M-type thioredoxins directly regulate the biogenesis of the photosystem II complex.
Abstract
Chloroplastic m-type thioredoxins (TRX m) are essential redox regulators in the light regulation of photosynthetic metabolism. However, recent genetic studies have revealed novel functions for TRX m in meristem development, chloroplast morphology, cyclic electron flow, and tetrapyrrole synthesis. The focus of this study is on the putative role of TRX m1, TRX m2, and TRX m4 in the biogenesis of the photosynthetic apparatus in Arabidopsis (Arabidopsis thaliana). To that end, we investigated the impact of single, double, and triple TRX m deficiency on chloroplast development and the accumulation of thylakoid protein complexes. Intriguingly, only inactivation of three TRX m genes led to pale-green leaves and specifically reduced stability of the photosystem II (PSII) complex, implying functional redundancy between three TRX m isoforms. In addition, plants silenced for three TRX m genes displayed elevated levels of reactive oxygen species, which in turn interrupted the transcription of photosynthesis-related nuclear genes but not the expression of chloroplast-encoded PSII core proteins. To dissect the function of TRX m in PSII biogenesis, we showed that TRX m1, TRX m2, and TRX m4 interact physically with minor PSII assembly intermediates as well as with PSII core subunits D1, D2, and CP47. Furthermore, silencing three TRX m genes disrupted the redox status of intermolecular disulfide bonds in PSII core proteins, most notably resulting in elevated accumulation of oxidized CP47 oligomers. Taken together, our results suggest an important role for TRX m1, TRX m2, and TRX m4 proteins in the biogenesis of PSII, and they appear to assist the assembly of CP47 into PSII.
Thioredoxins (TRXs), the ubiquitous small (approximately 12 kD) thiol:disulfide oxidoreductases, are essential redox regulatory elements in plant metabolism (Schürmann and Buchanan, 2008; Dietz and Pfannschmidt, 2011). All TRXs have a redox-active site that contains two conserved Cys residues in the peptide motif WC(G/P)C (Holmgren, 1989). In the reduced state, TRXs are able to reduce disulfide bridges in the target proteins, thereby modulating their functions and stability (Dietz and Pfannschmidt, 2011).
In contrast with other organisms, plants have a large number of TRXs. At least 20 TRX isoforms have been identified in Arabidopsis (Arabidopsis thaliana), and these are targeted to various cellular compartments, including the chloroplast, mitochondria, and cytosol (Meyer et al., 2005; Lemaire et al., 2007). Depending on their subcellular localization, TRXs are reduced by different electron donor systems. Both cytosolic and mitochondrial TRXs are reduced by compartment-specific NADPH:thioredoxin reductases (NTRs), whereas chloroplastic TRXs are reduced by the ferredoxin:thioredoxin reductase with electrons provided by photosynthetic electron transport (Schürmann and Jacquot, 2000). In addition, chloroplasts have a modified type of NTR, called NTRC, which contains a C-terminal TRX domain and acts as a bifunctional enzyme with NTR/TRX activity for the reduction of target proteins (Serrato et al., 2004).
The chloroplastic TRX system is quite complicated and includes five major groups (2f, 4m, 1x, 2y, and 1z) in Arabidopsis (Collin et al., 2003, 2004; Schürmann and Buchanan, 2008; Arsova et al., 2010). The diversity of TRXs implies the existence of an intricate TRX-modulated redox regulatory network in the chloroplast. Previously, in vitro studies using purified TRX proteins have shown that the f- and m-type TRXs are required for carbon metabolism mainly through the regulation of some Calvin cycle enzymes, while the x- and y-type TRXs seem to serve as hydrogen donors for antioxidant enzymes (Collin et al., 2003, 2004; Schürmann and Buchanan, 2008; Chibani et al., 2011). However, the physiological significance and functional specificity of these different TRX isoforms remain to be determined. Recently, the genetic characterization of TRXs in mutant plants has revealed their novel physiological functions in vivo. The inactivation of TRX f1 leads to decreased light activation of ADP-Glc pyrophosphorylase and altered diurnal starch turnover (Thormählen et al., 2013). The trx m3 mutation hampers meristem development, leading to a seedling-lethal phenotype (Benitez-Alfonso et al., 2009), and TRX y2 functions as an electron donor to Met sulfoxide reductases for protein repair (Laugier et al., 2013). Collectively, these data reveal the potential for functional diversity in chloroplastic TRXs.
The primary reactions of photosynthesis are mediated by three pigment-protein complexes, PSII, the cytochrome b6f (Cyt b6f) complex, and PSI, which are embedded in the thylakoid membranes of chloroplasts and connected in series by small, mobile electron carriers like plastoquinone and plastocyanin (Rascher and Nedbal, 2006; Eberhard et al., 2008). A characteristic feature of these photosynthetic apparatuses is that they all consist of multiple nucleus- and chloroplast-encoded subunits as well as numerous pigments, such as chlorophylls and xanthophylls. Hence, the biogenesis of the photosynthetic complexes depends upon a tight coordination between protein and pigment synthesis as well as the spatially and temporally coordinated assembly of the different subunits and the proper incorporation of various cofactors (Rochaix, 2011). Notably, mounting evidence suggests that TRXs play an essential role in the biogenesis of the photosynthetic apparatus. Global proteomic analyses have revealed that some photosynthetic apparatus subunits, such as D1 and PsbO in PSII, cytochrome f and Rieske FeS protein in the Cyt b6f complex, and PsaA, PsaF, and PsaN in PSI, may be TRX partners (Motohashi and Hisabori, 2006; Ströher and Dietz, 2008; Montrichard et al., 2009; Lindahl et al., 2011). TRX z has been shown to redox regulate chloroplastic gene expression and development (Arsova et al., 2010). NTRC participates in the posttranslational regulation of magnesium protoporphyrin methyltransferase in tetrapyrrole synthesis (Richter et al., 2013). In addition, HIGH CHLOROPHYLL FLUORESCENCE164 (HCF164), a lumenal TRX-like protein, has been shown to be involved in the assembly of the Cyt b6f complex (Lennartz et al., 2001; Motohashi and Hisabori, 2006, 2010). In spite of this, our knowledge of the regulatory function of TRXs in the biogenesis of the photosynthetic apparatus has been largely limited by the transient nature of interactions between TRXs and their target proteins or by the absence of detectable phenotypes in single TRX mutants that are presumably due to functional redundancy within TRX gene families.
In this study, we aimed to further investigate the role of chloroplastic TRXs in the biogenesis of the photosynthetic complexes. Among the numerous chloroplastic TRXs, the m-type TRX proteins have been suggested to be involved in leaf development, chloroplast morphology, cyclic electron flow, and tetrapyrrole synthesis (Ikegami et al., 2007; Chi et al., 2008; Benitez-Alfonso et al., 2009; Luo et al., 2012; Courteille et al., 2013). Besides these, the TRX m1, TRX m2, and TRX m4 proteins have been demonstrated to peripherally associate with the stroma-exposed thylakoid membranes (Peltier et al., 2002; Friso et al., 2004). All of these findings encouraged us to comprehensively investigate the impact of TRX m1, TRX m2, and TRX m4 deficiency on chloroplast development and the accumulation of the thylakoid protein complexes. Based on the pale-green leaf phenotype and the specifically impaired PSII complex in plants triply silenced for TRX m1, TRX m2, and TRX m4, we have established the redundant role of the TRX m1, TRX m2, and TRX m4 proteins in PSII accumulation. Furthermore, biochemical studies in vivo and in vitro indicate that the three TRX m proteins directly interact with PSII and modulate the reduction of intermolecular disulfide bonds in CP47.
RESULTS
Triple Inactivation of Three M-Type TRX Genes Causes a Pale-Green Leaf Phenotype in Arabidopsis
To examine the physiological functions of the TRX m1, TRX m2, and TRX m4 proteins in vivo, transfer DNA (T-DNA) insertional mutant libraries were screened, and homozygous trx m1 and trx m4 mutants were identified in previous studies (Courteille et al., 2013; Laugier et al., 2013). Although trx m4 mutants exhibited elevated cyclic electron flow capacity, all trx m1 and trx m4 mutant plants showed no obvious growth defects when compared with wild-type plants under normal growth conditions (Courteille et al., 2013; Laugier et al., 2013), implying that functional redundancy may exist between TRX m1, TRX m2, and TRX m4. To test this hypothesis and to avoid the lethality of multiple TRX m deficiency, the tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) approach was applied to efficiently silence one or more TRX m genes (Liu et al., 2002; Burch-Smith et al., 2006; Arsova et al., 2010; Luo et al., 2012).
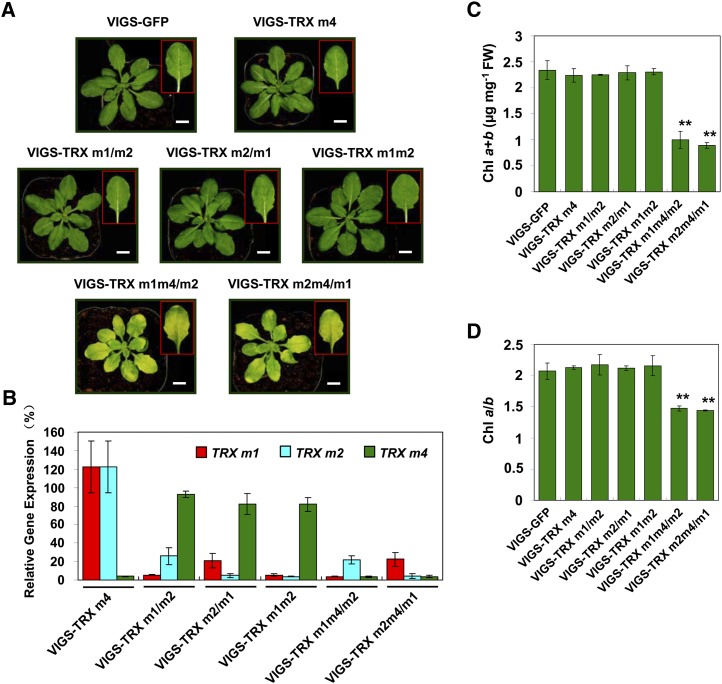
To verify the phenotypes in trx m single mutants, the coding regions of TRX m1, TRX m2, and TRX m4 were individually cloned into the pTRV2 vector (pTRV2-TRX m1, pTRV2-TRX m2, and pTRV2-TRX m4) for efficient silencing of single TRX m genes (Burch-Smith et al., 2006). Three weeks after infection, the time interval used for all biological experiments, the mRNA levels of TRX m genes in VIGS plants were analyzed by quantitative real-time reverse transcription (RT)-PCR. In comparison with VIGS-GFP plants (the negative control), the expression of TRX m4 was specifically reduced by more than 90% in the plants infiltrated with the pTRV2-TRX m4 vector (called VIGS-TRX m4 plants). Unexpectedly, we found that TRX m2 mRNA levels in plants infiltrated with the pTRV2-TRX m1 vector decreased by approximately 74%, and vice versa, whereas the expression of TRX m4 was not influenced in these plants (Fig. 1B; Supplemental Table S1). This off-target silencing effect on TRX m1 and TRX m2 could be attributed to the significantly higher coding sequence homology between TRX m1 and TRX m2 (76.1% identity) than between TRX m1 and TRX m4 (57.9% identity; Supplemental Fig. S1A; Jackson et al., 2003; Chibani et al., 2009). Therefore, plants infected with the pTRV2-TRX m1 and pTRV2-TRX m2 vectors exhibited double TRX m1 and TRX m2 silencing and were designated VIGS-TRX m1/m2 and VIGS-TRX m2/m1 plants, respectively. Similar to the single TRX m mutant plants, VIGS-TRX m4, VIGS-TRX m1/m2, and VIGS-TRX m2/m1 plants showed no obvious differences from VIGS-GFP plants (Fig. 1; Supplemental Table S1). Furthermore, we introduced the coding regions of TRX m1 and TRX m2 into the pTRV2 vector together (pTRV2-TRX m1m2) for simultaneous inactivation of these two TRX genes. Plants infected with the pTRV-TRX m1m2 vector showed more than 90% silencing efficiency for both TRX m1 and TRX m2 (called VIGS-TRX m1m2 plants) and also exhibited no visible phenotype (Fig. 1A; Supplemental Table S1). Therefore, we propose that silencing of individual m-type TRX genes and concurrent silencing of TRX m1 and TRX m2 have no obvious impact on leaf development.
