A newly identified cytochrome P450 isoform initiates the synthesis of valuable alkaloids in leaves of Catharanthus roseus by hydroxylating tabersonine.
Abstract
Hydroxylation of tabersonine at the C-16 position, catalyzed by tabersonine 16-hydroxylase (T16H), initiates the synthesis of vindoline that constitutes the main alkaloid accumulated in leaves of Catharanthus roseus. Over the last decade, this reaction has been associated with CYP71D12 cloned from undifferentiated C. roseus cells. In this study, we isolated a second cytochrome P450 (CYP71D351) displaying T16H activity. Biochemical characterization demonstrated that CYP71D12 and CYP71D351 both exhibit high affinity for tabersonine and narrow substrate specificity, making of T16H, to our knowledge, the first alkaloid biosynthetic enzyme displaying two isoforms encoded by distinct genes characterized to date in C. roseus. However, both genes dramatically diverge in transcript distribution in planta. While CYP71D12 (T16H1) expression is restricted to flowers and undifferentiated cells, the CYP71D351 (T16H2) expression profile is similar to the other vindoline biosynthetic genes reaching a maximum in young leaves. Moreover, transcript localization by carborundum abrasion and RNA in situ hybridization demonstrated that CYP71D351 messenger RNAs are specifically located to leaf epidermis, which also hosts the next step of vindoline biosynthesis. Comparison of high- and low-vindoline-accumulating C. roseus cultivars also highlights the direct correlation between CYP71D351 transcript and vindoline levels. In addition, CYP71D351 down-regulation mediated by virus-induced gene silencing reduces vindoline accumulation in leaves and redirects the biosynthetic flux toward the production of unmodified alkaloids at the C-16 position. All these data demonstrate that tabersonine 16-hydroxylation is orchestrated in an organ-dependent manner by two genes including CYP71D351, which encodes the specific T16H isoform acting in the foliar vindoline biosynthesis.
Catharanthus roseus displays an active specialized metabolism producing more than 100 monoterpene indole alkaloids (MIAs). MIAs are a major component of the chemical arsenal that likely protects the plant by acting against herbivores (Guirimand et al., 2010a; Roepke et al., 2010). These MIAs include highly valuable molecules that exhibit interesting pharmacological properties for human health. For instance, vinblastine, vincristine, and their derivatives are widely used in cancer chemotherapy, where they act as microtubule disruptors (Gigant et al., 2005). Due to highly complex structures, these therapeutic molecules are prepared by semisynthesis using natural precursors extracted from C. roseus leaves. Their extremely low level in planta explains their high cost of production and justifies the extensive study of the MIA biosynthetic pathway that has been carried out over the last three decades, making of C. roseus one of the most studied plants for specialized metabolism (Facchini and De Luca, 2008; Guirimand et al., 2010b).
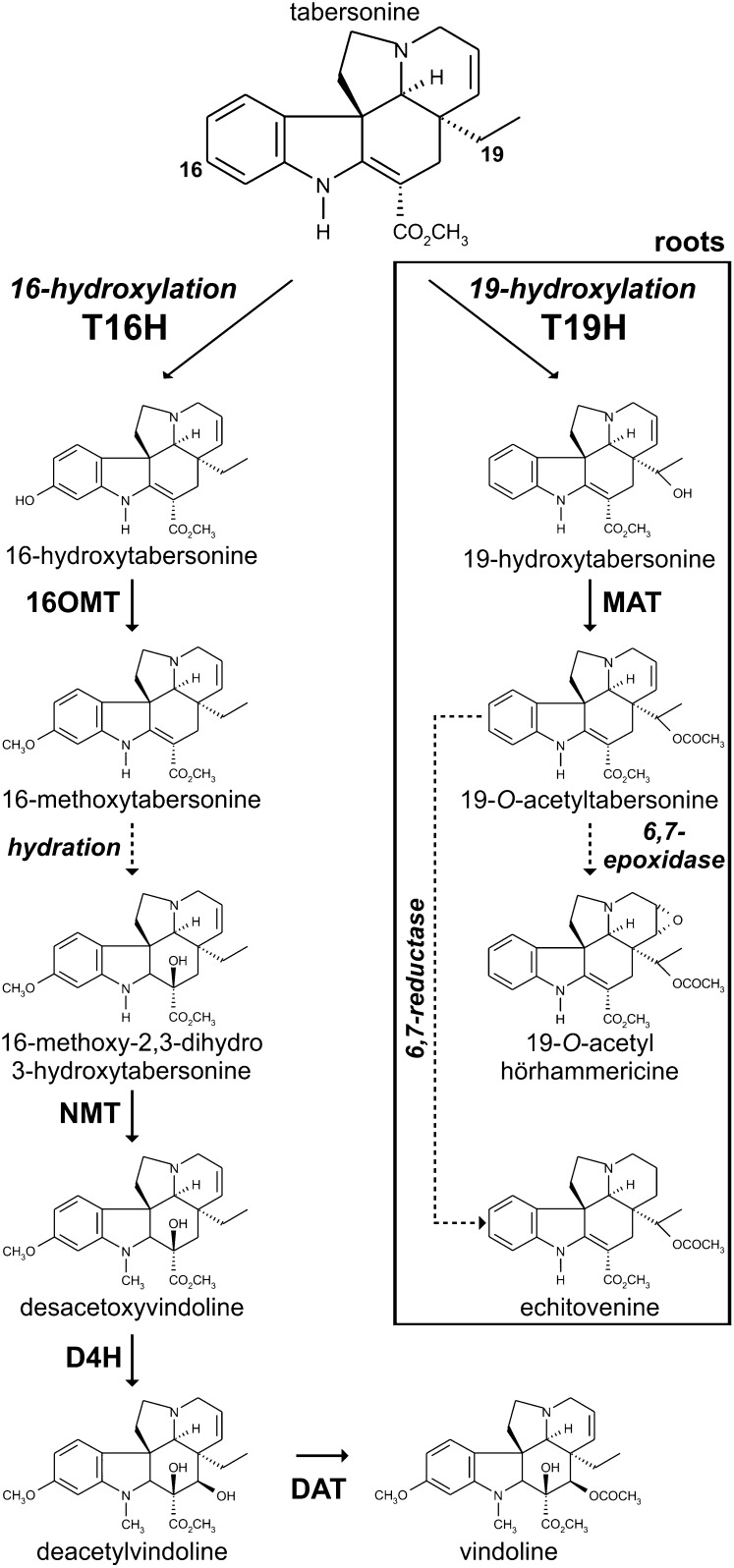
Vinblastine and vincristine are dimeric MIAs resulting from the oxidative coupling of two monomeric precursors, catharanthine (Iboga-type MIA) and vindoline (Aspidosperma-type MIA), which constitute the major MIAs that accumulate in C. roseus leaves as compared with the trace amounts of dimeric MIAs (Westekemper et al., 1980). Vindoline as well as dimeric MIAs are restricted to aerial organs such as leaves and stems, while catharanthine is accumulated equally throughout underground and aboveground organs (Deus-Neumann et al., 1987; Balsevich and Bishop, 1989), with a specific secretion of this MIA in leaf wax exudates (Roepke et al., 2010). In contrast to the limited available data regarding catharanthine biosynthesis, characterization of the in folio vindoline biosynthetic pathway is now approaching completion (Fig. 1). This pathway relies on a six-step conversion of tabersonine initiated by tabersonine 16-hydroxylase (T16H; EC 1.14.13.73), which is a cytochrome P450 (P450) that hydroxylates tabersonine at the 16 position (St-Pierre and De Luca, 1995). Accumulating evidence argues for a key role of T16H in vindoline biosynthesis, since, for instance, screening of low- and high-MIA-accumulating cultivars of C. roseus revealed a direct correlation of vindoline content with T16H activity (Magnotta et al., 2006). After hydroxylation, the resulting 16-hydroxytabersonine is successively O-methylated by 16-hydroxytabersonine 16-O-methyltransferase (16OMT; Levac et al., 2008), reduced at the C2-C3 double bond with concomitant hydroxylation at the C-3 position through an uncharacterized mechanism of hydration, and N-methylated at the indolic nitrogen by 16-methoxy-2,3-dihydro-3-hydroxytabersonine N-methyltransferase (NMT; De Luca et al., 1987; Liscombe et al., 2010) to form desacetoxyvindoline. The last two steps of this pathway involve an additional hydroxylation catalyzed by desacetoxyvindoline-4-hydroxylase (D4H; Vazquez-Flota et al., 1997) and a final O-acetylation performed by deacetylvindoline-4-O-acetyltransferase (DAT; St-Pierre et al., 1998), leading to the formation of vindoline. In roots, the fate of tabersonine is different and relies on its conversion into hörhammericine and echitovenine, where a recently characterized P450, CYP71BJ1, acts as 19-hydroxylase (T19H; Giddings et al., 2011) and a minovincinine-O-acetyltransferase (MAT; Laflamme et al., 2001) closely related to DAT catalyzes the subsequent reaction (Fig. 1).
Figure 1.
The position of tabersonine hydroxylation governs the biosynthesis of downstream MIAs. In aerial organs, tabersonine is hydroxylated at the C-16 position by T16H and subsequently metabolized by 16OMT, uncharacterized hydratase, NMT, D4H, and DAT to produce vindoline. In roots, tabersonine is hydroxylated at the C-19 position by T19H prior to acetylation catalyzed by MAT and additional reactions of reduction or epoxidation. Dashed lines represent uncharacterized enzymatic reactions.
