A pentatricopeptide repeat protein is involved in plastid gene expression required for normal chloroplast development.
Abstract
Chloroplasts are the site of photosynthesis and the biosynthesis of essential metabolites, including amino acids, fatty acids, and secondary metabolites. It is known that many seedling-lethal mutants are impaired in chloroplast function or development, indicating the development of functional chloroplast is essential for plant growth and development. Here, we isolated a novel transfer DNA insertion mutant, dubbed sel1 (for seedling lethal1), that exhibited a pigment-defective and seedling-lethal phenotype with a disrupted pentatricopeptide repeat (PPR) gene. Sequence analysis revealed that SEL1 is a member of the PLS subgroup, which is lacking known E/E+ or DYW domains at the C terminus, in the PLS subfamily of the PPR protein family containing a putative N-terminal transit peptide and 14 putative PPR or PPR-like motifs. Confocal microscopic analysis showed that the SEL1-green fluorescent protein fusion protein is localized in chloroplasts. Transmission electron microscopic analysis revealed that the sel1 mutant is impaired in the etioplast, as well as in chloroplast development. In sel1 mutants, plastid-encoded proteins involved in photosynthesis were rarely detected due to the lack of the corresponding transcripts. Furthermore, transcript profiles of plastid genes revealed that, in sel1 mutants, the transcript levels of plastid-encoded RNA polymerase-dependent genes were greatly reduced, but those of nuclear-encoded RNA polymerase-dependent genes were increased or not changed. Additionally, the RNA editing of two editing sites of the acetyl-CoA carboxylase beta subunit gene transcripts in the sel1 mutant was compromised, though it is not directly connected with the sel1 mutant phenotype. Our results demonstrate that SEL1 is involved in the regulation of plastid gene expression required for normal chloroplast development.
Chloroplasts have their own genome and gene expression machinery. The expression and regulation of plastid genes are very different from that of nuclear genes, and the coordinated expression of nuclear and plastid genes is important for the development of functional chloroplasts. Plastid genes are generally organized in polycistronic transcription units. Therefore, they are transcribed as long primary transcripts, which are extensively processed into mature transcripts by various posttranscriptional modifications, such as RNA splicing, RNA editing, RNA cleavage, and RNA stability. These posttranscriptional regulations of plastid gene expression are mediated by many nuclear-encoded proteins (Barkan and Goldschmidt-Clermont, 2000).
Recently, many chloroplast-localized pentatricopeptide repeat (PPR) proteins have emerged as primary nuclear factors that are involved in plastid gene expression and RNA metabolism in higher plants. The PPR proteins are characterized by a tandem array of degenerate 35-amino acid repeats, forming one of the largest protein families that is particularly prevalent in higher plants but not in other eukaryotes (Small and Peeters, 2000; Lurin et al., 2004; Schmitz-Linneweber and Small, 2008). Most of the PPR proteins are predicted to be targeted in the chloroplasts or mitochondria and involved in posttranscriptional regulation of organellar gene expression by interacting with RNA. The PPR protein family is divided into P and PLS subfamilies. The P subfamily usually contains only the classic PPR (P) motifs. By contrast, the PLS subfamily contains repeated blocks of PPR-like (P, L, and S) motifs, is specific to land plants (Lurin et al., 2004), and is also known as the plant combinatorial and modular protein (PCMP) subfamily (Aubourg et al., 2000; Rivals et al., 2006). Based on the presence of different C-terminal motifs, the PLS subfamily is further divided into the PLS, E, E+, and DYW subgroups (Lurin et al., 2004). Recent studies have shown that PPR proteins are sequence-specific RNA binding factors recruiting one or more effector proteins to the target RNA and are involved in posttranscriptional regulation of organellar gene expression (Schmitz-Linneweber and Small, 2008). Members of the P subfamily are involved in the transcription of plastid genes (Pfalz et al., 2006), RNA cleavage (Meierhoff et al., 2003; Nakamura et al., 2003), RNA stabilization (Beick et al., 2008; Pfalz et al., 2009), RNA splicing (Schmitz-Linneweber et al., 2006; de Longevialle et al., 2008), and translation (Yamazaki et al., 2004; Schmitz-Linneweber et al., 2005; Cai et al., 2011).
RNA editing is a posttranscriptional process that alters specific cytidine-to-uridine (C-to-U) conversion in the chloroplast and mitochondrial RNA of higher plants (Shikanai, 2006; Bentolila et al., 2008; Stern et al., 2010). In Arabidopsis (Arabidopsis thaliana), at least 34 sites are known to be edited in chloroplast transcripts (Chateigner-Boutin and Small, 2007), and more than 500 sites are specifically modified in mitochondrial transcripts (Bentolila et al., 2008; Takenaka et al., 2008). Thus far, more than 10 transfactors required for the editing of particular C targets in chloroplasts have been identified. They are all members of the E/E+ and DYW subgroups of the PLS subfamily of the PPR protein family. Members of the E/E+ subgroup proteins, CHLORORESPIRATORY REDUCTION4 (CRR4), CRR21, CHLOROPLAST BIOGENESIS19 (CLB19), and ORGANELLAR TRANSCRIPT PROCESSING80 (OTP80), and members of the DYW subgroup proteins, including CRR22, CRR28, YELLOW SEEDLING1 (YS1), EARLY CHLOROPLAST BIOGENESIS2 (AtECB2), PROTEIN REQUIRED FOR accD RNA EDITING1 (AtECB2), and OTP82, have been shown to be required for the editing of corresponding targets such as NADH dehydrogenase subunit4 (ndhB), ndhD, RNA polymerase alpha subunit (rpoA), acetyl-CoA carboxylase beta subunit gene (accD), and ndhG (Kotera et al., 2005; Okuda et al., 2006, 2007; Chateigner-Boutin et al., 2008; Hammani et al., 2009; Zhou et al., 2009). However, although the Arabidopsis genome has six PLS subgroup genes in the PLS subfamily of the PPR protein family (Lurin et al., 2004), the molecular function of members of the PLS subgroup proteins has not yet been reported.
The chloroplasts of higher plants contain two different types of RNA polymerases: nuclear-encoded RNA polymerase (NEP) and plastid-encoded RNA polymerase (PEP). NEP is a bacteriophage-type, single subunit RNA polymerase (Hedtke et al., 1997). PEP is a eubacterial-type, multisubunit RNA polymerase that consists of core subunits encoded by plastid genes (rpoA, rpoB, rpoC1, and rpoC2) and σ factors encoded by nuclear genes (Hess and Börner, 1999). It has been suggested that both NEP and PEP are responsible for the transcription of distinct types of plastid genes (Allison et al., 1996; Hajdukiewicz et al., 1997). Class I genes are predominantly transcribed by PEP, class II genes are transcribed by both NEP and PEP, and class III genes are transcribed exclusively by NEP. Whereas NEP transcribes nonphotosynthetic housekeeping genes, PEP transcribes photosynthesis-related genes (Hajdukiewicz et al., 1997; Swiatecka-Hagenbruch et al., 2007; Liere et al., 2011).
In addition to the core subunits, PEP complex contains many of the regulatory proteins encoded by nuclear genes that are essential for plastid gene expression (Pfalz et al., 2006; Steiner et al., 2011). Most of the loss-of-function mutants for these regulatory proteins show seedling-lethal and albino or pale-green phenotype. Though there have been numerous studies on the function of the regulatory proteins in plastid gene expression, the mechanism of the regulation has not yet been fully understood. Recent maize (Zea mays) nucleoid proteome analysis has shown that PEP and its regulatory proteins are found together with other molecules in plastid nucleoids. The nucleoid is a very large complex and composed of plastid DNA, RNA, and various proteins. It is also the site of DNA regulation, transcription, and translation. For example, RNA maturation and ribosome assembly occur cotranscriptionally in association with nucleoids. The function of the nucleoid-enriched proteome is subject to qualitative change from RNA metabolism to translation and homeostasis as proplastids convert to chloroplasts with assistance of regulatory proteins (Majeran et al., 2012).
In this study, we report the molecular characterization of seedling lethal1 (sel1) mutant, showing a pigment-defective and seedling-lethal phenotype. SEL1 encodes a novel chloroplast-localized PPR protein, which is a member of the PLS subgroup in the PLS subfamily of the PPR protein family. The mutation of SEL1 leads to defects in early chloroplast development. In the sel1 mutant, the transcription levels of PEP-dependent plastid genes are decreased, but those of NEP-dependent plastid genes are increased or not changed. Therefore, our results suggest that SEL1 is involved in the regulation of plastid gene expression required for normal chloroplast development.