Figure 1.
Characterization of the Arabidopsis VIGS-TRX m plants. A, Representative photographs of VIGS-GFP, VIGS-TRX m4, VIGS-TRX m1/m2, VIGS-TRX m2/m1, VIGS-TRX m1m2, VIGS-TRX m1m4/m2, and VIGS-TRX m2m4/m1 plants, which were infected with the pTRV2-GFP, pTRV2-TRX m4, pTRV2-TRX m1, pTRV2-TRX m2, pTRV2-TRX m1m2, pTRV2-TRX m1m4, and pTRV2-TRX m2m4 vectors, respectively, via agroinfiltration. Photographs of representative leaves from VIGS plants are shown in the insets. All the plants were observed 3 weeks after infiltration. At least three biological replicates were performed, and similar results were obtained. Bars = 1 cm. B, The suppression rate of TRX m genes in VIGS-TRX m plants was analyzed by quantitative real-time RT-PCR. The relative transcript levels of TRX m1, TRX m2, and TRX m4 in VIGS-TRX m4, VIGS-TRX m1/m2, VIGS-TRX m2/m1, VIGS-TRX m1m2, VIGS-TRX m1m4/m2, and VIGS-TRX m2m4/m1 plants were normalized to the level in VIGS-GFP plants (100%). The data represent means ± sd of three biological replicates. C and D, Total chlorophyll (Chl) content (C) and chlorophyll a/b ratio (D) in VIGS-TRX m and VIGS-GFP plants. FW, Fresh weight. The data represent means ± sd of three biological replicates. Statistical significance compared with the VIGS-GFP plants is indicated by asterisks (**P ≤ 0.01, Student’s t test).
To further investigate the influence of triple TRX m gene silencing on leaf development, we cloned the coding regions of TRX m4 with TRX m1 or TRX m2 into the pTRV2 vector (pTRV2-TRX m1m4 and pTRV2-TRX m2m4 vectors). Plants infected with either pTRV2-TRX m1m4 or pTRV2-TRX m2m4 showed the triple TRX m-silenced phenotype and were designated VIGS-TRX m1m4/m2 or VIGS-TRX m2m4/m1 plants, respectively (Fig. 1B; Supplemental Table S1). Interestingly, these plants had pale-green leaves with strongly reduced (approximately 45%) chlorophyll contents compared with the VIGS-GFP plants; the chlorophyll a/b ratio was 1.5 in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants versus 2.1 in VIGS-GFP plants (Fig. 1, C and D). Taken together, the phenotypic comparison between single, double, and triple TRX m-silenced plants indicates that the TRX m1, TRX m2, and TRX m4 proteins possess a functional redundancy in leaf greening, and only the inactivation of the three m-type TRX genes can result in pale-green leaves.
PSII Activity Is Dramatically Reduced in Plants Triply Silenced for TRX m1, TRX m2, and TRX m4
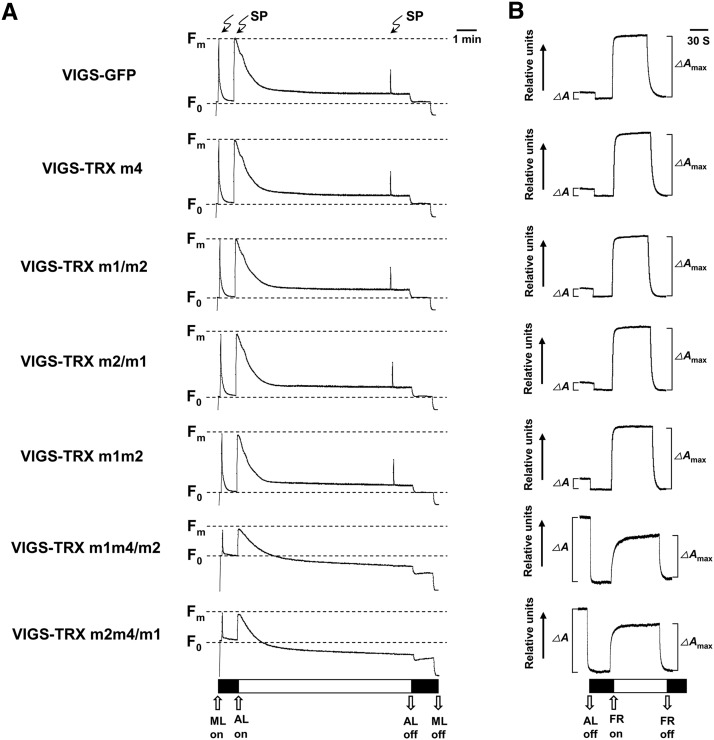
To gain insight into the primary target of the triple TRX m1, TRX m2, and TRX m4 gene silencing, noninvasive chlorophyll fluorometric analyses were performed to investigate photosynthetic electron transport in the VIGS plants (Baker, 2008). The ratio of variable fluorescence to maximal fluorescence (Fv/Fm), which is an indicator of the maximum efficiency of PSII photochemistry, was drastically decreased in the VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants (0.46 ± 0.02 versus 0.83 ± 0.01 in the VIGS-GFP controls; Fig. 2A; Table I). The decreased Fv/Fm below 0.5 indicates that the triple TRX m-silenced plants have primary defects in electron transfer within PSII or a partial loss of PSII capacity (Meurer et al., 1996). Consistently, the actual PSII photochemical efficiency (ΦPSII) and photochemical quenching (qP) could not be measured in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants (Table I). In contrast, nonphotochemical quenching (NPQ) in the VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants was higher than in the VIGS-GFP plants (Table I); NPQ is viewed as a compensatory mechanism in response to overexcited chlorophyll to prevent irreversible damage to PSII (Takahashi and Badger, 2011). Furthermore, inactive PSII always affects the light-induced redox kinetics of PSI (Meurer et al., 1996). The redox kinetics of P700 showed that P700 can be oxidized in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants, but the amplitude of changes in A820 induced by far-red light were lower than in the VIGS-GFP plants, and the ΔA/ΔAmax value was greater than 1 (Fig. 2B), implying that PSI in the triple TRX m-silenced plants is functional but with low activity. These spectroscopic analyses suggest that the photosynthetic changes in the triple TRX m-silenced plants primarily reflect a reduction in PSII activity.
Figure 2.
Spectroscopic characterization of VIGS-TRX m and VIGS-GFP plants. A, Chlorophyll a fluorescence induction analyses in VIGS plants. The minimal level of fluorescence (F0) and the maximal level of fluorescence (Fm) are indicated for each genotype. The lightning arrows indicate the application of saturating light pulses (SP). The white bar indicates exposure to actinic light (AL), and the blank bar indicates exposure to measuring light (ML). B, Redox kinetics of P700 at 820 nm induced by far-red light (FR; 720 nm) in VIGS plants. ΔA and ΔAmax are in vivo absorbance changes of P700 at 820 nm induced by actinic light and far-red light illumination, respectively. Three biological replicates were performed, and similar results were obtained.
Table I. Chlorophyll fluorescence parameters in VIGS-TRX m and VIGS-GFP plants.
Measurements of chlorophyll fluorescence parameters were done on VIGS plants after 15 min of dark adaptation. Actinic light intensity was 110 μmol photons m−2 s−1. 1 − qP represents excitation pressure. At least six leaves from VIGS plants were measured. Data are given as means ± se of three biological replicates. Statistical significance compared with the VIGS-GFP plants is indicated by asterisks (**P ≤ 0.01, Student’s t test).
| Parameter | VIGS-GFP | VIGS-TRX m4 | VIGS-TRX m1/m2 | VIGS-TRX m2/m1 | VIGS-TRX m1m2 | VIGS-TRX m1m4/m2 | VIGS-TRX m2m4/m1 |
|---|---|---|---|---|---|---|---|
| Fv/Fm | 0.830 ± 0.001 | 0.825 ± 0.003 | 0.824 ± 0.004 | 0.824 ± 0.006 | 0.835 ± 0.001 | 0.463 ± 0.021** | 0.461 ± 0.030** |
| ФPSII | 0.532 ± 0.014 | 0.553 ± 0.018 | 0.554 ± 0.015 | 0.589 ± 0.011 | 0.536 ± 0.005 | 0** | 0** |
| NPQ | 0.721 ± 0.071 | 0.753 ± 0.067 | 0.684 ± 0.035 | 0.601 ± 0.037 | 0.633 ± 0.076 | 1.877 ± 0.256** | 1.913 ± 0.226** |
| 1 − qP | 0.281 ± 0.118 | 0.242 ± 0.023 | 0.249 ± 0.021 | 0.209 ± 0.015 | 0.277 ± 0.015 | 1** | 1** |
Accumulation of the PSII Complex Is Specifically Impaired in Plants Triply Silenced for TRX m1, TRX m2, and TRX m4
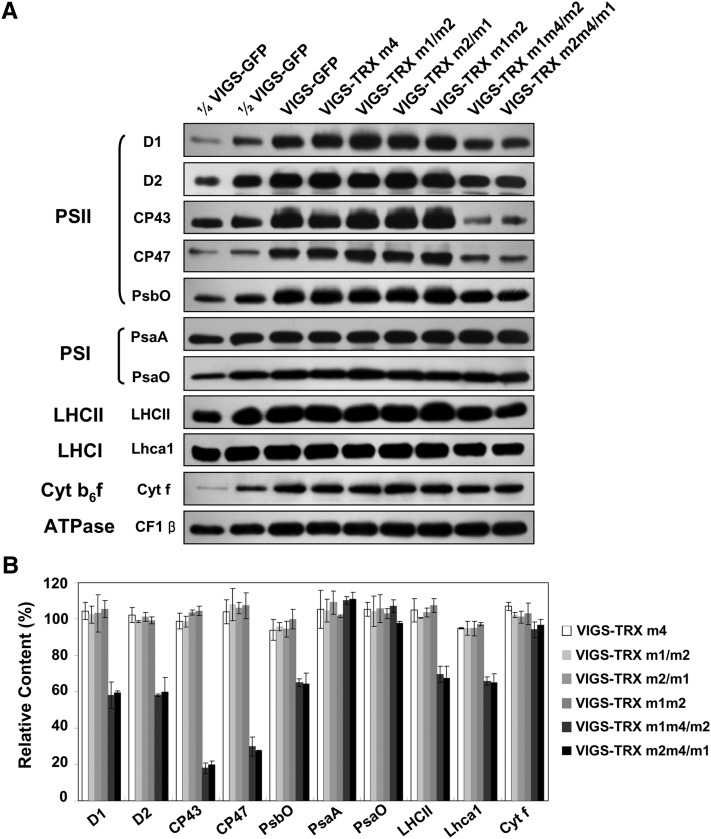
The defect in PSII photosynthetic electron transport could be associated with altered protein levels in the PSII complex. To examine the steady-state levels of thylakoid proteins in the VIGS plants, we performed immunoblot analyses using antibodies raised against diagnostic subunits from four thylakoidal membrane photosynthetic protein complexes (i.e. PSII, Cyt b6f complex, PSI, and ATP synthase). Our results showed that levels of the chloroplast-encoded PSII core subunits D1, D2, CP43, and CP47 in thylakoids isolated from VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants were reduced to approximately 58%, 58%, 18%, and 28% of VIGS-GFP control levels, respectively (Fig. 3). The nucleus-encoded 33-kD subunit of the oxygen-evolving complex (PsbO) accumulated to approximately 64% of VIGS-GFP levels, and the light-harvesting complex II (LHCII) and PSI antenna protein (Lhca1) were reduced to approximately 68% and 65% of control levels, respectively (Fig. 3). In contrast, the diagnostic subunits of PSI (PsaA), the Cyt b6f complex (cytochrome f), and ATP synthase (CF1 β) in the VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants accumulated to levels similar to those in the control VIGS-GFP plants (Fig. 3).
Figure 3.
