Biochemical, histological, and transcriptional characterization of lignification identifies substantial differences in two sugarcane genotypes.
Abstract
Sugarcane (Saccharum spp.) is currently one of the most efficient crops in the production of first-generation biofuels. However, the bagasse represents an additional abundant lignocellulosic resource that has the potential to increase the ethanol production per plant. To achieve a more efficient conversion of bagasse into ethanol, a better understanding of the main factors affecting biomass recalcitrance is needed. Because several studies have shown a negative effect of lignin on saccharification yield, the characterization of lignin biosynthesis, structure, and deposition in sugarcane is an important goal. Here, we present, to our knowledge, the first systematic study of lignin deposition during sugarcane stem development, using histological, biochemical, and transcriptional data derived from two sugarcane genotypes with contrasting lignin contents. Lignin amount and composition were determined in rind (outer) and pith (inner) tissues throughout stem development. In addition, the phenolic metabolome was analyzed by ultra-high-performance liquid chromatography-mass spectrometry, which allowed the identification of 35 compounds related to the phenylpropanoid pathway and monolignol biosynthesis. Furthermore, the Sugarcane EST Database was extensively surveyed to identify lignin biosynthetic gene homologs, and the expression of all identified genes during stem development was determined by quantitative reverse transcription-polymerase chain reaction. Our data provide, to our knowledge, the first in-depth characterization of lignin biosynthesis in sugarcane and form the baseline for the rational metabolic engineering of sugarcane feedstock for bioenergy purposes.
Sugarcane (Saccharum spp. hybrids) is an indigenous crop that originates from New Guinea (southeast Asia) and is grown in all tropical and subtropical regions of the world, on both sides of the equator (Lisboa et al., 2011). As a C4 plant, sugarcane is very efficient in converting solar radiation into biomass and also has the unique ability within the Poaceae family to accumulate up to 60% of its mature stem dry weight as Suc (Casu et al., 2004). At present, Brazil is the world’s largest sugarcane producer, with 612 million tons during the 2009/2010 crop season, from which 25 billion L of ethanol were generated (Cheavegatti-Gianotto et al., 2011). Due to the global interest in renewable energy, the production of sugar-based bioethanol has received increasing attention in recent years.
An emerging effort to increase the production of bioethanol from plants is based on the use of lignocellulosic biomass, resulting in the so-called second-generation biofuels (Dias et al., 2009). In tropical countries, sugarcane bagasse, the residue produced after Suc extraction, is the most plentiful lignocellulosic material. One ton of sugarcane generates 280 kg of bagasse, which is composed by 39% cellulose, 25% hemicelluloses, and 23% lignin, among other minor components (Carroll and Somerville, 2009; Rabelo et al., 2011). Currently, the remaining bagasse is used for the generation of heat and power for sugar processing into ethanol and also to produce electricity that is sold to the power grid (Carroll and Somerville, 2009). The production of second-generation ethanol could be advantageous for the sugarcane industry, as the cellulosic bioethanol could be coproduced by sharing part of the infrastructure that is used to produce the first-generation bioethanol (Dias et al., 2009, 2012; Rabelo et al., 2011).
Lignocellulose refers to plant biomass composed of the polymers cellulose, hemicellulose, and lignin. Cellulose is arranged in a tight network of microfibrils embedded in a matrix of hemicellulosic polysaccharides that are covalently cross linked with the heterogenous and complex structure of lignin (Vega-Sánchez and Ronald, 2010). The ability of lignin to resist degradation makes this phenolic polymer the major plant cell wall component responsible for biomass recalcitrance (Vanholme et al., 2010c; Van Acker et al., 2013). The fact that lignin is a nonlinear polymer built from different monomer units linked by chemically diverse and low-reactive bonds precludes the ability of any single enzyme to properly recognize and degrade it (Weng et al., 2008). Furthermore, lignin and some of its by-products, formed during the removal of lignin from biomass, can inhibit the enzymes that carry out the fermentation process, decreasing biofuel yield (Simmons et al., 2010). Consequently, the biomass-to-biofuel conversion process requires costly and harsh mechanical and chemical pretreatments to degrade lignin, to loosen up the rigid cell wall, and to provide access to the cell wall polysaccharides for saccharification (Vanholme et al., 2013a). In an attempt to facilitate the processing of plant biomass into biofuels, genetic engineering strategies have been targeted to produce plants that either deposit less lignin or that produce lignins that are more amenable to chemical degradation (Carroll and Somerville, 2009).
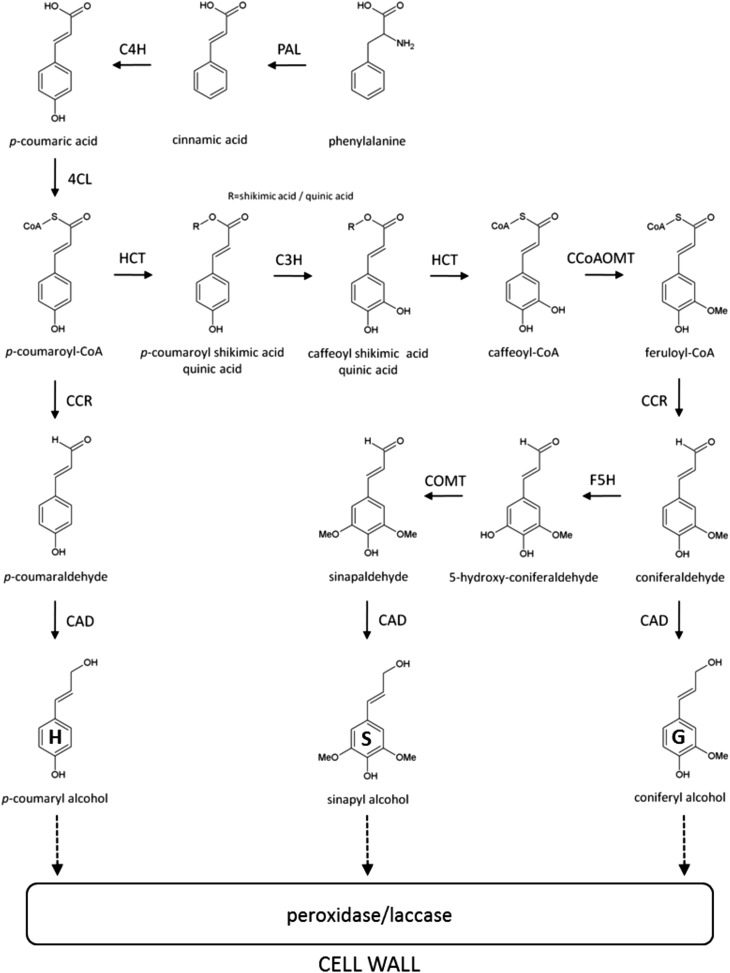
Lignins are complex aromatic heteropolymers produced by the oxidative combinatorial coupling of mainly three p-hydroxycinnamyl alcohol monomers differing in their degree of methoxylation, the p-coumaryl, coniferyl, and sinapyl alcohols (Fig. 1; Boerjan et al., 2003). When incorporated into the lignin polymer, these monomers are called p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively, and their individual contribution to lignin composition varies significantly among species, cell types, and tissues in the same plant (Bonawitz and Chapple, 2010). In addition to the three canonical monolignols, other phenylpropanoids can be incorporated into the polymer at varying levels, including hydroxycinnamates, hydroxycinnamyl aldehydes, and hydroxycinnamyl acetates (Boerjan, et al., 2003; Ralph et al., 2004). In general, the lignins from nonflowering vascular plants (i.e. ferns and gymnosperms) are composed mostly of G units with minor amounts of H units, whereas angiosperm dicotyledonous lignins are composed of G and S units and traces of H units. In monocots, both S and G units are present at similar levels and the amount of H units is higher than in dicots (Vanholme et al., 2010a).
Figure 1.
Schematic representation of the lignin biosynthetic pathway in plants. PAL, Phe ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; HCT, p-hydroxycinnamoyl-CoA:quinate shikimate p-hydroxycinnamoyl transferase; C3H, p-coumarate 3-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase.
The deposition of lignin is regulated in a spatiotemporal fashion and varies between primary and secondary cell walls and among tissues (Grabber et al., 2004). During the early stages of lignification, H and G units, but only a few S units, are incorporated into the polymer. Subsequently, coniferyl alcohol and increasing amounts of sinapyl alcohol are incorporated during secondary wall formation to form a mix of G and S units (Grabber, 2005). In grasses, significant amounts of the hydroxycinnamates ferulic acid and p-coumaric acid are also incorporated during secondary cell wall development and lignification (Vogel, 2008). While ferulic acid is mainly deposited in young grass cell walls, where it is esterified to arabinosyl residues of arabinoxylan chains, p-coumaric acid is esterified to the γ-position of the side chains of S units and, therefore, is considered as an indicator of cell wall maturity (Riboulet et al., 2009). Differential distribution of lignin units also occurs among distinct cell types. In most dicots, some secondary thickened cell walls, such as those in vessels, present in the xylem, typically have high levels of G units, whereas structural xylem fibers are rich in S units (Bonawitz and Chapple, 2010). In grasses, additional tissues of the stem, such as parenchyma, epidermis, and hypodermis, can become lignified, whereas xylary tissues and sclerenchyma are highly lignified and enriched in S lignins (Grabber et al., 2004). Finally, the deposition of lignin in plant cell walls is not only developmentally regulated but can also be affected by environmental conditions such as biotic and abiotic stresses (Moura et al., 2010).
The study of lignin biosynthesis and deposition in plant cell walls is one of the main foci in biofuel research, since several studies identified lignin as the main factor conferring recalcitrance of cell walls to saccharification in different plant species (Chen and Dixon, 2007; Van Acker et al., 2013). Despite the recent advances achieved with model plants such as Arabidopsis (Arabidopsis thaliana), poplar (Populus spp.), and maize (Zea mays), information on lignin metabolism in sugarcane is scarce, and only a few genes have been identified from large-scale transcript profiling studies (Ramos et al., 2001; Papini-Terzi et al., 2005). In this regard, a combination of coexpression analysis, tissue/cell type-specific expression analysis, and genetic complementation was recently used to correlate a laccase gene to the lignification process in sugarcane (Cesarino et al., 2013b).
Here, we present, to our knowledge, the first systematic study of lignin deposition in two genotypes from a sugarcane F1 forage breeding population contrasting for lignin content: IACSP04-529 (higher lignin content) and IACSP04-683 (lower lignin content). We have analyzed the deposition of lignin and its S/G composition throughout stalk development. In addition, we have used metabolite profiling to reveal phenolic metabolites and lignin oligomers that differentially accumulate in the same sample set. Furthermore, the Sugarcane EST Database (SUCEST) was extensively surveyed to identify phenylpropanoid and monolignol biosynthetic genes, and the expression of all identified genes was analyzed by quantitative PCR to pinpoint the most likely candidates to play a role in developmental lignin biosynthesis. Together, our data provide, to our knowledge, the first in-depth characterization of lignin biosynthesis in sugarcane.
RESULTS
A comprehensive study of lignin deposition and gene expression analysis was carried out in two sugarcane genotypes with contrasting lignin contents, named IACSP04-529 (with a relatively high lignin content) and IACSP04-683 (with a relatively low lignin content). Our choice for the two genotypes was based on a 3-year evaluation of agronomic traits (i.e. primarily Suc and fiber content) in an F1 population produced by the sugarcane breeding program of the Sugarcane Center of the Agronomic Institute of Campinas. Lignin of genotype IACSP04-683 averaged 4.56% for a 3-year evaluation (4.8%, 4.3%, and 4.7%), whereas that of genotype IACSP04-529 averaged 8.4% for a 3-year evaluation (9.2%, 8.0%, and 8.2%) on a dry weight basis.
