In meiosis I, the segregation of homologous chromosomes before pairing would be catastrophic. Kim et al. describe two mechanisms that prevent this. In early meiosis, Ipl1 triggers shedding of a kinetochore protein and prevents microtubule attachment. Ipl1 localizes to the spindle pole bodies (SPBs), where it blocks spindle assembly. These processes are reversed upon expression of Ndt80. CDK phosphorylates Ipl1, delocalizing it from SPBs and triggering spindle assembly. Ipl1 and Ntd80 coordinate chromosome and spindle behaviors to prevent the attachment of unpartnered chromosomes to the meiotic spindle.
Keywords: Ipl1, Clb4, Cdc28, spindle pole body, meiosis, kinetochores
Abstract
In meiosis I, homologous chromosomes pair and then attach to the spindle so that the homologs can be pulled apart at anaphase I. The segregation of homologs before pairing would be catastrophic. We describe two mechanisms that prevent this. First, in early meiosis, Ipl1, the budding yeast homolog of the mammalian Aurora B kinase, triggers shedding of a kinetochore protein, preventing microtubule attachment. Second, Ipl1 localizes to the spindle pole bodies (SPBs), where it blocks spindle assembly. These processes are reversed upon expression of Ndt80. Previous studies have shown that Ndt80 is expressed when homologs have successfully partnered, and this triggers a rise in the levels of cyclin-dependent kinase (CDK). We found that CDK phosphorylates Ipl1, delocalizing it from SPBs, triggering spindle assembly. At the same time, kinetochores reassemble. Thus, dual mechanisms controlled by Ipl1 and Ntd80 coordinate chromosome and spindle behaviors to prevent the attachment of unpartnered chromosomes to the meiotic spindle.
Meiosis I is a specialized division in which homologous chromosomes first become paired with one another and then attach to microtubules that will pull them toward opposite poles of the spindle. In humans, failures in this process are a leading cause of birth defects and infertility (Hassold and Hunt 2001). Prior to segregation, homologous chromosomes become partnered through a process of homologous pairing, synapsis, and recombination. Successful completion of these events in the pachytene stage of prophase satisfies a checkpoint (Xu et al. 1995; Hepworth et al. 1998; Wu and Burgess 2006). Pachytene exit is triggered through activation of the Ndt80 transcription factor (Chu and Herskowitz 1998). Ndt80 triggers the expression of Clb1 and Clb4, B-type cyclins that are responsible for the increase in cyclin-dependent kinase (CDK) levels that promotes the transition into metaphase (Grandin and Reed 1993; Carlile and Amon 2008).
Assembly of the meiotic spindle is restricted until homologous chromosomes are successfully partnered. In meiosis, the spindle pole body (SPB), which acts as a microtubule organizing center, is duplicated in S phase, but the duplicated SPBs remain side by side until pachytene exit (Byers and Goetsch 1975). In contrast, in mitotic cells, there is no period of homologous chromosome pairing, and spindle assembly occurs much earlier, coincident with DNA replication.
What are the mechanisms that protect meiotic chromosomes from segregating before they have become tethered to their partners? It has been shown that Ipl1 prevents spindle formation during meiotic prophase, but the mechanism is not known (Shirk et al. 2011). Additionally, it has been shown that Ndc80, an outer kinetochore protein required for attaching chromosomes to microtubules, is lost from kinetochores in early meiosis (Asakawa et al. 2005), but when Ndc80 reloads and whether this is coordinated with homologous pairing have not been determined. Finally, it has not been determined how the mechanisms that block spindle and kinetochore assembly are reversed once chromosomes are prepared to segregate. One candidate is CDK. CDK activity governs progression through the major transitions of the meiotic cell cycle (Grandin and Reed 1993; Carlile and Amon 2008; Miller et al. 2012). In budding yeast, this is done by the sole CDK (Cdc28) in association with nine cyclins. Cdc28 plays an essential role in spindle formation in both mitotic (Lim et al. 1996) and meiotic cells (Shuster and Byers 1989). Artificial overexpression of the cyclin genes CLB1 or CLB4 in prophase I can trigger precocious spindle assembly. This suggests that the induction of specific CDK/cyclin combinations (CDK/Clb1 and CDK/Clb4) at the correct time in meiosis may be the trigger for spindle assembly (Miller et al. 2012).
Here we explore the mechanisms used by meiotic cells to prevent the segregation of chromosomes before they become tethered to their eventual segregation partners. We found that Ipl1 blocks spindle assembly and kinetochore function, while chromosomes are in the pairing process. The completion of homolog partnering has been shown to activate Ndt80 expression, which allows pachytene exit (Xu et al. 1995; Hepworth et al. 1998; Wu and Burgess 2006). We found that Ndt80 activation reverses the actions of Ipl1, allowing segregation of the homologous pair.