RESULTS
Isolation and Characterization of sel1 Mutant
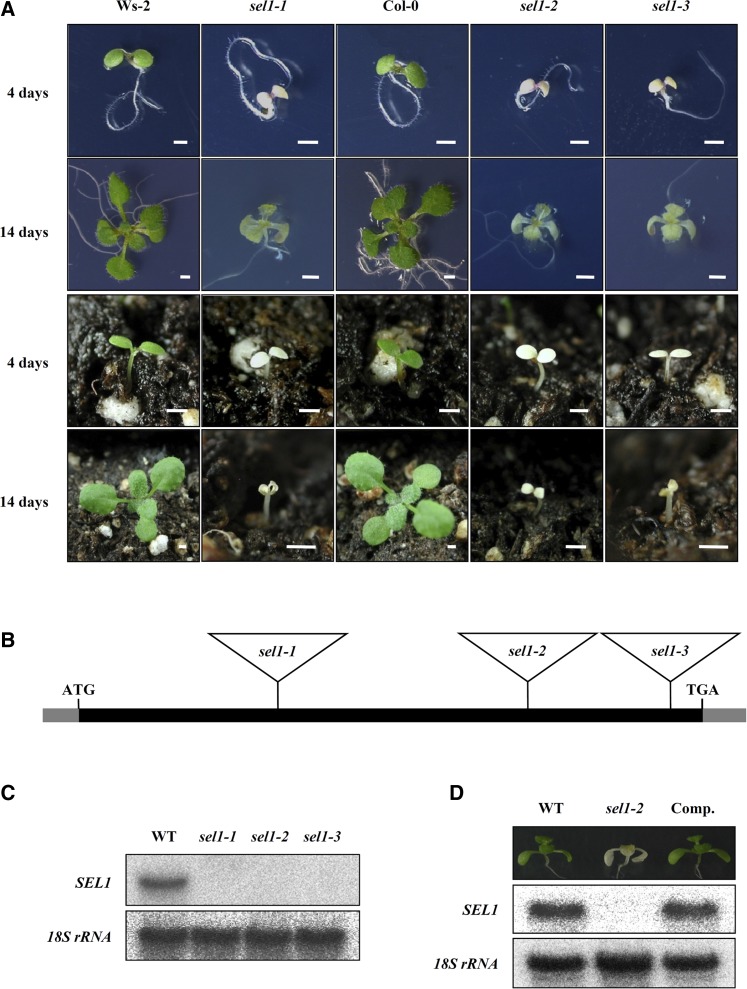
We generated a collection of T-DNA insertion mutants in Arabidopsis (ecotype Wassilewskija-2 [Ws-2] background) and isolated several seedling-lethal mutants with pigment-defective phenotypes, such as albino, pale green, and pale yellow. Among these mutants, we characterized a novel pigment-defective and seedling-lethal mutant grown under normal growth conditions. This mutant was named sel1-1 for the seedling-lethal phenotype. The germinated homozygous sel1-1 mutant on normal Murashige and Skoog (MS) medium containing Suc developed normally but failed to be pigmented (Fig. 1A). In the absence of Suc, the sel1-1 mutant was not pigmented and halted at the cotyledon stage (Supplemental Fig. S1). When grown on soil, the sel1-1 mutant stalled beyond the cotyledon stage and subsequently died (Fig. 1A). Pigment accumulations, including total chlorophyll and carotenoids, were compared between the wild type and sel1-1 mutant. The contents of total chlorophylls and carotenoids of the sel1-1 mutant were about 6 and 10 times lower, respectively, than those of the wild type (Supplemental Fig. S2).
Figure 1.
Isolation and characterization of the sel1 mutants. A, Pigment-defective and seedling-lethal phenotypes of the sel1 mutants. The wild-type (ecotypes Columbia and Ws-2) and sel1 mutant alleles were grown on MS medium with 1% (w/v) Suc or soil for 4 and 14 d, respectively. Bar = 1 mm. B, Gene structure of SEL1 (At4g18520). The black box represents an exon, and gray boxes represent the 5′ and 3′ untranslated regions. The positions of the T-DNA insertion in sel1-1, sel1-2 (SAIL_793_F11), and sel1-3 (SALK_054374) are represented by triangles. T-DNA is not drawn to scale. C, Northern-blot analysis of the SEL1 gene in wild-type (WT) and sel1 mutant alleles. Twenty micrograms of total RNA was isolated from 7-d-old seedlings grown on MS medium. 18S rRNA was used as a loading control. D, Molecular complementation of the sel1 mutant by pSEL1::SEL1 genomic DNA. The top row shows the 14-d-old wild type, sel1-2, and the complemented line (Comp.). The bottom row shows northern-blot analysis of SEL1 in the wild type, sel1-2, and the complemented line. 18S rRNA was used as a loading control. [See online article for color version of this figure.]
Genetic analysis indicated that the pigment-defective and seedling-lethal phenotype of the sel1-1 mutant was controlled by a recessive nuclear gene and cosegregated with the phosphinothricin resistance marker. Sequence analysis of the thermal asymmetric interlaced-PCR product revealed that the transfer DNA (T-DNA) was inserted 467 bp downstream from the start codon of the At4g18520 gene (Fig. 1B). Northern-blot analysis confirmed that At4g18520 transcript was detected in the wild type but not in the sel1-1 mutant (Fig. 1C). These results indicate that disruption of the At4g18520 gene leads to a pigment-defective and seedling-lethal phenotype of the sel1-1 mutant. To confirm that the sel1-1 mutant phenotype was resulted from the T-DNA insertion in the At4g18520 gene, we obtained two independent T-DNA insertion lines for the At4g18520 gene from the SAIL and SALK collections. Genotype and sequence analyses revealed that the position of the T-DNA insertion was 787 bp and 1,747 bp downstream from the start codon of the At4g18520 gene in SAIL_793_F11 (sel1-2) and SALK_054374 (sel1-3), respectively (Fig. 1B). The phenotype of sel1-2 and sel1-3 was identical to that of sel1-1 (Fig. 1A). Both the sel1-2 and sel1-3 mutants showed no SEL1 transcripts when using northern-blot analysis (Fig. 1C). Furthermore, we confirmed that the pigment-defective and seedling-lethal phenotype of sel1 mutants was associated with the T-DNA insertion into At4g18520 by molecular complementation. The genomic DNA of the At4g18520 gene and its putative promoter region was introduced into a heterozygous sel1-2 mutant. The subsequent genotype, phenotype, and northern-blot analyses confirmed that the introduction of At4g18520 genomic sequence restored the wild-type phenotype in the complemented lines (Fig. 1D). In addition, we generated complemented transgenic plants expressing FLAG-tagged SEL1 protein under the control of the SEL1 promoter. FLAG-tagged SEL1-complemented plants also grew normally like wild-type plants. Thus, we used the FLAG-tagged SEL1-complemented plants for further studies. Taken together, these results revealed that the phenotypes observed in sel1 mutants are caused by a loss of function of the At4g18520 gene.
SEL1 Gene Encodes a PPR protein That Is a Member of the PLS Subgroup
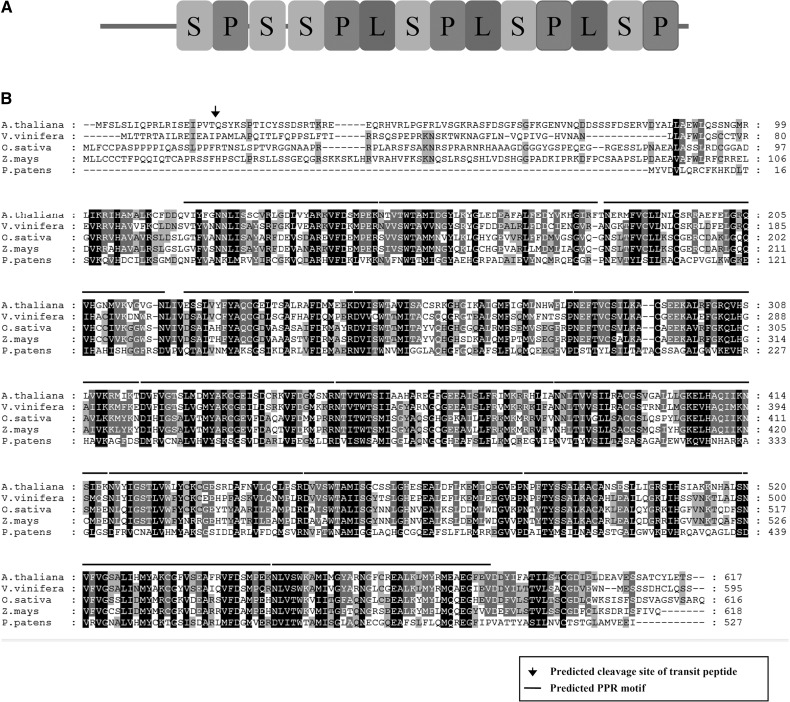
The SEL1 gene does not have an intron and encodes a putative protein of 617 amino acids with a predicted Mr of 69 kD. Sequence analysis revealed that SEL1 protein contains 14 PPR or PPR-like (P, L, and S) motifs but does not contain any other known domains (Fig. 2, A and B). The SEL1 protein is a member of the PLS subfamily and belongs to the PLS subgroup that does not have additional C-terminal domains (Lurin et al., 2004). Furthermore, SEL1 protein has been classified as AtPCMP A2, a member of the Plant Combinatorial Modular Protein subfamily, and belongs to the PCMP-A subgroup (Aubourg et al., 2000; Rivals et al., 2006). In Arabidopsis, the PLS or PCMP-A subgroup contains six members. Amino acid sequence analysis shows that SEL1 protein has significant sequence identity with uncharacterized proteins identified in grape (Vitis vinifera), rice (Oryza sativa), maize, and moss (Physcomitrella patens; Fig. 2B).
