Research into the systems biology of roots now encompasses transcriptomic and bioinformatic analysis combined with diverse modeling approaches, in order to relate gene regulatory networks to organ and rhizosphere scale processes, and will ultimately allow researchers to bridge the genotype-to-phenotype gap.
Abstract
Genetic and genomic approaches in model organisms have advanced our understanding of root biology over the last decade. Recently, however, systems biology and modeling have emerged as important approaches, as our understanding of root regulatory pathways has become more complex and interpreting pathway outputs has become less intuitive. To relate root genotype to phenotype, we must move beyond the examination of interactions at the genetic network scale and employ multiscale modeling approaches to predict emergent properties at the tissue, organ, organism, and rhizosphere scales. Understanding the underlying biological mechanisms and the complex interplay between systems at these different scales requires an integrative approach. Here, we describe examples of such approaches and discuss the merits of developing models to span multiple scales, from network to population levels, and to address dynamic interactions between plants and their environment.
Root architecture critically influences nutrient and water uptake efficiency and plays a central role in plant productivity (Lynch, 1995); selecting new crop varieties with improved root traits may produce a second Green Revolution (Lynch, 2007). Over the past several decades, reductionist approaches have pinpointed the individual genes or mechanisms that control root system architecture (for review, see Benfey et al., 2010). However, as our knowledge of biological systems has increased, researchers have realized that the underlying components (e.g. gene products, cells, tissues, and organs) function in highly complex, dynamic networks. The existence of emergent traits, robustness, and hierarchical organization of biological systems make them challenging to conventional reductionist experimental approaches (Bruggeman and Westerhoff, 2007). Understanding these properties requires studying the system rather than its individual components. Also, in contrast to the simple linear networks conveyed in textbooks, biological networks usually contain multiple branches, feedback and/or feed-forward loops, and other complex regulatory motifs. For example, most signal transduction pathways include negative feedback and/or positive feed-forward loops to switch off or amplify a response pathway. Hence, logic alone often fails to predict the output from such pathways. Systems approaches often employ mathematical or computational models to simulate the behaviors of these nonlinear networks and predict emergent behaviors (for review, see Middleton et al., 2012). Robust biological systems also have compensatory mechanisms that come into play when a key gene is removed. For example, 65% of knockout lines for Arabidopsis (Arabidopsis thaliana) transcription factors in the root stele gene regulatory network (GRN) showed molecular phenotypes, but only 16% showed morphological phenotypes (Brady et al., 2011). Thus, in the robust stele GRN, the network architecture or genetic redundancy can buffer or canalize changes in gene expression. Hierarchy is inherent to all biological systems. At scales relevant to agriculture, hierarchy starts at the field scale. Field-scale populations comprise individual plants, which are composed of organs, tissues, cells, organelles, and molecules. Each hierarchical level interacts with higher and lower scales. Hence, understanding the underlying biological mechanisms of the complex, critical interplay between these scales requires an integrative systems approach.
Root researchers currently use several distinct systems-based approaches. “Top-down” approaches automatically analyze large-scale data sets to uncover relationships between levels of transcripts and/or proteins (for review, see Bassel et al., 2012). This approach starts with experimental “omics” data, followed by data analysis and data integration to determine correlations, and ends with the formulation of hypotheses concerning the coregulation and interregulation of groups of molecules. These hypotheses predict new correlations that can be iteratively tested (Bruggeman and Westerhoff, 2007).
Another approach, termed “bottom up,” constructs detailed models of well-characterized networks that can be simulated mathematically and/or computationally. This approach starts with a manageable part of a system, such as a small GRN, and employs mechanism-based models (for review, see Middleton et al., 2012). Bottom-up studies take an integrative approach to look for emergent properties of a network with available quantitative information such as kinetic data, transcription rates, and protein stability. As information about GRN increases, so does the opportunity to implement mechanistic models for bottom-up studies.