The vindoline biosynthetic pathway displays a high degree of compartmentalization of both gene expression and enzymatic reactions, especially in C. roseus leaves. While transcripts and enzyme activities of both D4H and DAT have been localized to laticifers/idioblasts, adaxial and abaxial epidermal cells have been shown to host the starting steps of the biosynthetic pathway. The epidermal localization of 16OMT transcripts was first established using epidermis-specific mRNA enrichment procedures performed by carborundum abrasion or epidermis laser-capture microdissection/EST sequencing (Murata and De Luca, 2005; Levac et al., 2008; Murata et al., 2008) and was further confirmed by RNA in situ hybridization (Guirimand et al., 2011b). By contrast, the situation is less clear for T16H, whose only known coding sequence, CYP71D12, was cloned from C. roseus cell cultures (Schröder et al., 1999; Guirimand et al., 2011b). Evidence of the leaf epidermal localization of T16H transcripts was initially obtained by reverse transcription (RT)-PCR procedures and epidermis EST sequencing (Murata and De Luca, 2005; Murata et al., 2008). However, all our attempts to confirm these results by RNA in situ hybridization failed. This is surprising considering that the frequency of ESTs annotated as T16H in the C. roseus epidermome is similar to or higher than those of other MIA biosynthetic genes, such as strictosidine synthase and strictosidine β-d-glucosidase, both of which have previously been localized in leaf epidermis using RNA in situ hybridization (St-Pierre et al., 1999; Murata et al., 2008; Guirimand et al., 2010a). This discrepancy prompted us to investigate the 16-hydroxylation of tabersonine in more detail and led to the identification of a second P450 displaying T16H activity, CYP71D351. Comparative expression analysis of CYP71D351 and CYP71D12, cellular transcript distribution, as well as gene silencing in seedlings allowed us to conclude that the newly discovered CYP71D351, and not the previously cloned CYP71D12, initiates vindoline biosynthesis in C. roseus leaves. In contrast, CYP71D12 appears to be dedicated to vindoline biosynthesis in flowers.
RESULTS
Isolation of CYP71D351 Complementary DNA
While the coding sequence of the original full-length T16H (CYP71D12; GenBank accession no. FJ647194) could be efficiency amplified by PCR using complementary DNAs (cDNAs) prepared from C. roseus cell cultures (Guirimand et al., 2011b), all our attempts to carry out this amplification using reverse-transcribed RNAs extracted from young leaves failed. As a consequence, additional amplifications of a partial internal sequence were attempted using young leaf cDNAs and the previously described CrT16H-RT01 and CrRT16H-RT02 primers (Murata and De Luca, 2005). This allowed us to obtain a unique 242-bp cDNA fragment that displays 96% identity with the corresponding region of the CYP71D12 coding sequence (Supplemental Fig. S1). By combining 5′ and 3′ RACE with EST database analysis, we amplified a 1,698-bp-long cDNA that encompassed a 1,539-bp open reading frame (ORF) 86.5% identical to that of the CYP71D12 nucleic acid sequence. Interestingly, we noted that all four T16H-annotated ESTs identified within the C. roseus epidermome EST database were strictly identical to the new amplified cDNA and slightly different from CYP71D12 (Murata et al., 2008; Supplemental Table S1). The encoded 512-amino acid protein, which is 82% identical to CYP71D12 (Supplemental Fig. S2), was named CYP71D351 according to the standardized system of P450 nomenclature (Nelson, 2006). By contrast, CYP71D351 displays only low amino acid identity with other P450s associated with MIA biosynthesis, such as secologanin synthase (CYP72A1; 20%), geraniol 10-hydroxylase (CYP76B6; 33%), and the recently characterized tabersonine 19-hydroxylase (CYP71BJ1; 35%).
CYP71D351 Catalyzes the 16-Hydroxylation of Tabersonine
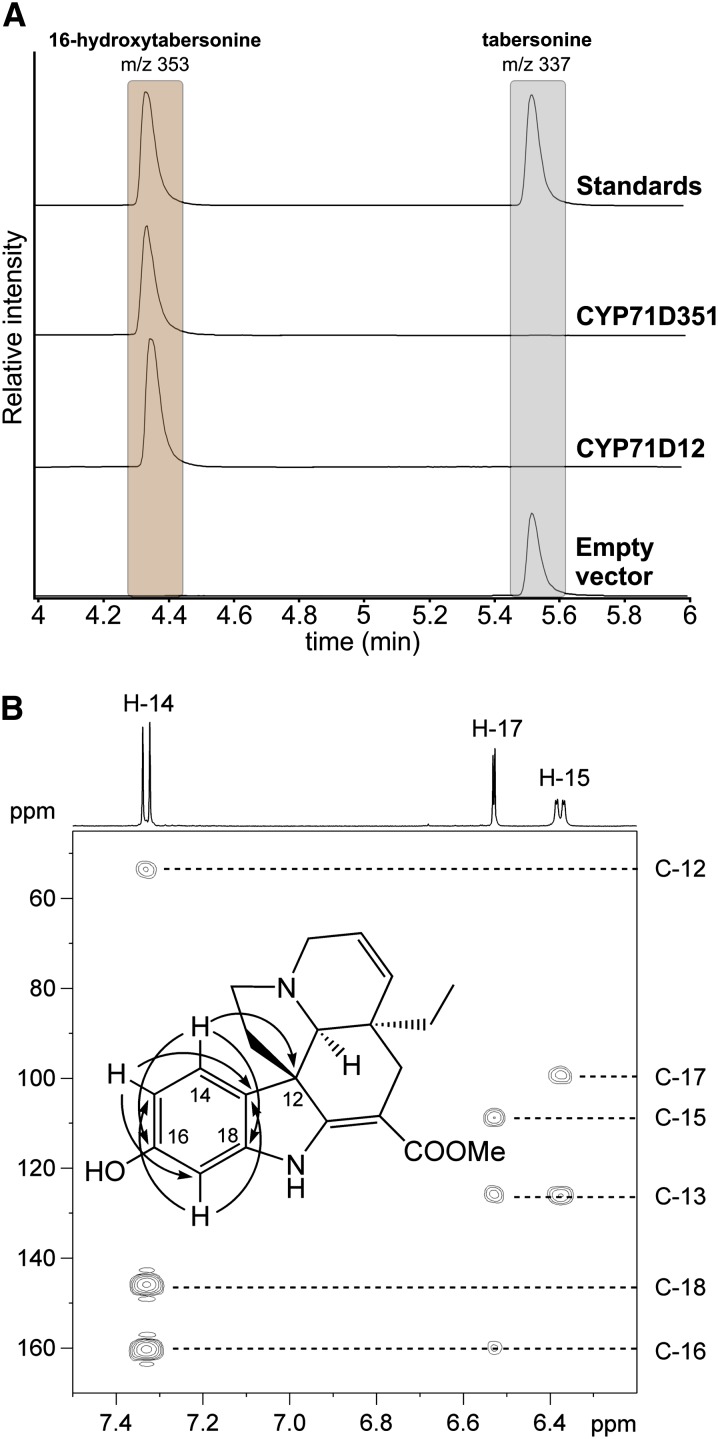
To analyze the activity of CYP71D351, both CYP71D12 (the gene originally assigned as T16H) and CYP71D351 were individually expressed in the Saccharomyces cerevisiae WAT11 strain, a yeast strain engineered for plant P450 protein studies via the coexpression of the Arabidopsis (Arabidopsis thaliana) NADPH P450 REDUCTASE1 (Pompon et al., 1996). While microsomes of the control yeast strain (carrying the empty expression vector) did not metabolize tabersonine, liquid chromatography (LC)-mass spectrometry (MS) and gas chromatography-MS analyses revealed that microsomes of both strains expressing the P450s could convert tabersonine into a more hydrophilic compound displaying a retention time and a mass-to-charge ratio consistent with hydroxylated tabersonine (Fig. 2A; Supplemental Fig. S3). No enzymatic products were observed when NADPH was omitted from the reactions, confirming that the hydroxylation occurred in a NADPH-dependent manner, as described previously for CYP71D12 (St-Pierre and De Luca, 1995; Schröder et al., 1999). To investigate the position at which CYP71D351 was hydroxylating tabersonine, the enzymatic product from a large-scale reaction was purified and analyzed by 1H-NMR, 1H-1H-COSY (for correlation spectroscopy), Heteronuclear Single Quantum Coherence (HSQC), and Heteronuclear Multiple Bond Correlation (HMBC) NMR. The 13C-NMR chemical shifts of the new metabolite, which were obtained from the HSQC and HMBC spectra, were similar to the ones reported previously for 16-hydroxytabersonine (He et al., 1994). Notably, in comparison with the 13C-NMR spectra reported for tabersonine (Wenkert et al., 1973), the signal from C-16 was shifted to a lower field (from δ 127.6 to δ 160.6), while the signals from C-15 and C-17 were shifted to a higher field (from δ 120.5 to δ 108.7 and from δ 109.2 to δ 99.6, respectively), clearly indicating an electronegative substituent, such as a hydroxyl group, at C-16. Also importantly, the three-spin system of H-14 (δ 7.32, d, J = 8.1 Hz), H-15 (δ 6.37, dd, J = 8.1, 2.1 Hz), and H-17 (δ 6.53, d, Jv= 2.1 Hz) and their HMBC correlations through three bonds (H-14 → C-12, C-16, C-18; H-15 → C-13, C-17; H-17 → C-13, C-15) confirmed the hydroxyl transfer at the C16 position of the aromatic ring (Fig. 2B). All these results demonstrate that CYP71D351 catalyzes the 16-hydroxylation of tabersonine and, in turn, prove the existence of two T16H isoforms in C. roseus. Therefore, we propose to extend the T16H nomenclature and to annotate CYP71D12 and CYP71D351 as T16H1 and T16H2, respectively.
Figure 2.
CYP71D351 and CYP71D12 catalyze the 16-hydroxylation of tabersonine. A, LC-MS results using selected ion monitoring (tabersonine, mass-to-charge ratio 337; 16-hydroxytabersonine, mass-to-charge ratio 353) of the reaction products of microsomes purified from yeast cell cultures expressing either CYP71D351 or CYP71D12 or containing the empty pYeDP60 vector. B, Partial HMBC spectrum for 16-hydroxytabersonine isolated from yeast culture expressing CYP71D351. The position of the new hydroxyl group was established from HMBC correlations through three bonds (H-14/C-12, H-14/C-16, H-14/C-18, H-15/C-13, H-15/C-17, H-17/C-13, and H-17/C-15) as indicated by arrows in the inset structure. [See online article for color version of this figure.]