Immunoblot analyses of thylakoid proteins from VIGS plants. A, Immunoblot analysis of thylakoid proteins loaded on the basis of equal total thylakoid proteins. Aliquots of 2.5 μg (¼ VIGS-GFP), 5 μg (½ VIGS-GFP), 10 μg (VIGS-GFP), and 10 μg (VIGS-TRX m) of total thylakoid proteins were loaded on the gels. Designations of thylakoid membrane protein complexes and their diagnostic components are labeled on the left. Three biological replicates were performed, and similar results were obtained. B, Semiquantitative analysis of thylakoid proteins. Immunoblots in A from three biological replicates were analyzed with Phoretix 1D software (Phoretix International). The protein contents (per unit of thylakoid protein) of the thylakoid membranes from VIGS plants were normalized to the level of the β-subunit of the ATP synthase (CF1 β). Protein levels in the VIGS-TRX m plants are shown relative to the levels in the VIGS-GFP leaves (100%). Data are given as means ± sd of three biological replicates.
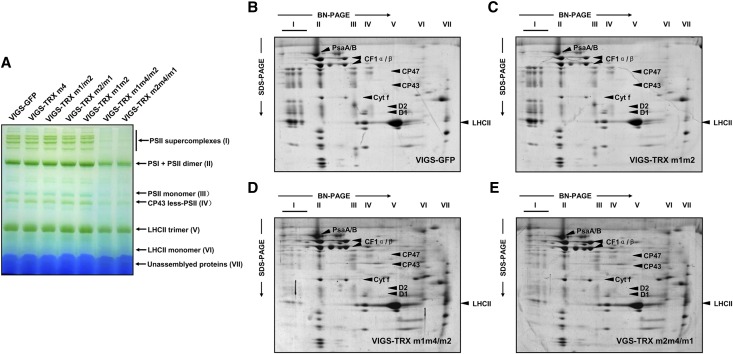
To further investigate the putative structural alterations of thylakoid membrane protein complexes in the triple TRX m gene-silenced plants, thylakoid membranes from VIGS plants were solubilized with n-dodecyl β-d-maltoside (DM), and the chlorophyll-protein complexes (with equal amounts of chlorophyll) were separated by blue native (BN)-PAGE (Schägger et al., 1994). Seven protein complexes were resolved in the first dimension in the presence of Coomassie blue dye; these represent PSII supercomplexes (band I), PSI monomers and PSII dimers (band II), PSII monomers (band III), CP43-minus PSII (band IV), trimeric (band V) and monomeric (band VI) LHCII, and unassembled proteins (band VII; Fig. 4A). The BN-PAGE analysis clearly showed that the amount of PSII complexes (bands I, II, III, and IV) per unit of chlorophyll was significantly lower in thylakoids isolated from the VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants than that from the VIGS-GFP control plants (Fig. 4A). Analyses of the second-dimension SDS-urea-PAGE gels after Coomassie blue staining confirmed that the relative amounts of the PSII core subunits D1, D2, CP43, and CP47 were considerably reduced in the VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants, while no significant changes were observed in the amounts of Cyt b6f and ATP synthase (Fig. 4, D and E). Collectively, our results indicate that the dramatically decreased PSII activity seen in plants silenced for the three TRX m genes could be attributed to the impaired accumulation of the PSII complex.
Figure 4.
Analyses of thylakoid protein complexes from VIGS plants. A, BN-PAGE analyses of thylakoid chlorophyll-protein complexes. Equal amounts of thylakoid membranes (8 µg of chlorophyll) from VIGS-TRX m and VIGS-GFP plants were solubilized with 1% (w/v) DM and separated by BN-PAGE. Assignments of the thylakoid membrane macromolecular protein complexes indicated at right were identified according to a previous study (Peng et al., 2006). B to E, Two-dimensional BN/SDS-PAGE fractionation of thylakoid protein complexes. Individual lanes from the BN-PAGE gels in A were subjected to second-dimension SDS-urea-PAGE followed by Coomassie blue staining. Identities of the relevant proteins are indicated by arrows. Three biological replicates were performed, and similar results were obtained. [See online article for color version of this figure.]
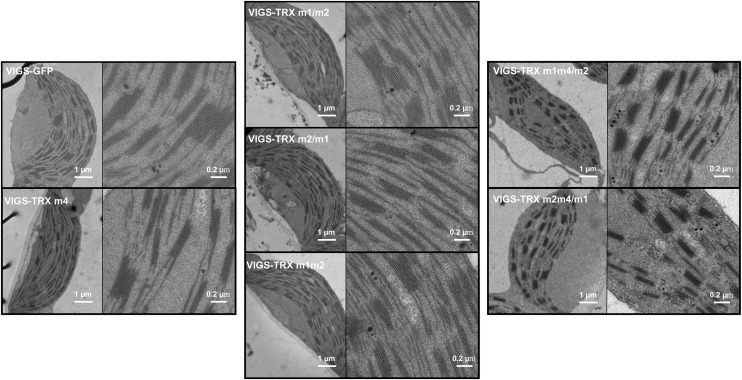
To assess whether the triple silencing of TRX m genes causes ultrastructural changes in chloroplasts, transmission electron micrographs of ultrathin sections of leaves from various VIGS plants were analyzed. In agreement with the phenotypic comparisons between VIGS-TRX m and VIGS-GFP plants, chloroplasts from VIGS-TRX m4, VIGS-TRX m1/m2, VIGS-TRX m2/m1, and VIGS-TRX m1m2 plants were similar in size to those from VIGS-GFP plants, and they contained well-developed membrane systems composed of grana connected by stroma lamellae (Fig. 5). In contrast, the thylakoid membranes from VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 chloroplasts were disrupted, such that the grana stacks in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 chloroplasts appeared to be less connected by stroma lamellae than those in VIGS-GFP control chloroplasts (Fig. 5).
Figure 5.
Electron micrographs of chloroplasts from VIGS plants. Images of intact chloroplasts and closeup views were obtained from VIGS-TRX m and VIGS-GFP plants. Leaves were collected 3 weeks after infection. Two biological replicates were performed, and similar results were obtained. Scale bars are as indicated.
Expression of Chloroplast-Encoded PSII Subunits Is Not Altered in Plants Triply Silenced for TRX m1, TRX m2, and TRX m4
The chloroplast transcriptional regulation system in Arabidopsis depends upon both the plastid-encoded plastid RNA polymerase (PEP) and the nucleus-encoded plastid RNA polymerase (NEP), which have different target genes (Shiina et al., 2005). Chloroplast genes, therefore, can be classified into three groups according to their promoter structures: most of the photosynthetic genes depend on PEP (class I), whereas RNA Polymerase Subunit Alpha (RpoA), RpoB, Acetyl-CoA Carboxylase Carboxyl Transferase Subunit Beta (AccD), and Uncharacterised Protein Family Ycf2 (Ycf2) genes are exclusively transcribed by NEP (class III), and the nonphotosynthetic housekeeping genes are generally transcribed by both PEP and NEP (class II; Hajdukiewicz et al., 1997).
The recently characterized TRX z participates in the PEP-dependent expression of chloroplast genes through the redox regulation of two fructokinase-like proteins (Arsova et al., 2010). To test whether the significant reduction in PSII core subunits observed in the triple TRX m-silenced plants resulted from impaired transcription of chloroplast genes, we used quantitative real-time RT-PCR to analyze mRNA levels of genes that encode the representative chloroplast proteins transcribed by NEP and/or PEP in the VIGS plants (Table II). The PsaA, PsbA, PsbB, PsbC, and PsbD genes that encode photosystem subunits were selected as representatives of the PEP-dependent genes (class I), RpoA, AccD, and Ycf2 were selected as NEP-dependent genes (class III), and Caseinolytic Protease P (ClpP) and NADH Dehydrogenase ND2 were chosen as class II genes. Quantitative real-time RT-PCR analysis revealed that the expression of class I genes in VIGS-TRX m plants was no different from that in VIGS-GFP control plants (Table II), suggesting that TRX m proteins are not involved in the regulation of PEP-dependent gene expression. In spite of this, triple silencing of TRX m1, TRX m2, and TRX m4 altered the mRNA levels of some chloroplast-encoded genes. The ClpP and Ycf2 transcript levels were significantly different in comparison with levels in the VIGS-GFP plants (Table II). In addition, expression of the nucleus-encoded genes PsaE, PsbO, and PSII Light Harvesting Complex Gene2.4 (Lhcb2.4), which encode proteins that are targeted to the chloroplast, was substantially reduced in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants (Table II). These data indicate that silencing the three TRX m genes does not result in a general inhibitory effect on the transcription of chloroplast protein genes.
Table II. Changes in transcript abundance of chloroplast-encoded and nucleus-encoded genes in VIGS-TRX m plants and VIGS-GFP plants.
The transcript levels of representative genes encoding chloroplast proteins were analyzed by quantitative real-time RT-PCR. For individual genes, relative mRNA levels were normalized with respect to ACTIN2 (At3g18780) and then to VIGS-GFP levels. The log2 (VIGS-TRX m/VIGS-GFP) value is given, where 3.32 corresponds to a 10-fold up-regulation and −3.32 corresponds to a 10-fold down-regulation in VIGS-TRX m plants relative to VIGS-GFP plants. UBIQUITIN4 (At5g20620) was also used as a reference gene with similar results (data not shown). N.D., Not detectable. The data represent means ± sd of three biological replicates. Statistical significance compared with the VIGS-GFP plants is indicated by asterisks (**P ≤ 0.01, Student’s t test).