Developmental Deposition of Lignin in Sugarcane Stems
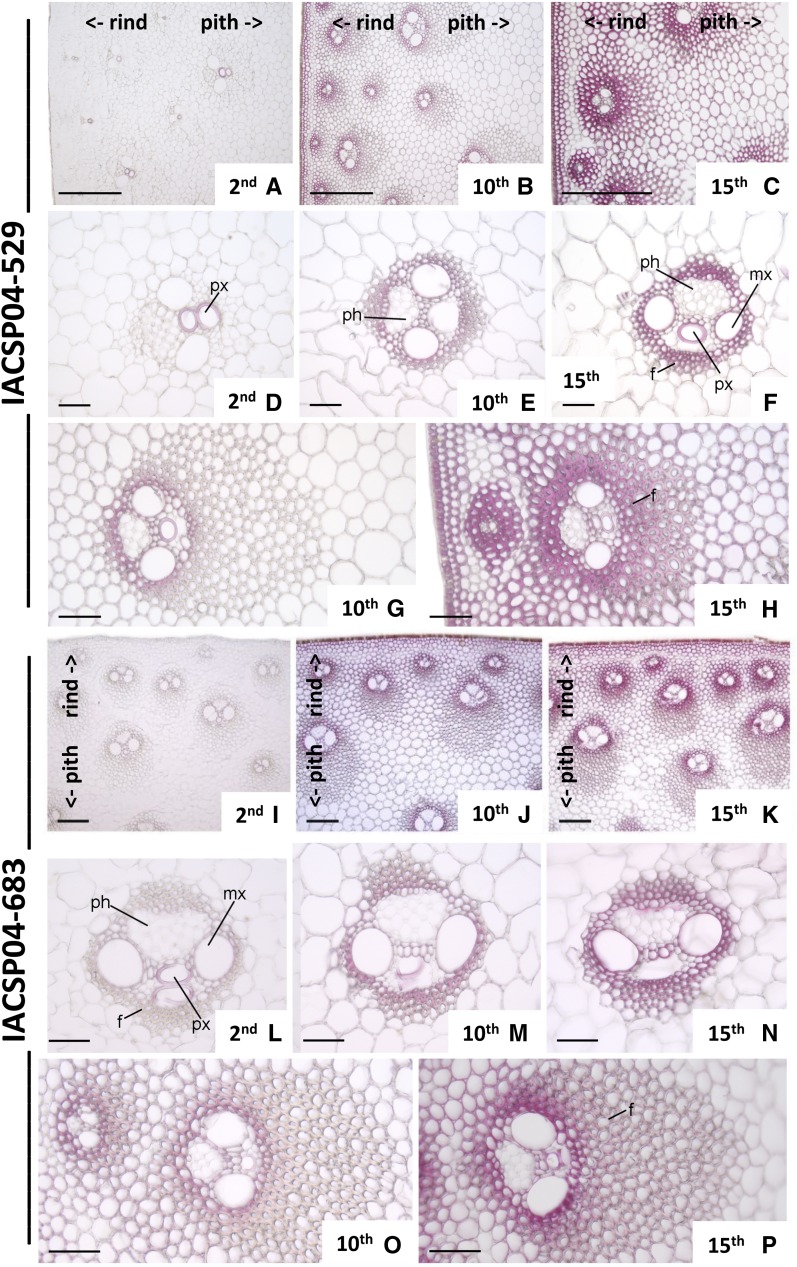
In order to evaluate the deposition of lignin during sugarcane stem development, histochemical analysis with phloroglucinol-HCl was performed. Apical (young), middle (intermediate), and basal (mature) internodes were analyzed (Fig. 2). Anatomically, the sugarcane stem has numerous vascular bundles distributed throughout the storage parenchyma. However, cross sections clearly showed that pith (internal tissue) and rind (periphery) are two distinct regions (Fig. 2, A–C and I–K). While the rind has a high number of vascular bundles with wide sclerenchymatic sheaths and minor amounts of parenchyma, the Suc-storing parenchyma in the pith harbors a smaller number of vascular bundles with less extensive sclerenchymatic sheaths. In addition, the differences between these two anatomic regions become more pronounced with stem maturity. Both genotypes studied here showed a similar structural tissue organization (Fig. 2).
Figure 2.
Transverse sections of internodes of two sugarcane genotypes stained for lignin with phloroglucinol. Genotypes IACSP04-529 (A–H) and IACSP04-683 (I–P) are shown. Details are as follows: second internode (A, D, I, and L); 10th internode (B, E, G, J, M, and O); 15th internode (C, F, H, K, N, and P); peripheral region (A–C and I–K); detail of the vascular bundle of the central region (D–F and L–N); and detail of the vascular bundle of the peripheral region (G, H, O, and P). f, Fibers; mx, metaxylem; ph, phloem; px, protoxylem. Numbers indicate the positions of the internode. <- rind and pith-> indicate their positions in the photographs. Bars = 200 μm (A–C) and 50 μm (D–M).
In young internodes (internode 2) of the high-lignin genotype IACSP04-529, lignification is restricted to tracheary elements of protoxylem in the vascular bundles of the rind and pith regions (Fig. 2, A and D), while in the low-lignin genotype IACSP04-683, lignin is also present in the tracheary elements of metaxylem and in the fibers of bundle sheaths (Fig. 2, I and L). Lignified cell types in intermediate and mature internodes (internodes 10 and 15, respectively) of both genotypes included the epidermis, hypodermis, parenchymatic cells, tracheary elements of metaxylem, and sclerenchyma surrounding the vascular bundles (Fig. 2, B, C, E–H, J, K, and M–P). Because the differential accumulation of lignin in rind and pith could help in identifying the gene family members specifically involved in developmental lignin biosynthesis, we separately sampled pith and rind for all further analyses, except for phenolic profiling.
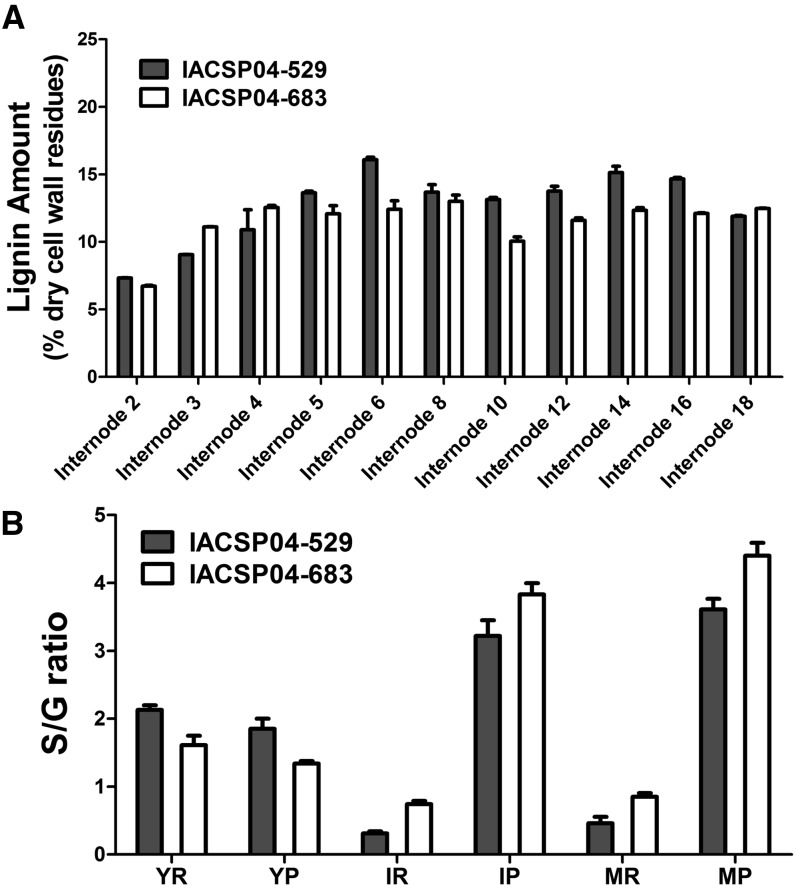
To investigate lignin deposition in sugarcane stems in more detail, lignin content was measured throughout the whole stalk, from internode 2 to internode 18 (Fig. 3A). Only the central part of individual internodes was used for the analysis, while the more lignified nodes were discarded. The Klason method showed that for most of the internodes, IACSP04-529 accumulated more insoluble lignin than IACSP04-683. This was particularly true for the older internodes.
Figure 3.
A, Percentage of insoluble lignin content in sugarcane internodes of genotypes IACSP04-529 and IACSP04-683 obtained with the Klason method. B, S-G ratio values in sugarcane in genotypes IACSP04-529 and IACSP04-683 obtained by thioacidolysis. YR, Rind of young internode; YP, pith of young internode; IR, rind of intermediate internode; IP, pith of intermediate internode; MR, rind of mature internode; MP, pith of mature internode. Error bars indicate se of three biological replicates (n = 3).
Thioacidolysis was used to determine the monomer yields of S and G units linked by 8-O-4-ether bonds in the lignin polymer. Thioacidolysis was carried out on pith and rind samples of internodes of different developmental stages for genotypes IACSP04-529 and IACSP04-683 (Table I; Fig. 3B). The S-G ratio was analyzed using a three-way ANOVA model with tissue type, developmental stage, and genotype as factors. Following the removal of nonsignificant interaction terms from the model, the reduced model still included two significant interactions. More specifically, variation in the S-G ratio over stem development showed a slightly different pattern in both genotypes (P = 0.014). However, the most significant interaction showed that the developmental changes in the S-G ratio were clearly different between pith and rind (P < 10−8). Further insight was obtained by considering Student’s t test and one-way ANOVA results (see “Materials and Methods”). In both genotypes, the S-G ratios were highest in young rind, followed by a significant decrease in later stages (P < 10−4), whereas intermediate and mature rinds showed similar values. On the other hand, S-G ratios in the pith increased throughout development (for each genotype, P < 0.01), whereas similar values were found between pith and rind in young tissues. The low S-G ratios in the peripheral regions of intermediate and mature tissues of both genotypes were mainly caused by higher levels of thioacidolysis-released G units (Table I). On the other hand, a higher abundance of thioacidolysis-released S units was observed in the mature pith internodes, suggesting a preferential biosynthesis of S in this tissue with maturity (Table I).
Table I. Monomeric composition of lignin determined by thioacidolysis.
YR, Rind of young internode; IR, rind of intermediate internode; MR, rind of mature internode; YP, pith of young internode; IP, pith of intermediate internode; MP, pith of mature internode. Data are means of three independent biological replicates.