Results
Ipl1 prevents formation of bipolar spindles during meiotic prophase
Previous studies have suggested that Ipl1 coordinates events in meiotic prophase by either promoting the disassembly of the synaptonemal complex (SC) or regulating spindle behavior (Jordan et al. 2009; Shirk et al. 2011). Cells enter meiotic prophase with duplicated SPBs that are positioned side by side and connected by a structure called the half-bridge (Byers and Goetsch 1975). The SPBs separate to form a spindle upon the exit from pachytene, concomitant with the disassembly of the SC (Dresser and Giroux 1988). Shirk et al. (2011) demonstrated that cells that were blocked in prophase by deleting the gene for the Ndt80 transcription factor were able to form spindles when Ipl1 was inactivated. We also found that prophase-arrested cells formed spindles when Ipl1 was depleted. (Supplemental Fig. S1). These ndt80Δ ipl1-mn cells ultimately developed tripolar and tetrapolar spindles (Supplemental Fig. S2). These results demonstrate that one role of Ipl1 is to coordinate meiotic events by preventing spindle formation in prophase. A previous report suggested a different role for Ipl1: mediating the disassembly of the SC such that loss of Ipl1 leads to cells with metaphase and anaphase spindles and persisting SC (Jordan et al. 2009). To re-examine the requirement for Ipl1 in SC disassembly, we monitored the presence of Zip1, a component of the central element of the SC, in IPL1 and ipl1-mn strains as they progressed through meiosis. In both IPL1 and ipl1-mn cells, Zip1 was present in prophase prior to SPB separation (Supplemental Fig. S3). In IPL1 metaphase cells (a single chromatin mass and short spindle), Zip1 is always gone (Supplemental Fig. S3A), consistent with the well-documented disassembly of the SC and loss of Zip1 as cells exit pachytene (Padmore et al. 1991; Jordan et al. 2009). However, in ipl1-mn cells with a single chromatin mass and separated SPBs (Supplemental Fig. S3B), Zip1 was frequently present, as reported previously (Jordan et al. 2009). If this simultaneous presence of SC and bipolar spindles in ipl1-mn mutants is due to a failure in SC disassembly, it should lead to cells in metaphase and beyond with intact SC (Jordan et al. 2009). To test this, in cells that contained both spindles and Zip1, we monitored Pds1, which is degraded at the metaphase-to-anaphase transition in mitosis and meiosis (Cohen-Fix et al. 1996; Salah and Nasmyth 2000). Anaphase cells—defined as cells with separated SPBs, dumbbell-shaped chromatin masses (DAPI), and no detectable Pds1—were identified and then scored for the presence or absence of Zip1. For both IPL1 and ipl1-mn strains, nearly all cells in anaphase were devoid of Zip1 (100% and 97%, respectively) (Supplemental Fig. S3A,B). We conclude that most ipl1-mn cells that simultaneously exhibit SC and spindles have precociously assembled spindles in prophase and are not anaphase cells with persisting SC.
Shedding of outer kinetochore components prevents chromosome–microtubule interactions in prophase
The formation of precocious spindles would be expected to result in attachment of chromosomes to the spindles in prophase. To test this, we analyzed the interactions of chromosomes with the precocious spindles formed in ipl1 mutants. To avoid potential phenotypes from the loss of Ipl1 activity at earlier steps in meiosis (Meyer et al. 2013), we allowed cells to proceed to pachytene with a functional version of Ipl1, Ipl1-as5, then inactivated this Ipl1-as5 (Pinsky et al. 2006) by the addition of 1-NA-PP1 to the medium. Pachytene cells were first accumulated by blocking expression of NDT80 (Chu and Herskowitz 1998), and then Ipl1 was inhibited to allow precocious spindle formation. During normal meiosis, following prophase exit, attachments of centromeres to the meiotic spindle typically result in poleward movement of the centromeres such that, in early metaphase, most centromeres are clustered at the poles (Fig. 1A; Meyer et al. 2013). We monitored attachment of chromosome I to precocious spindles by measuring the distance between CEN1 and the spindle pole. In ipl1 mutants, CEN1 did not colocalize to the poles in 86% of cells with precocious spindles (Fig. 1A,B), suggesting that the chromosomes may be unable to interact with the precocious spindles.
Figure 1.
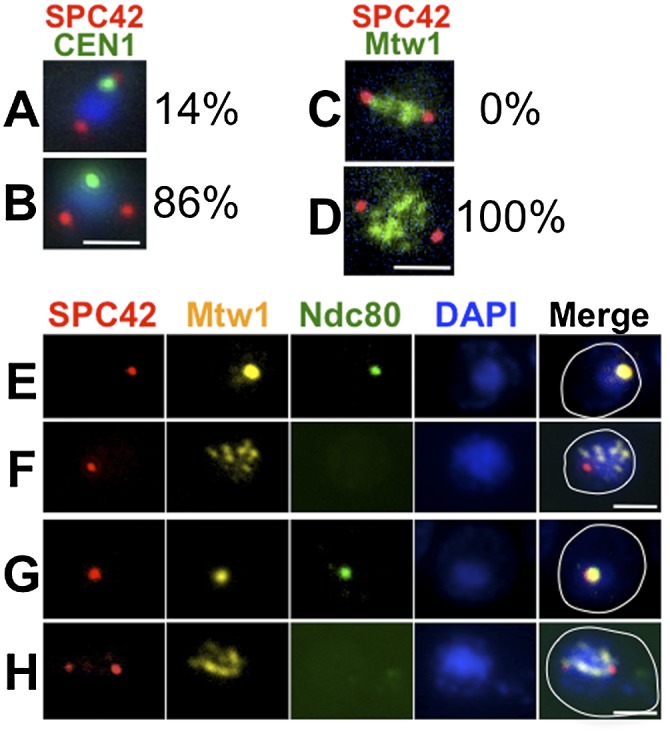
Kinetochore disassembly blocks precocious kinetochore–microtubule interactions. (A) ipl1-as5/ipl1-mn cells (DSY434 and DSY436) were sporulated. 1-NA-PP1 was added (T = 5 h) to allow spindle formation during the pachytene arrest (no β-estradiol added). The distance between CEN1 (CEN1 was tagged with GFP using the lacO/GFP-lacI system) (Straight et al. 1996; Obeso and Dawson 2010) and the closer SPB (Spc42-DsRed) was measured. (A) CEN1 at the pole (≤0.5 μm from the nearest pole). (B) CEN1 distant from the pole (>0.5 μm from the nearest pole) (n = 84). (C,D) ipl1-as5/ipl1-mn cells (DSY428) were sporulated, and 1-NA-PP1 was added (T = 5 h) to allow spindle formation during pachytene arrest (ndt80Δ). Kinetochores (Mtw1-GFP) and SPBs (Spc42-DSRed) were visualized. Examples of a wild-type cell with chromosomes aligned on the metaphase spindle (C) and an ipl1 cell with scattered chromosomes on a precocious spindle (D). In a sample of 44 ipl1 cells (at T = 9 h), 100% had scattered chromosomes. (E–H) ndt80Δ (DSY585) (E,F) and ndt80Δ ipl1-as5/ipl1-mn (DSY588, 1-NA-PP1 at 5 h) (G,H). Samples were prepared for whole-cell immunostaining for kinetochores (Mtw1-myc and Ndc80-GFP) and SPBs (Spc42-DsRed) at meiotic entry (E,G) and prophase (F,H). (E) Clustered Ndc80 was observed in 98% of the cells at meiotic entry (marked by clustered Mtw1; n = 55). (F) Ndc80 was absent in 100% of prophase cells (marked by dispersed Mtw1; n = 31). (G) Clustered Ndc80 was observed in 100% of cells at meiotic entry (marked by clustered Mtw1; n = 51). (H) Ndc80 was absent in 100% of prophase cells (marked by dispersed Mtw1; n = 31). Bar, 2 μm.