| Gene | Accession No. | VIGS-TRX m4 | VIGS-TRX m1/m2 | VIGS-TRX m2/m1 | VIGS-TRX m1m2 | VIGS-TRX m1m4/m2 | VIGS-TRX m2m4/m1 | |
|---|---|---|---|---|---|---|---|---|
| Class I genes | ||||||||
| PsaA | Atcg00350 | 0.215 ± 0.127 | −0.040 ± 0.134 | −0.145 ± 0.021 | −0.085 ± 0.021 | −0.228 ± 0.244 | −0.412 ± 0.087 | |
| PsbA | Atcg00020 | 0.350 ± 0.113 | 0.325 ± 0.085 | 0.235 ± 0.226 | 0.270 ± 0.014 | −0.150 ± 0.237 | −0.065 ± 0.239 | |
| PsbB | Atcg00680 | 0.068 ± 0.214 | −0.202 ± 0.427 | −0.188 ± 0.238 | −0.202 ± 0.427 | −0.105 ± 0.200 | −0.200 ± 0.075 | |
| PsbC | Atcg00280 | −0.308 ± 0.351 | 0.070 ± 0.627 | −0.152 ± 0.837 | −0.310 ± 0.113 | −0.478 ± 0.511 | −0.368 ± 0.046 | |
| PsbD | Atcg00270 | 0.285 ± 0.071 | 0.130 ± 0.177 | −0.153 ± 0.032 | 0.210 ± 0.156 | −0.367 ± 0.618 | −0.183 ± 0.154 | |
| Class II genes | ||||||||
| ClpP | Atcg00670 | −0.615 ± 0.163 | −0.245 ± 0.219 | −0.485 ± 0.120 | −0.225 ± 0.057 | −1.595 ± 0.054** | −1.627 ± 0.267** | |
| NADH | Atcg00890 | 0.230 ± 0.007 | 0.060 ± 0.177 | −0.120 ± 0.021 | −0.190 ± 0.120 | −0.065 ± 0.100 | −0.045 ± 0.072 | |
| Dehydrogenase ND2 | ||||||||
| Class III genes | ||||||||
| RpoA | Atcg00740 | 0.203 ± 0.247 | −0.113 ± 0.028 | 0.213 ± 0.049 | 0.183 ± 0.148 | −0.733 ± 0.208 | −0.606 ± 0.100 | |
| AccD | Atcg00500 | 0.140 ± 0.035 | 0.475 ± 0.001 | N.D. | 0.390 ± 0.205 | 0.290 ± 0.173 | 0.160 ± 0.064 | |
| Ycf2 | Atcg00860 | −0.295 ± 0.106 | −0.335 ± 0.099 | 0.145 ± 0.042 | 0.190 ± 0.078 | 1.885 ± 0.232** | 1.918 ± 0.206** | |
| Nucleus-encoded genes | ||||||||
| PsaE | At4g28750 | 0.115 ± 0.163 | −0.085 ± 0.106 | 0.090 ± 0.057 | 0.185 ± 0.014 | −1.212 ± 0.174** | −1.248 ± 0.215** | |
| PsbO | At5g66570 | 0.095 ± 0.163 | −0.050 ± 0.064 | −0.075 ± 0.014 | 0.140 ± 0.007 | −1.050 ± 0.228** | −0.868 ± 0.070** | |
| Lhcb2.4 | At3g27690 | −0.128 ± 0.061 | −0.337 ± 0.083 | −0.128 ± 0.061 | 0.480 ± 0.045 | −3.563 ± 0.191** | −3.310 ± 0.394** | |
VIGS-TRX m Plants Accumulate More Reactive Oxygen Species Than VIGS-GFP Plants
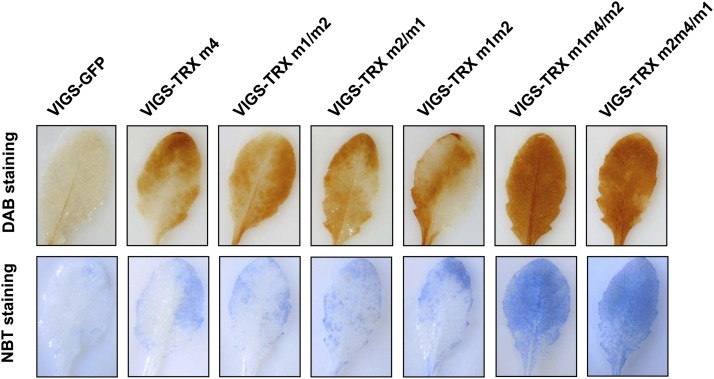
TRX m proteins function as electron donors for Cys-containing proteins and regulate the functions of target proteins by reducing their disulfide bonds. Among the target proteins of TRX m, 2-Cys peroxiredoxin is an important antioxidant for hydrogen peroxide (H2O2) detoxification in chloroplast (König et al., 2002; Dietz and Pfannschmidt, 2011; Tovar-Méndez et al., 2011). Previous studies suggest that TRX m proteins are able to reduce the oxidative state of 2-Cys peroxiredoxin to maintain the metabolic balance of reactive oxygen species (ROS; Collin et al., 2003; Chi et al., 2008). Therefore, we hypothesized that simultaneous inactivation of the Arabidopsis TRX m1, TRX m2, and TRX m4 genes could influence the ROS level in vivo. The results of histochemical staining demonstrated that elevated levels of H2O2 and superoxide anion radical (O2−) were detected in all VIGS-TRX m plants, especially in the triply silenced VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants (Fig. 6). ROS accumulation at similar levels has been detected in other TRX m gene-silenced plants, such as the Arabidopsis TRX m3 knockout mutant (Benitez-Alfonso et al., 2009) and in rice (Oryza sativa) OsTRX m RNA interference plants (Chi et al., 2008). Furthermore, it is widely accepted that ROS serve as an essential redox signal for the transcriptional regulation of photosynthesis-related nuclear genes (Nott et al., 2006; Ruckle et al., 2007). In line with this, our expression analysis showed that mRNA levels for the nucleus-encoded chloroplast protein genes PsaE, PsbO, and Lhcb2 were down-regulated (Table II). Taken together, our data suggest that triple silencing of TRX m1, TRX m2, and TRX m4 not only results in a defect in the PSII complex but also affects ROS metabolism.
Figure 6.
ROS accumulation in leaves of VIGS plants. Fully expanded leaves from VIGS-TRX m and VIGS-GFP plants grown under normal conditions were harvested for in situ histochemical staining using 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) to show the accumulated H2O2 (top row) and O2− (bottom row), respectively. Three biological replicates were performed, and similar results were obtained.
Confirmation of Impaired PSII Accumulation in Triple TRX m-Silenced Plants
The above results strongly indicated that the triple inactivation of TRX m1, TRX m2, and TRX m4 leads to pale-green leaves and impaired PSII accumulation. To further confirm these observations in TRX m mutant plants, the homozygous trx m1trx m4 double mutant was isolated by crossing and was found to have a similar phenotype to wild-type plants (Supplemental Fig. S2; Supplemental Tables S1 and S2). In combination with the phenotype observed in plants silenced for both TRX m1 and TRX m2, we propose that the simultaneous inactivation of TRX m1 and either TRX m2 or TRX m4 has no conspicuous effect on leaf development.
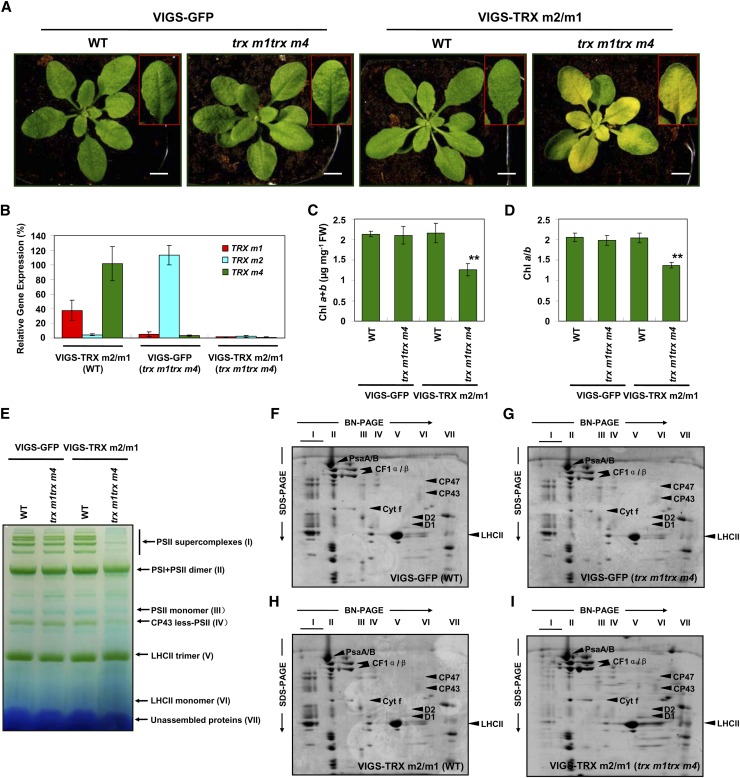
Furthermore, we introduced the TRV-TRX m2 vector into trx m1trx m4 double mutant plants to silence the TRX m2 gene. Three weeks after infection, plants infiltrated with the pTRV2-TRX m2 vector displayed evidently reduced expression of TRX m1 and TRX m2 genes in comparison with VIGS-GFP (wild-type) plants and were designated VIGS-TRX m2/m1 (WT) and VIGS-TRX m2/m1 (trx m1trx m4) plants (Fig. 7B; Supplemental Table S1). The VIGS-TRX m2/m1 (trx m1trx m4) plants also had pale-green leaves and sharply reduced chlorophyll contents (Fig. 7, A–D). The two-dimensional BN/SDS-PAGE analysis indicated that the accumulation of PSII complexes (bands I, II, III, and IV) and PSII core proteins was specifically and markedly reduced in thylakoid membranes isolated from VIGS-TRX m2/m1 (trx m1trx m4) plants (Fig. 7, E–I).
Figure 7.
Characterization of VIGS-TRX m2 (trx m1trx m4) plants. A, Representative photographs of the wild-type VIGS-GFP (WT), VIGS-GFP (trx m1trx m4), VIGS-TRX m1/m2 (WT), and VIGS-TRX m2/m1 (trx m1trx m4) plants, which were infected with the pTRV2-GFP and pTRV2-TRX m2 vectors via agroinfiltration in wild-type and trx m1trx m4 double mutant backgrounds, respectively. Representative leaves are shown in the insets. At least three biological replicates were performed, and similar results were obtained. Bars = 1 cm. B, The suppression rate of TRX m genes in VIGS-TRX m plants was analyzed by quantitative real-time RT-PCR. The relative mRNA levels of TRX m1, TRX m2, and TRX m4 in the VIGS-TRX m2/m1 (WT), VIGS-GFP (trx m1trx m4), and VIGS-TRX m2/m1 (trx m1trx m4) plants were normalized to the levels in VIGS-GFP (WT) plants (100%). The data represent means ± sd of three biological replicates. C and D, Total chlorophyll (Chl) contents (C) and chlorophyll a/b ratios (D) of the VIGS plants in wild-type and trx m1trx m4 double mutant backgrounds. FW, Fresh weight. The data represent means ± sd of three independent experiments. Statistical significance compared with the VIGS-GFP (WT) plants is indicated by asterisks (**P ≤ 0.01, Student’s t test). E, BN-PAGE analyses of thylakoid chlorophyll-protein complexes. Equal amounts of thylakoid membranes (8 µg of chlorophyll) from the VIGS plants in wild-type and trx m1trx m4 double mutant backgrounds 3 weeks after infiltration were solubilized with 1% (w/v) DM and separated by BN-PAGE. Assignments of the thylakoid membrane macromolecular protein complexes indicated at right were identified according to a previous study (Peng et al., 2006). F to I, Two-dimensional BN/SDS-PAGE fractionation of thylakoid protein complexes. Individual lanes from the BN gels in E were subjected to second-dimension SDS-urea-PAGE followed by Coomassie blue staining. Identities of the relevant proteins are indicated by arrowheads. Three biological repeats were performed, and similar results were obtained.
M-Type TRX Proteins Associate with Minor PSII Assembly Intermediates and Interact with PSII Core Subunits D1, D2, and CP47
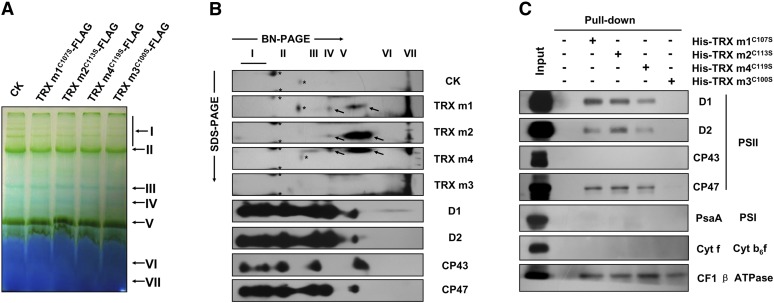
The observed reduction in the abundance of PSII complexes in the triple TRX m-silenced plants implies a direct regulatory function for m-type TRX proteins in the biogenesis of PSII. To test this hypothesis, we asked whether the TRX m1, TRX m2, and TRX m4 proteins are associated with the PSII complex. Because of the high degrees of similarity between the amino acid sequences of these three TRX proteins (Supplemental Fig. S1B), we were unable to prepare antibodies raised specifically against each of the three individual TRX m proteins. Alternatively, we combined 2× FLAG-tagged proteins at the C termini of TRX m amino acid sequences for immunoblot analysis. Moreover, to strengthen the interaction between TRX m and their target proteins, the C-terminal Cys residue in the TRX active-site motif (WCGPC) that catalyzes the dissociation of target proteins from TRXs was substituted with a Ser residue (Motohashi et al., 2001; Balmer et al., 2003). The TRX m1C107S-FLAG, TRX m2C113S-FLAG, and TRX m4C119S-FLAG fusion proteins were transiently expressed separately in Arabidopsis mesophyll protoplasts, and TRX m3C100S-FLAG was used as the reference protein (Supplemental Fig. S3A). Thylakoid membranes were then isolated from protoplasts and solubilized using 1% (w/v) DM for two-dimensional BN/SDS-PAGE analysis (Fig. 8A). After electrophoresis, immunoblot analyses were performed using antibodies raised against FLAG-tagged TRX m proteins and PSII subunits (D1, D2, CP43, and CP47). The results showed that the vast majority of the TRX m1, TRX m2, and TRX m4 proteins, but not TRX m3, could comigrate with minor PSII assembly intermediates such as the PSII reaction center (RC), the CP43 precomplex, and the CP47 precomplex, and the immunoblot signals for the TRX m1, TRX m2, and TRX m4 proteins were also detected in the CP43-minus PSII monomers (band IV; Fig. 8B). To confirm the association of TRX m with the PSII complex, we examined the interactions of TRX m with PSII subunits by means of pull-down assays. For this purpose, the immobilized His-TRX m1C107S, His-TRX m2C113S, His-TRX m4C119S, and His-TRX m3C100S recombinant proteins were used as baits to trap potential targets in the thylakoid membranes (Supplemental Fig. S3B). The trapped TRX m target proteins were eluted and subjected to immunoblot analysis. The results showed that TRX m1, TRX m2, and TRX m4, but not TRX m3, could directly interact with PSII core subunits D1, D2, and CP47, but not CP43. In addition, the four TRX m proteins could interact with the ATP synthase subunit (CF1 β), which is a well-known TRX target protein (Motohashi and Hisabori, 2006; Ströher and Dietz, 2008; Montrichard et al., 2009; Lindahl et al., 2011), but not with representative subunits of PSI (PsaA) and the Cyt b6f complex (cytochrome f; Fig. 8C).