Developmental Changes in the Phenolic Profile
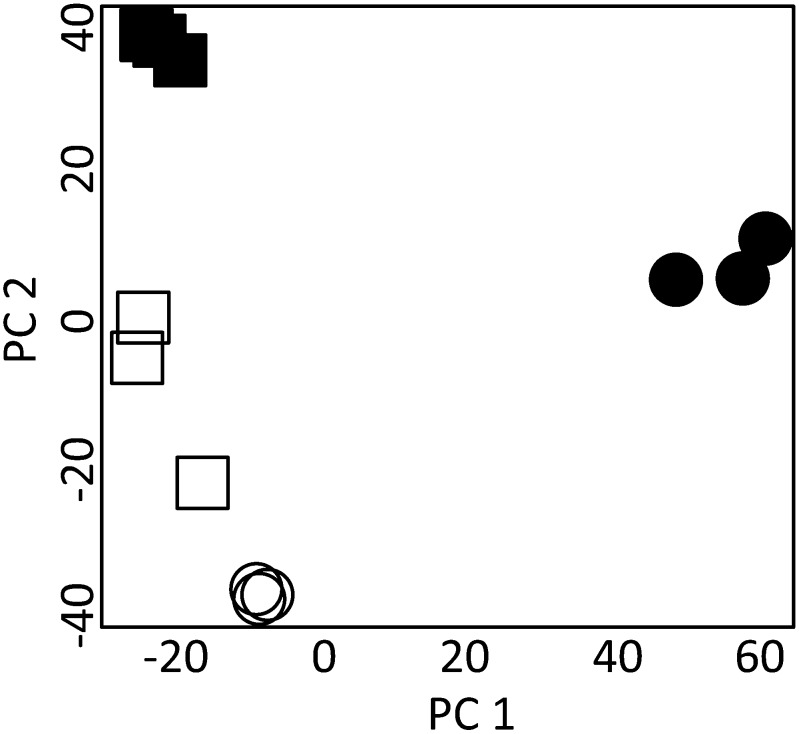
Methanol extracts were analyzed by liquid chromatography-mass spectrometry (LC-MS) to compare changes in the phenolic profiles during stem development of both sugarcane genotypes. Stem material at two developmental stages, young and mature, was used, while no separation into pith and rind was performed. In the analysis, only peaks that were present in all three biological replicates of at least one of the four genotype versus development groups were further analyzed. This procedure resulted in 2,904 peaks, which were used for principal component analysis to provide an initial overview of the phenolic metabolome of young and mature stem tissues from both sugarcane genotypes (Fig. 4). The first principal component (PC1), which explained most of the total variation within the data set (48%), allowed distinguishing mature from young tissues in genotype IACSP04-863. Separation of both genotypes was accomplished by PC2, which accounted for 26% of the total variation. Thus, principal component analysis showed that mature tissues can be differentiated from young tissues, but prominent differences can still be observed for the same tissue type between the two sugarcane genotypes. Phenolic compounds were structurally annotated by matching with in-house mass spectral libraries, taking into account the accurate mass-to-charge ratio value, the retention time, and the tandem mass spectrometry fragmentation spectrum. The identification procedure was focused on metabolites involved in lignin biosynthesis, from early steps in the pathway to final products of monomer polymerization, and related phenylpropanoid compounds such as flavonoids. Thirty-five compounds were structurally annotated and classified into four major groups (Table II): eight hydroxycinnamoyl esters, three hydroxycinnamic acids, 14 lignin oligomers, and 10 flavonoids. Within the hydroxycinnamoyl ester subgroup, cinnamate conjugates with quinic acid, including isomers of caffeoyl- and feruloyl-quinate, and shikimic acid, including isomers of caffeoyl-shikimate, were identified. The hydroxycinnamic acids p-coumaric acid, caffeic acid, and ferulic acid were also found, while no hydroxycinnamaldehydes were identified. A sequencing strategy for lignin oligomers (Morreel et al., 2010a, 2010b) allowed not only the sequence annotation of the monomer-derived units but also of the linkage types between the units within an individual oligomer. Dimers, trimers, and tetramers composed of G and S units linked by 8-8-, 8-5-, and 8-O-4-linkages were found. In addition, oligomers containing units derived from coniferaldehyde, sinapaldehyde, and ferulic acid, as well as oligomers conjugated with hexose, were annotated. Surprisingly, no oligomers containing H units were detected in the stem samples, despite the fact that grass lignin contains relatively higher levels of H units than lignins from dicots. In addition, 10 flavones were identified, all of them derived from tricin and tricin O-glycosides.
Figure 4.
Principal component analysis of the compounds identified in the phenolic profiling. Young (white squares) and mature (black squares) internodes of genotype IACSP04-529 (higher lignin levels) and young (white circles) and mature (black circles) internodes of genotype IACSP04-863 (lower lignin levels) are indicated. PC 1 and PC 2, Principal components 1 and 2.
Table II. Phenolic profiling of sugarcane stems.
Phenolic profiling was performed in young and mature stem regions of the varieties IACSP04-529 and IACSP04-683 with reverse-phase ultra-HPLC in connection with a Synapt HDMS quadrupole time-of-flight apparatus. Names of oligolignols are based on Morreel et al. (2004). The linkage type in the oligolignols is indicated in parentheses, and units derived from ferulic acid and coniferylaldehyde are designated FA and G′, respectively. The term “red” indicates reduced adjacent linkage. Ref indicates a bibliographic reference for mass spectrometry data as follows: Ref 1, Clifford (2003); Ref 2, Niggeweg et al. (2004); Ref 3, Duarte-Almeida et al. (2011); Ref 4, Morreel et al. (2010a); Ref 5, Morreel et al. (2010b); Ref 6, Colombo et al. (2006).
| Class | Compound (Reference) | Formula | Retention Time | Mass-to-Charge Ratio | Mass-to-Charge Ratio MS/MS | Δppm | Relative Abundance |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| IACSP04-529 |
IACSP04-683 |
|||||||||
| Young | Mature | Young | Mature | |||||||
| Hydroxycinnamate esters | Caffeoyl-quinate (Ref 1) | C16H17O9 | 2.94 | 353.0859 | 191, 179, 135 | −4.5 | 4.54 × 105 | 7.48 × 103 | 3.97 × 105 | 7.42 × 104 |
| Caffeoyl-quinate (Ref 1) | C16H17O9 | 4.20 | 353.0859 | 191, 173, 161, 133, 127 | −3.7 | 1.54 × 105 | 5.53 × 104 | 1.14 × 105 | 1.68 × 105 | |
| Caffeoyl-quinate (Ref 1) | C16H17O9 | 4.51 | 353.0859 | 191, 179, 173, 135, 133 | 1.7 | 1.36 × 105 | 1.49 × 104 | 7.29 × 104 | 1.03 × 105 | |
| Caffeoyl-quinate (Ref 1) | C16H17O9 | 5.59 | 353.0859 | 191, 173, 161, 133, 127 | 2.5 | 2.05 × 104 | 2.42 × 103 | 2.50 × 104 | 1.99 × 104 | |
| Feruloyl-quinate (Ref 1) | C17H19O9 | 4.68 | 367.1029 | 193, 134 | −1.6 | 8.11 × 104 | 1.07 × 104 | 1.92 × 105 | 1.02 × 105 | |
| Feruloyl-quinate (Ref 1) | C17H19O9 | 6.74 | 367.1029 | 193, 191, 173, 134 | −1.9 | 3.58 × 104 | 4.92 × 103 | 4.31 × 104 | 2.52 × 104 | |
| Caffeoyl-shikimate (Ref 2) | C16H15O8 | 5.58 | 335.0767 | 191, 179, 173, 135 | −5.1 | 1.45 × 104 | 2.69 × 10 | 1.80 × 104 | 1.47 × 102 | |
| Caffeoyl-shikimate (Ref 2) | C16H15O8 | 6.96 | 335.0767 | −1.8 | 3.42 × 103 | 1.13 × 10 | 6.97 × 102 | 3.01 × 10 | ||
| Hydroxycinnamates | p-Coumaric acid (Ref 3) | C9H7O3 | 6.78 | 163.0395 | 119 | 1.8 | 3.45 × 104 | 5.70 × 104 | 3.09 × 104 | 3.65 × 103 |
| Caffeic acid (Ref 3) | C9H7O4 | 4.90 | 179.0344 | 135 | −2.3 | 1.22 × 104 | 8.98 × 102 | 7.09 × 103 | 6.20 × 103 | |
| Ferulic acid (Ref 3) | C10H9O4 | 7.94 | 193.0501 | −35.7 | 1.03 × 103 | 2.32 × 102 | 5.46 × 102 | 5.75 × 10 | ||
| Oligolignols | S(8-8)S (Ref 4) | C22H25O8 | 10.89 | 417.1549 | 402, 387, 181, 166, 151 | 1.0 | 3.70 × 102 | 1.13 × 103 | 1.49 × 102 | 1.49 × 103 |
| S(8-O-4)S′ (Ref 4) | C22H25O9 | 12.56 | 433.1487 | 418, 403, 385, 373 | −2.8 | 6.05 × 102 | 1.45 × 104 | 3.38 | 9.38 × 102 | |
| G(red8-5)G (Ref 4) | C20H23O6 | 10.59 | 359.1495 | −4.5 | 1.07 × 103 | 6.92 × 102 | 3.49 × 102 | 4.49 × 102 | ||
| G(t8-O-4)G (Ref 4) | C20H23O7 | 8.18 | 375.1444 | −4.8 | 3.57 × 102 | 1.57 × 103 | 7.87 × 10 | 3.29 × 102 | ||
| G(8-O-4)FA (Ref 4) | C20H21O8 | 10.91 | 389.1236 | 2.8 | 1.40 × 103 | 1.11 × 104 | 5.00 × 102 | 1.06 × 102 | ||
| G(red8-8)G, lariciresinol + hexose (Ref 4) | C26H33O11 | 10.71 | 521.2023 | 503, 491, 488, 476, 341, 329 | 2.7 | 4.60 × 103 | 1.23 × 104 | 4.15 × 103 | 2.28 × 104 | |
| G(8-O-4)S(8-5)G (Ref 5) | C31H35O11 | 14.74 | 583.2179 | 535, 505, 369, 357, 195, 165, 150 | −1.0 | 9.29 × 103 | 2.39 × 104 | 4.84 × 103 | 5.98 × 103 | |
| G(8-O-4)S(8-5)G (Ref 5) | C31H35O11 | 15.56 | 583.2179 | 535, 369, 357, 195, 165, 150 | −1.2 | 5.14 × 103 | 1.60 × 104 | 2.55 × 103 | 3.73 × 103 | |
| G(8-O-4)S(8-5)G′ (Ref 5) | C31H33O11 | 16.26 | 581.2023 | 369, 193 | −1.9 | 7.52 × 102 | 4.03 × 103 | 4.52 × 102 | 8.20 × 102 | |
| G(8-O-4)S(8-5)G′ (Ref 5) | C31H33O11 | 16.76 | 581.2023 | 551, 533, 503, 367, 355, 165, 150 | 0.9 | 4.13 × 103 | 3.05 × 104 | 1.21 × 103 | 4.56 × 103 | |
| S(8-O-4)S(8-5)G (Ref 5) | C32H37O12 | 16.69 | 613.2285 | 467, 369, 325, 195, 163 | −11.6 | 5.62 × 102 | 1.14 × 103 | 2.92 × 102 | 6.13 × 102 | |
| S(8-O-4)S(8-8)S (Ref 5) | C33H39O13 | 16.43 | 643.2404 | 595, 417, 225 | 2.0 | 2.27 × 102 | 1.34 × 103 | 1.27 × 102 | 1.22 × 103 | |
| G(8-O-4)S(8-8)S(4-O-8)G (Ref 5) | C42H49O16 | 18.02 | 809.3021 | 791, 761, 613, 565, 417 | 3.1 | 1.73 × 103 | 7.28 × 103 | 6.24 × 102 | 2.76 × 103 | |
| G(8-O-4)S(8-8)S(4-O-8)G (Ref 5) | C42H49O16 | 18.70 | 809.3021 | 791, 761, 613, 565, 417 | 4.4 | 1.61 × 103 | 5.98 × 103 | 8.13 × 102 | 2.62 × 103 | |
| Flavonoids | Tricin | C17H13O7 | 17.09 | 329.0661 | 314, 299, 271, 227 | 18.5 | 1.54 × 104 | 8.30 × 103 | 5.45 × 104 | 1.76 × 104 |
| Tricin-7-O-neohesperoside (Ref 6) | C29H33O16 | 10.87 | 637.1769 | 329 | −3.8 | 4.07 × 103 | 2.11 × 103 | 1.98 × 103 | 2.05 × 103 | |
| Tricin-7-O-neohesperoside (Ref 6) | C29H33O16 | 11.47 | 637.1769 | 329, 314 | −0.5 | 7.87 × 103 | 1.15 × 104 | 1.42 × 104 | 1.32 × 104 | |
| Tricin-7-O-glycoside (Ref 6) | C23H23O12 | 7.33 | 491.119 | 9.8 | 2.29 × 102 | 1.69 × 103 | 1.58 × 102 | 5.97 × 102 | ||
| Tricin-7-O-glycoside (Ref 6) | C23H23O12 | 9.99 | 491.119 | 461, 329 | 7.9 | 1.31 × 103 | 1.32 × 103 | 1.97 × 104 | 6.29 × 103 | |
| Tricin-7-O-glycoside (Ref 6) | C23H23O12 | 11.43 | 491.119 | 476, 461, 343, 329, 314 | −5.7 | 9.41 × 103 | 5.15 × 103 | 7.29 × 103 | 9.00 × 103 | |
| Tricin-4′-O-guaiacyl ether (Ref 6) | C27H25O11 | 18.02 | 525.1397 | 477, 329, 314, 299, 195, 165, 150 | −0.4 | 1.32 × 104 | 1.32 × 104 | 3.19 × 104 | 3.00 × 104 | |
| Tricin-4′-O-guaiacyl ether (Ref 6) | C27H25O11 | 18.74 | 525.1397 | 329, 314, 299, 195, 165, 150 | 2.7 | 8.73 × 103 | 7.52 × 103 | 2.46 × 104 | 2.45 × 104 | |
| Tricin-4′-O-guaiacyl ether-7- O-glucopyranoside (Ref 6) | C33H35O16 | 12.86 | 687.1925 | 525, 491, 329, 314, 195, 165, 150 | −0.3 | 1.27 × 104 | 1.06 × 104 | 1.31 × 104 | 3.07 × 104 | |
| Tricin-4′-O-guaiacyl ether-7- O-glucopyranoside (Ref 6) | C33H35O16 | 13.60 | 687.1925 | 525, 491, 329, 195, 165, 150 | −1.7 | 8.36 × 103 | 6.56 × 103 | 4.74 × 103 | 1.24 × 104 | |
The abundances of the structurally characterized compounds were compared among the four genotype versus development groups by two-way ANOVA (Supplemental Table S1). A significant model (P < 0.05) was obtained for 23 of the 33 compounds. Among those 23 compounds, a significant interaction between genotype and development (P < 0.05) was observed for 15 compounds. For these compounds, the developmental abundance patterns were different in both genotypes. As a consequence, Student’s t tests (P < 0.05) had to be considered to obtain more insight into the statistical differences between both developmental stages within each genotype and between genotypes for a particular tissue type. Significant interaction effects were predominantly observed for the oligolignols (nine of the 14 oligolignols). These nine compounds had higher levels in genotype IACSP04-529 as compared with genotype IACSP04-683 in the mature tissues, in agreement with the higher lignin content in the former genotype. Furthermore, the abundances of these nine oligolignols showed significant developmental differences. However, in the case of the trilignols and S(8–O-4)S′, this developmental pattern was only significant for genotype IACSP04-529. Besides these nine oligolignols, the levels of two other oligolignols showed developmental differences. As expected, all 11 oligolignols but G(8–O-4)FA preferentially accumulated in mature internodes in both genotypes, because lignin amount increases with maturity. The dimer G(8-O-4)FA was more abundant in mature than in young tissues in genotype IACSP04-529, while the reverse was true in genotype IACSP04-683. The three remaining oligolignols for which no significant differences were observed between the two developmental stages or between the contrasting genotypes were S(8-8)S, Gred(8-5)G, and S(8-O-4)S(8-5)G. At least the former two might be (neo)lignans rather than lignin oligomers. Overall, the oligolignol amounts in the different tissues and genotypes corresponded with the lignin amounts in these four sample types.