The above result might also be explained if the chromosomes can interact with the microtubules but are prevented from moving to the poles. In this scenario, chromosomes would align on the precocious spindles but not migrate to the poles. To test this, we determined the location of all of the kinetochores in cells with precocious spindles. Whereas in wild-type metaphase cells kinetochores are located directly between the poles, in the ipl1 mutant cells with precocious spindles, the kinetochores were not aligned on the spindle but rather were scattered, indicating that they are not attached to the precocious spindles (Fig. 1C,D).
The failure of chromosomes to attach to the precocious spindles could be explained if kinetochores are not competent to attach to microtubules in these cells. Previous studies (Asakawa et al. 2005) demonstrated that the outer kinetochore component Ndc80, which is essential for chromosome–microtubule interactions (He et al. 2001), is shed from chromosomes upon meiotic entry. Therefore, we tested whether failure of chromosomes to interact with precocious spindles could be explained by the absence of Ndc80 from kinetochores in late prophase. Meiosis was induced, and the localization of Ndc80-GFP to the kinetochore (marked by Mtw1-MYC) was monitored by immunofluorescence microscopy. IPL1 cells began the meiotic process with their centromeres clustered at the spindle poles, as shown previously (Hayashi et al. 1998; Jin et al. 1998; Asakawa et al. 2005). In these cells, Ndc80-GFP and Mtw1-MYC colocalized (Supplemental Fig. 4A). In prophase, the kinetochores (Mtw1-myc) dispersed, and Ndc80-GFP was lost from the kinetochores (Supplemental Fig. 4B). The observed reduction in Ndc80 expression may also contribute to the observed reduction of kinetochore-associated Ndc80 (Brar et al. 2012; Miller et al. 2012). In metaphase and anaphase cells, Ndc80-GFP was again colocalized with Mtw1-myc (Supplemental Fig. 4C,D). To test whether the signal to reassemble kinetochores occurs during or after prophase, we assayed the association of Ndc80 with kinetochores in cells arrested in pachytene (ndt80Δ). Even after a prolonged pachytene arrest, the Ndc80 never reappeared on kinetochores (Fig. 1F). Consistent with this, when precocious spindles were allowed to form in pachytene-arrested ipl1 cells, Ndc80 could not be detected on the scattered kinetochores (Fig. 1H). We conclude that kinetochores are prevented from forming potentially dangerous microtubule attachments with microtubules throughout meiotic prophase by the shedding of Ndc80 (above) coupled with its diminished expression until metaphase (Brar et al. 2012; Miller et al. 2012).
Ipl1 localizes near unseparated SPBs
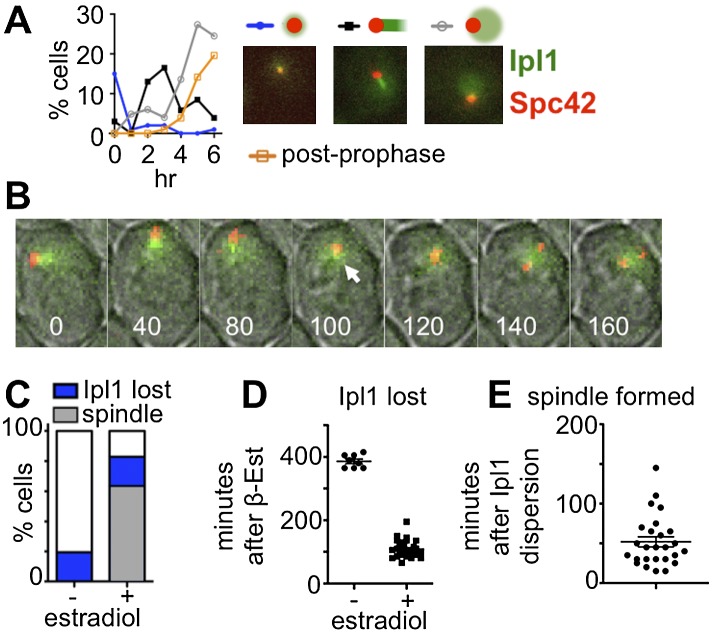
The mechanism by which Ipl1 inhibits spindle formation during meiotic prophase is not known. In mitotic metaphase cells, Ipl1 acts at kinetochores to regulate kinetochore–microtubule interactions (Biggins et al. 1999; Tanaka et al. 2002), and in anaphase, Ipl1 regulates microtubule dynamics at the spindle mid-zone (Kotwaliwale et al. 2007; Zimniak et al. 2009). These actions of Ipl1 are regulated by its localization to specific sites at specific times. By analogy, it might be that Ipl1 blocks meiotic spindle assembly by regulating microtubule dynamics at the prophase SPBs. If so, we would expect Ipl1 to localize to unseparated SPBs during prophase and dissipate prior to SPB separation. We tested this hypothesis. Cells expressing Ipl1-GFP, Spc42-DsRED to mark SPBs, and Mtw1-MYC were induced to enter meiosis, and as cells progressed through meiosis, samples harvested at timed intervals were analyzed to evaluate the localization of Ipl1-GFP (Fig. 2A). Most cells entering meiosis had either no detectable Ipl1-GFP signal or a faint signal near the single SPB. As cells moved into prophase, a population emerged with comet-like Ipl1-GFP near the SPBs. This was followed by the emergence of cells with dispersed nuclear Ipl1-GFP and less intense (or no detectable) staining near the SPBs. Cells with separated SPBs were observed after the appearance of cells with dispersed Ipl1-GFP. The above results are consistent with the model that spindle assembly follows the dispersal of Ipl1-GFP from the SPBs.
Figure 2.