“Middle-out” approaches construct models starting at the level for which we have the most information and combining predictive bottom-up and inference-based top-down approaches, thus building on the current body of knowledge (Noble, 2008). The middle-out approach can start at any scale. However, cells provide a natural level of organization for an organism, cell-based models can endow individual cells with considerable internal machinery (representing the subcellular networks), and the interactions of numerous individual cells can lead to the (emergent) tissue-scale properties. This makes cell-based models an attractive approach, as they naturally incorporate heterogeneity via cellular properties.
“Multiscale” approaches employ models that consider behaviors on two or more spatial scales, ranging from the subcellular up to the whole organism and beyond. Multiscale models must also incorporate temporal scales ranging over many orders of magnitude. Spatially, systems can be measured at the molecular scale (10−10 m) through the whole-plant scale (0.1–100 m) or up to the field level (more than 1,000 m). Likewise, temporal scales can range from nanoseconds (10−9 s) for enzymatic reaction to months and years (107 s) for a plant life cycle. Given this range of temporal and spatial scales, multiscale models are often challenging to construct, and relatively few examples involve plant roots (Band et al., 2012a).
Although our review focuses on roots, we do not ignore links to the whole-plant and field systems. We ultimately study roots because they take up water and nutrients, which influences crop yield and environmental adaptation. While the first point, that roots are fundamental to the whole plant, is obvious, the second point, that of the effect of the whole-plant context on the root, is often overlooked.
This review attempts to cover the plethora of activities that make up root systems research, employing the approaches detailed above. In recent years, “root systems biology” has become synonymous with the top-down approach pioneered by Benfey, Birnbaum, Coruzzi, and coworkers, involving transcriptomics and bioinformatic analysis (Poultney et al., 2007; Benfey et al., 2010; Katari et al., 2010). Here, we first describe the impressive progress made using this approach to build root GRNs and elucidate novel biological mechanisms. We also discuss how mathematical and computational modeling employing bottom-up, middle-out, and multiscale approaches helps us understand the mechanistic behavior of root systems from the GRN scale to whole-root system and rhizosphere scales, thereby enabling us to bridge the genotype-to-phenotype gap by integrating environmental information.
TOP-DOWN ROOT SYSTEMS BIOLOGY APPROACHES
Until recently, root-related gene and protein networks have been characterized using low-throughput molecular genetic and cell biological approaches (e.g. double mutants, fluorescence resonance energy transfer; Benfey et al., 2010). Although these approaches have enabled the discovery and characterization of many small GRNs controlling root developmental programs, they do not work for large-scale network identification, as they build GRNs in a gene-by-gene manner. In contrast, top-down systems biology approaches are ideally suited to construct large-scale root networks of genes or proteins (for review, see Bassel et al., 2012).
New omics profiling technologies such as transcriptomics have enabled explosive growth of top-down approaches in recent years, and the large-scale data sets required to conduct top-down approaches have increased almost exponentially since the sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000; for review, see Mardis, 2008; Egan et al., 2012; Hamilton and Buell, 2012). Expression analysis of specific cell types and developmental stages in Arabidopsis roots can be accomplished using techniques such as laser-capture microdissection (Kerk et al., 2003; Jiao et al., 2009). The most widely used technique uses fluorescence-activated sorting of marked cells, usually from transgenic lines with tissue-specific expression of GFP. This technique uses total RNA prepared from sorted protoplasts of these transgenic lines (Birnbaum et al., 2005). Another method involves expressing a tagged ribosomal protein behind a tissue-specific promoter (Zanetti et al., 2005). Immunoprecipitation of ribosomes from such transgenic plants allows researchers to profile only the actively translated transcripts.