Figure 2.
Sequence analysis of SEL1. A, Predicted motif structure of SEL1 protein. P or P-L-S block motifs are depicted as boxes with letters as previously proposed (Lurin et al., 2004); P, L, and S designate the PPR, PPR-like S, and PPR-like L motif, respectively. B, Amino acid sequence alignment of SEL1. The amino acid sequence of SEL1 was compared with the homologous proteins in other species using ClustalW 2.0 (Larkin et al., 2007). The putative cleavage site of the transit peptide in SEL1 is shown by an arrow. Lines above the sequences show the predicted P- or PPR-like (S and L) motifs. Black boxes indicate amino acid residues that are greater than 80% conserved, and gray boxes indicate amino acids that are greater than 60% conserved. The amino acid sequences are NP_193587.4 for Arabidopsis, XP_002284293.1 for grape, NP_001173562.1 for rice, ACG29369.1 for maize, and XP_001757146.1 for moss.
SEL1 Is Localized in the Chloroplast
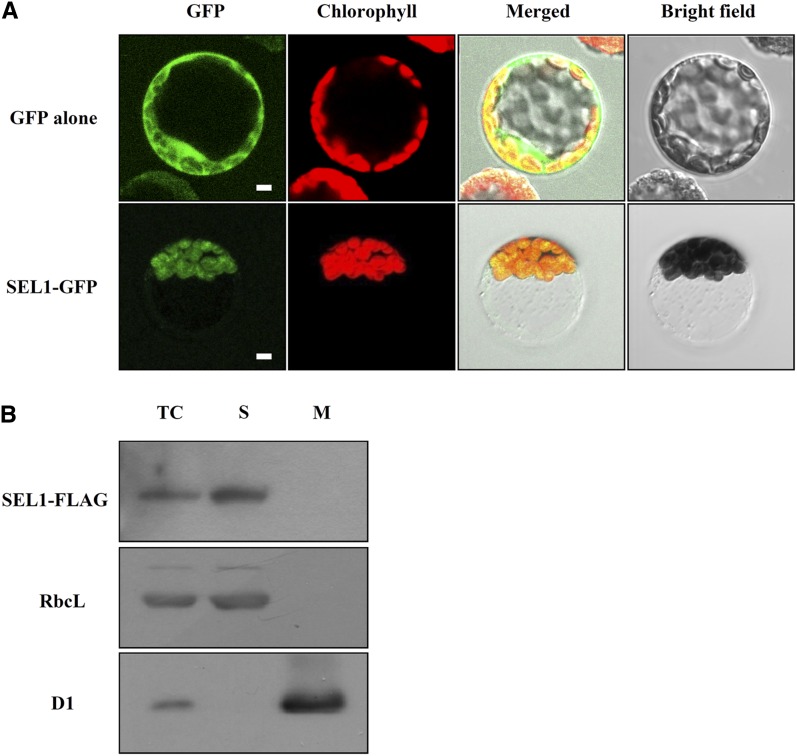
Many PPR proteins are predicted to be localized in the chloroplasts and mitochondria (Lurin et al., 2004). The ChloroP program predicted that the SEL1 protein is targeted to the chloroplast, and the N-terminal 19 amino acids of the SEL1 protein are a putative transit peptide to the chloroplast (Fig. 2B). To determine the subcellular localization of SEL1 protein, we made transgenic plants expressing full-length SEL1-GFP fusion protein under the control of a Cauliflower mosaic virus 35S promoter. When protoplasts were isolated and examined by confocal laser scanning microscopy, we observed the green fluorescence derived from the SEL1-GFP fusion protein exclusively in the chloroplasts (Fig. 3A). This green fluorescence was colocalized with the chlorophyll autofluorescence. Thus, this result indicates that SEL1 protein is localized and functions in the chloroplasts.
Figure 3.
SEL1 is localized in the chloroplasts. A, Subcellular localization of SEL1-GFP. Protoplasts were isolated from 35S::GFP and 35S::SEL1:GFP transgenic plants. The fluorescence of GFP and SEL1-GFP fusion protein in protoplasts was observed by confocal laser scanning microscope. Green fluorescence signals, chlorophyll red autofluorescence signals, merged images, and bright-field images are shown. Bar = 5 μm. B, Western-blot analysis of SEL1-FLAG fusion protein in chloroplast subfractions. Chloroplasts from pSEL1::SEL1:FLAG transgenic plants were purified and separated into soluble and membrane fractions by ultracentrifugation. The same proportion of each fraction was separated by SDS-PAGE and transferred to the nitrocellulose membrane. The blot was probed sequentially with antibody to FLAG and specific antibodies to RbcL and D1 as controls for the fractionation of soluble proteins and membrane proteins, respectively. TC, Total chloroplast protein; S, soluble protein; M, membrane protein. [See online article for color version of this figure.]
To further investigate the SEL1 protein localization in the chloroplasts, we examined the FLAG-tagged SEL1 protein in the chloroplast subfractions using western-blot analysis. As shown in Figure 3B, FLAG-tagged SEL1 protein was detected only in the soluble fraction but not in the membrane fraction of the chloroplasts. Therefore, this result indicates that SEL1 is a soluble protein located in the chloroplasts.
SEL1 Protein Is Part of Small and High Molecular Protein Complexes
Some PPR proteins are part of a high molecular protein complex that contains RNA molecules (Williams and Barkan, 2003; Schmitz-Linneweber et al., 2006; Beick et al., 2008). To determine whether SEL1 is a component of the RNA-protein complex in vivo, we examined the position of FLAG-tagged SEL1 protein in soluble fractions using sedimentation through a Suc gradient and western-blot analysis (Supplemental Fig. S3). FLAG-tagged SEL1 protein was broadly distributed in Suc gradient fractions and found predominantly in small Mr complexes, which are smaller than Rubisco, although small fractions of FLAG-tagged SEL1 protein were found in high Mr complexes that are larger than Rubisco. Ribonuclease A treatment prior to sedimentation leads to a reduced level of FLAG-tagged SEL1 protein in all fractions. These results suggest that SEL1 might be a part of a protein complex that is associated with RNA.
SEL1 Is Specifically Expressed in Green Aerial Tissues
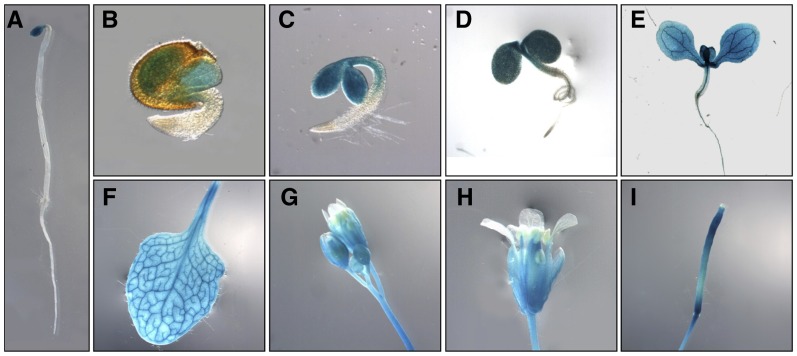
To investigate the expression pattern of the SEL1 gene in planta, we made transgenic plants expressing the GUS protein under the control of the SEL1 promoter and examined the GUS expression driven by the SEL1 promoter (Fig. 4). In germinating seeds and seedlings, GUS staining was observed in cotyledons but not in roots (Fig. 4, B–E). In adult plants, the GUS activity was strongly detected in rosette leaves, cauline leaves, stems, flowers, and siliques (Fig. 4, F–I). In flowers, GUS staining was observed exclusively in green tissues, such as sepals, stamens, and carpels, but not in petals (Fig. 4, G and H). GUS staining was also detected in all of the siliques but not in the stigma (Fig. 4I). Interestingly, GUS staining was also detected in the cotyledons of 3-d-old etiolated seedlings that were grown under dark (Fig. 4A). Therefore, these results indicate that SEL1 is specifically expressed in green aerial tissues and functions under light as well as dark conditions.
Figure 4.