Delocalization of Ipl1 from SPBs precedes spindle formation. (A,B) The localization of Ipl1 (IPL1-EGFP) relative to SPBs (SPC42-DsRed) was monitored (DSY461). (A) Samples were taken hourly (n ≥ 100 for all time points). Post-prophase cells were those with separated SPBs. (B) Cells (DSY461) were loaded into a microfluidic chamber 1.5 h after inducing sporulation and then imaged every 2 min. Selected frames are shown. Numbers indicate minutes since start of acquisition. (White arrow) Last frame in which Ipl1 focus could still be seen at SPB. (C–E) Cells (DSY670) expressing IPL1-EGFP and mCherry-TUB1 were sporulated, and pachytene cells were accumulated (no β-estradiol added). At 6.5 h, cells were loaded on a microfluidic plate, and images were collected (5-min intervals) to track the localization of Ipl1 (IPL1-EGFP) relative to microtubules (mCherry-TUB1). Following the first acquisition, medium with or without β-estradiol was introduced into the chamber to trigger NDT80 expression.
We tested this model by tracking Ipl1 in single cells from prophase to metaphase using live-cell imaging. If comet-like localization of Ipl1 prevents spindle formation, cells that progress into metaphase and form spindles should lose this pattern prior to spindle formation. A diploid strain carrying IPL1-EGFP, SPC42-DsRed, and MTW1-MYC was transferred to a microfluidic chamber 90 min after induction of meiosis, and images were collected every 2 min (Fig. 2B). In every case (n = 20), cells that formed metaphase spindles lost their Ipl1 localization to the SPBs before SPB separation by an average of 43 min.
Because spindle formation depends on pachytene exit, we tested whether the removal of Ipl1 from near the SPBs is coordinated with Ndt80 expression. Cells expressing Ipl1-GFP and mCherry-TUB1 were induced to enter meiosis and arrested in pachytene, and cells with comet-like Ipl1-GFP signal were imaged following their release from the pachytene arrest (Supplemental Fig. S5). Synchronous pachytene exit was accomplished by placing NDT80 under the control of the GAL1 promoter and expressing the Gal4 transcription factor as a hybrid protein with an estrogen receptor (Gal4-ER) moiety (Benjamin et al. 2003; Carlile and Amon 2008). Pachytene exit was triggered by the addition of β-estradiol to the medium. In control cells arrested in pachytene (no β-estradiol addition), the comet-like Ipl1-GFP persisted at SPBs for the duration of the experiment (Fig. 2C) in most cells, and spindles never formed. However, when pachytene exit was induced, Ipl1-GFP dispersed from the SPBs in 83% of the cells, and most of these (76%) went on to form spindles ∼46 min, on average, after losing the comet-like Ipl1 signal (Fig. 2D,E). We conclude that dispersion of Ipl1 from the SPBs in meiosis I follows the signal to exit pachytene and that loss of Ipl1 near the SPB precedes spindle assembly.
Ipl1 is concentrated on microtubules in prophase cells
The manner in which Ipl1 blocks spindle assembly is unknown, but Ipl1 could be imagined to act at the SPBs to prevent breakage of the half-bridge that holds the SPBs together or at the microtubules to prevent the formation of anti-parallel microtubule bundles. The comet-like pattern of Ipl1-GFP near the SPBs in prophase resembles the short prophase I microtubule array emanating from the unseparated SPBs (Supplemental Fig. S5), suggesting that a majority of the SPB-associated Ipl1 might be on the microtubules. To test this, we asked whether the Ipl1-GFP signal is diminished by the microtubule-destabilizing drug benomyl. Pachytene cells in a microfluidic chamber were flooded with benomyl-containing medium, and the patterns of Ipl1-GFP and mCherry-Tub1 were monitored by live-cell imaging (Fig. 3A). In control cells (no benomyl), the comet-like Ipl1-GFP pattern persisted >20 min, but when benomyl was added, both the overlapping mCherry-Tub1 and Ipl1-GFP signals were reduced from a comet shape to a punctate signal, and a dispersed GFP signal could be seen in most cells. Consistent with a mainly microtubule-associated localization of Ipl1-GFP, when the above experiment was repeated with Ipl1-GFP and SPC42-DsRED cells, the Ipl1-GFP signal clearly localized adjacent to, not with, the SPBs (Fig. 3B). The evaluation of Ipl1 localization using prophase chromosome spreads confirmed that the main localization of Ipl1 was not on kinetochores but adjacent to the SPBs (Supplemental Fig. S6). It has been previously demonstrated that Ipl1 localizes to the kinetochores in metaphase (Biggins and Murray 2001; Tanaka et al. 2002; Buvelot et al. 2003; Monje-Casas et al. 2007; Yu and Koshland 2007). Here, we demonstrate that in meiotic prophase, Ipl1 shows a localization near the SPBs that is microtubule-dependent.
Figure 3.
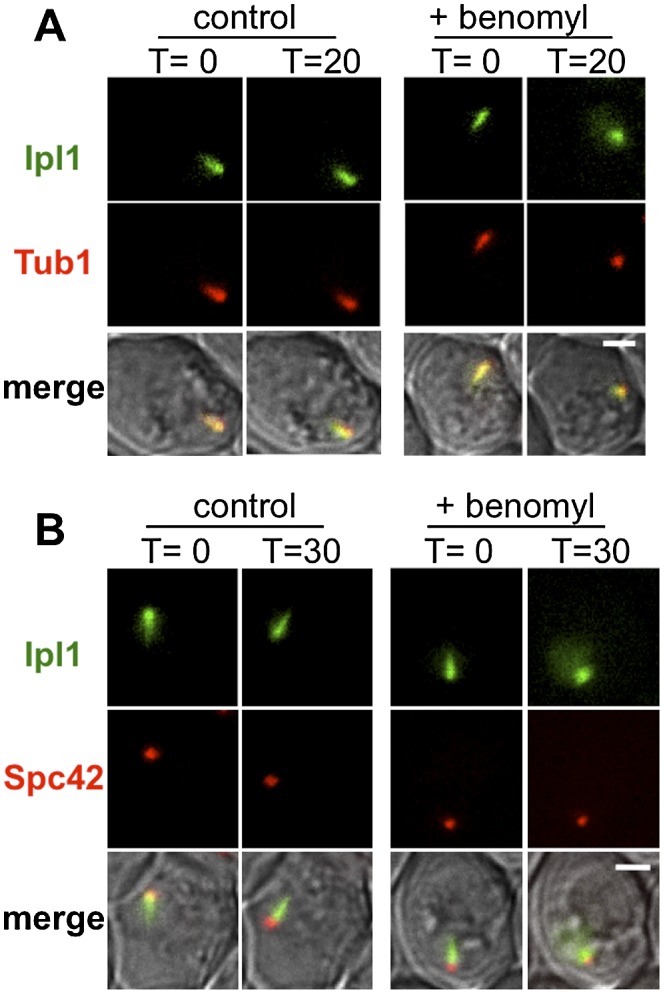
Ipl1 localizes to microtubule arrays in prophase of meiosis I. (A) The relative localization of Ipl1 (IPL1-GFP) and microtubules (DsRED-TUB1) was monitored before (T = 0 min) and after (T = 20 min) replacing the sporulation medium with fresh medium either without (control) or with (+benomyl) benomyl. Diploid cells (DSY670) were sporulated, and pachytene cells were accumulated (no β-estradiol added). At 5 h, cells were loaded onto a microfluidic plate, and images were acquired (5-min intervals). Following the first acquisition, medium with or without benomyl was introduced into the chamber. Bar, 2 μm. (B) Localization of Ipl1 (IPL1-EGFP) relative to SPBs (SPC42-DsRed) was monitored after microtubule depolymerization. Diploid cells (DSY673 and DSY674) were sporulated and imaged as described in A except that imaging was performed at 30 min after introduction of fresh medium. Bar, 5 μm.