The majority of current top-down examples in roots examine mRNA transcript abundance (transcriptomics), but transcript levels do not necessarily predict protein abundance. Directly examining proteins, a protein expression map of the Arabidopsis root provided the identity and cell type-specific localization of nearly 2,000 proteins, and colocalization data provided support for numerous protein-protein network interactions (Petricka et al., 2012). Other omics profiling techniques recently employed in roots include metabolomics, which measures the pool sizes of metabolites (molecules of less than 1,000 D) that collectively define the metabolome (Fiehn et al., 2000). Moussaieff et al. (2013) used a cell-sorting approach to perform nontargeted metabolomics assays on core cell types in the Arabidopsis root. This study provided a metabolic map of the key root cell types, identifying 50 metabolites.
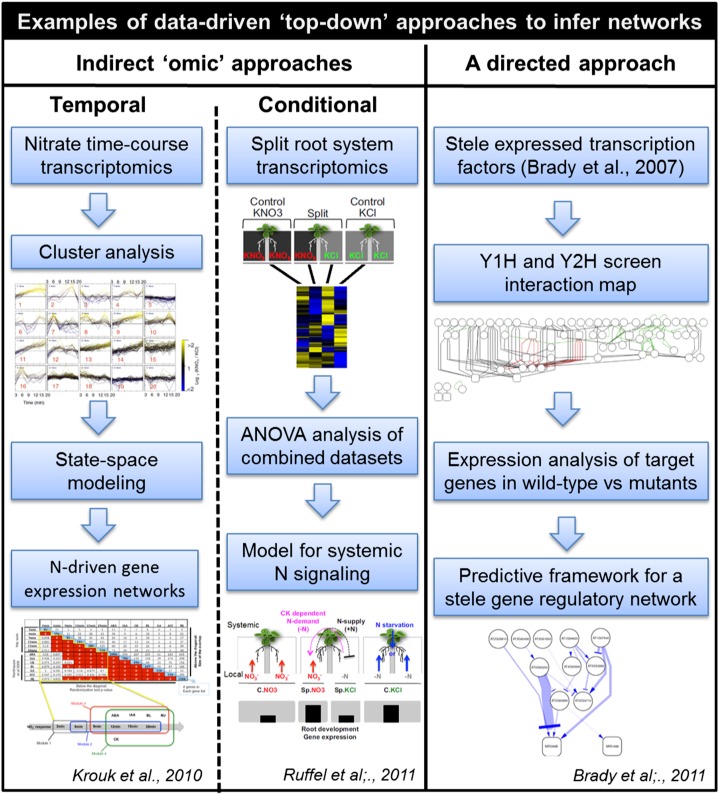
The challenge root researchers face is how to use these rich omics data sets to elucidate meaningful regulatory relationships between mRNAs, proteins, and other biological molecules. Two distinct approaches have been employed to construct large-scale root regulatory networks (summarized in Fig. 1 using two specific examples). The direct approach involves a systematic experimental screen of pairwise interactions among a carefully selected subset of potentially interacting root genes and/or proteins. The second, indirect approach exploits the availability of root omics (predominantly transcriptomic) data sets to infer GRNs through statistical analysis and the identification of regulatory relationships between tens to many thousands of genes. In the next section, we review both approaches using recent examples.
Figure 1.
Examples of top-down approaches to infer GRNs. Left column, Krouk et al. (2010) combined time-series transcriptomic data with published hormone treatment data sets (Nemhauser et al., 2006) to obtain a temporal model for nitrate-driven gene expression networks. They then used a machine-learning approach (state-space modeling) to infer a regulatory network governing nitrate response over time. Middle column, Ruffel et al. (2011) combined genetic and genomic approaches within a split-root system to study how systemic nitrate signaling affected genome-wide programming in root development. Right column, Brady et al. (2011) employed a modeling approach to predict the regulatory potential of each of the relationships they identified within the stele GRN. Stele-expressed transcription factors were selected from the GRN data set published previously (Brady et al., 2007). Protein-protein and protein-DNA maps were generated from yeast one-hybrid and two-hybrid screens of stele-expressed TFs and miRNAs. The key interactions between TFs and their target genes were measured by quantitative reverse transcription-PCR of target gene expression in the wild type and mutant lines. Regulatory interaction strength is represented by edge and arrow width, with thicker lines or arrowheads representing steeper slopes. P value strength is represented by edge opacity, with darker edges representing more significant interactions. Slopes were determined by plotting quantitative PCR expression values of the TF and its target and fitting a line using weighted least-squares regression. This predictive framework enables the identification of the most influential upstream activators or repressors to be manipulated to regulate the expression of a target.