Expression patterns of SEL1. GUS expression was analyzed in pSEL1::GUS transgenic plants. A, Three-day-old etiolated seedling. B, Germinating seed. C, Three-day-old seedling. D, Five-day-old seedling. E, Seven-day-old seedling. F, Rosette leaf. G, Flower buds. H, Flower. I, Silique. [See online article for color version of this figure.]
sel1 Mutants Have Defects Affecting the Development of Chloroplasts and Etioplasts
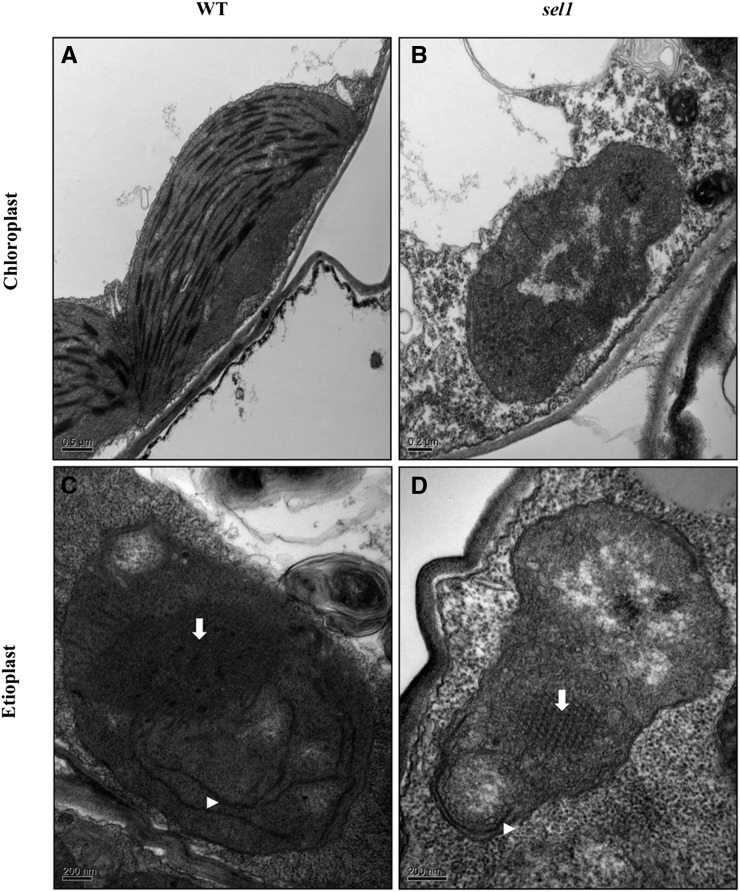
Because the sel1 mutant is pigment defective and SEL1 protein is localized in the chloroplasts, we examined the chloroplast morphology and ultrastructure in sel1 mutant by transmission electron microscopy (TEM). Under light conditions, chloroplasts in the wild type were lens shaped and contained well-organized thylakoid membrane systems composed of stromal and granal thylakoids (Fig. 5A). However, the plastids in the sel1 mutants were small and abnormally shaped, including round and amoeboid, and did not have thylakoid membranes or any membrane structures but contained a large number of small vesicles (Fig. 5B). In addition, densely stained globule aggregates were frequently observed in sel1 mutants.
Figure 5.
Ultrastructure of chloroplasts and etioplasts in sel1 mutant. A and B, Chloroplasts are from 7-d-old light-grown cotyledons of the wild type (WT) and sel1 mutant. C and D, Etioplasts are from 7-d-old dark-grown cotyledons of the wild type and sel1 mutant. Prolamellar body and prothylakoid are indicated by arrows and triangles, respectively.
Because SEL1 is expressed in the cotyledons of etiolated seedlings, we also examined the etioplasts ultrastructure. Etioplasts are defined by the presence of the paracrystalline prolamellar body (PLB). Under dark conditions, etioplasts in the wild type showed the typical structures, i.e. spherical or ellipsoidal shape enclosing PLBs and prothylakoid membranes (Fig. 5C). However, plastids in sel1 mutant did not have well-developed PLBs or prothylakoid membranes (Fig. 5D). Some plastids in sel1 mutant showed a PLB-like but disorganized structure. Taken together, these results indicate that the sel1 mutant is impaired in the development of chloroplast, as well as etioplasts, suggesting that SEL1 functions in the early stage of chloroplast development.
sel1 Mutant Fails to Accumulate Photosynthetic Protein Complexes
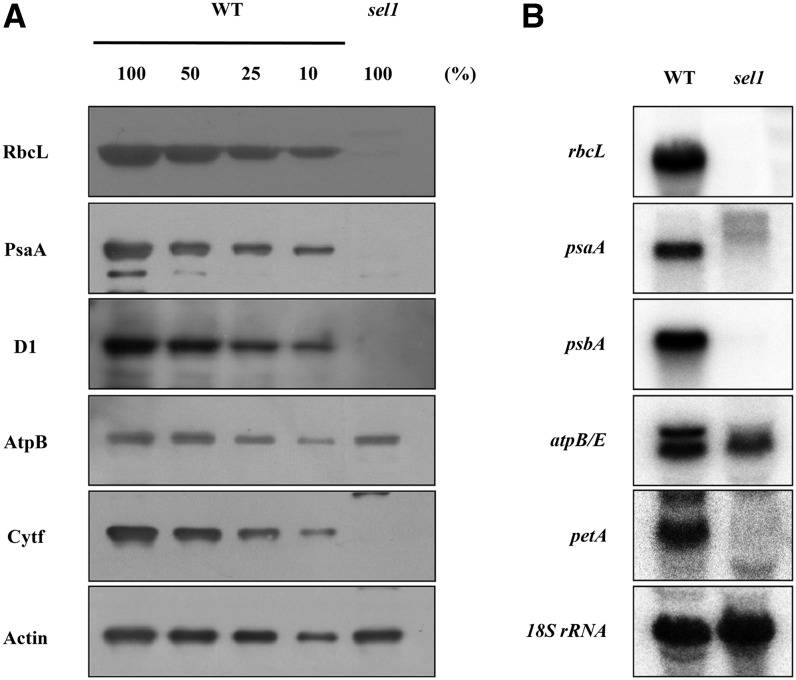
To compare the accumulation levels of chloroplast proteins of sel1 mutant with those of the wild type, protein amounts were evaluated comparably using western-blot analysis. As shown in Figure 6A, the sel1 mutant did not have detectable levels of subunits of photosynthetic protein complexes corresponding to the products of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL), PSI P700 chlorophyll a apoprotein A1 (psbA), PSII protein D1 (psbA), and cytochrome f (petA) genes. However, ATP synthase CF1 beta subunit (AtpB) was slightly reduced in the sel1 mutant when compared with the wild type. The reduction of plastid-encoded proteins could be due to the impaired accumulation of plastid transcripts. To determine whether reduced protein levels in sel1 mutants result from transcriptional defects in the expression of the plastid genes, we examined the expressions of corresponding genes encoding subunits of the photosynthetic protein complex in sel1 mutants by northern-blot analysis (Fig. 6B). Compared with the wild type, the levels of rbcL, psaA, psbA, and petA transcripts were not detected in sel1 mutants. However, transcript pattern of atpB gene in sel1 mutant were different from those in the wild type. While rbcL, psaA, psbA, and petA genes belong to class I plastid genes that are transcribed preferentially by PEP, the atpB gene is a class II plastid gene, which is transcribed by both NEP and PEP (Hajdukiewicz et al., 1997). In the wild type, there are two major atpB transcripts, in which 2.6- and 2-kb transcripts are transcribed by PEP and NEP, respectively (Loschelder et al., 2006; Schweer et al., 2006). However, in the sel1 mutant, the PEP-dependent transcript is strongly reduced, and the NEP-dependent transcript is not changed. In addition, the two long transcripts, which do not exist in the wild type, are accumulated in the sel1 mutant (Supplemental Fig. S4). This pattern of atpB transcripts in the sel1 mutant is similar to that observed in PEP-defective mutants such as clb19 and sigma factor6 mutants (Ishizaki et al., 2005; Chateigner-Boutin et al., 2008). This result suggests that the mutation of SEL1 may affect PEP activity but not NEP activity. Taken together, the deficiencies of the photosynthetic protein complexes in the sel1 mutant are caused by a defect in the transcription of corresponding plastid genes encoding subunits for photosynthetic protein complexes.
Figure 6.