Spindle assembly is triggered at pachytene exit by CDK/Clb4
CDK/Clb4 activity rises when cells exit pachytene (Grandin and Reed 1993; Carlile and Amon 2008), and precocious expression of CDK/Clb4 in prophase triggers spindle assembly (Miller et al. 2012). Could CDK/Clb4 trigger spindle assembly by causing the delocalization of Ipl1 from the microtubules? To test this, we induced expression of CLB4 in prophase using the copper-inducible PCUP1 promoter as described by others (Miller et al. 2012). We used live-cell imaging to monitor the localization of Ipl1-GFP and spindle formation (mCherry-Tub1) in pachytene-arrested cells (ndt80Δ) with and without induction of PCUP1-CLB4. Cells were induced to enter meiosis, and, at 4 h, copper was added to induce CLB4 expression. In control experiments (no copper), Ipl1-GFP maintained its comet-like shape, and spindles did not form (Fig. 4A,B). In contrast, following induction of CLB4 expression, all cells examined lost their comet-like Ipl1 pattern, and most of those cells went on to form spindles (Fig. 4). Ipl1 typically delocalized from the microtubules ∼50 min after Clb4 expression, and spindles formed ∼20 min later, on average—spindle formation never preceded Ipl1 delocalization (Fig. 4). Overexpression of CLB5, which is naturally expressed in meiotic prophase (Grandin and Reed 1993; Carlile and Amon 2008), was shown previously not to induce precocious spindle formation (Miller et al. 2012). If delocalization of Ipl1 from microtubules leads to spindle assembly, then Ipl1 should not delocalize efficiently from SPBs upon induction of PCUP1-CLB5. Consistent with this, much less Ipl1-GFP delocalization from spindles was observed following CLB5 expression (45% vs. 100%), and when it was observed, it occurred much later (∼111 min vs. 53 min). Thus, regulation of Ipl1 localization by CDK is specific to the version of cyclin B.
Figure 4.
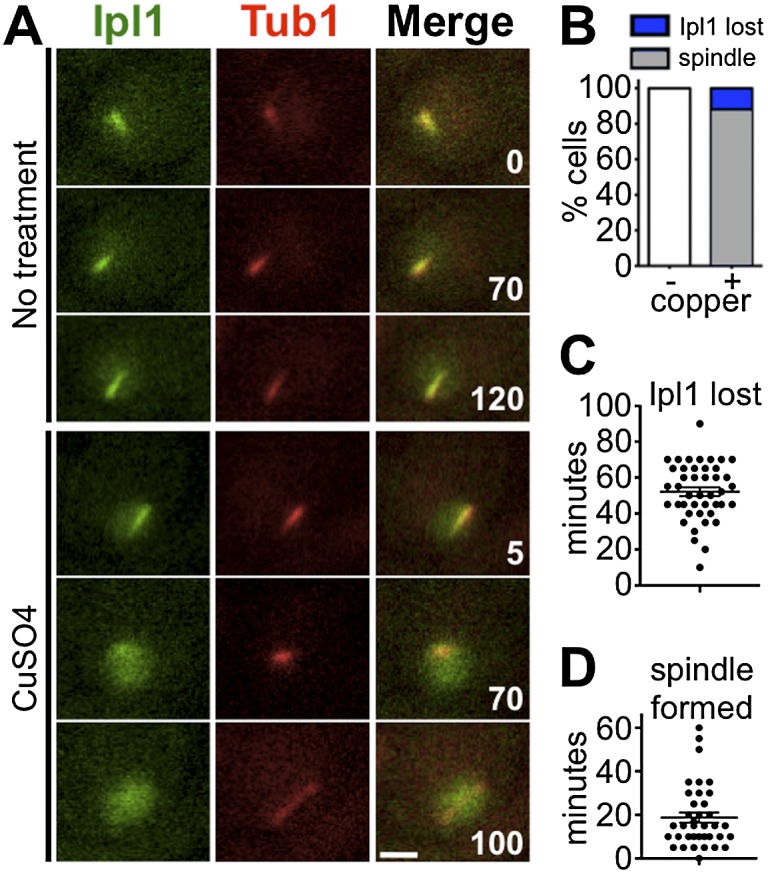
Ipl1 delocalizes from the prophase microtubule array before spindle formation. (A–D) Ipl1 localization (IPL1-EGFP) relative to the prophase microtubule array (mCherry-TUB1) was monitored after induced expression of CLB4 (PCUP1-CLB4) while in pachytene-arrested cells (ndt80Δ) (DHC244 and DHC245). (A) Meiosis was induced. At T = 4 h, cells were transferred to a microfluidic plate. Images were collected every 5 min for 120 min. After the first frame, sporulation medium with or without CuSO4 was introduced into the chamber. (B) Cells were scored for delocalization of Ipl1 from the SPBs and formation of spindles. All cells that formed spindles also lost SPB localization of Ipl1. (C) Cells were scored for the number of minutes after copper addition at which Ipl1 was lost from the SPBs. (D) Cells were scored for the number of minutes after Ipl1 delocalization at which a spindle formed. n > 40 for each sample. Bar, 2 μm.