Building Root Networks Using Interaction Screens
Brady et al. (2011) recently described an interaction-based screen to unravel the GRN controlling the identity of the stele, the innermost Arabidopsis root tissues. The authors initially used a high-resolution spatiotemporal root gene expression map (Brady et al., 2007) to select 167 genes encoding stele-enriched transcription factors (TFs). They carried out a yeast one-hybrid screen using these 167 TF genes as prey and the promoters of 65 out of the 167 genes as bait. They also included in their screen the promoters of 28 microRNAs (miRNAs) that target some of the selected TFs. Next, they completed their network with a yeast two-hybrid screen to identify protein-protein interactions between the 167 TFs. The two screens generated a network of 66 protein-DNA interactions and 25 protein-protein interactions. Fifty-nine percent of the protein-DNA interactions could be validated in planta by chromatin immunoprecipitation (ChIP) and genetic analysis. To determine whether the protein-DNA interactions detected by yeast one-hybrid screening and ChIP-PCR resulted in positive or negative effects on transcription, they next analyzed changes in transcript accumulation of the predicted targets in lines knocked down or overexpressing each putative regulator. This approach allowed them to quantify the strength of the interactions and identify the strongest regulators for each target. This systematic screening approach successfully recovered the core of the GRN controlling stele tissue identity and quantitatively measured the strength of the interactions in this GRN. This time-consuming method yields only a partial GRN, but it provides an excellent experimentally validated framework to undertake more detailed analysis of network topology through further laboratory-based and in silico modeling studies.
One of the key challenges to building GRNs lies in defining and confirming the cis-acting regulatory elements in the promoters of transcription factor targets. Recent genome-aided investigations of the developmental pathway have begun to provide insights into how useful this can be (Won et al., 2009). This approach depends on knowledge of the consensus binding motifs for the transcription factors, knowledge largely lacking in Arabidopsis. High-throughput genomic approaches such as ChIP sequencing and yeast one-hybrid screening have begun to fill in this knowledge gap, but the large number of known and putative transcription factors suggests that in silico scans with subsequent validation will remain the standard approach.
Building Root Networks Using Statistical Inference Algorithms
Network inference approaches typically look for correlative associations (e.g. sets of genes expressed under a particular condition) in genome-wide omics data sets, commonly using transcriptomic data. Initially, the approach identifies the components correlated with specific variables (e.g. time, developmental stage, genetic differences, and growth conditions). Conditional and temporal/developmental transcriptomic data can be used to infer GRNs. Conditional expression data compare variables such as the wild type versus root hair-defective mutants (Jones et al., 2006), treated versus mock treated (Paponov et al., 2008), or combinations of genetic and environmental variables (Okushima et al., 2005; Bruex et al., 2012). Time-series expression data can be collected following developmental or experimental induction of the system. Temporal and developmental transcriptomic data sets relevant to roots include those associated with lateral root formation (De Smet et al., 2008), root hair nodulation (Libault et al., 2010), or responses to hormone or nutrient treatments (Krouk et al., 2010; Bielach et al., 2012). For example, Ruffel et al. (2011) studied how systemic nitrate signaling affected genome-wide programming in root development (Fig. 1). This involved physically isolating root systems of the same plant using a “split root” and challenging them with different nitrate conditions. This system was used to probe systemic nitrate responses by comparative transcriptomic analysis of Arabidopsis mutants impaired in nitrate reduction and hormone synthesis and on decapitated plants. The authors performed three-way ANOVA to analyze the data as a whole, including the nitrogen and split-root effects. This analysis identified genes whose nitrate responses were affected by the split-root conditions and found at least two genetically independent systemic signaling mechanisms important for communicating the nitrate supply and demand of a plant (Ruffel et al., 2011).