Enzyme Kinetic Parameters
Both CYP71D12- and CYP71D351-enriched yeast microsomes were incubated with varying amounts of tabersonine to compare the kinetic parameters of the two enzymes (Supplemental Fig. S4). CYP71D12 (T16H1) displayed an apparent Km of 350 ± 100 nm and a Vmax of 1.8 ± 0.05 µm min−1. Interestingly, these values are of the same magnitude as those reported for T19H but much lower than the Km for tabersonine evaluated using a T16H/16OMT coupled system (11 µm; St-Pierre and De Luca, 1995; Giddings et al., 2011). By contrast, CYP71D351 (T16H2) exhibited a lower apparent Km of 70 ± 20 nm and a Vmax of 2.2 ± 0.25 µm min−1. This difference in apparent Km suggests that this new isoform has a higher affinity for tabersonine than CYP71D12.
Substrate Specificity Analysis
To analyze the substrate specificity of CYP71D351 and CYP71D12, microsomes from yeast strains expressing each enzyme were assayed against several different compounds, including MIAs, MIA precursors, and naringenin, which is a hydroxylatable flavonoid with aromatic ring, accumulated in leaf epidermis and substrate of geraniol 10-hydroxylase (Sung et al., 2011). LC-MS analysis of reaction mixtures revealed that, among all the tested compounds, only tabersonine (Aspidosperma-type MIA) and to a lesser extent the two closely related compounds, 2,3-dihydro-3-hydroxytabersonine and 2,3-dihydrotabersonine, were metabolized by the two enzymes (Table I). Interestingly, while hydroxylation of these last two compounds by CYP71D12 (T16H1) occurred at very low levels, CYP71D351 (T16H2) was more permissive toward these substrates. The lack of hydroxylating activity toward the other substrates tested, including naringenin, suggests that both CYP71D351 and CYP71D12 are specific enzymes acting in vivo in the conversion of tabersonine to 16-hydroxytabersonine in the vindoline pathway.
Table I. CYP71D351 and CYP71D12 substrate specificities.
+ and – denote capacity and incapacity of hydroxylation. The conversion of substrates (%) is given in parentheses.
| Substrate | CYP71D351 | CYP71D12 |
|---|---|---|
| Naringenin | – | – |
| Tryptamine | – | – |
| Secologanin | – | – |
| Strictosidine | – | – |
| Ajmalicine | – | – |
| Tabersonine | +++ (96.3 ± 0.6) | +++ (93.6 ± 0.3) |
| 2.3-Dihydrotabersonine | + (31.3 ± 1.7) | +/− (2.1 ± 0.2) |
| 2.3-Dihydro-3-hydroxytabersonine | + (18.2 ± 0.9) | +/− (2.6 ± 0.2) |
| Vindoline | – | – |
| Catharanthine | – | – |
CYP71D351 (T16H2) and CYP71D12 (T16H1) Display Different Transcript Distributions in C. roseus
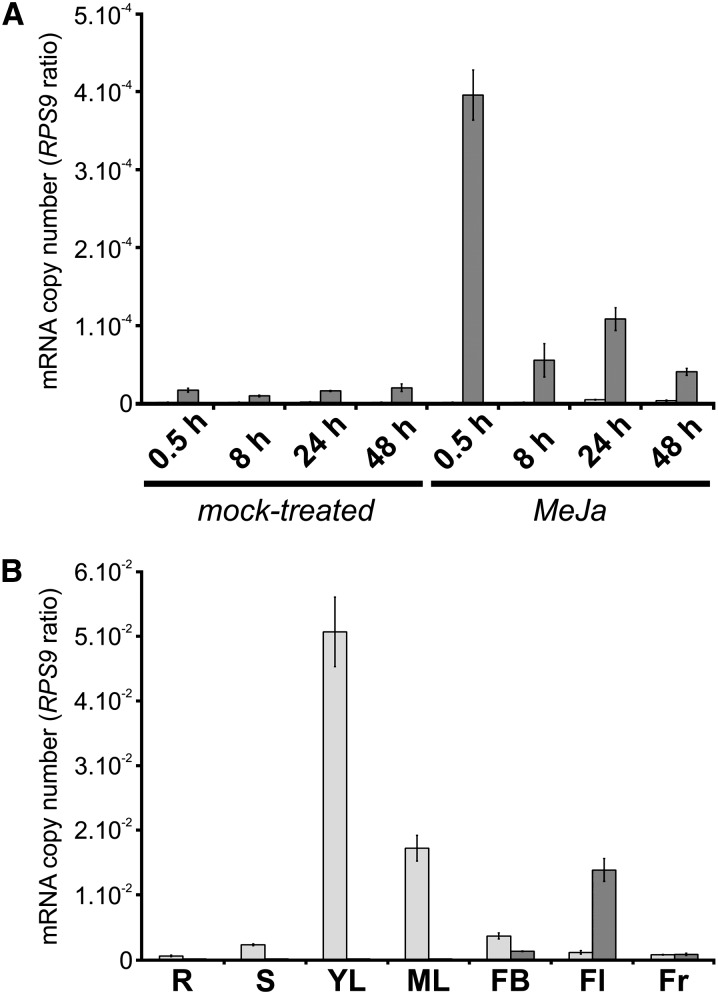
The identification of a second T16H isoform prompted us to investigate the distribution of CYP71D351 and CYP71D12 transcripts in C. roseus plant organs and cell cultures. Since CYP71D12 (T16H1) was initially isolated from C. roseus cell cultures, we first analyzed transcript levels of both enzymes in this system by quantitative real-time PCR. As expected, CYP71D12 transcripts were easily detectable in cell cultures and were responsive to methyl jasmonate (MeJa) treatment, as reported previously (Schröder et al., 1999; He et al., 2011; Fig. 3A). By contrast, transcripts of CYP71D351 (T16H2) were barely detectable (approximately 100- to 1,000-fold less abundant) and exhibited a weak response to MeJa treatment, suggesting that the T16H activity previously measured in cell cultures resulted mainly from CYP71D12 (Schröder et al., 1999). The situation was strikingly different in whole plants (cv Pacifica Pink), given that the CYP71D12 expression level was extremely low in most of the tested organs (Fig. 3B). CYP71D12 transcripts were barely detectable or undetectable in roots, internodes, young and mature leaves, and flower buds but were relatively abundant in fully developed flowers. The expression pattern of CYP71D351 was strikingly dissimilar, including very low transcript levels in roots and fruits, low levels in stems, flower buds, and flowers, and very high amounts in leaves, especially young leaves, where the highest levels of T16H enzymatic activity have been found (St-Pierre and De Luca, 1995). In this organ, CYP71D351 transcript level was also dramatically higher than the amount of CYP71D12 transcripts, which just reached the level of detection. These results suggest that CYP71D351 (T16H2) has a major role in the biosynthesis of vindoline in leaves, while CYP71D12 (T16H1) may play an analogous role in flowers, where vindoline does accumulate, albeit at lower levels (Supplemental Fig. S5).
Figure 3.
Analysis of CYP71D351 (T16H2) and CYP71D12 (T16H1) expression in C. roseus organs and cell cultures. CYP71D351 (light gray bars) and CYP71D12 (dark gray bars) transcript levels were determined by real-time RT-PCR analyses performed on total RNA extracted from C. roseus cells subjected to MeJa or mock treatment during 0.5, 8, 24, and 48 h (A) and from C. roseus organs (B). CYP71D351 and CYP71D12 transcript copy numbers were normalized using CrRPS9. R, Roots; S, stems; YL, young leaves; ML, mature leaves; FB, flower buds; Fl, flowers; Fr, fruits.
CYP71D351 (T16H2) Expression Follows the Expression of Other Vindoline Biosynthetic Genes
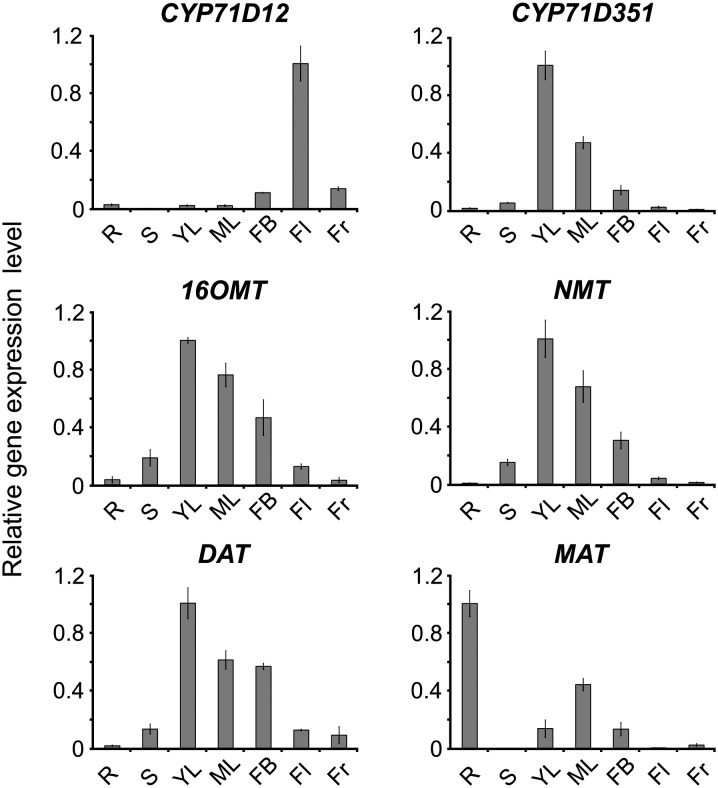
The specific distribution of CYP71D12 and CYP71D351 transcripts was also compared with that of other known vindoline biosynthetic pathway genes, including 16OMT, NMT, and DAT, together with MAT, which catalyzes the penultimate step in the biosynthesis of echitovenine in roots (Laflamme et al., 2001). As depicted in Figure 4, CYP71D351 (T16H2) displayed the same pattern of expression as 16OMT, NMT, and DAT, including highest transcript amounts in young leaves, lower levels in mature leaves and flower buds, and very low levels in roots, stems, flowers, and fruits. This contrasts with the expression profile of MAT, which had the highest mRNA levels in roots, consistent with its role in echitovenine/hörhammericine biosynthesis. In addition, CYP71D12 (T16H1) displayed an expression profile that was very different from the characteristic profile of the known vindoline biosynthetic genes. These expression profiles argue in favor of the involvement of CYP71D351 in leaf vindoline biosynthesis.