In mitotic cells, Ipl1 associates with the microtubules through multiple mechanisms. One mechanism is through a direct binding to Bim1, a protein that localizes to and stabilizes the plus ends of microtubules to promote net growth (Gardner et al. 2008). Ipl1 has six known target residues for CDK phosphorylation (Zimniak et al. 2012). Mitotic CDK phosphorylates two of these (Ser50 and Ser76) and diminishes the association of Ipl1 with Bim1, which promotes its delocalization from the plus ends of microtubules (Zimniak et al. 2012). We tested whether an analogous mechanism might be used in meiosis to trigger spindle assembly upon pachytene exit. If Cdk/Clb4 directly phosphorylates Ipl1 to mediate its displacement from microtubules, then a nonphosphorylatable version of Ipl1 should block prophase spindle formation following PCUP1-CLB4 overexpression. To test this, Ipl1 localization and spindle formation were monitored following CLB4 induction in cells expressing Ipl1-AA-GFP (in which the key CDK target serines are replaced by alanines). Three hours after meiotic induction, before CLB4 expression, both Ipl1-GFP and Ipl1-AA-GFP could be seen on microtubule arrays and dispersed in the nucleus (Fig. 5A,B). After inducing CLB4 expression, the Ipl1-GFP delocalized from microtubules, while in this same time frame, Ipl1-AA-GFP continued to accumulate on the microtubules (Fig. 5B). Consistent with this, the Ipl1-AA-GFP cells are refractory to spindle formation following CLB4 induction (Fig. 5C). These results strongly suggest that direct CDK phosphorylation of Ipl1 on Ser50 and Ser76 triggers delocalization of Ipl1 from microtubules and that this delocalization is necessary to permit spindle assembly. This is not to say that Ipl1 is the sole CDK target for triggering meiotic spindle assembly. For example, phosphorylation of Sli15 might also act to reduce Ipl1 on the microtubules, and multiple proteins involved in spindle dynamics could also be regulated by the increase in CDK activity following Ndt80 activation.
Figure 5.
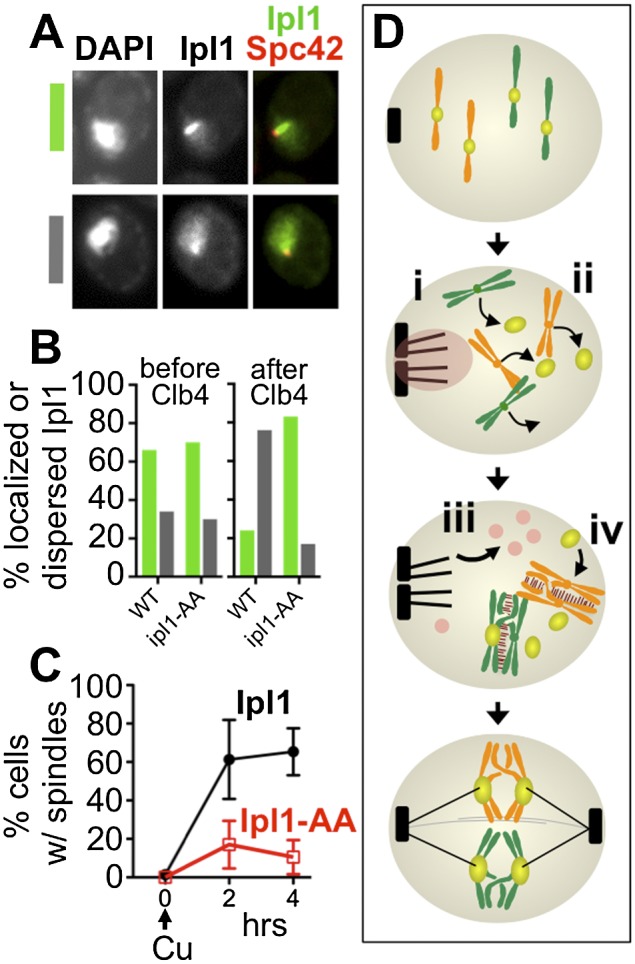
Phosphorylation of Ipl1 is necessary to trigger precocious spindle assembly in prophase. To test whether CDK phosphorylation of Ipl1 controls its localization in meiotic prophase, prophase-arrested cells (ndt80Δ) expressing either Ipl1-GFP (DHC257) or nonphosphorylatable Ipl1-AA-GFP (DHC258 and DHC259) were treated with copper to induce expression of CLB4 (PCUP1-CLB4) and then monitored to follow Ipl1 localization and spindle formation (Spc42-DSRed). (A) Three hours after meiotic entry (before CLB4 induction), some cells exhibited intense localization to the microtubule array (green), while others had a more dispersed nuclear distribution (gray). (B) Cells were assayed for their Ipl1 distribution at the time of copper addition (3 h after introduction to sporulation medium) or 4 h later. Following CLB4 induction, Ipl1-AA-GFP showed significantly higher levels of cells with Ipl1 at the microtubule array (n = 100 for both samples; Fisher's exact test, P > 0.0001). (C) Spindle formation (Spc42-DSRed) was monitored following CLB4 induction in cells expressing Ipl1-GFP (black) or Ipl1-AA-GFP (red) (average ± one standard deviation of three experiments). (D) Model for the coordination of microtubule and chromosome behaviors by Ipl1 and Ndt80 in meiosis. Upon meiotic entry, Ipl1 localizes to SPBs (i), preventing their separation, and triggers shedding of Ndc80 (ii). When homologous chromosomes have successfully partnered, Ndt80 activation triggers increased CDK activity (iii), which results in delocalization of Ipl1 from SPBs, and promotes kinetochore reassembly through an unknown mechanism (iv).