Clustering provides a useful first analysis of transcriptomic data sets (Eisen et al., 1998; Bansal et al., 2007). Hierarchical clustering uses a pairwise measure of distance between gene expression profiles to compute distance trees (dendrograms) that delimit clusters of coexpressed genes; genes showing similar expression profiles are likely to be coregulated. Clustering can help identify regulatory modules, but it does not reveal the regulatory relationships of the genes inside a cluster. If two genes cluster together, they could be targets of another gene or one gene could directly or indirectly regulate the other.
In contrast to clustering approaches, correlation network reconstruction algorithms aim to infer the regulatory relationships between correlated genes. The simplest way of building a correlation network consists of first calculating all pairwise Pearson’s correlations between expression profiles and then determining the edges of the network through a thresholding procedure. However, these simple correlation network analyses have the same drawbacks as clustering analyses.
Several algorithms based on various types of mathematical formalisms have been developed over the last decade to infer GRN state-space representation models; one of the earliest and widely used methods for modeling gene networks (Noor et al., 2013) attempts to capture the dynamic evolution of the gene network. Krouk et al. (2010) employed a state-space modeling approach termed machine learning to analyze time-series transcriptome data and generate testable hypotheses to probe potential mechanisms underlying the root nitrate signaling pathway. Out of 550 nitrogen-regulated genes, the authors extracted 67 that correspond to the predicted transcription factors plus nine nitrogen-regulated target genes belonging to the primary nitrogen assimilation pathway. The machine-learning approach proposed a regulatory network derived from the high-resolution transcriptome data set that was sampled at rapid time points after roots were treated with nitrate. Intriguingly, of the 76 studied genes, 60 have 500 significant connections, suggesting that nitrate regulation is highly complex. Indeed, the authors speculated that this high level of connectivity “may explain why, to date, (reductionist) experimental analyses have uncovered only few molecular actors specifically involved in the control of NO3−-induced gene expression” (Krouk et al., 2010). Nevertheless, by determining the topology of the GRN, the authors pinpointed a key node, the SBP BOX-LIKE9 (SPL9) transcription factor. The model identified SPL9 as the third most influential transcription factor regulating nitrogen assimilation genes, since it is induced minutes after nitrate treatment and controls at least six genes, including two highly important genes. Furthermore, SPL9 had the greatest magnitude and number of “in” connections controlling it. Hence, the authors concluded that “SPL9 constitutes a potential crucial bottleneck in the flux of information mediated by the proposed nitrogen regulatory network,” a prediction that was later validated experimentally through studies demonstrating that SPL9 overexpression modifies the characteristics of nitrate signal propagation.
Probabilistic graphical modeling techniques such as Bayesian, Gaussian, and qualitative probabilistic networks have also emerged as useful tools for reverse engineering GRNs (for review, see Noor et al., 2013). Bayesian approaches were initially developed to infer GRNs from steady-state expression data (Friedman et al., 2000). They were later extended to solve inference problems on time-series expression data (dynamic Bayesian network; Perrin et al., 2003; Yu et al., 2004). For example, to infer the GRN important for root hair differentiation, Bruex et al. (2012) examined the transcriptomes of 17 root epidermal mutants and two plant hormone treatments. They used a Bayesian approach to infer the regulatory relationships between 208 core genes and used expression information from a developmental time-series data set to position genes temporally within the network (Bruex et al., 2012).