The levels of four of the 10 flavonoids showed significant interaction effects between genotype and development. These flavonoids were tricin, two tricin-7-O-glycosides, and tricin-4′-O-guaiacyl ether-7-O-glucopyranoside. However, no coherent pattern of differences among the four sample types was evident for these four flavonoids. For two of the remaining six flavonoids, higher levels were present in genotype IACSP04-683 as compared with genotype IACSP04-529. Both flavonoids were tricin-4′-O-guaiacyl ether isomers.
Upstream of flavonoid and oligolignol metabolism (i.e. in phenylpropanoid metabolism), differences between both developmental stages and genotypes were not always coherent among the various metabolites. However, two general patterns emerged from analyzing the abundance of the hydroxycinnamoyl esters; in genotype IACSP04-529, hydroxycinnamoyl esters accumulated preferentially in young internodes but much less in mature internodes. In addition, the mature tissues of IACSP04-683 accumulated more hydroxycinnamoyl esters than those of IACSP04-529. Concerning the hydroxycinnamic acids, their abundances were generally higher in the young internodes of both genotypes. Furthermore, when compared with those in IACSP04-529, mature tissues of genotype IACSP04-683 contained higher amounts of caffeic acid but lower amounts of p-coumaric and ferulic acids.
Identification of Genes Associated with Monolignol Biosynthesis
The SUCEST database was surveyed to identify monolignol biosynthetic genes in sugarcane, based on the annotated genes from Arabidopsis, sorghum (Sorghum bicolor), rice (Oryza sativa), and maize. A total of 28 unigenes were identified, among them four PALs (EC 4.3.1.5), two C4Hs (EC 1.14.13.11), three 4CLs (EC 6.2.1.12), one HCT (EC 2.3.1.133), one HCT-like, two C3Hs (EC 1.14.13.36), three CCoAOMTs (EC 2.1.1.104), two CCRs (EC 1.2.1.44), one COMT (EC 2.1.1.68), one F5H (EC 1.14.13.-), and four CADs (EC 1.1.1.195; for definitions, see Fig. 1). The Sugarcane Assembled Sequences (SAS) tentative consensus number, the respective sugarcane gene names, and the primer sequences designed for quantitative reverse transcription-PCR analysis are shown in Supplemental Table S2. A phylogenetic analysis using amino acid sequences of monolignol biosynthesis enzymes from representative species of the Viridiplantae protein data set, including Populus trichocarpa, Selaginella moellendorffii, and Physcomitrella patens, and the identified homologs in sugarcane was performed to define classes within each gene family (Supplemental Figs. S1–S10). By including the sequences of previously characterized enzymes in other plant species, this phylogenetic analysis could be useful for the identification of the putative orthologs that might play a role in lignification in sugarcane stems. However, we were unable to unambiguously extract this information for most of the gene families, because the sugarcane sequences very often clustered into monocot-exclusive groups.
Dynamics of Phenylpropanoid/Monolignol Gene Expression during Stem Development
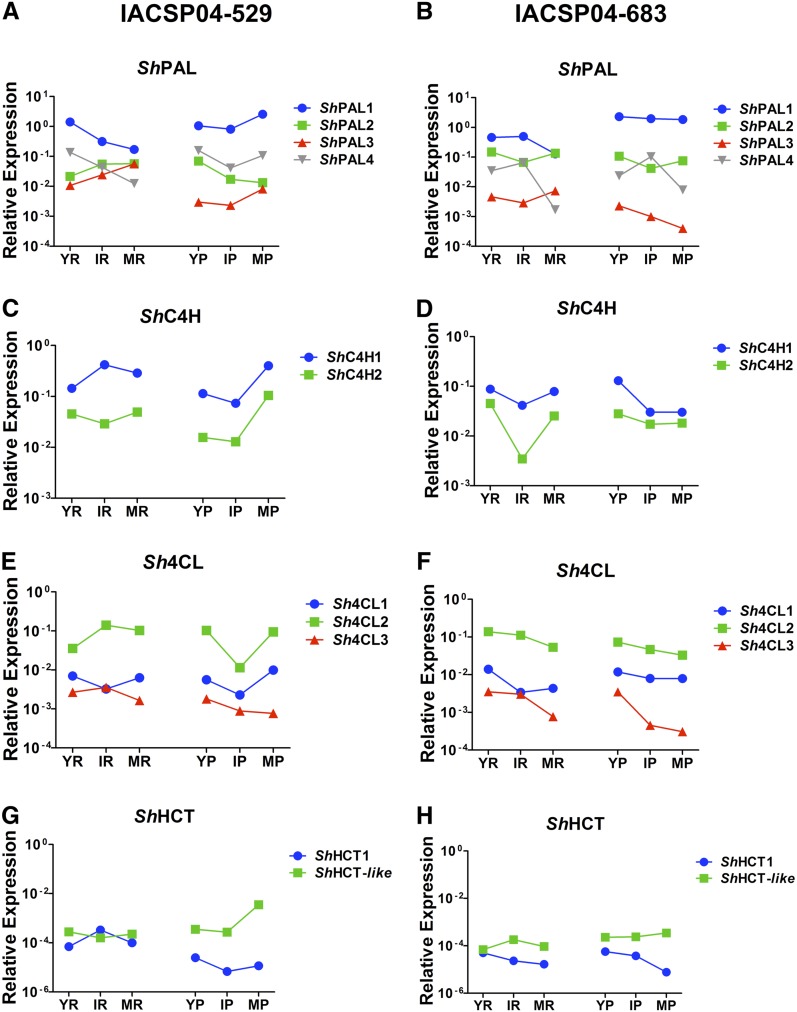
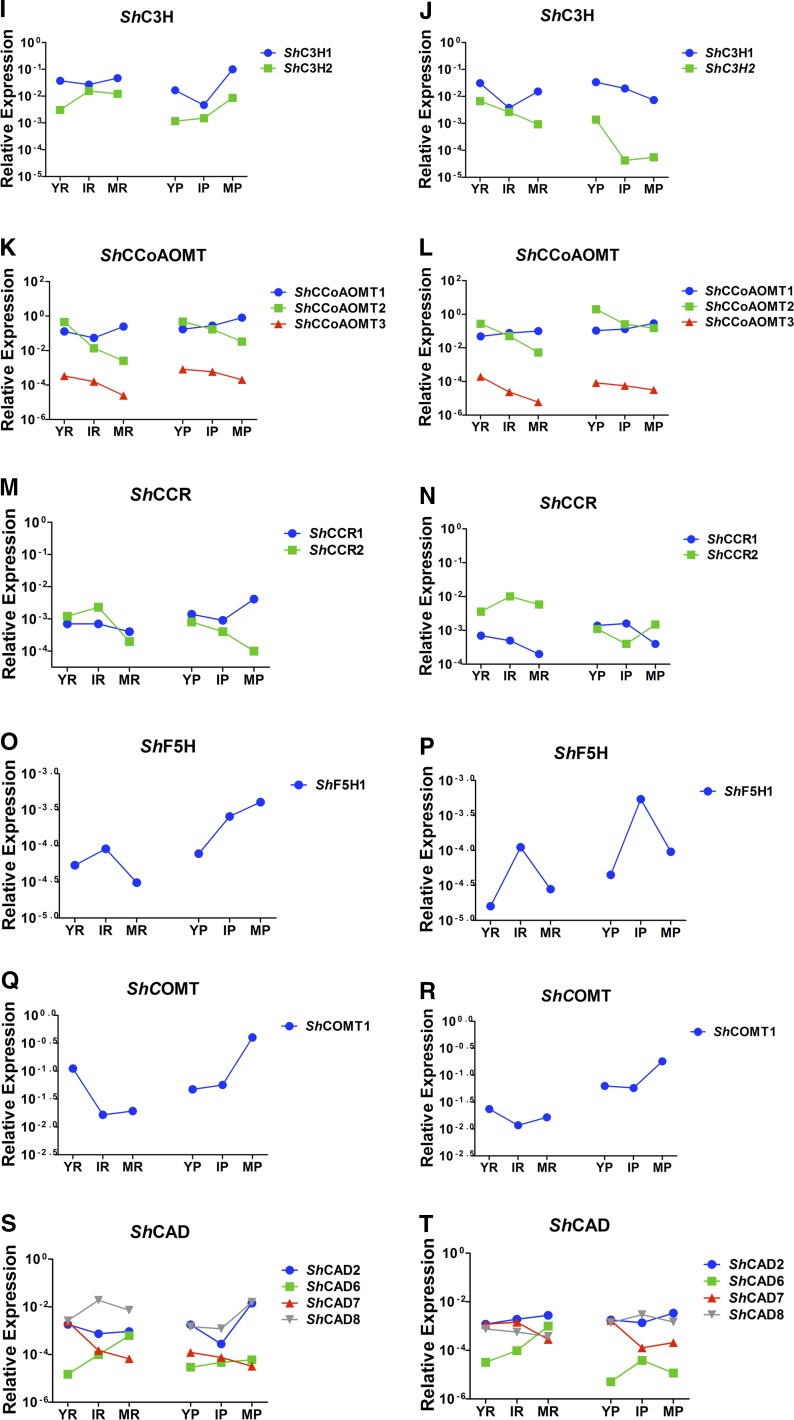
In an attempt to identify the gene family members that are likely involved in sugarcane stem lignification, we compared the transcript abundance of each phenylpropanoid gene during sugarcane stem development by quantitative reverse transcription-PCR. For both sugarcane varieties, three developmental stages of stem internodes (young, intermediate, and mature), divided into pith and rind, were used for RNA isolation. The gene members with the strongest expression levels in the stem samples and whose expression profile correlated with the increase of lignin content during stem development, as well as with the preferential deposition of lignin in the rind region when compared with the pith region, were considered the most qualified gene family members to play a role in the developmental lignification. Additionally, the phylogenetic relationships with previously characterized phenylpropanoid biosynthesis genes from other plant species were considered in the analysis.