Discussion
The results reported here demonstrate that there are dual mechanisms that prevent interactions of chromosomes with the spindle until pairing and recombination are complete (Fig. 5D); first, spindle formation is prevented, and second, Ndc80 is shed from kinetochores. The two mechanisms are controlled by the same regulators. Ipl1 blocks spindle assembly beginning in early prophase, concurrent with the loss of Ndc80 from kinetochores. The fact that kinetochores remain attached to microtubules throughout meiosis I in ipl1-mn mutants (Meyer et al. 2013) demonstrates that Ipl1 is also required for shedding the Ndc80. Just as both protective mechanisms require Ipl1 to be activated, they both require the expression of the Ndt80 transcription factor for their reversal. Because Ndt80 is only activated once the pachytene checkpoint is satisfied, this ensures that the machinery used to segregate homologous partners will remain inactive until the partners are fully prepared to be segregated.
The roles for Ipl1 reported here are related to but distinct from roles played by Aurora kinases in vertebrates. Outer kinetochores are lost and then reassembled upon mitotic entry in vertebrate cells. Aurora B is involved in this process but has been implicated mainly in promoting the reassembly (Emanuele et al. 2008), rather than the shedding, of kinetochores. Aurora B has not been strongly implicated in the spindle assembly process. Instead, its homolog, Aurora A, localizes to centrosomes and, in many organisms, has been found to regulate mitotic spindle assembly (for review, see Mardin and Schiebel 2012; Hochegger et al. 2013). In contrast to Ipl1, Aurora A promotes, rather than restricts, spindle assembly. Despite the fact that they are operating through different regulatory mechanisms, it is interesting to note that in spindle assembly, Ipl1 is functioning more like Aurora A than Aurora B.
The manner in which Ipl1 is regulated to perform its functions in budding yeast meiosis, but apparently not in mitosis, remains an intriguing question. These mechanisms do not appear to be in place in mitotic cells, and perhaps there is no need for them, as mitotic chromosomes are immediately prepared for segregation when replication is complete. In meiosis, shedding of the kinetochores and blocking spindle assembly clearly prevent meiotic chromosomes from beginning to segregate before they have completed the prolonged process of becoming tethered to their homologous partners. One aspect of the meiotic chromosome pairing involves dynamic movements of the chromosomes in the prophase nucleus (Conrad et al. 2008). A second possible important role for the shedding of the outer kinetochores is that this liberates the centromeres from the SPBs and allows centromeres to move freely as chromosomes become repositioned in prophase.
The roles for Ipl1 described here can be added to a long list of functions for this cell cycle regulator. One previously ascribed role for Ipl1, as the main regulator for synaptonemal disassembly, was based in part on the observation that ipl1-mn meiotic cells simultaneously exhibit spindles and SC (Jordan et al. 2009), leading to the suggestion that ipl1 cells often proceed through meiosis I, and sometimes meiosis II, without disassembling their SCs. We found that Zip1 is undetectable by the end of anaphase I in ipl1 mutants. This finding plus the discovery that ipl1-mn mutants form spindles in prophase (Shirk et al. 2011) demonstrate that Ipl1 is not essential for SC disassembly. Instead, Ipl1 is necessary for the regulation of multiple meiotic processes, including the release of centromeres from the SPB at meiotic entry (Meyer et al. 2013), spindle assembly (Shirk et al. 2011), the timing of the metaphase-to-anaphase transition (Meyer et al. 2013), and likely others. We suggest that rather than acting as the main regulator of SC disassembly, Ipl1 is necessary for the coordination of events such that, in its absence, the relative timing of landmark events is altered.
The manner in which Ipl1 is causing kinetochores to disassemble is unclear. We followed the shedding of the Ndc80 protein from the kinetochores, but the full complement of proteins that are shed is not known. The kinetochore contains multiple proteins that are known targets of Ipl1, and it will be of interest to determine how this shedding process is regulated. Interestingly, although the outer kinetochore is not shed in yeast mitotic cells, it is in mammalian mitotic and meiotic cells (Maiato et al. 2004; Emanuele et al. 2008; Parra et al. 2009).
Similarly, the manner in which Ipl1 prevents spindle assembly remains a mystery. Our results support the model in which the association of Ipl1 with the plus end microtubule-binding protein Bim1 is essential to block spindle assembly. One possibility is that Ipl1 directly regulates Bim1 function at the side-by-side SPBs. The interaction of Ipl1 and Bim1 is important for destabilization of the anaphase spindle in mitosis (Zimniak et al. 2009). Thus, it is plausible that an analogous mechanism is in place to destabilize the microtubules at the unseparated SPBs in meiotic prophase to prevent spindle formation. Alternatively, it may be that the high local concentration of Ipl1 around the SPBs and its actions on other targets are key for blocking spindle assembly.
When homologs have completed their preparations for segregation, Ndt80 promotes the release of cells into prometaphase. This includes the reassembly of functional kinetochores—although the regulation of this process remains a mystery—and the assembly of spindles, which is triggered by the Ndt80-promoted increase in CDK activity. Our results strongly support the model in which CDK promotes spindle assembly by directly phosphorylating Ipl1, releasing it from prophase microtubules. These linked dual mechanisms promote segregation fidelity by preventing the catastrophic segregation of homologous chromosomes that have not become tethered to their partners.
Materials and methods
Yeast strains and culture conditions
All strains are XY derivatives (Dresser et al. 1994). We used standard yeast culture methods (Burke et al. 2000). To induce meiosis, cells were grown in YP-acetate and then shifted to 1% potassium acetate at 108 cells per milliliter. 1-NA-PP1 (10 mM stock in dimethylsulfoxide; Tocris) and estradiol (5 mM stock in ethanol; Sigma) were added when indicated.
Strain construction
PCR-based methods were used to create gene modifications (Longtine et al. 1998; Janke et al. 2004). GAL4-ER constructs are described in Benjamin et al. (2003). ipl1-as5 (Pinsky et al. 2006) was crossed into the XY strains (five backcrosses). The strain genotypes are reported in Supplemental Table S1.