Tools and Resources Available for Root System Biology Studies
A wealth of available tools and databases can be used for data collection, data mining, analysis, and meta-analysis. The high spatial resolution of transcriptome data sets has proved particularly valuable for root studies (Brady et al., 2011). For example, 85% of the 3,656 differentially expressed genes identified by a cell-sorting analysis were not detected in whole-root experiments (Gifford et al., 2008). This illustrates the sensitivity of these approaches; also, transcriptional responses to environmental cues are likely to be tissue specific and masked in whole-root assays.
As the capacity to generate high-throughput large-scale data sets increases, so does the need for tools designed to collect and examine this wealth of new information. Despite the focus on aboveground tissues and whole-plant expression, many tools and resources are already available for root biologists to adopt for their studies. The publicly available Arabidopsis eFP Browser visualizes relative and absolute gene expression data across approximately 22,000 genes from Arabidopsis, as represented on the ATH1 GeneChip from Affymetrix (Winter et al., 2007). This platform includes root-expressed genes profiled using 19 marker lines and 13 developmental stages, corresponding to cell types and tissues at progressive developmental stages, and transcript profiles from quiescent center cells of the root meristem (Brady et al., 2007). The updated Arabidopsis eFP Browser incorporates gene expression data from a variety of sources, including roots under abiotic stresses such as high salinity, iron deprivation, nitrogen availability, low pH, and sulfur and phosphate deficiency (Dinneny et al., 2008; Gifford et al., 2008; Iyer-Pascuzzi et al., 2011; Lin et al., 2011).
The majority of root transcriptome data sets contain microarray data, but many future data sets will contain next-generation sequencing data, from RNA analysis through complementary DNA sequencing at a massive scale (RNA-seq; Ozsolak and Milos, 2011). Indeed, RNA-seq overcomes many of the limitations of microarray technologies. For example, RNA-seq can detect alternative isoforms of RNA transcripts and discover miRNAs (Fahlgren et al., 2007) or small, interfering RNAs (Kasschau et al., 2007; for review, see Ghildiyal and Zamore, 2009; Ozsolak and Milos, 2011). Next-generation sequencing-based investigation of Arabidopsis root system architecture in response to nitrate availability discovered a nitrate-responsive miRNA and its target (Vidal et al., 2010). The authors found that nitrate induces the expression of both miRNA393 and its target, the auxin receptor AUXIN SIGNALING F-BOX3 (AFB3), with a time delay, creating a negative feedback loop of regulation; this module regulates primary and lateral roots by modifying auxin responses via AFB3 (Vidal et al., 2010). Vidal et al. (2013) recently extended their studies to dissect regulatory networks activated by nitrate in roots and acting downstream of AFB3. Employing the Sungear tool (for details, see Table I), they found that the NAC4 transcription factor controls a nitrate-responsive network. Intriguingly, the nac4 mutant exhibits altered lateral root growth but normal primary root growth in response to nitrate. The authors conclude that AFB3 activates two independent pathways to control root system architecture.
Table I. Databases, data mining, and analysis tools for root systems biology.
Freely available databases and mining tools for root systems biology are listed. Many of these databases also may be useful for the study of other plant organs and systems.