Several distinct spatiotemporal expression profiles were found among the analyzed sugarcane genes. In addition, transcript levels of a given gene were often different between the two contrasting sugarcane varieties (Fig. 5). Therefore, the association between gene expression and lignin deposition was not always obvious. Nevertheless, the expression profiles of some family members were in agreement with their expected roles in lignification. For instance, the most likely genes involved in the developmental lignification of the sugarcane stem are ShPAL1, ShC4H1, Sh4CL2, ShC3H1, ShCCoAOMT1, ShF5H, ShCOMT, ShCAD2, and ShCAD8. Based on our criteria, ShPAL1, ShC4H1, Sh4CL2, ShC3H1, and ShCAD2 were chosen because they were the most highly expressed members of their gene family, while the expression of ShCCoAOMT1 and ShCAD8 significantly increased with stem maturity (Fig. 5). Transcript levels of ShHCT1 and ShHCT-like genes were low in all analyzed tissues and in both sugarcane genotypes (Fig. 5, G and H), which is not expected for genes involved in lignification. In addition, the ShCCR transcript abundances were not conclusive to pinpoint a ShCCR gene involved in lignification, as both ShCCR1 and ShCCR2 were relatively lowly expressed, and also due to significant differences in expression between the two genotypes and the tissues analyzed (Fig. 5, M and N). The expression of ShF5H1 appeared low in all analyzed tissues (Fig. 5, O and P), making it doubtful to be involved in lignification. However, ShF5H1 was relatively more expressed in pith compared with rind, which might explain the higher S-G ratio in intermediate and mature pith as compared with rind (Fig. 3). Interestingly, the latter observation was also true for ShCOMT1, which had much higher expression levels than ShF5H1 (Fig. 5, Q and R). Finally, the lower expression levels of some family members could be due to their preferential expression in limited tissues or cell types.
Figure 5.
Expression profile of monolignol biosynthetic genes analyzed by quantitative reverse transcription-PCR. The sequences were retrieved from the SUCEST database based on the annotated genes from Arabidopsis, sorghum, rice, and maize. YR, Young internode rind; IR, intermediate internode rind; MR, mature internode rind; YP, young internode pith; IP, intermediate internode pith; MP, mature internode pith. IACSP04-529 is the genotype with higher lignin levels, and IACSP04-683 is the genotype with lower lignin levels. Statistical analysis is found in Supplemental Table S3.
Bayesian Network of the Lignin Biosynthesis Genes
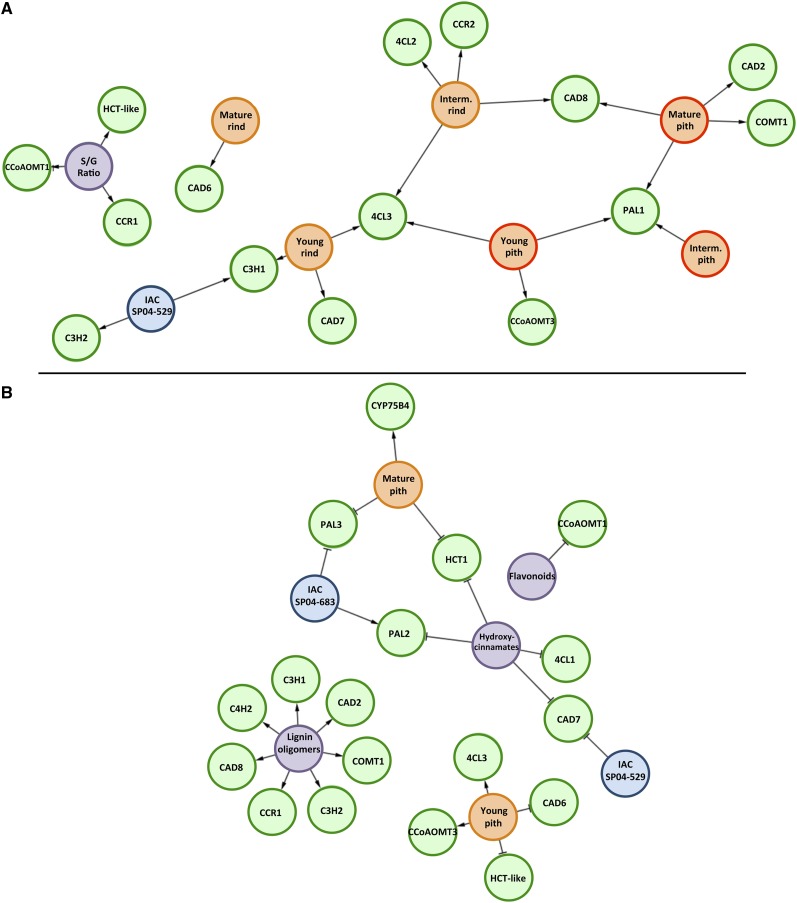
Complex biological processes can be investigated by inferring regulatory networks based on relationships between transcriptional profiles and genetic, chemical, and physiological parameters. This approach can also be used to capture the behavior of the system under further perturbations. We currently have a preliminary understanding of how genes are organized in complex networks to generate the flexibility in many biological processes (Bruex et al., 2012). To obtain a systems-level view of monolignol biosynthesis in sugarcane, we applied a Bayesian network approach to exploit relationships in gene expression and to generate a graph of nodes that best predicts downstream nodes. In an attempt to get insight into the dependencies among all the variables obtained, the expression data were integrated with the biochemical data (i.e. lignin amount and composition and metabolite levels) from the two sugarcane genotypes to construct two different regulatory networks. Accordingly, it is possible to identify the genes that best predict these characteristics, providing insight that cannot be achieved from simple transcript profile comparisons. The first network included expression data, lignin content, and S-G ratio of the samples of pith and rind of all internodes (young, intermediate, and mature; Fig. 6A). In the second network, the data of the intermediate internode were excluded and compounds identified by phenolic profiling were included (Fig. 6B). Two different regulatory networks were constructed because the plant material used for phenolic profiling was obtained only from young and mature internodes and was not separated into pith and rind samples.
Figure 6.
Bayesian networks based on the relationships between gene expression and biochemical data. A, The network was inferred based on gene expression, lignin composition (S-G ratio), and lignin content data from internodes at three developmental stages (young, intermediate, and mature) and different tissue types (pith and rind) of IACSP04-529 (high lignin) and IACSP04-683 (low lignin). B, The phenolic profiling data were included to infer the network, but the biochemical data for intermediate internodes were excluded. In A, colored circles are as follows: green = genes; blue = sugarcane genotypes; purple = biochemical data; orange with orange boarder = rind tissue; orange with red boarder = pith tissue. In B, colored circles are as follows: green = genes; blue = sugarcane genotypes; purple = biochemical data; orange = pith tissue. In the obtained networks, it is assumed that the expression of genes can depend on conditions (genetic, morphological, and biochemical characteristics) but not the opposite, and the type of interaction can be a positive (arrows) or negative (blockers) correlation between variables.
The network presented in Figure 6A only identified positive correlations. The expression of ShHCT-like, ShCCoAOMT1, and ShCCR1 was intimately linked with S-G ratio, whereas that of ShC3H2 was only linked to IACSP04-529, indicating a very specific relationship. In some cases, gene expression was typically correlated with specific tissue types: ShCAD6 in mature rind; Sh4CL2 and ShCCR2 in intermediate rind; ShCAD2 and ShCOMT1 in mature pith; ShCCoAOMT3 in young pith; ShCAD7 in young rind. The expression of Sh4CL3, ShPAL1, ShCAD8, and ShC3H1 linked different tissues, suggesting a general role.
When data of the phenolic profiling were included in the analysis, a more complex network was obtained and negative relationships were found (Fig. 6B). The accumulation of the hydroxycinnamates caffeic acid, p-coumaric acid, and ferulic acid was negatively correlated with the expression of ShHCT1, ShPAL2, Sh4CL1, and ShCAD7. Except for ShCAD7, the other three genes were not displayed in the first network. On the other hand, the abundance of lignin oligomers (lignols in Table II) was positively correlated with the expression of multiple genes (ShCAD2, ShCAD8, ShC4H2, ShC3H1, ShC3H2, ShCCR1, and ShCOMT1). Similar to the first network, young pith was positively correlated with ShCCoAOMT3 expression, but also with Sh4CL3, and negatively correlated with the expression of ShCAD6 and ShHCT-like. Curiously, the expression of some genes was negatively correlated with either one or the other genotype, as was the case for ShCAD7 and IACSP04-529 and ShC3H1 and IACSP04-683.
DISCUSSION
Lignification starts early in sugarcane stem development in the vascular bundles of the young internodes, whereas deposition of lignin in other tissues such as epidermis, hypodermis, and storage parenchyma is achieved later in development (Jacobsen et al., 1992; Casu et al., 2007). The lignin content measured with the Klason method showed a continuous increase until the fifth internode for IACSP04-683 and until the sixth internode for IACSP04-529, after which much less variation was observed during later stages of stem development. This pattern of lignification is in agreement with previous reports (Lingle and Thomson, 2012). A similar developmental pattern of lignin deposition was observed in switchgrass (Panicum virgatum), a C4 perennial forage grass that has been selected for development as a biofuel crop in North America, and for which a fast increase in lignin content was observed during the progressive elongation stage (Shen et al., 2009). Similarly, a high increase in Klason lignin content was found in early stages of maize stem development (i.e. between tassel emergence and silking stages), while constant amounts were observed during the following stages (Jung and Casler, 2006; Riboulet et al., 2009). This lignin accumulation pattern is coherent with the fact that lignification begins before the cessation of the elongation process, with the deposition of secondary cell walls in specific cell types. In addition, it has been shown that the top part of individual elongating internodes in maize is more lignified than the bottom part of the same internode (Morrison et al., 1994). In agreement, Shen et al. (2009) showed that the variation in lignin content within one internode is lower in more mature stages than in earlier stages of switchgrass stem development.
The differences in magnitude between the lignin quantification data presented in this paper and those of the 3-year evaluation of an F1 sugarcane population can be explained by technical differences between the methods used. First, for the 3-year evaluation, lignin content was indirectly derived from the determination of acid and neutral detergent fibers (Van Soest, 1967), a method that underestimates lignin, especially in grasses (Hatfield et al., 1994), while in this study, lignin was measured with the Klason method, a classical method commonly used in wood research. Second, due to the low Suc levels and the presence of components that interfere with the ethanol yield (i.e. high levels of polyphenol oxidase activity), the top internodes and leaves were discarded during the harvest for the 3-year evaluation.
The values for lignin content presented in our study are significantly lower than those previously reported for sugarcane bagasse (Lopes et al., 2011; Masarin et al., 2011; Rezende et al., 2011). Sugarcane bagasse is the residue produced after Suc extraction and is composed of 39% cellulose, 25% hemicelluloses, and 23% lignin, among other minor components (Carroll and Somerville, 2009). The methodology for lignin analysis in sugarcane bagasse is different from that used here. Normally, whole culms without leaves are pressed to remove the juice, and the resulting bagasse, containing nodes and internodes, is subsequently dried and ground to a powder that is used for wet chemistry analyses. Here, only the internode, the region between two adjacent nodes, was used for lignin determination, while the highly lignified and fibrous nodes were discarded. Therefore, the high lignin values previously reported for sugarcane bagasse (Lopes et al., 2011; Masarin et al., 2011; Rezende et al., 2011) reflect the lignin content of a mixture of internodes and nodes and, therefore, cannot be directly compared with the values obtained here.