Analysis of photosynthetic protein complexes in the sel1 mutant. A, Accumulation of representative subunits of photosynthetic protein complexes determined by western-blot analysis with specific antibodies to proteins indicated to the left; RbcL is the large subunit of Rubisco. PsaA and D1 are subunits of PSI and PSII, respectively. AtpB and Cytf (for cytochrome subunit f) are subunits of ATP synthase and cytochrome b6f complex, respectively. Total proteins (10 μg or the indicated dilution of the wild-type sample) from 7-d-old seedlings were loaded per lane. Actin was used as a loading control. B, Northern-blot analysis for the accumulation of photosynthesis-related transcripts. Five micrograms of total RNA was isolated from 7-d-old seedlings of the wild type (WT) and sel1 mutant and hybridized with the gene-specific probes indicated. 18S rRNA was used as a loading control.
sel1 Mutants Have Global Defects in Plastid Gene Expression
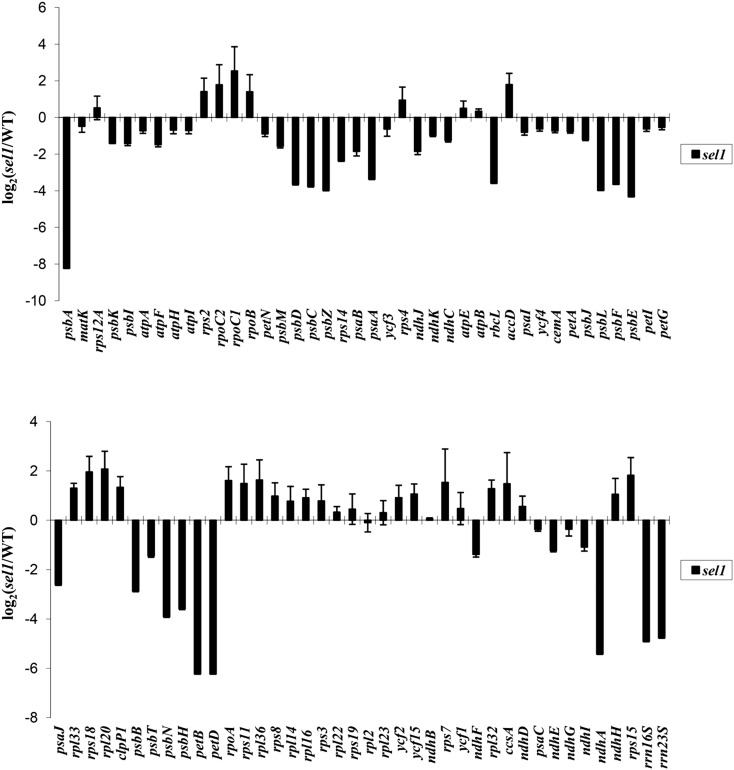
To investigate whether the SEL1 mutation specifically affects transcription by PEP but not NEP, we examined the transcript abundance of various plastid genes in the sel1 mutant by quantitative reverse transcription (qRT)-PCR analysis. As shown in Figure 7, there were significant differences in plastid gene expression between the wild type and sel1 mutant. Compared with the wild type, the expressions of the plastid genes that are transcribed by PEP, including the psbA, psbB, psbD, psbE, petB, ndhA, and rbcL genes, were strongly reduced in the sel1 mutant. In addition, plastid ribosomal RNAs (rRNAs) genes (rrn16s [for16S ribosomal RNA] and rrn23s), which are transcribed by both NEP and PEP, were also significantly reduced in sel1 mutant. However, transcript levels of the plastid genes, including the accD, ribosomal protein L33 (rpl33), rps18, hypothetical chloroplast open reading frames2, and rpo genes, which are transcribed by NEP, were increased or unchanged in sel1 mutant (Fig. 7). These results indicate that SEL1 is required for proper expression of plastid genes.
Figure 7.
RNA levels of plastid-encoded genes in sel1 mutant. The transcript abundance of protein-encoding genes and rRNAs of the plastid genome were measured from the wild type (WT) and sel1 mutant by quantitative RT-PCR. The graph shows the log2 ratio of transcript levels in the sel1 mutant compared with levels in the wild type. The genes are sorted according to physical location on the plastid genome. Error bars indicate sd.
sel1 Mutant Is Defective in RNA Editing of accD Transcripts
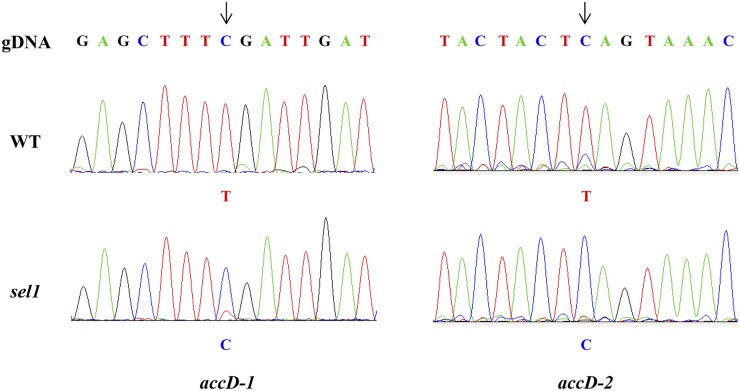
The impairment in RNA editing has been reported in some PPR mutants, such as clb19, ys1, atecb2, and vanilla cream1 (vac1), leading to PEP-defective transcription (Chateigner-Boutin et al., 2008; Yu et al., 2009; Zhou et al., 2009; Tseng et al., 2010). These mutants showed the similar pigment-defective and seedling-lethal phenotype to sel1 mutants. To examine whether the SEL1 functions in RNA editing, we tested the 34 RNA editing sites of plastid transcripts in the sel1 mutant using high-resolution melting (HRM) analysis (Chateigner-Boutin and Small, 2007). The sel1 mutant shows altered editing extent at most of RNA editing sites compared with the wild type, although they were edited correctly, as in the wild type (Supplemental Fig. S5). Interestingly, among 34 RNA editing sites known to be edited in the wild-type chloroplast, the RNA editing of accD-2 was severely impaired in the sel1 mutant, indicating that SEL1, as a member of the PLS subgroup, might be required for RNA editing of accD-2 site in Arabidopsis. To confirm this defect, we examined the RNA editing of accD transcript by direct sequencing of reverse transcription (RT) product and identified defects of RNA editing of accD-1 as well as accD-2 in the sel1 mutant (Fig. 8). The RNA editing of accD-1 is significantly reduced and that of accD-2 is completely abolished in the sel1 mutant. We also evaluated the editing rates of these two editing sites in the wild type and sel1 mutant. The editing rates of the two sites in the wild type were 100% and 91%, respectively, whereas they were 16% and 0%, respectively, in the sel1 mutant (Table I). Furthermore, in the sel1 mutant, RNA editing of some plastid transcripts (ATP-dependent Clp protease proteolytic subunit, rpoA, rpoB, rpoC1, and rps14) is mildly enhanced relative to the wild type (Supplemental Fig. S6). Taken together, these results suggest that SEL1 might be required for RNA editing of plastid transcripts, including most notably accD transcripts.
Figure 8.
The sel1 mutant is defective in RNA editing of accD transcripts. Sequence analysis for the accD transcripts from the wild type (WT) and sel1 mutant. Nucleotide sequences including the RNA editing sites of accD-1 and accD-2 are shown as chromatograms. Editing sites of accD-1 and accD-2 are indicated by arrows pointing to the corresponding peaks. [See online article for color version of this figure.]
Table I. RNA editing rate for the editing sites of accD transcripts in the wild type and sel1 mutant.
The editing rate was determined by analysis of approximately 25 independent clones from RT-PCR products including accD-1 and accD-2 in the wild type and sel1 mutant.
|
accD-1 |
accD-2 |
|||||
|---|---|---|---|---|---|---|
| Genotype | Unedited | Edited | Editing Rate | Unedited | Edited | Editing Rate |
| % | % | |||||
| Wild type | 0 | 24 | 100 | 2 | 20 | 91 |
| sel1 | 21 | 4 | 16 | 25 | 0 | 0 |
Plastid Ribosomes Are Functional But Reduced in Amount in the sel1 Mutant
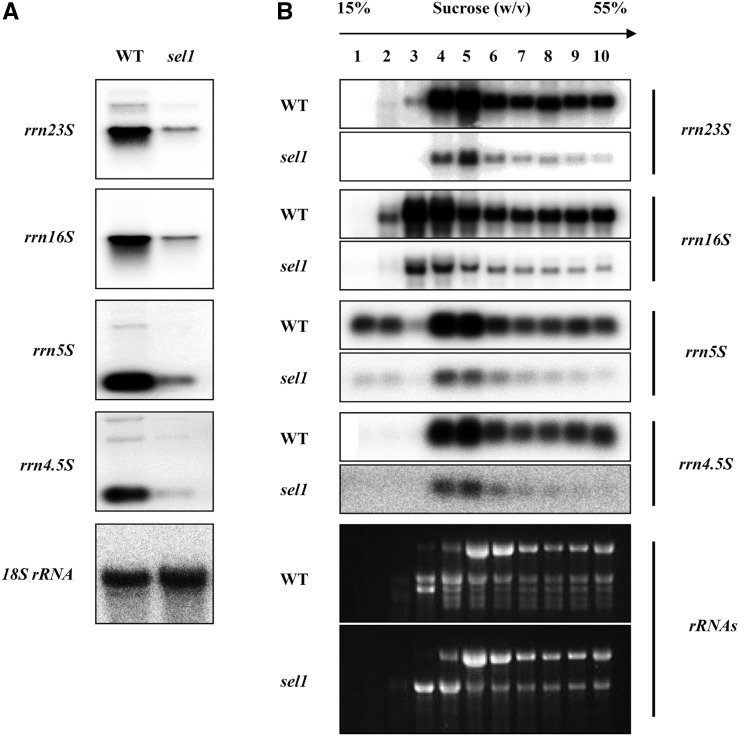
In mutants with defects in the plastid ribosome, pleiotropic effects in plastid gene expression have been reported, in which they could not translate plastid-encoded proteins, including PEP core subunits (Hess et al., 1993). Therefore, the reduced levels of PEP-dependent transcripts in the sel1 mutant might be caused by functional defects of the plastid ribosome. To ensure the link of the less PEP activity to sel1 mutant phenotype, the transcript levels and patterns of plastid rRNAs in sel1 mutant were investigated using northern-blot analysis (Fig. 9A). Compared with the wild type, the expression levels of all plastid rRNAs were significantly reduced in the sel1 mutant. However, the transcript patterns of plastid rRNAs in the sel1 mutant were similar to those of the wild type.