Fluorescence microscopy
The analysis of spindle formation and Ipl1 localization with fixed cells was performed by fixing cells for 5 min in 5% formaldehyde and then mounting the cells on agarose pads for viewing. Indirect immunofluorescence was preformed using published methods (Pringle et al. 1991) to detect the following epitopes: Mtw1-13MYC, mouse anti-MYC (9E10; gift from Susannah Rankin), and Alexa Fluor 647 goat anti-mouse IgG (Invitrogen); Pds1-13XMYC, mouse anti-MYC, and Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen); Zip1, goat anti-Zip1 (Santa Cruz Biotechnology), and Cy5 donkey anti-goat IgG (Jackson ImmunoResearch); and Ndc80-GFP, chicken anti-GFP (Chemicon), and Alexa Fluor 488 goat anti-chicken IgG. Images were collected using a Zeiss AxioImager microscope with Chroma filters, a Roper HQ2 CCD, and Axiovision software. Images were processed and analyzed using Axiovision software. Time-lapse imaging was performed with CellAsic microfluidic flow chambers (Y04D) with a flow rate of 5 psi. Images were collected with a Nikon Eclipse TE2000-E equipped with Perfect Focus system and NIS software. Images were processed and analyzed using NIS software.
Copper induction of CLB4 and CLB5 in meiotic cultures
Diploid cells of IPL1-GFP or ipl1-AA-GFP carrying PCUP1-CLB4, ndt80Δ, and SPC42-DSRED were induced to sporulate at 30°C. CuSO4 (50 μM) was added 3 h after cells were shifted into sporulation medium to induce the expression of CLB4. Cells were harvested every hour after induction. They were fixed with 3.7% formaldehyde for ∼7 min and then washed once in buffer B. DAPI-stained cells were then imaged using Zeiss Axiovision software. Two SPC42-Dsred dots were defined as spindle formed and one dot was defined as no spindle formed. Fifty cells were counted for each time point.
Acknowledgments
We thank Kirsten Benjamin, Sue Biggins, Dan Burke, Michael Conrad, Michael Dresser, David Morgan, Susannah Rankin, and Aaron Straight for reagents. We thank Jane Song for help with Western blots. We thank members of the Dawson laboratory for the use of shared strains and reagents, Susannah Rankin for the use of the Nikon Perfect Focus system, and members of the Dawson laboratory, past and present, and the Program in Cell Cycle and Cancer Biology for their many contributions to the development of this project. This project was supported by NIH grant R01GM087377.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.227454.113.
References
- Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y 2005. Dissociation of the Nuf2–Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell 16: 2325–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17: 1524–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev 15: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev 13: 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D, Dawson DS, Stearns T 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Buvelot S, Tatsutani SY, Vermaak D, Biggins S 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol 160: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L 1975. Electron microscopic observations on the meiotic karyotype of diploid and tetraploid Saccharomyces cerevisiae. Proc Natl Acad Sci 72: 5056–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Amon A 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell 1: 685–696 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10: 3081–3093 [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133: 1175–1187 [DOI] [PubMed] [Google Scholar]
- Dresser ME, Giroux CN 1988. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol 106: 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Harwell SN, Coody D, Conrad MN 1994. Nonhomologous synapsis and reduced crossing over in a heterozygous paracentric inversion in Saccharomyces cerevisiae. Genetics 138: 633–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol 181: 241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Haase J, Mythreye K, Molk JN, Anderson M, Joglekar AP, O'Toole ET, Winey M, Salmon ED, Odde DJ, et al. 2008. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol 180: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI 1993. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol 13: 2113–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P 2001. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet 2: 280–291 [DOI] [PubMed] [Google Scholar]
- Hayashi A, Ogawa H, Kohno K, Gasser SM, Hiraoka Y 1998. Meiotic behaviours of chromosomes and microtubules in budding yeast: Relocalization of centromeres and telomeres during meiotic prophase. Genes Cells 3: 587–601 [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK 2001. Molecular analysis of kinetochore–microtubule attachment in budding yeast. Cell 106: 195–206 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Friesen H, Segall J 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol 18: 5750–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Hegarat N, Pereira-Leal JB 2013. Aurora at the pole and equator: Overlapping functions of Aurora kinases in the mitotic spindle. Open Biol 3: 120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Jin Q, Trelles-Sticken E, Scherthan H, Loidl J 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol 141: 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Copsey A, Newnham L, Kolar E, Lichten M, Hoffmann E 2009. Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis. Genes Dev 23: 2237–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale CV, Frei SB, Stern BM, Biggins S 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev Cell 13: 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HH, Goh PY, Surana U 1996. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol 16: 6385–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC 2004. The dynamic kinetochore–microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Mardin BR, Schiebel E 2012. Breaking the ties that bind: New advances in centrosome biology. J Cell Biol 197: 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS 2013. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science 339: 1071–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP, Unal E, Brar GA, Amon A 2012. Meiosis I chromosome segregation is established through regulation of microtubule–kinetochore interactions. eLife 1: e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeso D, Dawson DS 2010. Temporal characterization of homology-independent centromere coupling in meiotic prophase. PLoS ONE 5: e10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66: 1239–1256 [DOI] [PubMed] [Google Scholar]
- Parra MT, Gomez R, Viera A, Llano E, Pendas AM, Rufas JS, Suja JA 2009. Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet 5: e1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S 2006. The Ipl1–Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol 8: 78–83 [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK 1991. Immunofluorescence methods for yeast. Methods Enzymol 194: 565–602 [DOI] [PubMed] [Google Scholar]
- Salah SM, Nasmyth K 2000. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma 109: 27–34 [DOI] [PubMed] [Google Scholar]
- Shirk K, Jin H, Giddings TH Jr, Winey M, Yu HG 2011. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J Cell Sci 124: 2891–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster EO, Byers B 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW 1996. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol 6: 1599–1608 [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K 2002. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Wu HY, Burgess SM 2006. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr Biol 16: 2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol 15: 6572–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HG, Koshland D 2007. The Aurora kinase Ipl1 maintains the centromeric localization of PP2A to protect cohesin during meiosis. J Cell Biol 176: 911–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T, Stengl K, Mechtler K, Westermann S 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol 186: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T, Fitz V, Zhou H, Lampert F, Opravil S, Mechtler K, Stolt-Bergner P, Westermann S 2012. Spatiotemporal regulation of ipl1/aurora activity by direct cdk1 phosphorylation. Curr Biol 22: 787–793 [DOI] [PubMed] [Google Scholar]