Figure 8.
Evidence for the interaction of TRX m1, TRX m2, and TRX m4 proteins with PSII protein complexes. A, Analysis of thylakoid protein complexes by BN-PAGE. Freshly isolated thylakoid membranes from protoplasts transformed with pUC19-TRX m1C107S-FLAG, pUC19-TRX m2C113S-FLAG, pUC19-TRX m4C119S-FLAG, and pUC19-TRX m3C100S-FLAG plasmids were solubilized with 1% (w/v) DM at a chlorophyll concentration of 15 μg, and the protein samples were separated by BN-PAGE. Protoplasts transformed with the empty transient expression vector were the negative control (CK). Assignments of the thylakoid membrane macromolecular protein complexes indicated at right were identified according to a previous study (Peng et al., 2006). B, Two-dimensional BN/SDS-PAGE fractionation and immunoblot analysis of PSII protein complexes. Thylakoid membrane complexes separated in A were further subjected to second-dimension SDS-urea-PAGE, and the proteins were immunodetected with specific antibodies against FLAG-tagged TRX m proteins, D1, D2, CP43, and CP47. The asterisks indicate nonspecific signals on the immunoblots. The arrows indicate the FLAG-tagged TRX m proteins. Two biological repeats were conducted, and similar results were obtained. C, Pull-down assay. His-TRX m1C107S, His-TRX m2C113S, His-TRX m3C100S, and His-TRX m4C119S recombinant proteins were immobilized on cyanogen bromide-activated Sepharose 4B resin and incubated with total thylakoidal membrane proteins solubilized with 1% (w/v) DM; empty cyanogen bromide-activated Sepharose 4B resin with no coupled His-TRX m protein was used as the negative control. After incubating and washing, proteins interacting with the His-TRX m mutant proteins were eluted with 10 mm DTT and detected by immunoblot analysis. Identities of thylakoid membrane protein complexes and their subunits are indicated at right. Three biological repeats were performed, and similar results were obtained. [See online article for color version of this figure.]
The Deficiency of TRX m1, TRX m2, and TRX m4 Interrupts the Redox Status of PSII Core Subunits
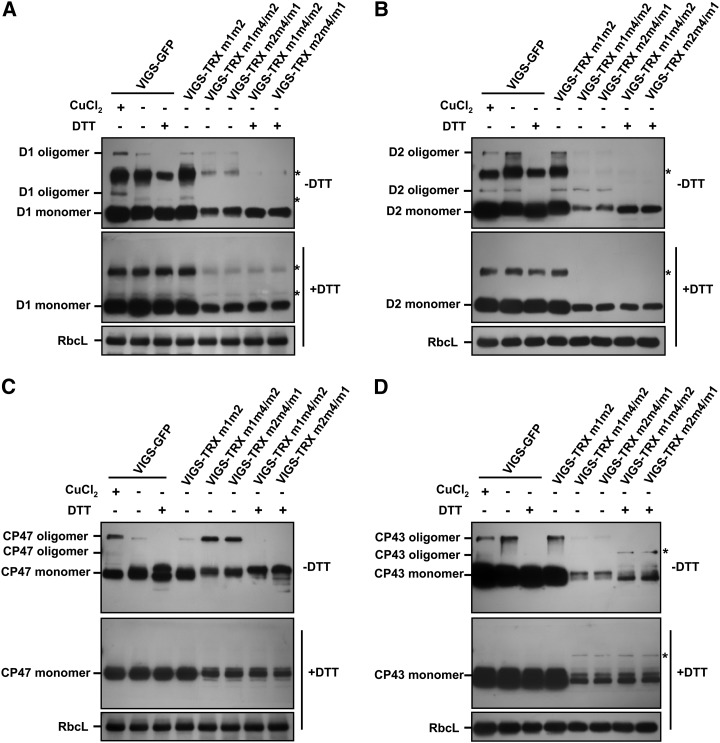
Based on the interactions between TRX m and PSII, we wondered whether the triple inactivation of TRX m1, TRX m2, and TRX m4 could have an effect on the redox status of the PSII complex in vivo. Amino acid sequence analyses showed that many PSII subunits, including D1, D2, CP43, CP47, and PsbO, have at least two conserved Cys residues that can form the intramolecular or intermolecular disulfide bridges (Shimada et al., 2007). Therefore, we investigated the redox states of intramolecular and intermolecular disulfide bonds in PSII proteins isolated from VIGS plants.
To analyze the redox status of intramolecular disulfide bridges in PSII subunits, 4-acetoamido-4-maleimidylstilbene-2,2-disulfonate (AMS; Invitrogen) was used to bind the reduced Cys residues (thiol group/sulfhydryl group) in reduced PSII proteins. In contrast, oxidized proteins with disulfide bridges or sulfenic acids cannot bind AMS; thus, the AMS-labeled reduced proteins migrate slower on nonreducing SDS-PAGE gels than do the oxidized proteins (Ikegami et al., 2007; Karamoko et al., 2011; Richter et al., 2013). To comprehensively analyze the exchanges of intramolecular disulfide bonds in PSII subunits, protein extracts from VIGS-GFP plants were treated with either the oxidant CuCl2, to induce disulfide bond formation, or the reducing agent dithiothreitol (DTT), for reduction of Cys residues, prior to AMS treatment. The mobility of AMS-labeled PSII subunits on nonreducing SDS-PAGE gels was then analyzed by immunoblotting. For D1, D2, CP43, and CP47, only one band was detected in the VIGS-GFP protein extracts, and the protein mobility was unaltered following CuCl2 or DTT treatment, whereas the mobility of AMS-labeled PsbO proteins showed a response to different redox conditions in that two oxidized PsbO bands were detected in the CuCl2-treated and nontreated VIGS-GFP protein extracts, and DTT could reduce partially oxidized PsbO to reduced PsbO (Supplemental Fig. S4). This suggested that only PsbO proteins are capable of forming intramolecular disulfide bridges and is supported by the evidence that LUMEN THIOL OXIDOREDUCTASE1 (LTO1) mediates the formation of intramolecular disulfide bonds in PsbO, which is essential for the assembly of PSII (Karamoko et al., 2011). However, the mobility of AMS-labeled PsbO in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants was not different from that in VIGS-GFP and VIGS-TRX m1m2 plants (Supplemental Fig. S4). These results suggest that the triple inactivation of TRX m genes does not influence the reduction of intramolecular disulfide bonds in PSII subunits.
Furthermore, we also examined the role of TRX m proteins in reducing the intermolecular disulfide bridges in PSII core subunits. With the formation of intermolecular disulfide bonds, two or more proteins can be aggregated to form homooligomeric/heterooligomeric complexes (Buchanan and Balmer, 2005; Dietz and Pfannschmidt, 2011). To improve the mobility and visualization of intermolecular disulfide bond-mediated macromolecular complexes, protein extracts from VIGS plants were separated by nonreducing SDS-PAGE without AMS labeling (Kirchsteiger et al., 2009; Arsova et al., 2010; Richter et al., 2013). On nonreducing SDS-PAGE gels, the monomers and oligomers of PSII core proteins D1, D2, CP43, and CP47 were observed in VIGS-GFP protein extracts, and the vast majority of PSII core subunits were present as monomeric proteins (Fig. 9). Moreover, we found that the oligomerization of D1 and CP47 was redox sensitive. CuCl2 treatment could significantly increase the aggregation of D1 and CP47 proteins, while DTT could reduce the oligomers to monomers (Fig. 9, A and C), suggesting that D1 and CP47 can form redox-regulated intermolecular disulfide bridges. One possible explanation for the observed oligomeric D2 and CP43 proteins could be the physical interactions between the four PSII core subunits (Wollman et al., 1999; Iwata and Barber, 2004; Nelson and Yocum, 2006). Interestingly, more oxidized CP47 oligomers were detected in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants in comparison with VIGS-GFP and VIGS-TRX m1m2 plants. In contrast, the levels of oligomers and monomers of D1, D2, and CP43 detected in VIGS-TRX m1m4/m2 and VIGS-TRX m2m4/m1 plants were significantly lower than in VIGS-GFP and VIGS-TRX m1m2 plants (Fig. 9C). These data suggest that TRX m1, TRX m2, and TRX m4 are required for modulating the redox status of intermolecular disulfide bridges in PSII core subunits, and the simultaneous inactivation of the three TRX m genes leads to elevated levels of oxidized CP47 oligomers.
Figure 9.
Visualization of the redox status of intermolecular disulfide bonds in PSII subunits from VIGS plants. Proteins from VIGS-TRX m1m2, VIGS-TRX m1m4/m2, VIGS-TRX m2m4/m1, and VIGS-GFP leaves were extracted under nonreducing conditions and were treated with either 1 mm CuCl2 or 10 mm DTT. The treated and untreated samples were separated on 12% nonreducing (−DTT) or reducing (+DTT) SDS-PAGE gels (15 μg of total leaf proteins per lane) and probed with antibodies raised against D1 (A), D2 (B), CP47 (C), and CP43 (D). RbcL (the large subunit of Rubisco) was used as a loading control. The asterisks indicate a nonspecific signal. Oxidized oligomers and reduced monomers of PSII subunits are labeled. Three biological replicates were performed, and similar results were obtained.
DISCUSSION
PSII is an integral membrane, multisubunit protein complex that catalyzes the light-driven water-splitting reaction and reduction of plastoquinone to initiate photosynthetic electron flow. In addition to structural studies of PSII, an emphasis has been placed on the identification of auxiliary proteins involved in the biogenesis and maintenance of the PSII complex (Mulo et al., 2008; Nixon et al., 2010; Komenda et al., 2012). Understandably, the identification of PSII accessory factors and the characterization of their functions will greatly improve our understanding of regulatory mechanisms by which the PSII complex accumulates and is maintained. Here, we report previously unknown functions for the classical chloroplastic TRX m1, TRX m2, and TRX m4 in the regulation of PSII biogenesis in Arabidopsis.
TRX m1, TRX m2, and TRX m4 Are Required for the Accumulation of PSII in Arabidopsis
To investigate the functions of TRX m1, TRX m2, and TRX m4 in the accumulation of the photosynthetic apparatus, we comprehensively characterized the phenotypes of plants silenced for one, two, or three of these TRX m genes. Interestingly, single deficiencies of either TRX m1 or TRX m4, and double inactivation of TRX m1 along with either TRX m2 or TRX m4, resulted in no macroscopic alteration in phenotype, whereas plants silenced for all three genes had pale-green leaves (Figs. 1 and 7; Supplemental Fig. S2; Supplemental Table S1), suggesting the existence of mutual functional compensation between TRX m1, TRX m2, and TRX m4. This could be associated with their similar protein structures and analogous thylakoid membrane localization (Supplemental Fig. S1; Peltier et al., 2002; Friso et al., 2004). In addition, our hypothesis is supported by the fact that the expression of TRX m1, TRX m2, or TRX m4 in yeast can rescue the oxidant-hypersensitive phenotype of the trx null strain (Issakidis-Bourguet et al., 2001). Functional compensation between TRX isoforms has also been observed in tetrapyrrole biosynthesis (Ikegami et al., 2007; Luo et al., 2012).