Figure 9.
Chloroplast rRNAs are significantly decreased in the sel1 mutant. A, Northern-blot analysis of chloroplast rRNAs in the wild type (WT) and sel1 mutant. One microgram of total RNA was isolated from 7-d-old seedlings and analyzed by hybridization to probe for the plastid 23S, 16S, 5S, and 4.5S rRNA. 18S rRNA was used as a loading control. B, Polysome analysis of the wild type and sel1 mutant. Total polysome isolation and RNA purification from 7-d-old seedlings were performed as described previously (Kwon and Cho, 2008). The ethidium bromide-stained gel represents equal proportional loading of the 10 gradient fractions.
Although the accumulation of plastid rRNAs is strongly reduced, the function of the plastid ribosome may not be impaired in the sel1 mutant. To test this possibility, we examined plastid rRNAs associated with the polysome in the sel1 mutant by polysomal RNA gel-blot analysis. As shown in Figure 9B, there was no difference in the distribution of plastid rRNAs between the wild type and sel1 mutant, although the amount of plastid rRNAs associating with polysomes was reduced in the sel1 mutant. Because the sel1 mutant contains significantly fewer plastid rRNAs than the wild type, this is likely a consequence of plastid rRNAs deficiency, which could be either due to a lack of functional PEP activity or to an abnormal rRNA metabolism. Likewise, the majority of the psaA, psbA, and rbcL transcripts were associated with polysomes in the wild type but were not detected in the sel1 mutant (Supplemental Fig. S7). These results indicate that in sel1 mutant, the functional plastid ribosomes exist, but the content of functional plastid ribosomes is significantly reduced.
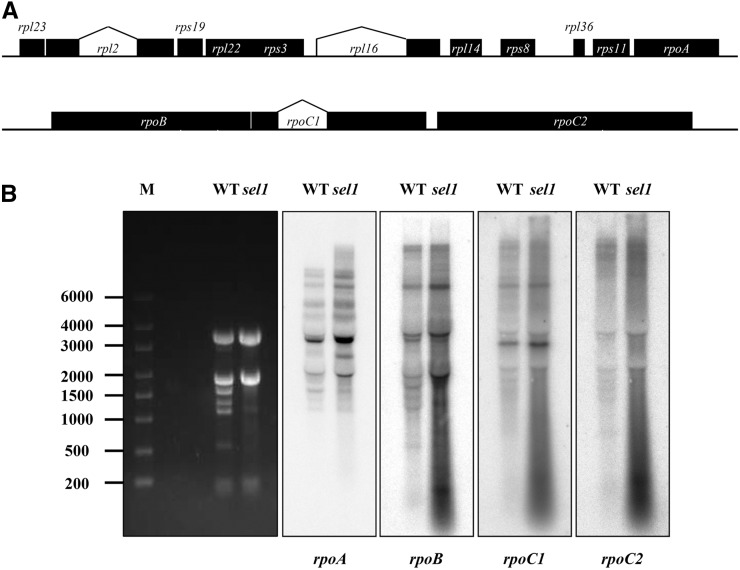
SEL1 Mutation Affects the Transcript Amount But Not the Transcript Pattern of rpo Genes
Because the visible and molecular phenotype of sel1 mutant is similar to those of mutants that have a defect in PEP function (Ishizaki et al., 2005; Pfalz et al., 2006; Chateigner-Boutin et al., 2008, 2011; Zhou et al., 2009; Gao et al., 2011; Steiner et al., 2011), it is likely that sel1 mutant is a PEP-defective mutant. PEP is composed of plastid-encoded core subunits and nuclear-encoded σ factors. The core subunits of the PEP are encoded by the plastid rpoA, rpoB, rpoC1, and rpoC2 genes, which are NEP-dependent genes (class III). In sel1 mutant, the transcript levels for rpo genes showed multiple times greater than those of the wild type (Fig. 7). To probe any possibility of abnormal rpo transcripts in sel1 mutant, the amount and pattern of rpo transcripts were compared with those of the wild type using northern-blot analysis. As shown in Figure 10A, rpoA is cotranscribed with several ribosomal protein genes as part of rpl23 operon, whereas rpoB, rpoC1, and rpoC2 are cotranscribed as part of rpoB operon. In sel1 mutant, the transcript accumulation for all rpo genes were significantly increased compared with the wild type, but the transcript patterns were similar in the wild type and sel1 mutant (Fig. 10B). This result indicates that SEL1 mutation causes a significant increase in the amount of rpo transcripts with no change of transcript patterns of rpo genes. To investigate whether the increased transcripts led to increase the corresponding proteins comparably, RpoB protein in sel1 mutant was compared with that of the wild type using western-blot analysis. The sel1 mutant showed similar level of RpoB to the wild type, though the rpoB transcript level of sel1 mutant is much higher than that of the wild type (Supplemental Fig. S8B). This result suggests that sel1 mutant may have comparable PEP level to the wild type, but PEP activity of sel1 mutant is not parallel to the wild type. Taken together, these results suggest that the mutation of SEL1 disturbed the PEP function, which decreased the overall level of functional rRNAs in plastids. Eventually, the nonoperational transcription and translation in sel1 mutant impeded the proper chloroplast biogenesis, resulting in seedling-lethal phenotype.
Figure 10.
Northern-blot analysis of the rpo genes encoding the PEP core subunits in the wild type (WT) and sel1 mutant. A, Schematic representation of chloroplast rpl23 and rpoB operons. rpoA is the last gene of the rpl23 operon. rpoB, rpoC1, and rpoC2 are part of the rpoB operon. The bent lines indicate introns. B, Five micrograms of total RNA from 7-d-old seedlings was analyzed by hybridization to the probe for rpoA, rpoB, rpoC1, and rpoC2. The size marker (M) is a RNA ladder, and ethidium bromide staining is shown as loading control.
DISCUSSION
SEL1 Gene Is Essential for Early Chloroplast Development
It has been known that a large portion of the seedling-lethal mutants is defective in chloroplast function or development, suggesting that the development of functional chloroplasts is tightly coordinated with plant growth and development (Meinke et al., 2003, 2008; Waters and Langdale, 2009). The sel1 mutants show seedling-lethal phenotype accompanied with white cotyledons under autotrophic conditions; however, on Suc-supplemented medium, primary leaves of sel1 mutants show pale yellow color (Fig. 1A; Supplemental Fig. S1). This ivory phenotype implicates that the plastids are still active and able to synthesize carotenoids (Steiner et al., 2011). In sel1 mutants, chlorophyll content was strongly reduced, while carotenoids content was moderately reduced (Supplemental Fig. S2). The visible phenotype of sel1 mutants and complementation analysis with the SEL1 gene under the control of its own promoter indicate that the disruption of the SEL1 causes the pigment-defective and seedling-lethal phenotype and that SEL1 is essential for normal plant growth and development.
Electron microscopy analysis revealed that the pigment-defective and seedling-lethal phenotypes of sel1 mutants resulted from defects of chloroplast development. In the sel1 mutant, the ultrastructure of the chloroplasts exhibited abnormal morphology and had no thylakoid membrane structure (Fig. 5B). Interestingly, etioplasts of sel1 mutants also showed abnormal morphology and failed to develop normal prolamellar body (Fig. 5D). Similar phenotypes were observed in previous studies related to PEP-associated proteins (PAPs) and plastid transcriptionally active chromosome proteins (pTACs; Pfalz et al., 2006; Steiner et al., 2011). Homozygote pap or ptac knockout mutants, such as pap5/ptac12 and pap8/ptac6 (Pfalz et al., 2006), pap7/ptac14 (Gao et al., 2011; Steiner et al., 2011), pap4/fad3 and pap9/fsd2 (Myouga et al., 2008), pap6/fln1 (Arsova et al., 2010; Steiner et al., 2011), pap10/trxz (Arsova et al., 2010), and pap11/atmurE (Garcia et al., 2008) mutants, showed white cotyledon and yellowish primary leaves on Suc-containing medium, while they are lethal when grown with no supply of an exogenous carbon source. Chloroplasts of these mutants do not contain thylakoid membrane structure, which is replaced by oval-shaped vesicles. Therefore, sel1 mutants are very similar to pap or ptac mutants regarding chloroplast ultrastructure as well as visible phenotype. These results suggest that SEL1 plays crucial role in early stage of chloroplast development. In agreement with an essential function in chloroplast development, SEL1 is localized in chloroplasts (Fig. 3) and the SEL1 gene is expressed only in green tissues, which was shown in the SEL1 promoter-GUS transgenic plant analysis (Fig. 4).