Histochemical analysis suggested that pith and rind are two distinct regions within the sugarcane stem, the latter being more heavily lignified than the pith region (Cesarino et al., 2012). Not only lignin amount but also lignin composition is an important factor influencing saccharification. It has been suggested that S-rich lignin is more easily degraded than G-rich lignin because the methoxy group at the C-5 position of S units prevents the formation of a degradation-resistant carbon-carbon bond at this position (Ziebell et al., 2010). A progressive increase in the S-G ratio during stem development has been shown in several grasses such as tall fescue (Festuca arundinacea; Chen et al., 2002), maize (Jung and Casler, 2006; Riboulet et al., 2009), and switchgrass (Shen et al., 2009). However, here, we show that lignin composition also varies in a radial fashion within the sugarcane stem, being differentially regulated between the two anatomically distinct regions, pith and rind. In the rind of both sugarcane genotypes, there was a marked decrease in the S-G ratio in intermediate and mature tissues compared with young tissues, caused by an increase in the G unit content. Conversely, the S-G ratio in the pith regions increased during stem maturation. The increase in S lignin in the intermediate and mature central regions is probably due to the lignification of parenchyma cells, which are higher in number than the vascular bundles. Interestingly, a combination of histological and biochemical studies defined a maize ideotype for cell wall digestibility as containing reduced amounts of lignin, higher levels of S units, and preferential localization of lignin in the rind (Méchin et al., 2005). These authors suggested that, compared with the rind, a less lignified pith would favor cell wall degradability by increasing the accessible and potentially degradable area. Similarly, Siqueira et al. (2011) observed that sugarcane pith samples were promptly hydrolyzed by cellulases, while the conversion of cellulose in rind samples was dependent on chlorite pretreatments. The significantly higher cell wall recalcitrance of sugarcane rind samples was mainly attributed to the massive presence of lignin-rich fibers and vessels, but the presence of a more condensed G-rich lignin in this anatomical region (this study) of the stem might also be implicated. Altogether, our data show that not only lignin content but also lignin composition is differentially regulated throughout sugarcane stem development, and significant variation exists between different cell types within the same organ. Additionally, the data show the existence of genetic variability in the S-G ratio between sugarcane genotypes, opening perspectives for breeding.
A total of 35 compounds related to the phenylpropanoid pathway, and especially to lignin biosynthesis, were identified in sugarcane stems by LC-MS. To our knowledge, this is the first work in which a “lignomics” approach was applied to sugarcane. Previous studies on the sugarcane metabolome were focused on primary metabolism related to Suc accumulation (Glassop et al., 2007) or performed simple phenolic compositional analysis by comparing chromatographic profiles of sugarcane culm extracts with HPLC of a few known phenolic compounds (Duarte-Almeida et al., 2011). Our data showed that hydroxycinnamoyl esters were the most abundant phenolic metabolites in both sugarcane genotypes, while the contents of hydroxycinnamates and flavonoids were similar. Differential accumulation of phenolics was also observed between the same tissue types of the different sugarcane genotypes. Additionally, while Duarte-Almeida et al. (2011) could only detect ferulic acid in sugarcane by-products (i.e. raw juice and molasses) but not in plant tissues, we could identify low amounts of this hydroxycinnamic acid in young and mature internodes of both genotypes, despite the fact that its content was much lower than that of caffeic and p-coumaric acids.
The investigation of flavonoid content in sugarcane bagasse and leaves demonstrated that the major flavonoids accumulating in these tissues are flavone O- and C-glycosides, most of them being derivatives of apigenin, luteolin, and tricin (Colombo et al., 2005, 2006). Our mass spectrometry data corroborate this observation, since all flavonoids identified in our data were tricin and tricin O-glycosides. It has been recently shown that tricin, a flavone widely distributed in grasses, is probably incorporated into lignin in wheat (Triticum aestivum) straw, which suggest that this alternative monomer is exported to the apoplast, where it undergoes radical coupling with canonical monolignols (Del Río et al., 2012). Due to their hybrid structure, derived from both the phenylpropanoid and the acetate/malonate-derived polyketide pathways, flavones are chemically capable of forming polyphenols by radical homocoupling reactions, which suggests that they might also be able to form copolymers with normal monolignols (Grabber et al., 2010). In fact, tricin was shown to form flavonolignan derivatives, such as tricin 4′-O-β-guaiacylglycerol ether, in which the tricin skeleton is linked to a G moiety through a β-O-4-bond (Wenzig et al., 2005). These observations highlight the metabolic flexibility of cell wall lignification and suggest that the concept of “normal monolignols” might vary significantly between taxa.
In agreement with the developmental program of lignin deposition in sugarcane stem, monolignol biosynthesis pathway intermediates and oligomers could be detected in both young and mature internode samples, whereas the differences were limited to the relative abundance of each compound. Hydroxycinnamates and hydroxycinnamoyl esters were detected in higher concentrations in young internodes, which supports their role as intermediates of monolignol biosynthesis (Boerjan et al., 2003; Bonawitz and Chapple, 2010; Vanholme et al., 2013b). On the other hand, lignin oligomers clearly showed a trend to accumulate in mature internodes, which is in line with the higher deposition of lignin with increasing maturity. Interestingly, the content of nearly all of the lignin oligomers was higher in the genotype IACSP04-529, which contained the higher lignin content, while no obvious trend was observed for the other groups of identified compounds. However, a correlation between oligomer abundance and lignin deposition is not obvious, because lignin is mainly produced by the addition of monolignols to the growing polymer, resulting in a polymer enriched in 8-O-4-linkages. Conversely, monolignol-monolignol coupling is more prevalent during oligomer formation, resulting in relatively more 8-8-linkages. Furthermore, (neo)lignans will also contribute to the oligomer pool (Morreel et al., 2004). Nevertheless, the presence of several lignin oligomers in a tissue that undergoes extensive lignification still suggests that these compounds are monolignol-coupling products formed under oxidative conditions in the cell wall (Morreel et al., 2004).
Grass cell walls are different from those of dicots in terms of the content and composition of polysaccharides, pectins, enzymatic and structural proteins, and phenolic compounds (Vogel, 2008). Grass lignin also contains relatively higher amounts of H units (Grabber et al., 2004) and monolignol-hydroxycinnamate conjugates as primary building blocks (Ralph, 2010). Interestingly, lignin oligomers containing H units were not found in our phenolic profiling data, similar to the findings of Kiyota et al. (2012), who found only the dimer G(8-5)H in the soluble lignin of sugarcane. In addition, several other oligomers detected in the phenolic profiles of Arabidopsis stems (Vanholme et al., 2010b) and poplar xylem (Morreel et al., 2010a) could not be detected in sugarcane stem. Due to the above mentioned differences between grass and dicot lignins, it is possible that sugarcane contains several oligomers composed of subunits and linkages not present in dicot plants and, therefore, not previously reported, despite the variety of oligomers available for identification in the in-house library (Morreel et al., 2010a, 2010b). Furthermore, several other metabolites, such as coumarins, procyanidins, lignans, neolignans, flavonol glycosides, and hydroxycinnamate esters conjugated with sugars, that were already published for Arabidopsis and poplar wild-type and mutant plants, were not found in sugarcane. The fact that the mass spectrometry libraries based on data from Arabidopsis and poplar were not very efficient for the identification of phenolic compounds in sugarcane stem highlights the importance of studies in a broad variety of plant species to fully understand lignin composition, polymerization, and regulation.
The enzymes responsible for the sequential reactions of the monolignol biosynthesis pathway are normally encoded by multigene families in most dicots and monocots (Bonawitz and Chapple, 2010). However, the different isoforms for a given enzyme may have different kinetic properties and may be preferentially expressed in a particular organ or developmental stage. Therefore, only a limited number of phenylpropanoid genes may play a role in lignification of the sugarcane stem and consequently contribute to biomass recalcitrance. Here, a combination of phylogenetic and transcriptomic data were used to determine the member(s) of each gene family that is most likely involved in monolignol biosynthesis in sugarcane stem as well as in different anatomical regions within the internode (i.e. pith and rind). Extensive analysis of the SUCEST identified several members for each phenylpropanoid gene family, with the exception of HCT, COMT, and F5H, which apparently are encoded by single genes. Nevertheless, since the searches for phenylpropanoid gene homologs in sugarcane were performed against an EST database instead of a fully sequenced genome database, we cannot exclude the possibility that additional members exist for a given gene family. The determination of gene number within each family and their phylogenetic relationships have already been made in other pioneering studies in Arabidopsis (Raes et al., 2003), maize (Guillaumie et al., 2007), and poplar (Shi et al., 2010).
For many of the lignin biosynthetic pathway genes, similar expression patterns and transcript abundances were found for members of the same gene family, suggesting functional redundancy. Interestingly, different expression profiles were found for some genes when comparing pith and rind, which also differed in lignin amount and composition. Noteworthy, S branch-specific genes (i.e. F5H and COMT) were up-regulated with maturity and were more highly expressed in the pith than in the rind, which is in agreement with the increased S-G ratio found in intermediate and mature pith when compared with young tissues. Conversely, the same genes were down-regulated in rind samples, in which the deposition of G units was favored. However, in several examples, there was no obvious correlation between lignin content and gene expression. Riboulet et al. (2009) used the correlation between variation in gene expression and lignin content during stem development to identify the phenylpropanoid genes most likely to participate in lignin biosynthesis in maize. Interestingly, most of the analyzed phenylpropanoid genes showed maximum expression in young ear internodes, followed by a decrease during the later stages of development, whereas lignin content remarkably increased with maturity. A possible explanation is that, although the lignification process is highly active in developing and mature internodes, cells of young internodes already deposit secondary cell walls in protoxylem and metaxylem elements and inner and outer portions of sclerenchymatic bundle sheaths (Cesarino et al., 2012a). Therefore, it is possible that higher gene expression occurs in early stages of stem development and that, even with subsequent down-regulation, the lignification process is still maintained due to the stability of lignin biosynthesis enzymes. The identification of genes potentially involved in the developmental lignification of sugarcane might be facilitated by our Bayesian analysis. From this analysis, ShCCoAOMT1, ShCCR1, and ShHCT-like, whose transcript abundances were positively correlated with S-G ratio, and ShCAD2, ShCAD8, ShC4H2, ShC3H1, ShC3H2, ShCCR1, and ShCOMT1, whose expression levels were positively correlated with the accumulation of lignin oligomers, turned out to be the best candidates.
Although phylogenetic and expression analyses are widely used to link a gene with a specific biological process, transgenic approaches are more reliable to characterize gene functions. In this regard, the use of genetic engineering has allowed identifying functional orthologs of lignin biosynthesis genes in grasses (for review, see Harrington et al., 2012). However, until recently, none of the lignin biosynthetic genes had been functionally characterized through forward or reverse genetics in sugarcane. Genetic transformation is still a challenge due to transgene instability, since silencing of the transgenes seems to occur very often in genetically modified sugarcane (Arruda, 2012). Nevertheless, significant progress has been reported. RNA interference-mediated silencing of COMT in sugarcane reduced the lignin content by 3.9% to 13.7% and the S-G ratio from 1.47 in the wild type to values between 1.27 and 0.79 (Jung et al., 2012). Moreover, these changes in the cell wall resulted in an increase of up to 34% in the fermentable Glc yield. To our knowledge, this study not only can be considered as the first functional characterization of a lignin biosynthetic gene in sugarcane by reverse genetics but also confirms that RNA interference is a promising tool for the genetic engineering of sugarcane biomass for second-generation biofuels.
In conclusion, we describe, to our knowledge, the first systematic study of lignin deposition during sugarcane stem development. Sugarcane is the most prominent bioenergy crop and is currently used for the production of sugar-based bioethanol on a commercial scale in developing countries. The remarkable biomass yield potential of sugarcane makes this grass an attractive lignocellulosic feedstock. However, the production of cellulosic biofuels from sugarcane still requires considerable research, especially because of its complex genome and lack of extensive genomic resources. The finding that rind and pith are not only distinct at the anatomical level but also have different lignin content and composition (S-G ratio) must be taken into account in future genetic engineering strategies to tailor sugarcane as a bioenergy crop with reduced recalcitrance and without affecting plant yield and architecture. Therefore, we believe that this work can be considered as a reference study for future applications of sugarcane as a biomass feedstock for renewable biofuels and other bio-based materials made from fermentable sugars.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Two sugarcane (Saccharum spp. hybrids) genotypes were used, IACSP04-529 and IACSP04-683, both derived from an F1 population originating from the breeding program carried out by the Sugarcane Center of the Agronomic Institute of Campinas, at Ribeirão Preto, Brazil. They were selected based on 3 years of evaluation for several technological traits used by the Brazilian sugarcane industry, including lignin content. The two genotypes contained the highest (IACSP04-529) and the lowest (IACSP04-683) lignin contents among 66 individuals of the population. The plants were maintained in the field under usual fertilization and agronomical procedures for the first sugarcane ratoon. The analyzed material was harvested from the first ratoon.