Furthermore, triple inactivation of the three TRX m genes has pleiotropic effects on photosynthetic electron transport and chloroplast development. Chlorophyll fluorescence measurements showed that Fv/Fm was dramatically reduced below 0.5 in the triple-silenced plants (Fig. 2; Table I). The decrease in Fv/Fm is due to specific impairment of the PSII complexes (Figs. 4 and 7). In line with this, no ΦPSII and elevated NPQ were detected, indicating that the electron flow within PSII is severely inhibited in the triple-silenced plants (Table I). In addition, determination of absorbance changes of P700 at 820 nm demonstrates that PSI in the triple-silenced plants is functional but has low activity (Fig. 2B). These spectroscopic properties also have been observed in a number of mutants with defects in PSII, such as low PSII accumulation1 (lpa1; Peng et al., 2006), lpa2 (Ma et al., 2007), lpa3 (Cai et al., 2010), photosynthesis affected mutant68 (pam68; Armbruster et al., 2010), and hcf243 (Zhang et al., 2011). Moreover, immunoblot analysis confirmed that essential subunits of the PSII complex are severely reduced in plants silenced for TRX m1, TRX m2, and TRX m4 (Fig. 3). Finally, the specifically impaired PSII complex in the triple-silenced plants had a strong effect on the organization of the thylakoid membranes. The amount of stroma lamellae and the stacking of grana thylakoids in the triple-silenced plants are obviously impaired in comparison with that in VIGS-GFP plants (Fig. 5). Taken together, our data on spectroscopy measurements, biochemical analyses, and chloroplast ultrastructure reveal a specific defect in PSII accumulation in the triple TRX m-silenced plants, thus providing a straightforward explanation for the observed pale-green leaf phenotype (Figs. 1 and 7).
The analyses described above indicate that TRX m1, TRX m2, and TRX m4 together participate in the accumulation of the PSII complex. However, the mechanism involved remains to be elucidated. Considering the hydrophilic nature of these three TRX m proteins, the possibility that they may be integral constituents of PSII can be excluded. Thus, these three TRX m proteins probably interact transiently with the PSII complex and some PSII subunits, in a manner similar to several PSII auxiliary factors that have been shown to interact with specific PSII subunits and assist their assembly, but are absent in the functional complexes (Peng et al., 2006; Ma et al., 2007; Armbruster et al., 2010; Cai et al., 2010). Indeed, TRX m1, TRX m2, and TRX m4, but not TRX m3, comigrate with minor PSII assembly intermediates and interact with PSII core subunits D1, D2, and CP47 (Fig. 8). All of these data suggest that the three TRX m proteins participate directly in the biogenesis of the PSII core complex.
Assembly of the PSII Core Complex Is Redox Dependent and Can Be Modulated by m-Type TRXs
Thiol-disulfide chemistry plays an essential role in the biogenesis of the photosynthetic apparatus and in the maintenance of photosynthetic efficiency (Dietz and Pfannschmidt, 2011; Rochaix, 2011; Kieselbach, 2013). While the formation of disulfide bonds in the oxygen-evolving complex has been shown to be essential for the assembly of the PSII complex (Burnap et al., 1994; Karamoko et al., 2011), relatively few studies have examined the mechanisms of disulfide bridge exchanges in PSII core proteins. This is hampered by multiple limitations such as the hydrophobic nature of PSII subunits and the lack of PSII subunits in mutant plants. Our biochemical analyses in vivo show that two PSII core subunits, D1 and CP47, can form redox-sensitive intermolecular, but not intramolecular, disulfide bridges (Fig. 9; Supplemental Fig. S4), implying that the assembly of the PSII core complex also requires proper redox status of the PSII core subunits and the involvement of redox regulators. This hypothesis is supported by several lines of evidence. The assembly of D1 proteins into the PSII RC and PSII core complex depends on proper redox conditions. The thiol reactants DTT, N-ethylmaleimide, and iodosobenzoic acid could reduce the stability of early PSII assembly intermediates, inhibit C-terminal processing of the D1 precursor, and prevent the reassembly of CP43 into PSII (Zhang et al., 2000). Moreover, SNOWY COTYLEDON2 and LOW QUANTUM YIELD OF PHOTOSYSTEM II1 (LQY1), with protein disulfide isomerase activity, interact with PSII core subunits CP43 and CP47 and are suggested to function in the biogenesis of thylakoid membranes in cotyledons and in PSII disassembly and/or reassembly during the PSII repair cycle, respectively (Shimada et al., 2007; Lu et al., 2011; Tanz et al., 2012). Intriguingly, the triple silencing of TRX m1, TRX m2, and TRX m4 disrupted the redox state of PSII subunits and led to elevated levels of oxidized CP47 oligomers (Fig. 9C), indicating the important role of TRX m proteins in modulating the redox status of PSII core subunits.
Biogenesis of the PSII complex is a highly ordered process in which a series of assembly steps occur sequentially and are well coordinated in different compartments of the thylakoid membranes (Komenda et al., 2012; Nickelsen and Rengstl, 2013). Thus, it is interesting to consider which step(s) of PSII assembly might involve TRX m1, TRX m2, and TRX m4. In the early stages of PSII assembly, PSII core subunits always combine with distinct low-molecular-mass proteins to form minor PSII assembly intermediate subcomplexes, such as the PSII RC, the CP47 precomplex, and the CP43 precomplex. In two-dimensional BN/SDS-PAGE, TRX m1, TRX m2, and TRX m4 can associate with these PSII assembly intermediates (Fig. 8B). Similar combinations with small PSII complexes have been observed for some PSII accessory factors that are involved in the early steps of PSII biogenesis; HCF136 and YCF48 localize to the thylakoid lumen and participate in the stabilization of newly synthesized D1 precursor and mediate the formation of the PSII RC (Plücken et al., 2002; Komenda et al., 2008; Rengstl et al., 2011). The intrinsic thylakoid proteins PAM68 and SII0933 play an essential role in the conversion of the PSII RC into larger PSII complexes (Armbruster et al., 2010; Rengstl et al., 2011). Furthermore, the chloroplast thylakoid membrane system is particularly complex in plants, because it is organized into nonappressed stroma lamellae and appressed grana stacks. The various PSII complexes are characterized in distinct thylakoid membrane domains. The functionally dimeric PSII, including PSII supercomplexes and PSII core dimers, dominate in the thylakoid grana, whereas the PSII assembly intermediate subcomplexes, such as PSII monomers, the CP47-containing PSII RC, and the PSII RC, prevail in the stroma lamellae (Danielsson et al., 2006). Thus, it is reasonable that the triple silencing of the three TRX m genes resulted in the conspicuous impaired stroma thylakoid membranes and grana (Fig. 5). Taken together, these results suggest that TRX m1, TRX m2, and TRX m4 may play an essential role in the early steps of PSII biogenesis for proper assembly of the PSII core complex.
Previous studies of PSII mutants, mainly performed in Chlamydomonas reinhardtii, indicated that D2 and CP47 serve a central function in the stable assembly of the PSII complex (Erickson et al., 1986; Vermaas et al., 1986; Lang and Haselkorn, 1989). However, the regulatory mechanism underlying the integration of CP47 into the PSII complex is an open question. At present, only two auxiliary proteins, PAM68 in Arabidopsis and Psb28 in cyanobacterium (Synechocystis PCC 6803), have been identified as being involved in the assembly of CP47 into PSII (Dobakova et al., 2009; Armbruster et al., 2010). Considering that TRX m1, TRX m2, and TRX m4 interact with PSII assembly intermediates and CP47 proteins, and that their deficiency leads to drastically reduced CP47 protein levels and the accumulation of redox-sensitive CP47 oligomers (Figs. 3, 8, and 9), we tentatively propose that these three m-type TRX proteins play a role in assisting the proper assembly of CP47 into the PSII core complex.
Are There Other Relevant Targets of m-Type TRX Disulfide-Reducing Activity in PSII Biogenesis?
Despite evidence supporting the disulfide-reducing activity of TRX m1, TRX m2, and TRX m4 in modulating the redox state of PSII subunits, which may in turn correlate with the reduced accumulation of PSII core subunits and PSII complexes (Figs. 3, 4, 7, and 9), the physiological significance of redox-sensitive CP47 oligomerization seems rather unclear. In principle, three scenarios for CP47 oligomerization can be envisioned: (1) several CP47 proteins can form CP47-CP47 homooligomers through intermolecular disulfide bridges; (2) CP47 combines with other PSII subunits such as D1 to form CP47-D1 heteroligomers; and (3) CP47 transiently associates with PSII assembly factors by means of intermolecular disulfide bridges. Based on crystalline structure analyses of the PSII complex and the low amounts of PSII protein oligomers in comparison with their monomers (Fig. 9; Iwata and Barber, 2004; Nelson and Ben-Shem, 2004; Dekker and Boekema, 2005), the first two possibilities seem unlikely. Interestingly, as shown in Table III, many PSII auxiliary proteins that are involved in the early steps of PSII biogenesis, such as Albino3 (Alb3), C-Terminal Processing Protease for D1 Protein of PSII, HCF243, LPA1, PAM68, and LPA3, all have conserved Cys residues. Strikingly, Degradation of Periplasmic Proteins1 and FK-506 BINDING PROTEIN20-2 have been suggested to be target proteins of chloroplastic TRXs (Lima et al., 2006; Ströher and Dietz, 2008). Therefore, in order to clarify the m-type TRX-mediated redox regulation of PSII biogenesis, we will further identify the potential reducing acceptors that are essential for PSII biogenesis and assembly of the PSII complex in a future study.
Table III. Summary of conserved Cys residues in described PSII assembly factors in Arabidopsis.
The PSII assembly factors containing conserved Cys residues are indicated in boldface.
| PSII Assembly and PSII Repair Processa | Assembly Factor | Accession No. | Sizeb | Localizationc | No. of Conserved Cys Residuesd | Site of Conserved Cys Residuese | References |
|---|---|---|---|---|---|---|---|
| kDa | |||||||
| PSII RC assembly | Alb3f | At2g28800 | 44 | Intrinsic TM | 1 | 154 | Bellafiore et al. (2002); Ossenbühl et al. (2004); Göhre et al. (2006) |
| C-Terminal Processing Protease for D1 Protein of PSII (CtpA) | At4g17740 | 46 | Extrinsic L | 5 | 224, 299, 410, 412, 420 | Anbudurai et al. (1994) | |
| HCF136 | At5g23120 | 38 | Extrinsic L | 0 | – | Plücken et al. (2002) | |
| HCF243 | At3g15095 | 65 | Intrinsic TM | 19 | 19, 61, 78, 90, 94, 98, 116,126, 198, 210, 261, 379, 427, 433, 478, 455, 512, 546, 562 | Zhang et al. (2011) | |
| LPA1 | At1g02910 | 40 | Intrinsic TM | 3 | 28, 29, 42 | Peng et al. (2006) | |
| PSII RC47 assembly | PAM68 | At4g19100 | 20 | Intrinsic TM | 1 | 13 | Armbruster et al. (2010) |
| Incorporation of CP43 | LPA2 | At5g51545 | 15 | Intrinsic TM | 0 | – | Ma et al. (2007) |
| LPA3 | At1g73060 | 34 | Extrinsic S | 3 | 64, 140, 242 | Cai et al. (2010) | |
| LPA19g | At1g03600 | 12 | Extrinsic L | 0 | – | Chen et al. (2006); Wei et al. (2010) | |
| Assembly of the oxygen-evolving Mn4CaO5 cluster | CYCLOPHILIN38 (CYP38) | At3g01480 | 39 | Extrinsic L | 1 | 174 | Fu et al. (2007); Sirpiö et al. (2008); Vasudevan et al. (2012) |
| LTO1 | At4g35760 | 35 | Intrinsic TM | 10 | 1, 64, 71, 150, 153, 185, 248, 251, 271, 286 | Karamoko et al. (2011) | |
| PSII dimmer and PSII supercomplex assembly | Degradation of Periplasmic Proteins1 (Deg1) | At3g27925 | 42 | Extrinsic L | 2 | 40, 365 | Kapri-Pardes et al. (2007); Sun et al. (2010) |
| FK-506 BINDING PROTEIN20-2 (FKBP20-2) | At3g60370 | 24 | Extrinsic L | 5 | 1, 11, 26, 96, 210 | Lima et al. (2006) | |
| Psb29/Thylakoid Formation1 (Thf1)g | At2g20890 | 27 | Extrinsic S | 2 | 159, 217 | Wang et al. (2004); Keren et al. (2005) | |
| PSII repair | LQY1 | At1g75690 | 12 | Intrinsic TM | 8 | 44, 47, 55, 58, 78, 81, 89, 92 | Lu et al. (2011) |
| PSBP-LIKE PROTEIN 1 (PPL1) | At3g55330 | 18 | Extrinsic L | 0 | – | Ishihara et al. (2007) | |
| THYLAKOID LUMEN PROTEIN18.3 (TLP18.3)/Psb32f | At1g54780 | 23 | Intrinsic TM | 0 | – | Sirpiö et al. (2007) |
PSII assembly factors are classified according to their proposed functions in the biogenesis and repair of the PSII complex. bPredicted molecular mass of mature proteins. cL, Lumen; S, stroma; TM, thylakoid membrane. dThe number of conserved Cys residues in the mature proteins. eThe site of conserved Cys residues in amino acid sequences of mature proteins. fAlb3 is involved in the integration of LHCII into PSII supercomplexes; THYLAKOID LUMEN PROTEIN18.3/Psb32 functions in the assembly of PSII supercomplex. gLPA19 and Psb29/Thylakoid Formation1 play roles in PSII repair.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana ecotype Columbia) was grown in soil in a growth chamber (110 μmol photons m−2 s−1, 16-h photoperiod, 21°C, and 60% relative humidity). The T-DNA insertion mutant lines for two m-type TRX genes (trx m1 [Salk_087118C] and trx m4 [Salk_032538]) were obtained from the Arabidopsis Biological Resource Center as described by Courteille et al. (2013). The homozygous T-DNA insertion lines were confirmed by PCR using the gene-specific and T-DNA-specific primers described in Supplemental Table S3. The trx m1trx m4 double mutant was obtained by crossing single mutants of trx m1 and trx m4 and screening the F2 population by PCR using the primers described above.