SEL1 Is Required for RNA Editing of the accD Transcripts
PPR proteins have been shown to be involved in the RNA editing of the chloroplasts or mitochondria (Kotera et al., 2005; Okuda et al., 2006, 2007; Chateigner-Boutin et al., 2008; Cai et al., 2009; Hammani et al., 2009; Kim et al., 2009; Okuda et al., 2009; Robbins et al., 2009; Yu et al., 2009; Zehrmann et al., 2009; Zhou et al., 2009). So far, all of these proteins belong to the E/E+ and DYW subgroup of the PPR protein family. In this study, we found that sel1 mutant is also defective in RNA editing of plastid transcripts, suggesting that SEL1 is required for the proper RNA editing process in chloroplast. Interestingly, among 34 known RNA editing sites in chloroplasts (Tillich et al., 2005; Chateigner-Boutin and Small, 2007), the RNA editing of accD-2 (C1568) sites was completely abolished and that of the accD-1 (C794) is severely impaired in the sel1 mutant (Fig. 8; Supplemental Fig. S5). The RNA editing defects in sel1 mutant are similar to those in atecb2 and vac1 mutants, which are independent mutant alleles of AtECB2/VAC1 encoding a DYW subgroup PPR protein that is required for the editing of accD and ndhF transcripts (Yu et al., 2009; Tseng et al., 2010). However, it has been reported that the loss of RNA editing at the accD-1 (C794) site was not associated with a pigment-defective and seedling-lethal phenotype; because Protein Required for accD RNA Editing1 mutant showed wild type-like phenotype, even the RNA editing of the accD-1 (C794) site is completely abolished (Robbins et al., 2009). Therefore, the editing defect in the sel1 mutant is unlikely to be a primary effect by the loss of SEL1 protein. The decreased editing extent is probably caused by the greatly increased level of accD transcripts in sel1 mutant as shown in Figure 7 and Supplemental Figure S8A. Too many transcripts may saturate the editing activity and consequently leads to lower editing extent as shown in previous studies on the relation between the transcript abundance and the level of editing extent (Lu and Hanson, 1992). However, we cannot exclude the possibility that SEL1 is involved in RNA editing process of accD transcript, because the editing events on the ATP-dependent Clp protease proteolytic subunit, rpoA, rpoB, rpoC1, and rps14 transcripts are not significantly affected (Supplemental Fig. S5), although they are highly expressed in the sel1 mutant compared with the wild type (Figs. 7 and 10).
SEL1 Plays an Important Role in the Regulation of Plastid Gene Expression
Analysis of plastid gene expression revealed that, in sel1 mutants, the transcript levels of PEP-dependent genes were dramatically reduced, whereas those of NEP-dependent genes were increased or unchanged (Fig. 7), suggesting that sel1 mutant is severely impaired in PEP activity and SEL1 plays an important role in the regulation of plastid gene expression. Thus, it seems likely that the early arrest of chloroplast development in sel1 mutants is due to the reduced PEP activity. Although the sel1 mutant shows many similarities in molecular phenotypes, including transcript pattern and accumulation, and expression profiles of plastid genes to other PEP-related mutants (Pfalz et al., 2006; Chateigner-Boutin et al., 2008, 2011; Zhou et al., 2009; Steiner et al., 2011), the molecular function of SEL1 currently remains elusive. However, it is obvious that SEL1 is required for the proper expression of plastid genes. From the recent study, maize ortholog of SEL1 has been identified in nucleoid-enriched protein fractions (Majeran et al., 2012). Thus, it is possible that SEL1 might be involved in the control of PEP activity as a component of nucleoid proteins like pTACs or PAPs, and required for the proper plastid gene expression and normal chloroplast development. In addition to the global reduction of PEP-dependent transcripts, several lines of evidence suggest that sel1 mutant is defective in PEP activity. When we investigated the accumulation of plastid-encoded proteins in sel1 mutants, the accumulation of Rubisco and thylakoid membrane protein complexes, except for ATP synthase, was severely decreased or not detected in the sel1 mutant due to the lack of corresponding transcripts (Fig. 6A). The amount of PEP-dependent transcript of atpB decreased (upper band, approximately 2.6 kb) in the RNA gel blot (Fig. 6B; Supplemental Fig. S4), even though the level of AtpB protein did not change significantly. As seen in Figure 5B, the existence of mitochondria was observed in sel1 mutant. Thereby, the amount of AtpB protein could be overestimated due to the cross reactivity of antibody used against the mitochondrial ATPase (Kwon and Cho, 2008). The amount of NEP-dependent transcript of atpB is not significantly altered and novel 4.8-kb transcript, which is not detected in the wild type, accumulated in sel1 mutant (Fig. 6B; Supplemental Fig. S4). A similar transcription pattern was also observed in the sigma factor6 (Loschelder et al., 2006; Schweer et al., 2006) and other PEP-defective mutants, such as ptac2/ptac6/ptac12/ptac14 (Pfalz et al., 2006; Gao et al., 2011), clb19 (Chateigner-Boutin et al., 2008), delayed green1 (Chi et al., 2008), and ys1 (Zhou et al., 2009).
In terms of the molecular phenotypes, sel1 mutants show the similarity to those of mutants with defects in plastid ribosome as well (Bang et al., 2012). In ribosome-deficient mutants, the expression of PEP-dependent genes is decreased, whereas that of NEP-dependent genes is increased. In sel1 mutant, steady-state levels of all plastid rRNAs were significantly decreased (Fig. 9). This may lead to a global defect in the expression of plastid genes due to the lack of functional plastid ribosomes, which causes the level of PEP to be much less than required to synthesize plastid proteins controlled by PEP. In sel1 mutant, protein levels of AccD and RpoB are not significantly changed, even though their transcripts are highly up-regulated (Figs. 7 and 10B; Supplemental Fig. S8). Thus, reduced translation of accD and rpoB could be a consequence of the decreased level of plastid rRNAs. Global defects in plastid gene expression in sel1 mutants are similar to those of plastid ribosome-deficient mutants, suggesting that the SEL1 may be involved in the regulation of plastid ribosome assembly or function. So, the loss of SEL1 leads to overall declines of all PEP-dependent transcriptions caused by the reduced translation of PEP. However, we could not rule out the possibility that the reduction in the plastid ribosome is also caused by defects of PEP activity.
In sel1 mutant, the expression of PEP-dependent genes is decreased, whereas that of NEP-dependent genes is increased. This increase in NEP-dependent transcripts in sel1 can be indirect and possibly explained by a recent study on the division of labor between PEP and NEP during plastid development and in mature chloroplasts using the primary transcriptome analysis in barley (Hordeum vulgare; Zhelyazkova et al., 2012). In this study, authors found that the extremely high number of promoters, at least 222 NEP promoters, can be used in white leaves in the absence of PEP and that NEP is able to generate virtually the same mRNAs as PEP in several genes, suggesting that activation of transcription by NEP can be a compensatory mechanism for the absence of PEP in higher plants.
In this work, we report a novel pigment-defective and seedling-lethal mutant, sel1 mutant, that disrupted genes encoding a member of the PLS subgroup of the PPR protein family. Our results demonstrate that SEL1 is required for the proper plastid gene expression and thus essential for normal chloroplast development. To discover the more detailed molecular function of SEL1, the identification of target RNA and interacting proteins of SEL1 in the future should be helpful to provide the insight into its function in plastid gene expression and chloroplast development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds were surface sterilized and incubated at 4°C for 3 d to synchronize germination and plated on MS medium (Duchefa Biochemie) containing 1% (w/v) Suc and 0.7% (w/v) agar in a growth chamber at 22°C under 16-h-light/8-h-dark condition. Plants were grown in soil (Sunshine Mix #1; SunGro) in a growth room at 22°C under continuous light. The sel1-1 mutant was isolated from T-DNA insertion mutant lines, which are generated in Ws-2 background with simple T-DNA vector pDAP101 (Sessions et al., 2002), whereas seeds of sel1-2 (SAIL_793_F11) and sel1-3 (SALK_054374) were obtained from the Arabidopsis Biological Resource Center.
Complementation Analysis
For the complementation test of the sel1 mutant, the 2,629-bp wild-type genomic DNA fragment of the At4g18520 gene, containing 645 bp of a putative promoter sequence and 130 bp of 3′ untranslated region, was amplified using Pfu-X polymerase (SolGent) and gene-specific primers. After verification by sequencing, the amplified fragment was cloned into a pBIN19 binary vector and subsequently introduced into heterozygous sel1-3 mutant plants using the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). For the generation of FLAG-tagged SEL1 complementation line, DNA encoding three copies of FLAG was amplified and cloned into pCB308 binary vector (Xiang et al., 1999), producing pCB308-FLAG vector. The SEL1 genomic DNA without stop codon, including 645-bp upstream region from ATG, was amplified and cloned into pCB308-FLAG vector and subsequently introduced into heterozygous sel1-2 mutant plants.