Figure 4.
CYP71D351 (T16H2) has a similar expression pattern to other vindoline biosynthetic genes. Relative expression of CYP71D12, CYP71D351, 16OMT, NMT, and DAT was determined by real-time RT-PCR analyses performed on total RNA extracted from various C. roseus organs. MAT, a root-specific hydroxylase, was also included for comparison. CrRPS9 was used as a reference gene. R, Roots; S, stems; YL, young leaves; ML, mature leaves; FB, flower buds; Fl, flowers; Fr, fruits.
CYP71D351 (T16H2) Transcripts Are Localized in the Epidermis of C. roseus Leaves
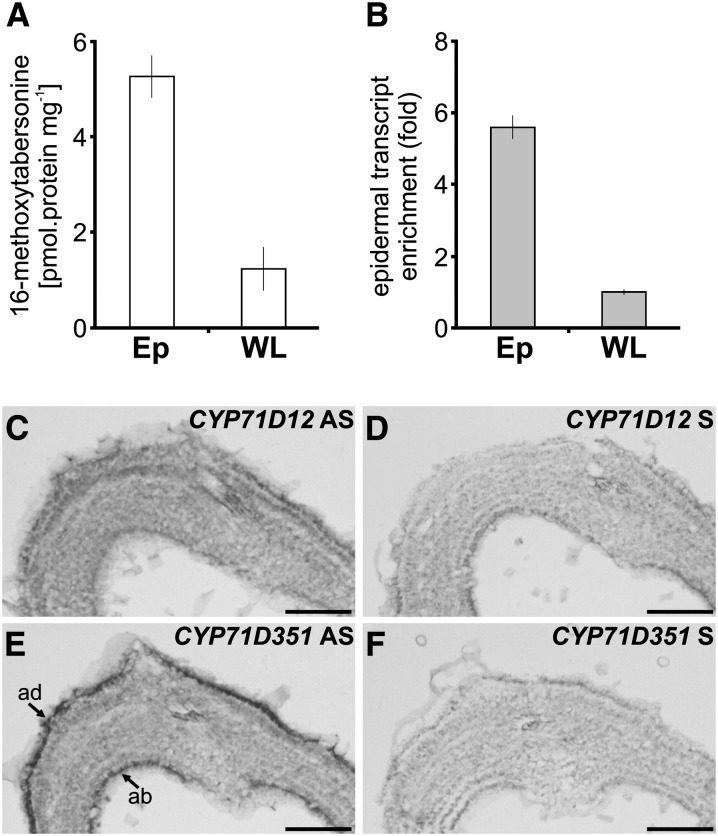
Previous work suggested an epidermis-specific localization of CYP71D12 (T16H1) transcripts in leaf (Murata and De Luca, 2005). However, we found that the primer pair used for this analysis (CrT16H-RT01 and CrRT16H-RT02) was not isoform specific and allowed the amplification of either CYP71D12 or CYP71D351, depending on the source of the transcripts. Since T16H transcripts detected in leaves are almost exclusively encoded by CYP71D351 (T16H2), we reinvestigated the spatial distribution of both transcripts in this organ. By comparing T16H activity in an epidermis-enriched fraction of leaves obtained by carborundum abrasion versus T16H activity in whole leaves (Fig. 5A), we first confirmed that the epidermis hosts most of the corresponding enzymatic activity, as described previously by Murata and De Luca (2005). Quantification of CYP71D351 expression in the same extracts using isoform-specific primers revealed that these gene transcripts followed a similar enrichment in the leaf epidermal fraction (Fig. 5B). The carborundum abrasion approach was complemented by localization studies performed by in situ RNA hybridization. Using moderate riboprobe hydrolysis and high-stringency conditions to distinguish between the two highly similar genes (86.5% nucleotide identity; Supplemental Fig. S1), no hybridization signals were detected in young leaves using a CYP71D12-specific probe (Fig. 5, C and D). Under the same conditions, CYP71D351 mRNA was readily detected in adaxial and abaxial epidermis cells (Fig. 5, E and F). These results are consistent with the occurrence of four CYP71D351 ESTs in the C. roseus EST epidermome database that were previously incorrectly annotated as CYP71D12 (Supplemental Table S1; Murata et al., 2008) and are also in agreement with the postulated localization of the first two steps of vindoline biosynthesis in the epidermis (Murata and De Luca, 2005; Murata et al., 2008; Guirimand et al., 2011b).
Figure 5.
T16H activity and CYP71D351 (T16H2) transcripts are specifically located in epidermis of C. roseus leaves. A and B, Analysis of T16H enzymatic activity (A) and relative expression of CYP71D351 in leaf epidermis-enriched protein/transcript extracts produced by carborundum abrasion compared with that found in whole leaves (B). Cr60S was used as a reference gene. Ep, Epidermis; WL, whole leaf. C to F, Analysis of CYP71D351 and CYP71D12 transcript distribution performed by RNA in situ hybridization. Serial sections of young developing leaves were hybridized either with CYP71D12 antisense (AS) probes (C), CYP71D351 antisense probes (E), CYP71D12 sense (S) probes (D), or CYP71D351 sense probes (F) used as negative controls. ab, Abaxial epidermis; ad, adaxial epidermis. Bars = 100 µm.
CYP71D351 (T16H2) Is an Endoplasmic Reticulum-Anchored Enzyme
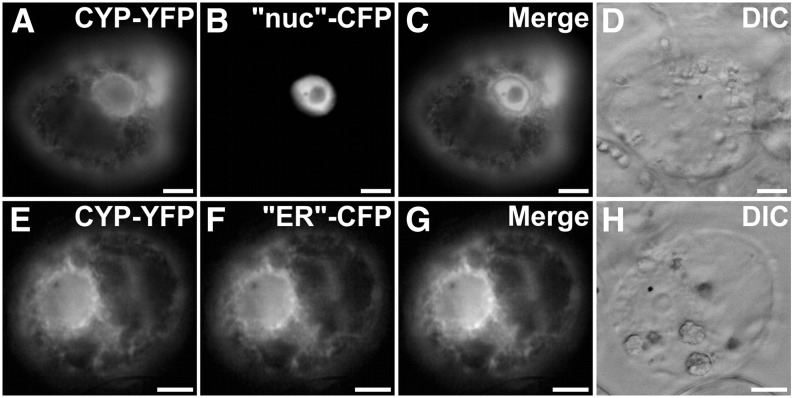
Most plant P450s characterized to date function as endoplasmic reticulum (ER)-anchored enzymes, as exemplified in C. roseus for CYP71D12, CYP72A1, and CYP76B6 (Guirimand et al., 2009, 2011a, 2011b). However, the recent localization of two carotenoid-hydroxylating P450s to plastids (Quinlan et al., 2012) prompted us to analyze the subcellular localization of CYP71D351 (T16H2). A predictive sequence analysis of this enzyme, which identified a putative 19-residue transmembrane N-terminal helix (residues 4–22; Supplemental Fig. S6), suggested that CYP71D351 (T16H2) was a traditional ER-anchored P450. The precise subcellular localization of CYP71D351 was investigated using a C-terminal yellow fluorescent protein (YFP) fusion protein (CYP71D351-YFP) that ensured the accessibility of the predicted transmembrane helix located at the N terminus of the P450. In transiently transformed C. roseus cells, the fusion protein displayed a fluorescence signal appearing as a network structure surrounding the nucleus and branching all across the cell (Fig. 6, A–D). This signal also colocalized perfectly with the signal of the “ER”-cyan fluorescent protein (CFP) marker (Fig. 6, E–H), which indicates that the protein is anchored to the ER membrane and probably releases the 16-hydroxytabersonine product in the cytosol according to the classical topology of P450s.
Figure 6.
CYP71D351 (T16H2) is located to the ER. C. roseus cells were transiently transformed with CYP71D351-YFP-expressing vector (CYP-YFP; A and E) in combination with plasmids expressing a nucleus-CFP marker (“nuc”-CFP; B) or an ER-CFP marker (“ER”-CFP; F). Colocalization of the two fluorescence signals appears on the merged images (C and G). Cell morphology (D and H) was observed with differential interference contrast (DIC). Bars = 10 µm.
CYP71D351 Expression Correlates with Vindoline Accumulation in C. roseus Cultivars
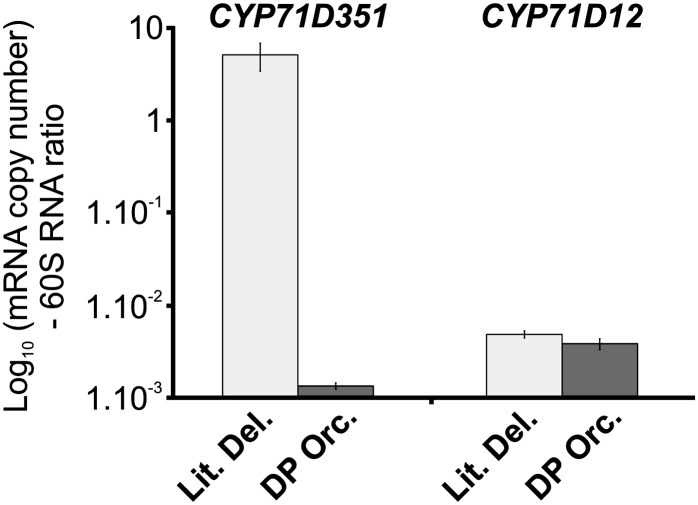
Since T16H activity is reduced or missing in the low-vindoline-accumulating cv Vinca Mediterranean DP Orchid, the expression levels of CYP71D12 and CYP71D351 were analyzed in the young leaves of this cultivar with respect to cv Little Delicata, a cultivar with an average level of vindoline (Magnotta et al., 2006). Interestingly, CYP71D351 (T16H2) transcript levels were 4,000-fold lower in cv Vinca Mediterranean DP Orchid as compared with cv Little Delicata (Fig. 7). By contrast, CYP71D12 (T16H1) transcript levels were similar in both cultivars and also 1,000-fold lower than those of CYP71D351 in cv Little Delicata. This provides compelling evidence that only the expression of CYP71D351 (T16H2) is directly correlated with foliar vindoline accumulation.