VIGS Assay
Plasmids pTRV1 and pTRV2 are VIGS vectors based on Tobacco rattle virus as described by Liu et al. (2002). To construct the pTRV2-TRX m vectors, TRX m1, TRX m2, and TRX m4 complementary DNAs (cDNAs) were generated by RT-PCR with the primers described in Supplemental Table S3 and inserted into the pTRV2 vector. The pTRV2-GFP vector was used as a negative control. pTRV1 and pTRV2 derivatives were introduced into Arabidopsis plants by Agrobacterium tumefaciens infiltration as described by Burch-Smith et al. (2006).
Transmission Electron Microscopy
Transmission electron microscopy was performed as described by Yao and Greenberg (2006). Fully expanded leaves from the VIGS-TRX m and VIGS-GFP plants were collected 3 weeks after infection. Micrographs were taken using a transmission electron microscope (JEM1400; JEOL).
Pigment Analysis and Chlorophyll Fluorescence Measurements
Chlorophyll was extracted from whole leaves with 80% acetone in 25 mm HEPES-KOH (pH 7.5), and the chlorophyll content was determined as described by Wellburn (1994).
Chlorophyll fluorescence was measured using Maxi-Imaging PAM and Dual-PAM 100 (Walz; http://www.walz.com). Arabidopsis leaves were dark adapted for 15 min prior to fluorescence measurements. The minimum fluorescence yield was measured under a weak measuring light. The maximum fluorescence yield was determined by applying a saturating light pulse (3 × 103 μmol photons m−2 s−1). Leaves were illuminated for 6 min under actinic light illumination (110 μmol photons m−2 s−1). Thereafter, the maximal fluorescence of light-adapted leaves was measured after applying a second saturating pulse. The ΦPSII, excitation pressure (1 – qP), and NPQ were recorded. Measurement of light-induced P700 absorbance changes at 820 nm was performed as described by Meurer et al. (1996). Absorbance changes induced by saturating far-red light represent the relative amounts of photooxidizable P700.
Thylakoid Membrane and Total Protein Preparations
Thylakoid membranes were prepared according to Robinson et al. (1980). Isolated thylakoid membranes were quantified based on total chlorophyll as described by Porra et al. (1989). Total proteins extracted from leaves of VIGS plants or from thylakoid membrane preparations were prepared according to Martinez-Garcia et al. (1999). Protein concentrations were determined using the Bio-Rad detergent-compatible colorimetric protein assay according to the manufacturer’s protocol.
BN/SDS-PAGE and Immunoblot Analyses
BN-PAGE was performed essentially according to Schägger et al. (1994) with the modifications described by Peng et al. (2006). For two-dimensional analysis, the excised BN-PAGE lanes were soaked in SDS sample buffer (100 mm Tris-HCl [pH 6.8], 2% [w/v] SDS, 15% [v/v] glycerol, and 2.5% [v/v] β-mercaptoethanol) for 15 min at room temperature and then layered onto 1.5-mm-thick 12% SDS-PAGE gels containing 6 m urea. The gels were stained with Coomassie Brilliant Blue G250 and scanned using a GS-800 densitometer (Bio-Rad).
For immunoblot analysis, total proteins extracted from VIGS leaves or thylakoid membrane preparations were separated on 12% SDS-PAGE gels containing 6 m urea. After electrophoresis, proteins were transferred to polyvinylidene difluoride membranes (Millipore) and probed using antibodies specifically directed against individual PSII subunits (D1, D2, CP43, CP47, and PsbO), PSI subunits (PsaA and PsaO), the light-harvesting antenna proteins (Lhcb1 and Lhca1), the Cyt b6/f complex (cytochrome f), and the ATP synthase β-subunit (CF1 β; Agrisera). Signals were detected with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Gene Expression Analysis
Total RNA was extracted from Arabidopsis plants with the Plant RNeasy Mini kit (Omega). Five-microgram samples of total RNA were used for cDNA synthesis using the PrimeScript RT reagent kit (Takara) according to the manufacturer’s instructions. Quantitative real-time RT-PCR was performed with SYBR Premix Ex Taq (Takara), and real-time amplification was monitored on the LightCycler480 system (Roche). Expression of ACTIN2 (At3g18780) and UBIQUITIN4 (At5g20620) was relatively stable in VIGS plants, and the genes were amplified as internal controls in this study. Primer sequences are listed in Supplemental Table S3.
Determination of ROS
In situ detection of H2O2 and O2− was performed mainly according to Lu et al. (2011) with minor modifications. For H2O2 visualization, leaves from VIGS plants were vacuum infiltrated with a solution containing 1 mg mL−1 3,3′-diaminobenzidine (pH 3.8) at room temperature for 1 h and incubated in the same solution under growth light illumination for 1 h. The stained leaves were then boiled in a solution of ethanol:acetic acid:glycerol (3:1:1, v/v/v) for 10 min. For O2− detection, leaves were vacuum infiltrated for 30 min with 50 mm phosphate buffer (pH 7.5) containing 1 mg mL−1 nitroblue tetrazolium and subsequently boiled in 90% (v/v) ethanol for 10 min.
Transient Expression of Mutant TRX m Proteins in Arabidopsis Protoplasts
The full-length cDNAs for TRX m1, TRX m2, TRX m3, and TRX m4 were amplified from Arabidopsis cDNA by RT-PCR. Site-directed mutagenesis of the C-terminal Cys residues in the TRX m active-site motif (WCGPC) were introduced by overlap-extension PCR. Full-length cDNA fragments of TRX m1C107S, TRX m2C113S, TRX m3C100S, and TRX m4C119S were then individually cloned into the pUC19-FLAG transient expression vector. Primer sequences are listed in Supplemental Table S3. Protoplasts were isolated from 14-d-old Arabidopsis seedlings according to Zhai et al. (2009) and transformed with the pUC19-TRX m1C107S, pUC19-TRX m2C113S, pUC19-TRX m3C100S, and pUC19-TRX m4C119S-FLAG plasmids according to Yoo et al. (2007). Transformed protoplasts were collected by centrifugation for 2 min at 100g, resuspended in W5 solution, and kept in the dark for 16 h prior to further analysis.
Expression and Purification of Recombinant His-Tagged Mutant TRX m Proteins
The cDNA fragments encoding the mature TRX m mutant proteins were amplified from pUC19-TRX m1C107S, pUC19-TRX m2C113S, pUC19-TRX m3C100S, and pUC19-TRX m4C119S-FLAG plasmids by RT-PCR and individually cloned into the pET28a expression vector (Novagen). Primer sequences are listed in Supplemental Table S3. The pET28a-TRX m1C107S, pET28a-TRX m2C113S, pET28a-TRX m3C100S, and pET28a-TRX m4C119S plasmids were transformed into Escherichia coli BL21 (DE3; Stratagene) and induced to express with 0.6 mm isopropyl β-d-thiogalactoside. Purification of four mutant His-tagged TRX m proteins was carried out with Ni+ affinity chromatography according to the manufacturer’s protocol (GE Healthcare). The imidazole in the elution buffer was removed by fast ultrafiltration using 3-kD-limit Amicon Ultra-15 centrifugal filter units (Millipore).
Pull-Down Assay
The pull-down assay was performed essentially according to the method of Tada et al. (2008) with some modifications. Purified His-TRX m1C107S, His-TRX m2C113S, His-TRX m3C100S, and His-TRX m4C119S recombination proteins were immobilized on Sepharose 4B resin according to the manufacturer’s protocol (GE Healthcare) and incubated with thylakoid membrane proteins solubilized in 1% (w/v) DM overnight at 4°C. The empty resin without TRX m protein was used as the negative control. The resins were then extensively washed with washing buffer containing 0.5 m NaCl until the A280 reached almost zero. The TRX m target proteins linked by the newly formed heterodisulfide bonds were eluted with buffer containing 10 mm DTT and 0.5 m NaCl and analyzed by immunoblotting.
Determination of the in Vivo Redox State of PSII Proteins
The in vivo redox state of PSII proteins was determined as in previous studies (Ikegami et al., 2007; Tada et al., 2008) with some modifications. Total leaf proteins were extracted from VIGS plants 3 weeks after infiltration without reducing agents. The extracts were divided into three equal parts and incubated with the oxidizing agent (1 mm CuCl2), the reducing agent (10 mm DTT), or the corresponding amount of distilled, deionized water at 23°C for 30 min, and the proteins were then precipitated with 5% (w/v) TCA. The denatured protein pellets were washed twice with 100% acetone and resuspended in 1 volume of 1% (w/v) SDS buffer and 1 volume of SDS-PAGE gel loading buffer without DTT. One part of each sample was incubated with AMS (Invitrogen) for 20 min at room temperature. Both AMS-treated and untreated samples were separated by nonreducing SDS-PAGE and immunoblotted with antibodies against PSII subunits. For a loading control, the third part of the leaf protein extracts was boiled in reducing SDS-PAGE gel loading buffer containing DTT and probed with antibodies against PSII subunits and the large subunit of Rubisco.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree and amino acid sequence alignment of TRX m proteins in Arabidopsis and rice.
Supplemental Figure S2. Phenotype and identification of trx m1trx m4 double mutant plants.
Supplemental Figure S3. Expression of TRX m-FLAG proteins in Arabidopsis protoplasts and purification of His-TRX m proteins.
Supplemental Figure S4. Visualization of the redox status of intramolecular disulfide bonds in PSII subunits from VIGS plants.
Supplemental Table S1. Summary of TRX m silencing plants and TRX m mutant plants in this study.
Supplemental Table S2. Chlorophyll content and chlorophyll fluorescence parameters in TRX m mutant plants.
Supplemental Table S3. List of primers used for vector construction in this study.
Supplementary Material
Acknowledgments
We thank Yule Liu (Tsinghua University) for the kind gift of TRV-based VIGS vectors, Lianwei Peng (Institute of Botany, Chinese Academy of Sciences) for assistance in BN/SDS-PAGE and for discussing the manuscript, Bernhard Grimm (Humboldt University Berlin) for critical reading of the manuscript, and Xia Feng (Plant Protection Research Institute, Guangdong Academy of Agricultural Science) for supporting Dual-PAM 100.