Northern-Blot Analysis
Total RNA was extracted from 7-d-old seedlings using the easy-BLUE Total RNA Extraction Kit (iNtRON Biotechnology), separated on MOPS-formaldehyde agarose gel, and transferred to a nitrocellulose membrane (Whatman). The 32P-labeled probes were prepared using a DecaLabel DNA Labeling Kit (Fermentas). Prehybridization, hybridization, washes, and signal detection were performed as described previously (Kwon and Cho, 2008).
Subcellular Localization of GFP Fusion Proteins in Protoplasts
Analysis of the subcellular localization of GFP fusion proteins in protoplasts was performed as described previously (Kwon and Cho, 2008).
Chloroplast Isolation and Fractionation
Chloroplasts were isolated and separated into soluble and membrane fractions as described previously (Kwon and Cho, 2008).
Histochemical GUS Staining
Tissues were incubated in cold 90% (v/v) acetone and placed in a staining buffer (50 mm sodium phosphate buffer, pH 7.2, 0.2% Triton-X, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, and 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid). After a vacuum infiltration, samples were incubated overnight at 37°C. GUS-stained tissues were cleared with an ethanol series and fixed in formaldehyde-acetic acid solution (50% [v/v] ethanol, 10% [v/v] acetic acid, and 5% [v/v] formaldehyde). Following fixation, samples were stored in 70% (v/v) ethanol and examined under the dissecting microscope.
TEM Analysis
TEM analysis was performed as described previously (Kwon and Cho, 2008), with minor modification. For analysis of the etioplast ultrastructure, wild-type and sel1 mutant seedlings were grown on MS agar plates in the dark for 7 d. Under dim green light, cotyledons of the wild type were directly detached from seedlings and then immediately soaked in fixation solution. For the sel1 mutant, cotyledons were detached and soaked in numbered 1.5-mL tubes containing fixation solution. The genotype of the sample was determined by PCR using the remaining part of the seedling. The detached cotyledons in the corresponding tubes confirmed that homozygous sel1 mutant was used for TEM analysis.
Western-Blot Analysis
Total proteins were isolated from 7-d-old seedlings as described previously (Barkan, 1998), separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, immunoblotted with various antibodies, and detected using the enhanced chemiluminescence kit (Santa Cruz Biotechnology). Antibodies were obtained from Agrisera (RbcL, PsaA, D1 protein of PSII, AtpB, and cytochrome subunit f) and from Uniplastomic (AccD and RpoB).
Analysis of Plastid Transcript Abundance
Total RNA was extracted from 7-d-old wild-type and sel1 mutant seedlings by using the NucleoSpin RNA Plant Kit (Macherey-Nagel). DNA-free RNA (2 μg) was then reverse transcribed with random hexamers by using a RevertAid First-Strand cDNA Synthesis Kit (Fermentas). Quantitative real-time PCR was carried out in an IQ5 Light Cycler (Bio-Rad) using SYBR Premix Ex Taq II (Takara). The relative quantification of gene expression data were analyzed using the IQ5 Optical System Software (Bio-Rad). The data set was normalized using cytosolic 18S rRNA as a reference and setting the median value to 0. The primer sequences used in quantitative RT-PCR analysis are described in Okuda et al. (2009).
Analysis of RNA Editing
HRM analysis was performed as previously described (Chateigner-Boutin and Small, 2007; Okuda et al., 2009). Genomic DNA for HRM analysis was isolated with plant I genomic DNA (iNtRON Biotechnology). RNA isolation and RT were performed as above. The sequence including the accD RNA editing sites (accD-1 and accD-2) from the wild type and sel1 mutant was amplified by PCR with gene-specific primer pairs, and then the RT-PCR products were sequenced directly or cloned into a T-blunt vector using a T-blunt PCR Cloning Kit (SolGent). Plasmids prepared from 25 independent colonies of each sample were sequenced to determine the RNA editing efficiency of accD-1 and accD-2.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At4g18520.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S2. The content of chlorophyll and carotenoids in sel1 mutant.
Supplemental Figure S3. Sucrose gradient sedimentation analysis of SEL1-containing particles in chloroplast.
Supplemental Figure S4. The transcript accumulation and pattern of atpB gene in sel1 mutant.
Supplemental Figure S5. High-resolution melting analysis of plastid RNA editing in sel1 mutant.
Supplemental Figure S6. RNA editing of plastid transcripts in sel1 mutant.
Supplemental Figure S7. Polysome association of photosynthesis-related mRNAs in sel1 mutant.
Supplemental FigureS8. Analysis of the plastid-encoded genes in sel1 mutant.
Supplementary Material
Acknowledgments
We thank Allen Sessions (Syngenta Biotechnology, Torrey Mesa Research Institute) and Syngenta Biotechnology for providing the pDAP101 vector.
Glossary
- NEP
nuclear-encoded RNA polymerase
- PEP
plastid-encoded RNA polymerase
- MS
Murashige and Skoog
- T-DNA
transfer DNA
- TEM
transmission electron microscopy
- PLB
paracrystalline prolamellar body
- RT
reverse transcription
- rRNA
ribosomal RNA
- HRM
high-resolution melting
- Ws-2
ecotype Wassilewskija-2
- qRT
quantitative reverse transcription
References
- Allison LA, Simon LD, Maliga P. (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg S, Boudet N, Kreis M, Lecharny A. (2000) In Arabidopsis thaliana, 1% of the genome codes for a novel protein family unique to plants. Plant Mol Biol 42: 603–613 [DOI] [PubMed] [Google Scholar]
- Bang WY, Chen J, Jeong IS, Kim SW, Kim CW, Jung HS, Lee KH, Kweon HS, Yoko I, Shiina T, et al (2012) Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J 71:122–134 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Photosynthesis. Molecular Biology of Energy Capture 297: 38–57 [Google Scholar]
- Barkan A, Goldschmidt-Clermont M. (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Elliott LE, Hanson MR. (2008) Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics 178: 1693–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, Lu C, Zhang L. (2009) LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol 150: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Okuda K, Peng L, Shikanai T. (2011) PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J 67: 318–327 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I. (2011) OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J 65: 532–542 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. (2007) A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res 35: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. (2008) The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I. (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN. (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53: 924–934 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A. (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277: 809–811 [DOI] [PubMed] [Google Scholar]
- Hess WR, Börner T (1999) Organellar RNA polymerases of higher plants. Int Rev Cytol 190: 1–59 [DOI] [PubMed] [Google Scholar]
- Hess WR, Prombona A, Fieder B, Subramanian AR, Börner T. (1993) Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J 12: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. (2005) A nuclear-encoded σ factor, Arabidopsis SIG6, recognizes σ-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Kim SR, Yang JI, Moon S, Ryu CH, An K, Kim KM, Yim J, An G. (2009) Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J 59: 738–749 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Kwon KC, Cho MH. (2008) Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J 55: 428–442 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T. (2011) The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. (2006) Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol 142: 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Hanson MR. (1992) A single nuclear gene specifies the abundance and extent of RNA editing of a plant mitochondrial transcript. Nucleic Acids Res 20: 5699–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. (2012) Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved Sucrose-dependent growth phenotype of sel1 mutant. in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Meinke LK, Showalter TC, Schissel AM, Mueller LA, Tzafrir I. (2003) A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol 131: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K. (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Meierhoff K, Westhoff P, Schuster G. (2003) RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur J Biochem 270: 4070–4081 [DOI] [PubMed] [Google Scholar]
- Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA 104: 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. (2006) A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem 281: 37661–37667 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivals E, Bruyère C, Toffano-Nioche C, Lecharny A. (2006) Formation of the Arabidopsis pentatricopeptide repeat family. Plant Physiol 141: 825–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR. (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15: 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2005) RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Loschelder H, Link G. (2006) A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett 580: 6617–6622 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2006) RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol Life Sci 63: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N. (2000) The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M, Liere K, Börner T. (2007) High diversity of plastidial promoters in Arabidopsis thaliana. Mol Genet Genomics 277: 725–734 [DOI] [PubMed] [Google Scholar]
- Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Tillich M, Funk HT, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, Maier RM. (2005) Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J 43: 708–715 [DOI] [PubMed] [Google Scholar]
- Tseng CC, Sung TY, Li YC, Hsu SJ, Lin CL, Hsieh MH. (2010) Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant. Plant Mol Biol 73: 309–323 [DOI] [PubMed] [Google Scholar]
- Waters MT, Langdale JA. (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PM, Barkan A. (2003) A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J 36: 675–686 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tasaka M, Shikanai T. (2004) PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J 38: 152–163 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. (2009) A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. (2012) The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang J. (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J 58: 82–96 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.