Figure 7.
Comparison of CYP71D351 (T16H2) and CYP71D12 (T16H1) transcript levels in low- and high-vindoline-accumulating C. roseus cultivars. CYP71D12 and CYP71D351 transcript levels were measured by real-time RT-PCR analyses performed on total RNA extracted from C. roseus leaves of the low-vindoline-accumulating cv Vinca Mediterranean DP Orchid (DP Orc.; dark gray bars) and of the high-vindoline-accumulating cv Little Delicata (Lit. Del.; light gray bars). Transcript copy numbers were normalized using the ribosomal 60S RNA.
Silencing of CYP71D351 (T16H2) Reduces Vindoline Levels and Alters the Levels of Related Metabolites
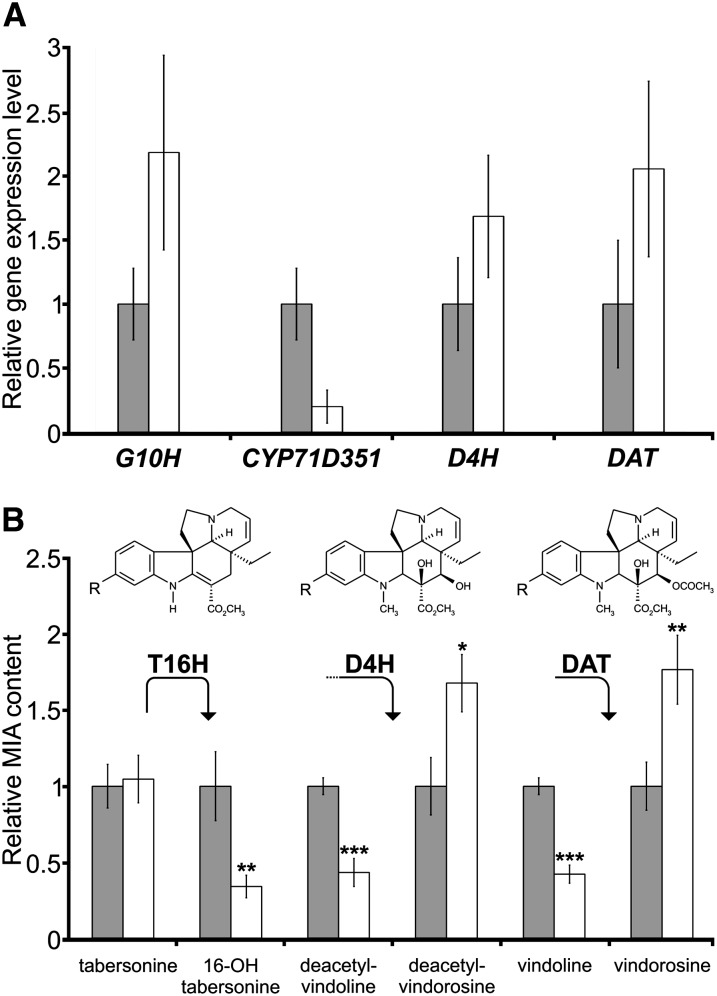
To confirm the involvement of CYP71D351 (T16H2) in vindoline synthesis in young leaves of C. roseus, a virus-induced gene silencing (VIGS) approach was implemented. In our preferred cultivar for VIGS studies, cv Sunstorm Apricot, foliar transcript levels of CYP71D12 (T16H1) were below real-time RT-PCR detection levels. However, we designed a specific down-regulation construct for CYP71D351 including its 3′-untranslated region, which presented the highest dissimilarity with respect to CYP71D12 (T16H1) to avoid potential cross-silencing reactions (Supplemental Fig. S1). Compared with the empty-vector control, the CYP71D351-silencing construct caused a decrease of approximately 80% of the corresponding transcript in young C. roseus leaves (Fig. 8A), while CYP71D12 mRNAs still remained below the detection level. We also measured transcript levels of other MIA biosynthetic genes acting both upstream (G10H) and downstream (D4H and DAT) of the T16H step and found that these were not down-regulated (Fig. 8A). Concomitant with the silencing of CYP71D351, we observed a reduction in vindoline of about 60% as compared with empty-vector controls. Tabersonine levels, on the other hand, were seemingly unaffected (Fig. 8B). Further investigation into the metabolite profiles of silenced leaves identified the significant increase of a metabolite with a mass corresponding to that of vindorosine, a leaf alkaloid that is structurally identical to vindoline except for the absence of the methoxy group at the 16 position. Notably, the increase in vindorosine content corresponded roughly to the decrease in vindoline content. Furthermore, putative biosynthetic intermediates in the tabersonine-to-vindoline pathway have been shown to accumulate in C. roseus leaves, albeit at low levels (Liscombe et al., 2010). Therefore, we also investigated their fate with respect to the silencing of CYP71D351. We observed the significant decrease of a metabolite with the mass of 16-hydroxytabersonine, the direct product of T16H. We also identified metabolites with masses corresponding to those of the last two intermediates in the vindoline and vindorosine pathways. For the two late vindoline intermediates (desacetoxyvindoline and deacetylvindoline; Fig. 1), a reduction was observed, while for the two late vindorosine intermediates (desacetoxyvindorosine and deacetylvindorosine), an increase was observed. In order to verify the plausibility of our assignment of these metabolites as vindoline/vindorosine pathway intermediates, we carried out an accurate mass analysis of control and silenced leaf extracts, which revealed that the measured masses were within 3 ppm of the theoretical masses of the proposed intermediates (Supplemental Table S2).
Figure 8.
Down-regulation of CYP71D351 (T16H2) affects vindoline biosynthesis. A, CYP71D351 transcript down-regulation by VIGS. Relative expression of G10H (encoding an enzyme involved in secologanin biosynthesis), CYP71D351, D4H, and DAT was determined by real-time RT-PCR analyses performed on total RNA extracted from C. roseus leaves of CYP71D351-silenced plants (CYP-VIGS; white bars) or plants transformed with an empty vector control (gray bars). CrRPS9 was used as a reference gene. Data correspond to average values (n = 4) ± sd of independent transformant plants. B, Relative MIA content in CYP-VIGS plants (white bars) as compared with empty vector plants (gray bars). The accumulation of tabersonine and 16-hydroxytabersonine (first molecule; R = H and R = OH, respectively), deacetylvindoline and deacetylvindorosine (second molecule; R = OCH3 and R = H, respectively), vindoline and vindorosine (last molecule; R = OCH3 and R = H, respectively) was quantified by LC-MS. The amount of each MIA in CYP-VIGS plants was expressed relative to that measured in empty vector plants (normalized to 1). Asterisks denote statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test). The results shown are representative of eight CYP-VIGS plants and eight empty vector plants.
DISCUSSION
The initial hydroxylation step of tabersonine determines the fate of the Aspidosperma-type alkaloid branch in C. roseus. In aerial organs, 16-hydroxylation by T16H directs the flux of tabersonine toward the biosynthesis of vindoline, which is a direct precursor of the valuable dimeric MIAs. In underground organs, hydroxylation at the 19 position by T19H leads to the production of hörhammericine and echitovenine in roots. T19H has recently been cloned and characterized biochemically and was shown to act stereoselectively on tabersonine and to a lesser extent on lochnericine, an oxygenated tabersonine derivative (Giddings et al., 2011). Recently, the enzymatic characterization of low- and high-vindoline-accumulating cultivars has revealed the close correlation between T16H activity and vindoline biosynthesis, highlighting the crucial role that T16H plays in controlling the levels of valuable dimeric MIAs (Magnotta et al., 2006). However, no in-depth characterization of T16H has been published since the cloning of a T16H isoform from cell suspension cultures in the late 1990s (Schröder et al., 1999).
Within all tested C. roseus organs, young leaves have been shown to possess the highest levels of T16H activity, which mirrors the vindoline accumulation profile (St-Pierre and De Luca, 1995). Interestingly, while not able to synthesize vindoline, C. roseus cell cultures also display relatively high levels of T16H activity and are a convenient system for laboratory studies. Therefore, Schröder et al. (1999) could readily use cell suspension cultures to clone a T16H isoform (CYP71D12-T16H1). However, there are inconsistencies regarding the CYP71D12 expression profile in comparison with that of other vindoline biosynthetic genes, especially regarding cellular localization. Most notably, the 16OMT transcripts that encode the enzyme acting immediately downstream of T16H have been located to leaf epidermis by complementary technical approaches, including epidermis enrichment with carborundum abrasion, cell-specific preparation by laser-capture microdissection, epidermis EST sequencing, and RNA in situ hybridization (Murata and De Luca, 2005; Levac et al., 2008; Murata et al., 2008; Guirimand et al., 2011b). For CYP71D12, however, only the first three approaches have suggested epidermal localization, while RNA in situ hybridization assays always failed in our hands (data not shown). These inconsistencies prompted us to clone T16H using C. roseus leaves rather than cell suspension cultures as starting material.