Glossary
- TRX
thioredoxin
- NTR
NADPH:thioredoxin reductase
- Cyt b6f
cytochrome b6f
- T-DNA
transfer DNA
- VIGS
virus-induced gene silencing
- RT
reverse transcription
- Fv/Fm
ratio of variable fluorescence to maximal fluorescence
- qP
photochemical quenching
- ΦPSII
PSII photochemical efficiency
- NPQ
nonphotochemical quenching
- DM
n-dodecyl β-d-maltoside
- BN
blue native
- PEP
plastid-encoded plastid RNA polymerase
- NEP
nucleus-encoded plastid RNA polymerase
- ROS
reactive oxygen species
- H2O2
hydrogen peroxide
- O2−
superoxide anion radical
- RC
reaction center
- AMS
4-acetoamido-4-maleimidylstilbene-2,2-disulfonate
- cDNA
complementary DNA
References
- Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB. (1994) The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc Natl Acad Sci USA 91: 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB. (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S, Ferris P, Naver H, Göhre V, Rochaix JD. (2002) Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell 14: 2303–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D. (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y. (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap RL, Qian M, Shen JR, Inoue Y, Sherman LA. (1994) Role of disulfide linkage and putative intermolecular binding residues in the stability and binding of the extrinsic manganese-stabilizing protein to the photosystem II reaction center. Biochemistry 33: 13712–13718 [DOI] [PubMed] [Google Scholar]
- Cai W, Ma J, Chi W, Zou M, Guo J, Lu C, Zhang L. (2010) Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis. Plant Physiol 154: 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen H, Zhang D, Guo J, Wu H, Jin M, Lu Q, Lu C, Zhang L. (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61: 567–575 [DOI] [PubMed] [Google Scholar]
- Chi YH, Moon JC, Park JH, Kim HS, Zulfugarov IS, Fanata WI, Jang HH, Lee JR, Lee YM, Kim ST, et al. (2008) Abnormal chloroplast development and growth inhibition in rice thioredoxin m knock-down plants. Plant Physiol 148: 808–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani K, Tarrago L, Schürmann P, Jacquot JP, Rouhier N. (2011) Biochemical properties of poplar thioredoxin z. FEBS Lett 585: 1077–1081 [DOI] [PubMed] [Google Scholar]
- Chibani K, Wingsle G, Jacquot JP, Gelhaye E, Rouhier N. (2009) Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol Plant 2: 308–322 [DOI] [PubMed] [Google Scholar]
- Collin V, Issakidis-Bourguet E, Marchand C, Hirasawa M, Lancelin JM, Knaff DB, Miginiac-Maslow M. (2003) The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J Biol Chem 278: 23747–23752 [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. (2004) Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiol 136: 4088–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courteille A, Vesa S, Sanz-Barrio R, Cazalé AC, Becuwe-Linka N, Farran I, Havaux M, Rey P, Rumeau D. (2013) Thioredoxin m4 controls photosynthetic alternative electron pathways in Arabidopsis. Plant Physiol 161: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson R, Suorsa M, Paakkarinen V, Albertsson PA, Styring S, Aro EM, Mamedov F. (2006) Dimeric and monomeric organization of photosystem II. Distribution of five distinct complexes in the different domains of the thylakoid membrane. J Biol Chem 281: 14241–14249 [DOI] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ. (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Pfannschmidt T. (2011) Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 155: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobakova M, Sobotka R, Tichy M, Komenda J. (2009) Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA. (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Erickson JM, Rahire M, Malnoe P, Girard-Bascou J, Pierre Y, Bennoun P, Rochaix JD. (1986) Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J 5: 1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A, He Z, Cho HS, Lima A, Buchanan BB, Luan S. (2007) A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Ossenbühl F, Crèvecoeur M, Eichacker LA, Rochaix JD. (2006) One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas. Plant Cell 18: 1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 264: 13963–13966 [PubMed] [Google Scholar]
- Ikegami A, Yoshimura N, Motohashi K, Takahashi S, Romano PG, Hisabori T, Takamiya K, Masuda T. (2007) The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J Biol Chem 282: 19282–19291 [DOI] [PubMed] [Google Scholar]
- Ishihara S, Takabayashi A, Ido K, Endo T, Ifuku K, Sato F. (2007) Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol 145: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issakidis-Bourguet E, Mouaheb N, Meyer Y, Miginiac-Maslow M. (2001) Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. Plant J 25: 127–135 [DOI] [PubMed] [Google Scholar]
- Iwata S, Barber J. (2004) Structure of photosystem II and molecular architecture of the oxygen-evolving centre. Curr Opin Struct Biol 14: 447–453 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Kapri-Pardes E, Naveh L, Adam Z. (2007) The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19: 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamoko M, Cline S, Redding K, Ruiz N, Hamel PP. (2011) Lumen Thiol Oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23: 4462–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB. (2005) Psb29, a conserved 22-kD protein, functions in the biogenesis of photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17: 2768–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach T. (2013) Oxidative folding in chloroplasts. Antioxid Redox Signal 19: 72–82 [DOI] [PubMed] [Google Scholar]
- Kirchsteiger K, Pulido P, González M, Cejudo FJ. (2009) NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Mol Plant 2: 298–307 [DOI] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichý M, Prásil O, Eichacker LA, Nixon PJ. (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ. (2012) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz KJ. (2002) The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc Natl Acad Sci USA 99: 5738–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JD, Haselkorn R. (1989) Isolation, sequence and transcription of the gene encoding the photosystem II chlorophyll-binding protein, CP-47, in the cyanobacterium Anabaena 7120. Plant Mol Biol 13: 441–457 [DOI] [PubMed] [Google Scholar]
- Laugier E, Tarrago L, Courteille A, Innocenti G, Eymery F, Rumeau D, Issakidis-Bourguet E, Rey P. (2013) Involvement of thioredoxin y2 in the preservation of leaf methionine sulfoxide reductase capacity and growth under high light. Plant Cell Environ 36: 670–682 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Michelet L, Zaffagnini M, Massot V, Issakidis-Bourguet E. (2007) Thioredoxins in chloroplasts. Curr Genet 51: 343–365 [DOI] [PubMed] [Google Scholar]
- Lennartz K, Plücken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K. (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima A, Lima S, Wong JH, Phillips RS, Buchanan BB, Luan S. (2006) A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc Natl Acad Sci USA 103: 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Mata-Cabana A, Kieselbach T. (2011) The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal 14: 2581–2642 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Lu Y, Hall DA, Last RL. (2011) A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Fan T, Liu Y, Rothbart M, Yu J, Zhou S, Grimm B, Luo M. (2012) Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiol 159: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L. (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez-Garcia JF, Monte E, Quail PH. (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P. (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F. (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86: 419–433 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. (2009) Thioredoxin targets in plants: the first 30 years. J Proteomics 72: 452–474 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Hisabori T. (2006) HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J Biol Chem 281: 35039–35047 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Hisabori T. (2010) CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid Redox Signal 13: 1169–1176 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T. (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P, Sirpio S, Suorsa M, Aro EM. (2008) Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth Res 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Nelson N, Ben-Shem A. (2004) The complex architecture of oxygenic photosynthesis. Nat Rev Mol Cell Biol 5: 971–982 [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF. (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B. (2013) Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Ossenbühl F, Göhre V, Meurer J, Krieger-Liszkay A, Rochaix JD, Eichacker LA. (2004) Efficient assembly of photosystem II in Chlamydomonas reinhardtii requires Alb3.1p, a homolog of Arabidopsis ALBINO3. Plant Cell 16: 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Söderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plücken H, Müller B, Grohmann D, Westhoff P, Eichacker LA. (2002) The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett 532: 85–90 [DOI] [PubMed] [Google Scholar]
- Porra R, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rascher U, Nedbal L. (2006) Dynamics of photosynthesis in fluctuating light. Curr Opin Plant Biol 9: 671–678 [DOI] [PubMed] [Google Scholar]
- Rengstl B, Oster U, Stengel A, Nickelsen J. (2011) An intermediate membrane subfraction in cyanobacteria is involved in an assembly network for photosystem II biogenesis. J Biol Chem 286: 21944–21951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamäki E, Grimm B. (2013) Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiol 162: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson HH, Yocum CF. (1980) Cyclic photophosphorylation reactions catalyzed by ferredoxin, methyl viologen and anthraquinone sulfonate: use of photochemical reactions to optimize redox poising. Biochim Biophys Acta 590: 97–106 [DOI] [PubMed] [Google Scholar]
- Rochaix JD. (2011) Assembly of the photosynthetic apparatus. Plant Physiol 155: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Buchanan BB. (2008) The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal 10: 1235–1274 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot JP. (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279: 43821–43827 [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. (2005) Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol 244: 1–68 [DOI] [PubMed] [Google Scholar]
- Shimada H, Mochizuki M, Ogura K, Froehlich JE, Osteryoung KW, Shirano Y, Shibata D, Masuda S, Mori K, Takamiya K. (2007) Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19: 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S, Allahverdiyeva Y, Suorsa M, Paakkarinen V, Vainonen J, Battchikova N, Aro EM. (2007) TLP18.3, a novel thylakoid lumen protein regulating photosystem II repair cycle. Biochem J 406: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S, Khrouchtchova A, Allahverdiyeva Y, Hansson M, Fristedt R, Vener AV, Scheller HV, Jensen PE, Haldrup A, Aro EM. (2008) AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J 55: 639–651 [DOI] [PubMed] [Google Scholar]
- Ströher E, Dietz KJ. (2008) The dynamic thiol-disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiol Plant 133: 566–583 [DOI] [PubMed] [Google Scholar]
- Sun X, Ouyang M, Guo J, Ma J, Lu C, Adam Z, Zhang L. (2010) The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. Plant J 62: 240–249 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Badger MR. (2011) Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Kilian J, Johnsson C, Apel K, Small I, Harter K, Wanke D, Pogson B, Albrecht V. (2012) The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J 69: 743–754 [DOI] [PubMed] [Google Scholar]
- Thormählen I, Ruber J, von Roepenack-Lahaye E, Ehrlich SM, Massot V, Hümmer C, Tezycka J, Issakidis-Bourguet E, Geigenberger P. (2013) Inactivation of thioredoxin f1 leads to decreased light activation of ADP-glucose pyrophosphorylase and altered diurnal starch turnover in leaves of Arabidopsis plants. Plant Cell Environ 36: 16–29 [DOI] [PubMed] [Google Scholar]
- Tovar-Méndez A, Matamoros MA, Bustos-Sanmamed P, Dietz KJ, Cejudo FJ, Rouhier N, Sato S, Tabata S, Becana M. (2011) Peroxiredoxins and NADPH-dependent thioredoxin systems in the model legume Lotus japonicus. Plant Physiol 156: 1535–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D, Fu A, Luan S, Swaminathan K. (2012) Crystal structure of Arabidopsis cyclophilin38 reveals a previously uncharacterized immunophilin fold and a possible autoinhibitory mechanism. Plant Cell 24: 2666–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas WF, Williams JG, Rutherford AW, Mathis P, Arntzen CJ. (1986) Genetically engineered mutant of the cyanobacterium Synechocystis 6803 lacks the photosystem II chlorophyll-binding protein CP-47. Proc Natl Acad Sci USA 83: 9474–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL. (2004) Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol 136: 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Guo J, Ouyang M, Sun X, Ma J, Chi W, Lu C, Zhang L. (2010) LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J Biol Chem 285: 21391–21398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. (1994) The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144: 307–313 [Google Scholar]
- Wollman FA, Minai L, Nechushtai R. (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim Biophys Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT. (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhai Z, Sooksa-nguan T, Vatamaniuk OK. (2009) Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol 149: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhou G, Liu B, Kong Y, Chen N, Qiu Q, Yin H, An J, Zhang F, Chen F. (2011) HCF243 encodes a chloroplast-localized protein involved in the D1 protein stability of the Arabidopsis photosystem II complex. Plant Physiol 157: 608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM. (2000) Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.