Internodes 2 to 18 were used for Klason lignin analyses in the stem. Internodes were numbered downward on the stalk as described previously (Moore, 1987). Only the region between two adjacent nodes was used, while the nodes themselves were discarded. For the youngest analyzed internode (internode 2), the region subtending the top visible dewlap leaf was harvested. Anatomical and histochemical analyses were carried out on internodes 2, 10, and 15, while phenolic profiling was performed on a pool (n = 5 biological replicates) of young (internodes 1–3) and mature (internodes 15–17) internodes only. For gene expression (quantitative PCR) and analysis of lignin composition (S-G ratio), the sugarcane materials were separated into three pools of internodes, the first made with young internodes (internodes 1–3), the second with intermediate internodes (internodes 5–7), and the third with mature internodes (internodes 15–17). Except for Klason lignin analysis and the phenolic profiling, each pool of internodes was separated into central (pith) and peripheral (rind) regions. A blade was used to obtain the rind, which was removed by cutting thin strips along the internodes (Cesarino et al., 2012). Immediately after the separation of pools and tissues (rind and pith), the plant materials were frozen in liquid N2 and stored at −80°C until use. Noteworthy, for all of the above-mentioned analyses, fresh material was used, in which no Suc extraction was performed previously. The materials are referred to as young pith, young rind, intermediate pith, intermediate rind, mature pith, and mature rind.
Lignin Determinations
The Klason method was used to estimate lignin content, as described by Hatfield and Fukushima (2005). Klason lignin was expressed as the percentage of dry cell wall residue, obtained after the extraction procedure, in relation to the initial dry mass.
In order to determine the S-G ratio, thioacidolysis was carried out according to Lange et al. (1995), and the reaction products were analyzed by gas chromatography-mass spectrometry (Lapierre et al., 1991) using a Shimadzu GCMS-QP Plus 2010, equipped with a moving needle-type injector and a flame ionization mass detector. Silylated samples were injected onto a polydimethylsiloxane capillary column (DB5; 30 m, 0.32 mm i.d., film thickness of 0.25 pm; J&W Scientific), and helium was the gas carrier, using a temperature program from 160°C to 260°C at 2°C min−1. Mass spectra were recorded at 70 eV.
Histochemical Analysis
Stem samples were collected from young (internode 2), intermediate (internode 10), and mature (internode 15) internodes. Sample fixation, embedding, and sectioning were performed according to Cesarino et al. (2012a). Sections were stained for lignin with phloroglucinol (Johansen, 1940). Photomicrographs were taken with an Olympus BX 51 photomicroscope equipped with an Olympus DP71 camera.
Phenolic Profiling
Lyophilized stem tissues were ground to powder, and an aliquot of 10 mg was extracted with 1 mL of 90% methanol at 70°C for 10 min while shaking. After centrifugation, 900 µL of supernatant was freeze dried, and the pellet was resuspended in 200 µL of 1:1 (v/v) cyclohexane:MilliQ water. After centrifugation, the aqueous phase was recovered, and a 15-µL aliquot was subjected to LC-MS analysis with an Acquity ultra-performance liquid chromatography system (Waters) connected to a Synapt HDMS quadrupole time-of-flight mass spectrometer (Micromass). Chromatographic separation was performed on a Waters Acquity BEH C18 column (2.1 mm × 150 mm, 1.7 µm) with a gradient elution, with the mobile phase composed of water containing 1% acetonitrile and 0.1% formic acid (A) and acetonitrile containing 1% water and 0.1% formic acid (B). During the gradient elution, a flow rate of 350 µL min−1 was applied, with initialization at time 0 min, 5% B, 30 min, 50% B, and 33 min, 100% B. The mass spectrometry parameters were used as described by Grunewald et al. (2012).
The identity of phenolic compounds was confirmed by accurate mass measurements and fragmentation patterns using Masslynx software (Waters). For relative quantification of each identified compound, chromatograms were integrated and aligned with the MzMine 2.2 software (Katajamaa and Oresic, 2005). The final peak list contained only peaks that were present in all three biological replicates of at least one sample type (i.e. genotype × tissue × development). Principal component analysis was performed with the prcomp function available in R version 2.6.2. The abundance of the identified compounds was calculated as the peak area normalized to the dry weight.
Analysis of the Brazilian Sugarcane EST Database
The SAS were retrieved from the SUCEST database (http://sucest-fun.org). A BLASTx search of SAS against the sequences of proteins putatively involved in lignin biosynthesis from sorghum (Sorghum bicolor), rice (Oryza sativa), maize (Zea mays), and Arabidopsis (Arabidopsis thaliana) was performed (e-value cutoff less than e−5). In a second step, a new BLASTx alignment also included representative species of the Viridiplantae protein data set, including Populus trichocarpa, Selaginella moellendorffii, and Physcomitrella patens. The SAS coding sequences were deduced from the multiple amino acid sequence alignment with the BLASTx best-match hit.
The translated SAS was then aligned with the 40 first BLASTx hits by MAFFT (Katoh et al., 2005) using default parameters. Comparative analysis of sugarcane lignin genes was done by constructing phylogenetic trees containing the corresponding most similar Viridiplantae sequences. The phylogenetic relationship of the aligned protein sequences was then inferred by maximum likelihood using PhyML (Guindon et al., 2010) with the Whelan and Goldman (WAG) plus gamma substitution model and Aligment Transformation Environment test. This process allowed the identification of the most probable lignin ortholog gene for each identified SAS. As a complementary step to identify orthologs from each gene family, the previously identified SAS were queried by BLASTp (Altschul et al., 1997) into the nonredundant protein databases (UNIPROT [http://www.uniprot.org/], The Arabidopsis Information Resource [http://www.arabidopsis.org/], and National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/]) and by tBLASTx into the MAIZEWALL nucleotide database (http://www.polebio.scsv.ups-tlse.fr/MAIZEWALL/index.html/). The amino acid sequences were aligned with the ClustalW program included in MEGA 5 (Tamura et al., 2011) and then used to construct a phylogenetic tree using the neighbor-joining algorithm (Saitou and Nei, 1987). Gap regions were excluded by manual adjustments. Bootstrap values were obtained after 5,000 replications.
Expression Analysis
Plant material from three biological replicates was ground to powder in liquid N2 using a precooled mortar and pestle and stored at −80°C until use. Total RNA isolation, complementary DNA synthesis, and quantitative reverse transcription-PCR were performed as described previously (Cesarino et al., 2013a). In order to obtain an accurate internal normalization, the expression stability of three reference genes, UBIQUITIN (forward, 5′-TATCAACAGCAATGGGAGCA-3′; reverse, 5′-GTCAGAAGGGAGCAGATGGA-3′), TUBULIN (forward, 5′-CTCCACATTCATCGGCAACTC-3′; reverse, 5′-TCCTCCTCTTCTTCCTCCTCG-3′), and GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (GAPDH; forward, 5′-TTGGTTTCCACTGACTTCGTT-3′; reverse, 5′-CTGTAGCCCCACTCGTTGT-3′), was investigated using the NormFinder software (Andersen et al., 2004). Since GAPDH was the most stable, transcript expression levels were standardized to transcripts of GAPDH as the reference gene. Each PCR was performed in triplicate, and PCR in the absence of template was also performed as a negative control for each primer pair. Relative quantification was determined comparing the transcript level between the target genes and the reference gene by the formula 2−ΔCt, where ΔCt = (Cttag − Ctref), Ct = threshold cycle, tag = tag gene, and ref = reference gene. This method was derived from the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Bayesian Network Analysis
Bayesian network analysis was performed using the BNFinder software (Wilczyński and Dojer, 2009). A total of 35 data points were collected from the network that had samples from young, intermediate, and mature internodes, while 11 data points were collected from the network including biochemical data. In the obtained networks, it is assumed that the expression of genes can depend on conditions (genetic, morphological, and biochemical characteristics) but not the opposite. Also, the orientation of the regulatory interactions was inferred. It was specified that the quantitative PCR data were continuous measurements and that other data (genotype, tissue type, lignin composition, lignin content, and other biochemical information) were discretized. The Bayesian-Dirichlet equivalence scoring criterion was used to learn the network structure for these discrete and continuous variables. In this study, the interactions represented the fact that one variable depends on some other variable, and the type of interaction can be a positive or negative correlation between variables. The network topology was properly reconstructed using Cytoscape (Smoot et al., 2011). Because the data on phenolic profiling were obtained only for mature and young internodes, a network specific for this particular analysis was produced, excluding the data obtained in the intermediate internodes.
Statistical Analysis
Student’s t tests were used to assess differences in lignin content between samples. Three technical replicates were used for each of the three biological replicates (three different plants). For thioacidolysis analysis, only one technical replicate was used for each of three biological replicates. Data were expressed as means ± se. A one-way ANOVA was used to assess differences in gene expression among the analyzed stem tissues. When a significant difference was found, Tukey’s test was used as a post hoc comparison. Statistical significance was considered at an α of 5%. Analyses were carried out using the software GraphPad Prism 5.00 for Windows (GraphPad Software). Phenolic profiling data were analyzed by a two-way ANOVA model to take into account the effects of genotype, development, and the interaction between genotype and development. In the case of a nonsignificant interaction, the two-way ANOVA model was reduced by eliminating the interaction term. Whenever the interaction was significant, Student’s t tests were computed to evaluate the main effects. Student’s t tests were computed between both genotypes within each developmental stage and also between both developmental stages within each genotype. Whenever necessary, Box-Cox transformations (root, natural logarithm, square root, or inverse transformation) were performed to ensure homoscedasticity (Supplemental Table S1). To correct for multiple testing, the Benjamini-Hochberg Q value was computed using the qvalue (lambda = 0) function present in the R-based qvalue package (Storey, 2002). All computations were performed in R version 2.14.1, and ANOVA models were obtained using the lm function.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1–S10. Phylogenetic analysis of amino acid sequences of monolignol biosynthesis enzymes retrieved from NCBI, TAIR, Uniprot, and MAIZEWALL databases and the identified homologs in sugarcane.
Supplemental Table S1. Statistical analysis of the metabolomic data.
Supplemental Table S2. Gene names, SAS identification, primers, and size of amplicons used for quantitative PCR analysis.
Supplemental Table S3. Values of the quantitative reverse transcription PCR expression analysis of 29 lignin biosynthesis-related genes.
Supplementary Material
Acknowledgments
We thank Viviane Gabriela Santarosa, Kellen Cristine Hisatugo, Laerti Reis Roque, and Eduardo Kiyota for technical assistance.