The full-length cDNA that we cloned from young leaves encoded a protein sharing 82% identity with CYP71D12 (T16H1), and we named it CYP71D351 or T16H2. Biochemical characterization of recombinant CYP71D12 and CYP71D351 revealed that both enzymes catalyze the regiospecific hydroxylation of tabersonine at the C16 position (Fig. 2A; Supplemental Fig. S3). Except for the close tabersonine analogs 2,3-dihydro-3-hydroxytabersonine and 2,3-dihydrotabersonine, no other tested compounds were hydroxylated by either of the enzymes, demonstrating a high degree of substrate specificity. In addition, CYP71D351 and CYP71D12 exhibit apparent Km values for tabersonine in the nanomolar range (70 ± 20 nm and 350 ± 100 nm, respectively). Thus, both enzymes have a very high affinity for the tabersonine substrate, suggesting a high efficiency of tabersonine hydroxylation in vivo. These values are similar to the ones reported for T19H (Km of 300 ± 50 nm; Giddings et al., 2011) but are much lower than the Km value initially determined for T16H in crude leaf extracts (11 µm), which was established using a T16H/16OMT coupled reaction (St-Pierre and De Luca, 1995). However, since the Km value of 16OMT (for 16-hydroxytabersonine) was later shown to be 2.6 µm (Levac et al., 2008), it is clear that the initial estimation of the T16H Km value for tabersonine was limited by the second step of the coupled assay.
The occurrence of two homologous proteins displaying T16H activity is in agreement with a genomic Southern-blot analysis carried out using CYP71D12 (T16H1) cDNA as a probe, which suggested the presence of at least two closely related genes for T16H in C. roseus (Schröder et al., 1999). Notably, T16H constitutes the only known MIA biosynthetic enzyme encoded by two distinct genes in C. roseus. However, the recent availability of massive transcriptomic data from different research groups will make possible a thorough characterization of other biosynthetic genes for homologs. The high level of amino acid sequence conservation of these two isoforms suggests that they share a common origin with T16H activity and resulted from a relatively recent gene duplication event leading to subfunctionalization or specialization. At this point, not enough orthologs and paralogs from Apocynaceae and related plant species are available to further evaluate the history of this gene duplication. However, relaxation of substrate specificity in CYP71D351 (T16H2) suggests divergent selection pressure compared with CYP71D12 (T16H1). By contrast, T19H, which also hydroxylates tabersonine, displays only 35% identity with both CYP71D12 and CYP71D351 and probably resulted from the recruitment of a phylogenetically distinct P450-encoding gene.
High levels of T16H activity and vindoline production have been reported previously in young leaves (St-Pierre and De Luca, 1995). Even though the biochemical assays indicate that both CYP71D12 and CYP71D351 could potentially be involved in the biosynthesis of vindoline, our analysis of gene expression suggests that CYP71D351 is the major T16H isoform in C. roseus leaves, particularly young leaves. Transcripts of CYP71D12 were only barely detectable in these organs (Fig. 3B). The flower-specific expression of CYP71D12 is surprising but might reflect either a specialization of this isoform to nonchlorophyllous tissues, including etiolated cell cultures, or defense mechanisms dedicated to flowers, as observed for pathogenesis-related proteins (Liu and Ekramoddoullah, 2006). The flower bud expression of both CYP71D351 and CYP71D12 might also reflect the differential expression of these P450s in chlorophyllous and nonchlorophyllous tissues. Floral synthesis of vindoline reaches 6% of leaf level and was, as expected, supported by low levels of NMT, D4H, and DAT gene expression in flowers (Fig. 4; Supplemental Fig. S5; St-Pierre et al., 1998). These initial observations about the preferential expression of CYP71D351 in leaves were confirmed by the specific detection of CYP71D351 transcripts in leaf epidermis, while no CYP71D12 transcripts were detected anywhere in leaf cross sections (Fig. 5, A–C). In addition, the four T16H ESTs annotated as T16H in the C. roseus leaf epidermome database (Murata et al., 2008) correspond strictly to CYP71D351 and not to CYP71D12 (Supplemental Table S1). Therefore, the initial report of CYP71D12 mRNA within leaf epidermis, obtained by RT-PCR performed on epidermis-enriched RNA extracts, likely resulted from priming to CYP71D351 (Murata and De Luca, 2005). Indeed, in our hands, the pair of primers used in the former study to amplify CYP71D12 leads to the amplification of CYP71D351 from leaf cDNA and, therefore, is not isoform specific. Furthermore, the specific expression of CYP71D351 mRNA in leaf epidermis definitively demonstrates that genes encoding T16H and 16OMT are coexpressed in this tissue. In addition, the anchoring of CYP71D351 to the ER, as revealed by subcellular localization studies, most likely exposes the catalytic site toward the cytosol and permits the release of 16-hydroxytabersonine into the cytosol, where 16OMT resides, thus facilitating the following methoxylation reaction (Fig. 6; Guirimand et al., 2011b). The comparison of gene expression in low- and high-vindoline-accumulating cultivars provided further evidence for the key role of CYP71D351 in foliar vindoline biosynthesis. Both types of cultivars were shown to have similar levels of activity of vindoline biosynthetic enzymes except for T16H, whose activity is strongly reduced in the low-vindoline-accumulating cultivar (Magnotta et al., 2006). Our experiments showed that this deficiency in T16H activity can be directly correlated to the lack of CYP71D351 transcripts.
Final proof of the crucial role of CYP71D351 (T16H2) in foliar vindoline biosynthesis was provided by down-regulation experiments using VIGS. As expected, the down-regulation of CYP71D351 led to a strong decrease of vindoline accumulation. This effect was accompanied by a significant reduction of metabolites with masses corresponding to vindoline pathway intermediates (16-hydroxytabersonine, desacetoxyvindoline, and deacetylvindoline). Simultaneously, metabolites with masses related to the vindorosine pathway (deacetoxyvindorosine, desacetylvindorosine, and vindorosine itself) were significantly increased. Vindorosine is the demethoxylated analog of vindoline, and the downstream enzymes NMT, D4H, and DAT are known to accept both the 16-methoxylated and the demethoxylated tabersonine derivatives for the production of either vindorosine or vindoline (Fig. 1; De Luca et al., 1987; Liscombe et al., 2010). Therefore, down-regulation of T16H seems to have caused the metabolic flux to be reoriented toward the synthesis of vindorosine rather than toward the production of other MIAs, with no detectable increase of tabersonine (Fig. 8B). This also implies that the yet unknown enzyme catalyzing the hydration of 16-methoxytabersonine in the vindoline pathway can accept tabersonine as a substrate, a fact that might facilitate its discovery.
In conclusion, our results show that CYP71D351 (T16H2) is the main isoform in the biosynthesis of vindoline in C. roseus by catalyzing the 16-hydroxylation of tabersonine in young leaves. By contrast, the previously cloned CYP71D12 (T16H1) does not play a role in this foliar biosynthesis and is likely dedicated to the production of vindoline in flowers. Therefore, vindoline biosynthesis is subject to an extraordinary degree of spatial control in C. roseus. The existence of such isoforms of MIA biosynthetic enzymes acting in an organ-dependent manner makes the architecture and the regulation of secondary metabolism in C. roseus even more complex.
MATERIALS AND METHODS
Plant and Cell Culture Growth
Mature Catharanthus roseus ‘Pacifica Pink’ plants were used for microscopy fixation (RNA in situ hybridization experiments) and RNA extraction (cloning experiments and gene expression measurements in various organs). A low-vindoline-accumulating C. roseus cultivar (cv Vinca Mediterranean DP Orchid) and a vindoline-accumulating control cultivar (cv Little Delicata; lines 49 and 50, respectively, in Magnotta et al., 2006) were used for gene expression analysis. VIGS assays were performed on the C. roseus cv Sunstorm Apricot. The C. roseus C20D cell suspension culture used for subcellular localization studies and gene expression analysis was propagated in Gamborg B5 medium (Duchefa) at 24°C under continuous shaking (100 rpm) for 7 d as described previously (Guirimand et al., 2009).
RNA Extraction and RT
For cDNA cloning, extraction of total RNA from young leaves was performed using the NucleoSpin RNA Plant kit (Macherey-Nagel). First-strand cDNA was synthesized from 5 µg of total RNA using oligo(dT)18 primers (0.5 µm) and 15 units of Thermoscript reverse transcriptase (Invitrogen). Following RT, complementary RNA was removed by treatment with Escherichia coli RNase H (Invitrogen) for 20 min at 37°C. For 5′ RACE, similar reactions were carried out except that total RNA was subsequently removed by treatment with RNase A (Sigma-Aldrich) and the reaction product was purified using the NucleoSpin Extract II kit. For real-time PCR, total RNA was extracted from different C. roseus organs and cells with the RNeasy Plant mini kit (Qiagen) and treated (1.5 µg) with RQ1 RNase-free DNase (Promega) before being used for first-strand cDNA synthesis by priming with oligo(dT)18 (0.5 µm). RT was carried out using SuperScript III reverse transcriptase (Invitrogen) at 50°C.
Cloning of CYP71D351
The first rounds of amplification were performed on cDNA obtained by RT of young leaf RNA using primers CrT16H-RT01 and CrT16H-RT02 (primer sequences are given in Supplemental Table S3). The amplification product was cloned into the pSC-A plasmid (Stratagene), and 15 resulting clones were sequenced. On the basis of the resulting sequences and EST sequence analysis, the sense 2T16H-A primer was designed to perform 3′ RACE-PCR according to Oudin et al. (2007). Similarly, the reverse primers 2T16H-51 and 2T16H-52 were used to perform a nested 5′ RACE-PCR as described previously (Guirimand et al., 2012). Following sequence assembly, the two specific primers 2T16H-YFPfor and 2T16H-YFPrev were used to amplify a full-length ORF using Phusion DNA polymerase (New England Biolabs). The resulting amplicon was cloned into the pSC-A vector following A-tailing and sequenced before deposition at the National Center for Biotechnology Information under GenBank accession number JF742645.
Subcellular Localization Studies
To study subcellular localization, the full-length CYP71D351 ORF was amplified by PCR using primers 2T16H-YFPfor and 2T16H-YFPrev (Supplemental Table S3) and cloned into the BglII and SpeI restriction sites of the pSC-A cassette YFPi plasmid in frame with the 5′ extremity of the YFP coding sequence. This recombinant plasmid was used for transient transformation of C. roseus cells by particle bombardment and GFP imaging according to Guirimand et al. (2009, 2010a), in combination with plasmids expressing the ER-CFP marker (CD3-954; Nelson et al., 2007) or the nucleus-CFP marker (Guirimand et al., 2011a).
Heterologous Expression of CYP71D351 and CYP71D12 in Yeast
Full-length CYP71D12 and CYP71D351 cDNAs were amplified using the primer pairs T16H-BglII/T16H-Bgl2stop and 2T16H-YFPfor/2T16H-Bgl2-stop, respectively (Supplemental Table S3), carrying BglII restriction sites at both extremities, and shifted into the BamHI restriction site of the pYeDP60 vector. Both recombinant plasmids as well as the empty vector were independently transformed into the Saccharomyces cerevisiae strain WAT11 engineered to express the Arabidopsis (Arabidopsis thaliana) NADPH P450 REDUCTASE1 (Pompon et al., 1996). Yeast strains were grown in YPGE medium (1% bacto peptone, 1% yeast extract, 0.5% Glc, and 3% ethanol), and recombinant expression was induced by Glc starvation in YPI medium (1% bacto peptone, 1% yeast extract, and 2% Gal) according to Pompon et al. (1996) from a single colony. Cells were harvested by centrifugation, and proteins were isolated with microsomal membranes as described by Heitz et al. (2012).
Enzyme Assays
CYP71D351 and CYP71D12 activities, including kinetic parameter determination and substrate specificities, were analyzed by ultra-performance liquid chromatography-MS in a final volume of 50 μL containing 100 mm Tris-HCl, pH 8.0, 4 mm dithiothreitol, 1 mm NADPH, various amounts of substrate, and 2 and 4 µL of microsomes for CYP71D351 and CYP71D12, respectively. The reactions were initiated by the addition of NADPH, incubated at room temperature, and quenched by the addition of 50 µL of methanol. Assays without NADPH or with microsomes from yeast harboring the empty expression vector were used as controls. For Km and Vmax measurements of CYP71D12 and CYP71D351, tabersonine was used at concentrations ranging from 0.12 to 7.5 µm and from 0.25 to 3 µm, respectively. Km and Vmax values were calculated from Lineweaver-Burk plots. For substrate specificity determination, enzymatic reactions were carried out at 30°C during 30 min with the following compounds (commercially available or prepared according to De Luca et al. [1987]): catharanthine (300 µm), vindoline (220 µm), ajmalicine (280 µm), secologanin (260 µm), naringenin (370 µm) tryptamine (620 µm), strictosidine (20 µm), 2,3-dihydrotabersonine (50 µm), 2,3-dihydro-3-hydroxytabersonine (50 µm), and tabersonine (50 µm). The ultra-performance liquid chromatography system was coupled to a SQD mass spectrometer equipped with an electrospray ionization source controlled by Masslynx 4.1 software (Waters). Sample separation was performed on a Waters Acquity HSS T3 C18 column (150 mm × 2.1 mm i.d., 1.8 µm) with a flow rate of 0.4 mL min−1 at 55°C. 16-Hydroxytabersonine was analyzed in electrospray ionization-positive mode using the following gradient: from 10:90 to 50:50 acetonitrile:water acidified with 0.1% formic acid over 5 min. The capillary and sample cone voltages were 3,000 and 30 V, respectively. The cone and desolvation gas flow rates were 60 and 800 L h−1.
CYP71D351 Reaction Product Characterization
For product characterization, the CYP71D351 enzymatic reaction was performed as described in the previous section but in a final volume of 60 mL containing 2 mg of tabersonine (100 µm), 50 mg NADPH, and 14 mL of microsomes. Following evaporation, the viscous sample (3 mL) was separated into four portions of 750 µL. Each portion was passed through an RP-18 cartridge (500 mg; preconditioned with methanol and water). Each cartridge was flushed with 3 mL of water and eluted with 3 mL of methanol. The methanol eluates were combined and evaporated to dryness, reconstituted in 1 mL of methanol, and subjected to HPLC purification using an Agilent HP1100 LC system (Agilent Technologies) and a Zorbax SB-C18 column (150 × 4.6 mm i.d., 3.5 µm) with a flow rate of 1 mL min−1. The injected volume was 100 µL per run, and the gradient was as follows: from 10:90 to 30:70 acetonitrile:water acidified with 0.1% trifluoroacetic acid over 30 min. UV absorption was monitored at 330 nm. The peak at retention time 22.8 min was subjected to NMR analysis. 1H-NMR, 1H-1H-COSY, HSQC, and HMBC spectra were recorded on an Avance 500 NMR spectrometer (Bruker-Biospin) at 300 K using a 5-mm TCI CryoProbe. The sample was dissolved in methanol-d4 (120 µL) and measured in a 2.5-mm NMR tube. Chemical shifts (δ) are given relative to tetramethylsilane as an internal standard, and coupling constants are in Hz.
NMR Data of 16-Hydroxytabersonine
1H-NMR (500 MHz, methanol-d4): δ 7.32 (1H, d, J = 8.1 Hz, H-14), 6.53 (1H, d, J = 2.1 Hz, H-17), 6.37 (1H, dd, J = 8.1, 2.1 Hz, H-15), 6.05 (1H, d, J = 10.2 Hz, H-6), 6.00 (1H, dd, J = 10.2, 4.1 Hz, H-7), 4.21 (1H, d, J = 15.8 Hz, H-8a), 4.01 (1H, brd, J = 15.8 Hz, H-8b), 3.89 (1H, s, H-19), 3.80 (3H, s, OCH3), 3.66 (1H, m, H-10a), 3.51 (1H, m, H-10b), 2.92 (1H, d, J = 16.7 Hz, H-4a), 2.31 (1H, d, J = 16.7 Hz, H-4b), 2.16 (2H, m, H2-11), 1.23 (1H, m, H-20a), 1.18 (1H, m, H-20b), 0.71 (3H, t, J = 7.2 Hz, H3-21).
13C-NMR (125 MHz, methanol-d4): δ 169.2 (CO), 164.4 (C-2), 160.0 (C-16), 145.9 (C-18), 135.4 (CH-6), 126.5 (C-13), 122.5 (CH-7), 122.4 (CH-14), 108.7 (CH-15), 99.6 (CH-17), 92.6 (C-3), 72.0 (CH-19), 52.6 (C-12), 51.7 (OCH3), 51.4 (CH2-10), 51.2 (CH2-8), 44.4 (CH2-11), 41.6 (C-5), 28.5 (CH2-4), 27.4 (CH2-20), 7.6 (CH3-21).
Gene Expression Analysis (Real-Time RT-PCR)
The expression of CYP71D351, CYP71D12, G10H, 16OMT, NMT, MAT, D4H, and DAT was analyzed by real-time RT-PCR using the primers given in Supplemental Table S3. Depending on the experiments, gene expression measurements were performed on reverse-transcribed RNA extracted from different C. roseus organs of cv Pacifica Pink (such as roots, first internodes, young and mature leaves, flower buds, flowers, and fruits), young leaves of C. roseus of cv Vinca Mediterranean, Little Delicata, and Sunstorm Apricot, epidermis-enriched fractions of young leaves (cv Little Delicata), and also from C. roseus cells treated during 0.5, 8, 24, or 48 h with 100 µm MeJa or mock treated with ethanol. Real-time PCR was run on a CFX96 Touch Real-Time PCR System (Bio-Rad) using the SYBR Green I technology. Each reaction was performed in a total reaction volume of 25 µL containing an equal amount of cDNAs, 0.2 µm forward and reverse primers, and 1× SsoAdvanced SYBR Green Supermix (Bio-Rad). The reaction was initiated by a denaturation step at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Melting curves were used to determine the specificity of the amplifications. Relative quantification of gene expression was calculated according to the delta-delta cycle threshold method using the 40S ribosomal protein S9 (RPS9) or 60S ribosomal protein as reference gene (Figs. 4, 5, and 8). Alternatively, absolute quantification of transcript copy number was performed with calibration curves and normalization with RPS9 or 60S reference gene ratios (Figs. 3 and 7). In Figures 3, 4, 5, and 7, each assay was performed in triplicate, and expression measurements were performed at least twice with independent experimental replicates, although only one experiment is shown. In Figure 8, each assay was performed in duplicate, and the data correspond to average values of four (for G10H, D4H, and DAT expression) or eight (for CYP71D351 expression) independently silenced plants ± sd.
Preparation of Epidermis-Enriched Fractions of C. roseus Leaves
The epidermis of young C. roseus leaves was abraded with carborundum to allow RNA extraction and crude protein extract preparation according to Murata and De Luca (2005). CYP71D351 gene expression was analyzed by real-time RT-PCR, and T16H activity was estimated by measuring the amount of 16-methoxytabersonine produced by crude protein extract from the same fractions as described by St-Pierre and De Luca (1995).
RNA in Situ Hybridization of C. roseus Leaves
The cellular localization of gene transcripts was performed by RNA in situ hybridization according to Mahroug et al. (2006). Briefly, full-length ORFs of CYP71D351 and CYP71D12 were amplified with Phusion DNA polymerase and cloned into the pSC-A plasmid for use in the synthesis of sense and antisense digoxigenin-labeled RNA probes. Paraffin-embedded serial longitudinal sections of young developing leaves were subsequently hybridized with digoxigenin-labeled probes and localized with anti-digoxigenin alkaline phosphatase.