Glossary
- H
p-hydroxyphenyl
- G
guaiacyl
- S
syringyl
- LC-MS
liquid chromatography-mass spectrometry
- SAS
Sugarcane Assembled Sequences
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250 [DOI] [PubMed] [Google Scholar]
- Arruda P. (2012) Genetically modified sugarcane for bioenergy generation. Curr Opin Biotechnol 23: 315–322 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Somerville C. (2009) Cellulosic biofuels. Annu Rev Plant Biol 60: 165–182 [DOI] [PubMed] [Google Scholar]
- Casu RE, Dimmock CM, Chapman SC, Grof CP, McIntyre CL, Bonnett GD, Manners JM. (2004) Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Mol Biol 54: 503–517 [DOI] [PubMed] [Google Scholar]
- Casu RE, Jarmey JM, Bonnett GD, Manners JM. (2007) Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip Sugarcane Genome Array expression profiling. Funct Integr Genomics 7: 153–167 [DOI] [PubMed] [Google Scholar]
- Cesarino I, Araújo P, Sampaio Mayer JL, Paes Leme AF, Mazzafera P. (2012) Enzymatic activity and proteomic profile of class III peroxidases during sugarcane stem development. Plant Physiol Biochem 55: 66–76 [DOI] [PubMed] [Google Scholar]
- Cesarino I, Araújo P, Leme AFP, Creste S, Mazzafera P. (2013a) Suspension cell culture as a tool for the characterization of class III peroxidases in sugarcane. Plant Physiol Biochem 62: 1–10 [DOI] [PubMed] [Google Scholar]
- Cesarino I, Araújo P, Sampaio Mayer JL, Vicentini R, Berthet S, Demedts B, Vanholme B, Boerjan W, Mazzafera P. (2013b) Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J Exp Bot 64: 1769–1781 [DOI] [PubMed] [Google Scholar]
- Cheavegatti-Gianotto A, de Abreu HM, Arruda P, Bespalhok Filho JC, Burnquist WL, Creste S, di Ciero L, Ferro JA, de Oliveira Figueira AV, de Sousa Filgueiras T, et al. (2011) Sugarcane (Saccharum × officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Trop Plant Biol 4: 62–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA. (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen L, Auh C, Chen F, Cheng X, Aljoe H, Dixon RA, Wang Z. (2002) Lignin deposition and associated changes in anatomy, enzyme activity, gene expression, and ruminal degradability in stems of tall fescue at different developmental stages. J Agric Food Chem 50: 5558–5565 [DOI] [PubMed] [Google Scholar]
- Clifford MN, Johnston KL, Knight S, Kuhnert N. (2003) Hierarchical scheme for LC-MSn identification of chlorogenic acids. J Agric Food Chem 51: 2900–2911 [DOI] [PubMed] [Google Scholar]
- Colombo R, Yariwake JH, Queiroz EF, Ndjoko K, Hostettmann K. (2005) On-line identification of sugarcane (Saccharum officinarum L.) methoxyflavones by liquid chromatography-UV detection using post-column derivatization and liquid chromatography-mass spectrometry. J Chromatogr A 1082: 51–59 [DOI] [PubMed] [Google Scholar]
- Colombo R, Yariwake JH, Queiroz EF, Ndjoko K, Hostettmann K. (2006) On-line identification of further flavone C- and O-glycosides from sugarcane (Saccharum officinarum L., Gramineae) by HPLC-UV-MS. Phytochem Anal 17: 337–343 [DOI] [PubMed] [Google Scholar]
- Del Río JC, Rencoret J, Prinsen P, Martínez AT, Ralph J, Gutiérrez A. (2012) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60: 5922–5935 [DOI] [PubMed] [Google Scholar]
- Dias MO, Ensinas AV, Nebra SA, Maciel R, Rossell CE, Maciel MR. (2009) Production of bioethanol and other bio-based materials from sugarcane bagasse: integration to conventional bioethanol production process. Chem Eng Res Des 87: 1206–1216 [Google Scholar]
- Dias MO, Junqueira TL, Cavalett O, Cunha MP, Jesus CD, Rossell CE, Maciel Filho R, Bonomi A. (2012) Integrated versus stand-alone second generation ethanol production from sugarcane bagasse and trash. Bioresour Technol 103: 152–161 [DOI] [PubMed] [Google Scholar]
- Duarte-Almeida JM, Salatino A, Genovese MI, Lajolo FM. (2011) Phenolic composition and antioxidant activity of culms and sugarcane (Saccharum officinarum L.) products. Food Chem 125: 660–664 [Google Scholar]
- Glassop D, Roessner U, Bacic A, Bonnett GD. (2007) Changes in the sugarcane metabolome with stem development: are they related to sucrose accumulation? Plant Cell Physiol 48: 573–584 [DOI] [PubMed] [Google Scholar]
- Grabber JH. (2005) How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci 45: 820–831 [Google Scholar]
- Grabber JH, Ralph J, Lapierre C, Barrière Y. (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. C R Biol 327: 455–465 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Schatz PF, Kim H, Lu F, Ralph J. (2010) Identifying new lignin bioengineering targets. 1. Monolignol-substitute impacts on lignin formation and cell wall fermentability. BMC Plant Biol 10: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, De Smet I, Lewis DR, Löfke C, Jansen L, Goeminne G, Vanden Bossche R, Karimi M, De Rybel B, Vanholme B, et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109: 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie S, San-Clemente H, Deswarte C, Martinez Y, Lapierre C, Murigneux A, Barrière Y, Pichon M, Goffner D. (2007) MAIZEWALL: database and developmental gene expression profiling of cell wall biosynthesis and assembly in maize. Plant Physiol 143: 339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Harrington MJ, Mutwil M, Barrière Y, Sibout R. (2012) Lignins: biosynthesis, biodegradation and bioengineering. Adv Bot Res 61: 77–112 [Google Scholar]
- Hatfield R, Fukushima RS. (2005) Can lignin be accurately measured? Crop Sci 45: 832–839 [Google Scholar]
- Hatfield RD, Jung HJ, Ralph J, Buxton DR, Weimer PJ. (1994) A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J Sci Food Agric 65: 51–58 [Google Scholar]
- Jacobsen KR, Fisher DG, Maretzki A, Moore PH. (1992) Developmental changes in the anatomy of the sugarcane stem in relation to phloem unloading and sucrose storage. Bot Acta 105: 70–80 [Google Scholar]
- Johansen DA. (1940) Plant Microtechnique. McGraw-Hill, New York [Google Scholar]
- Jung HG, Casler MD. (2006) Maize stem tissues: cell wall concentration and composition during development. Crop Sci 46: 1793–1800 [Google Scholar]
- Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F. (2012) RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol J 10: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Katajamaa M, Oresic M. (2005) Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics 6: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma Ki, Toh H, Miyata T. (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyota E, Mazzafera P, Sawaya ACHF. (2012) Analysis of soluble lignin in sugarcane by ultrahigh performance liquid chromatography-tandem mass spectrometry with a do-it-yourself oligomer database. Anal Chem 84: 7015–7020 [DOI] [PubMed] [Google Scholar]
- Lange BM, Lapierre C, Sandermann H., Jr (1995) Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol 108: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Monties B. (1991) Thioacidolysis of spruce lignin: GC-MS analysis of the main dimers recovered after Raney nickel desulphuration. Holzforschung 45: 61–68 [Google Scholar]
- Lingle SE, Thomson JL. (2012) Sugarcane internode composition during crop development. BioEnergy Res 5: 168–178 [Google Scholar]
- Lisboa CC, Butterbach-Bahl K, Mauder M, Kiese R. (2011) Bioethanol production from sugarcane and emissions of greenhouse gases: known and unknowns. Glob Change Biol Bioenergy 3: 277–292 [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopes FJ, Silvério FO, Baffa DC, Loureiro ME, Barbosa MH. (2011) Determination of sugarcane bagasse lignin S/G/H ratio by pyrolysis GC/MS. J Wood Chem Technol 31: 309–323 [Google Scholar]
- Masarin F, Gurpilhares DB, Baffa DC, Barbosa MH, Carvalho W, Ferraz A, Milagres AM. (2011) Chemical composition and enzymatic digestibility of sugarcane clones selected for varied lignin content. Biotechnol Biofuels 4: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchin V, Argillier O, Rocher F, Hébert Y, Mila I, Pollet B, Barriére Y, Lapierre C. (2005) In search of a maize ideotype for cell wall enzymatic degradability using histological and biochemical lignin characterization. J Agric Food Chem 53: 5872–5881 [DOI] [PubMed] [Google Scholar]
- Moore PH. (1987) Anatomy and morphology. In D Heinz, ed, Sugarcane Improvement through Breeding. Elsevier, Amsterdam, pp 85–142 [Google Scholar]
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inzé D, Messens E, et al (2010a) Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol 153: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Kim H, Lu FC, Dima O, Akiyama T, Vanholme R, Niculaes C, Goeminne G, Inzé D, Messens E, et al (2010b) Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal Chem 82: 8095–8105 [DOI] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W. (2004) Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TA, Kessler JR, Buxton DR. (1994) Maize internode elongation patterns. Crop Sci 34: 1055–1060 [Google Scholar]
- Moura JC, Bonine CA, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P. (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52: 360–376 [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C. (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Papini-Terzi FS, Rocha FR, Vêncio RZ, Oliveira KC, Felix JdM, Vicentini R, Rocha CdS, Simões AC, Ulian EC, di Mauro SM, et al. (2005) Transcription profiling of signal transduction-related genes in sugarcane tissues. DNA Res 12: 27–38 [DOI] [PubMed] [Google Scholar]
- Rabelo SC, Carrere H, Maciel Filho R, Costa AC. (2011) Production of bioethanol, methane and heat from sugarcane bagasse in a biorefinery concept. Bioresour Technol 102: 7887–7895 [DOI] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W. (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxy phenyl propanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Ramos RL, Tovar FJ, Junqueira RM, Lino FB, Sachetto-Martins G. (2001) Sugarcane expressed sequences tags (ESTs) encoding enzymes involved in lignin biosynthesis pathways. Genet Mol Biol 24: 235–241 [Google Scholar]
- Rezende CA, de Lima MA, Maziero P, Deazevedo ER, Garcia W, Polikarpov I. (2011) Chemical and morphological characterization of sugarcane bagasse submitted to a delignification process for enhanced enzymatic digestibility. Biotechnol Biofuels 4: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboulet C, Guillaumie S, Mechin V, Bosio M, Pichon M, Goffner D, Lapierre C, Pollet B, Lefevre B, Martinant JP, et al (2009) Kinetics of phenylpropanoid gene expression in maize growing internodes: relationships with cell wall deposition. Crop Sci 49: 211–223 [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Shen H, Fu C, Xiao X, Ray T, Tang Y, Wang Z, Chen F. (2009) Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. BioEnergy Res 2: 233–245 [Google Scholar]
- Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL. (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Simmons BA, Loqué D, Ralph J. (2010) Advances in modifying lignin for enhanced biofuel production. Curr Opin Plant Biol 13: 313–320 [DOI] [PubMed] [Google Scholar]
- Siqueira G, Milagres AM, Carvalho W, Koch G, Ferraz A. (2011) Topochemical distribution of lignin and hydroxycinnamic acids in sugar-cane cell walls and its correlation with the enzymatic hydrolysis of polysaccharides. Biotechnol Biofuels 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. (2002) A direct approach to false discovery rates. J R Stat Soc B 64: 479–498 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W. (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B, Desmet T, Ronsse F, Rabaey K, Breusegem FV, Mey MD, Soetaert W, Boerjan W. (2013a) Towards a carbon-negative sustainable bio-based economy. Front Plant Sci 4: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013b) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. (2010a) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010b) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Van Acker R, Boerjan W. (2010c) Potential of Arabidopsis systems biology to advance the biofuel field. Trends Biotechnol 28: 543–547 [DOI] [PubMed] [Google Scholar]
- Van Soest PJ. (1967) Development of a comprehensive system of feed analysis and its application to forage. J Anim Sci 26: 119–120 [Google Scholar]
- Vega-Sánchez ME, Ronald PC. (2010) Genetic and biotechnological approaches for biofuel crop improvement. Curr Opin Biotechnol 21: 218–224 [DOI] [PubMed] [Google Scholar]
- Vogel J. (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11: 301–307 [DOI] [PubMed] [Google Scholar]
- Weng JK, Li X, Bonawitz ND, Chapple C. (2008) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr Opin Biotechnol 19: 166–172 [DOI] [PubMed] [Google Scholar]
- Wenzig E, Kunert O, Ferreira D, Schmid M, Schühly W, Bauer R, Hiermann A. (2005) Flavonolignans from Avena sativa. J Nat Prod 68: 289–292 [DOI] [PubMed] [Google Scholar]
- Wilczyński B, Dojer N. (2009) BNFinder: exact and efficient method for learning Bayesian networks. Bioinformatics 25: 286–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell A, Gracom K, Katahira R, Chen F, Pu Y, Ragauskas A, Dixon RA, Davis M. (2010) Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. J Biol Chem 285: 38961–38968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.