VIGS
Due to high sequence identity with CYP71D12, the CYP71D351-silencing fragment was designed in the 3′ end and 3′ untranslated region of the corresponding gene (Supplemental Fig. S1) and amplified with primers 2T16H fw and 2T16H rev (Supplemental Table S3). The resulting fragment (252 bp) was cloned into the pTRV2u vector described by Geu-Flores et al. (2012). The resulting plasmid and the empty vector were used to perform the VIGS assays on C. roseus seedlings as described by Liscombe and O’Connor (2011). Leaves from the first two leaf pairs to emerge following inoculation were harvested from eight plants transformed with each construct and subjected to gene expression analysis by real-time RT-PCR (primers are given in Supplemental Table S3). The alkaloid content of silenced leaves was determined by LC-MS as described previously (Liscombe and O’Connor, 2011; Geu-Flores et al., 2012).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JF742645.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleic acid sequence alignment of CYP71D12 and CYP71D351.
Supplemental Figure S2. Amino-acid sequence alignment of CYP71D351 and CYP71D12.
Supplemental Figure S3. Characterization of the reaction product of CYP71D351 and CYP71D12.
Supplemental Figure S4. Effect of tabersonine concentration on T16H activity of CYP71D12 and CYP71D351.
Supplemental Figure S5. Analysis of MIAs accumulation in flowers and young leaves of C. roseus.
Supplemental Figure S6. Detection of a putative transmembrane helix at the N-terminal end of CYP71D351.
Supplemental Table S1. Nucleic acid sequence identity of T16H-annotated EST with CYP71D351 and CYP71D12.
Supplemental Table S2. Measured masses of the intermediates of vindoline/vindorosine biosynthetic pathways.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. David Nelson (University of Tennessee, Memphis) for annotation of CYP71D351, Dr. Christian Paetz (Max-Planck-Institute for Chemical Ecology) and Dr. Lionel Hill (John Innes Centre) for mass spectrometric analysis, and Dr. Renate Ellinger (Max-Planck-Institute for Chemical Ecology) for technical support. We also thank Marie-Antoinette Marquet, Marie-Françoise Aury, Evelyne Danos, Cédric Labarre, and Emelyne Marais (EA2106 Biomolécules et Biotechnologies Végétales) for help in maintaining cell cultures and plants.
Glossary
- MIA
monoterpene indole alkaloid
- T16H
tabersonine 16-hydroxylase
- P450
cytochrome P450
- 16OMT
16-hydroxytabersonine 16-O-methyltransferase
- NMT
16-methoxy-2,3-dihydro-3-hydroxytabersonine N-methyltransferase
- D4H
desacetoxyvindoline-4-hydroxlyase
- DAT
deacetylvindoline-4-O-acetyltransferase
- MAT
minovincinine-O-acetyltransferase
- RT
reverse transcription
- cDNA
complementary DNA
- LC
liquid chromatography
- MS
mass spectrometry
- MeJa
methyl jasmonate
- ER
endoplasmic reticulum
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- VIGS
virus-induced gene silencing
- ORF
open reading frame
- T19H
tabersonine 19-hydroxylase
References
- Balsevich J, Bishop GJ. (1989) Distribution of catharanthine, vindoline and 3′4′-anhydrovinblastine in the aerial parts of some Catharanthus roseus plants and the significance thereof in relation to alkaloid production in cultured cells. In WGW Kurz, ed, Primary and Secondary Metabolism of Plant Cell Cultures. Springer Verlag, Berlin, pp 149–153 [Google Scholar]
- De Luca V, Balsevich J, Tyler RT, Kurz WGW. (1987) Characterization of a novel N- methyltransferase (NMT) from Catharanthus roseus plants. Plant Cell Rep 6: 458–461 [DOI] [PubMed] [Google Scholar]
- Deus-Neumann B, Stöckigt J, Zenk MH. (1987) Radioimmunoassay for the quantitative determination of catharanthine. Planta Med 53: 184–188 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54: 763–784 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE. (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492: 138–142 [DOI] [PubMed] [Google Scholar]
- Giddings LA, Liscombe DK, Hamilton JP, Childs KL, DellaPenna D, Buell CR, O’Connor SE. (2011) A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J Biol Chem 286: 16751–16757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B, Wang C, Ravelli RB, Roussi F, Steinmetz MO, Curmi PA, Sobel A, Knossow M. (2005) Structural basis for the regulation of tubulin by vinblastine. Nature 435: 519–522 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Burlat V, Oudin A, Lanoue A, St-Pierre B, Courdavault V. (2009) Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep 28: 1215–1234 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Courdavault V, Lanoue A, Mahroug S, Guihur A, Blanc N, Giglioli-Guivarc’h N, St-Pierre B, Burlat V. (2010a) Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”? BMC Plant Biol 10: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G, Courdavault V, St-Pierre B, Burlat V (2010b) Biosynthesis and regulation of alkaloids. In EC Pua, M Davey, eds, Plant Developmental Biology: Biotechnological Perspectives, Vol 2. Springer-Verlag, Berlin, pp 139–160 [Google Scholar]
- Guirimand G, Guihur A, Ginis O, Poutrain P, Héricourt F, Oudin A, Lanoue A, St-Pierre B, Burlat V, Courdavault V. (2011a) The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J 278: 749–763 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Guihur A, Phillips MA, Oudin A, Glévarec G, Melin C, Papon N, Clastre M, St-Pierre B, Rodríguez-Concepción M, et al. (2012) A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol Biol 79: 443–459 [DOI] [PubMed] [Google Scholar]
- Guirimand G, Guihur A, Poutrain P, Héricourt F, Mahroug S, St-Pierre B, Burlat V, Courdavault V. (2011b) Spatial organization of the vindoline biosynthetic pathway in Catharanthus roseus. J Plant Physiol 168: 549–557 [DOI] [PubMed] [Google Scholar]
- He L, Yang L, Tan R, Zhao S, Hu Z. (2011) Enhancement of vindoline production in suspension culture of the Catharanthus roseus cell line C20hi by light and methyl jasmonate elicitation. Anal Sci 27: 1243–1248 [DOI] [PubMed] [Google Scholar]
- He YL, Chen WM, Feng XZ. (1994) Melomorsine, a new dimeric indoline alkaloid from Melodinus morsei. J Nat Prod 57: 411–414 [Google Scholar]
- Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, et al. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287: 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme P, St-Pierre B, De Luca V. (2001) Molecular and biochemical analysis of a Madagascar periwinkle root-specific minovincinine-19-hydroxy-O-acetyltransferase. Plant Physiol 125: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levac D, Murata J, Kim WS, De Luca V. (2008) Application of carborundum abrasion for investigating the leaf epidermis: molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Liscombe DK, O’Connor SE. (2011) A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry 72: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, Usera AR, O’Connor SE. (2010) Homolog of tocopherol C methyltransferases catalyzes N methylation in anticancer alkaloid biosynthesis. Proc Natl Acad Sci USA 107: 18793–18798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Ekramoddoullah AKM. (2006) The family 10 of plant pathogenesis-related proteins: their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68: 3–13 [Google Scholar]
- Magnotta M, Murata J, Chen J, De Luca V. (2006) Identification of a low vindoline accumulating cultivar of Catharanthus roseus (L.) G. Don by alkaloid and enzymatic profiling. Phytochemistry 67: 1758–1764 [DOI] [PubMed] [Google Scholar]
- Mahroug S, Courdavault V, Thiersault M, St-Pierre B, Burlat V. (2006) Epidermis is a pivotal site of at least four secondary metabolic pathways in Catharanthus roseus aerial organs. Planta 223: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Murata J, De Luca V. (2005) Localization of tabersonine 16-hydroxylase and 16-OH tabersonine-16-O-methyltransferase to leaf epidermal cells defines them as a major site of precursor biosynthesis in the vindoline pathway in Catharanthus roseus. Plant J 44: 581–594 [DOI] [PubMed] [Google Scholar]
- Murata J, Roepke J, Gordon H, De Luca V. (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nelson DR. (2006) Cytochrome P450 nomenclature, 2004. Methods Mol Biol 320: 1–10 [DOI] [PubMed] [Google Scholar]
- Oudin A, Mahroug S, Courdavault V, Hervouet N, Zelwer C, Rodríguez-Concepción M, St-Pierre B, Burlat V. (2007) Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol Biol 65: 13–30 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Quinlan RF, Shumskaya M, Bradbury LM, Beltrán J, Ma C, Kennelly EJ, Wurtzel ET. (2012) Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol 160: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke J, Salim V, Wu M, Thamm AM, Murata J, Ploss K, Boland W, De Luca V. (2010) Vinca drug components accumulate exclusively in leaf exudates of Madagascar periwinkle. Proc Natl Acad Sci USA 107: 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder G, Unterbusch E, Kaltenbach M, Schmidt J, Strack D, De Luca V, Schröder J. (1999) Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett 458: 97–102 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V. (1995) A cytochrome P-450 monooxygenase catalyzes the first step in the conversion of tabersonine to vindoline in Catharanthus roseus. Plant Physiol 109: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, Laflamme P, Alarco AM, De Luca V. (1998) The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J 14: 703–713 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Vazquez-Flota FA, De Luca V. (1999) Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung PH, Huang FC, Do YY, Huang PL. (2011) Functional expression of geraniol 10-hydroxylase reveals its dual function in the biosynthesis of terpenoid and phenylpropanoid. J Agric Food Chem 59: 4637–4643 [DOI] [PubMed] [Google Scholar]
- Vazquez-Flota F, De Carolis E, Alarco AM, De Luca V. (1997) Molecular cloning and characterization of desacetoxyvindoline-4-hydroxylase, a 2-oxoglutarate dependent-dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L.) G. Don. Plant Mol Biol 34: 935–948 [DOI] [PubMed] [Google Scholar]
- Wenkert E, Cochran DW, Hagaman EW, Schell FM, Neuss N, Katner AS, Potier P, Kan C, Plat M, Koch M, et al. (1973) Carbon-13 nuclear magnetic resonance spectroscopy of naturally occurring substances. XIX. Aspidosperma alkaloids. J Am Chem Soc 95: 4990–4995 [DOI] [PubMed] [Google Scholar]
- Westekemper P, Wieczorek U, Gueritte F, Langlois N, Langlois Y, Potier P, Zenk MH. (1980) Radioimmunoassay for the determination of the indole alkaloid vindoline in Catharanthus. Planta Med 39: 24–37 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.