| Resource | Description | Reference |
|---|---|---|
| VirtualPlant 1.3 (http://www.virtualplant.org) | Integrates genomic data and provides visualization and analysis tools to aid the generation of biological hypotheses; also includes predicted and validated protein-DNA interactions | Katari et al. (2010) |
| Sungear (http://virtualplant.bio.nyu.edu/cgi-bin/sungear/index.cgi) | Enables rapid, visually interactive exploration of large sets of genomic data; allows browsing of gene sets by experiment, gene annotation, and ontological term; makes otherwise complicated queries quick and visually intuitive | Part of the VirtualPlant suite of tools |
| Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) | Enables the visualization of relative and absolute gene expression data from approximately 22,000 Arabidopsis genes in different tissues and under a variety of conditions | Winter et al. (2007) |
| GENIUS (Gene Network Inference Using Signatures; http://networks.bio.puc.cl/genius/) | Uses large-scale gene expression data to identify signatures that characterize and discriminate the process of interest, and then uses these signatures to predict a functional gene network | |
| VisuaLRTC | Lateral root transcriptome compendium | Parizot et al. (2010) |
| iRootHair (http://www.iroothair.org/) | A comprehensive database of root hair genomics information | Kwasniewski et al. (2013) |
| SUBA3 (http://suba.plantenergy.uwa.edu.au) | Combines manual literature curation of large-scale subcellular proteomics, fluorescent protein visualization, and protein-protein interaction data sets from Arabidopsis with subcellular targeting calls from 22 prediction programs | Tanz et al. (2013) |
| Arabidopsis Interaction Viewer (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi) | Queries a database of 70,944 predicted and 28,556 confirmed Arabidopsis interacting proteins against the protein(s) of interest; includes subcellular localization from SUBA | Geisler-Lee et al. (2007) |
| PlantMetabolomics.org (http://plantmetabolomics.vrac.iastate.edu/ver2/) | A National Science Foundation-funded multi-institutional project that is developing metabolomics as a functional genomics tool for elucidating the functions of Arabidopsis genes; the consortium has established metabolomic platforms that detect approximately 1,800 metabolites | Bais et al. (2010) |
| Ionomics Hub (http://www.ionomicshub.org/home/PiiMS) | Contains curated ionomic data on many thousands of plant samples (from Arabidopsis, Brassica napus, rice [Oryza sativa], maize, and soybean [Glycine max]) |
The Arabidopsis Interactions Viewer searchable database includes experimental and predictive information from protein-protein interactions (Geisler-Lee et al., 2007). This freely available and curated resource (Table I) queries a database of 70,944 predicted and 28,556 confirmed Arabidopsis interacting proteins against a protein(s) of interest. The experimentally confirmed and predicted interactions come from BIND, the Biomolecular Interaction Network Database (Willis and Hogue, 2006), from high-density Arabidopsis protein microarrays (Popescu et al., 2007, 2009), from the Arabidopsis Interactome Mapping Consortium (2011), the Membrane protein INteractome Database MIND (Lalonde et al., 2010), the Arabidopsis G-signaling Interactome Database (Klopffleisch et al., 2011), and over 1,190 other literature sources. The subcellular localization data in the Arabidopsis Interactions Viewer comes from an older version of SUBA (which does not include predicted localization from SUBA3). Other freely available tools include user-friendly platforms designed to facilitate hypothesis formulation by depositing information on high-throughput profiling information relating to the plant tissue metabolome (Bais et al., 2010) and mineral composition (termed the ionome; Baxter et al., 2007).
Resources for mining high-throughput profiling data sets increasingly include user-friendly platforms, but the ever-increasing information contributed by low-throughput studies remains less well annotated. A wealth of information relevant to root biology remains buried within various repositories, including the scientific literature. Text mining can retrieve these data, but as the body of information grows, the feasibility of manual annotation decreases. However, advanced text-mining methodology has recently been implemented (Van Landeghem et al., 2013), including work to collate information about specific tissues or processes in a readily accessible format. For example, the more than 1,300 articles that describe processes related to root hair cells would require extensive time and effort to extract relevant information by a traditional literature review (Kwasniewski et al., 2013). iRootHair addresses this problem and provides a freely available resource that collects, presents, and shares the available information in a systematic, curated fashion (Kwasniewski et al., 2013). Features of iRootHair include four searchable sections: Genes, Processes of root hair formation, Root hair mutants, and References. The database also integrates bioinformatic tools and can be used to identify particular genes of interest or to find broader information about root hair genomics.
Bottom-Up Root Systems Biology Approaches
In contrast to the phenomenological-type models used in top-down studies, the hypothesis-driven bottom-up approach uses predictive models that can be tested experimentally (for review, see Middleton et al., 2012). This requires quantitative data on key components of the steady-state system to build mechanism-based models to predict emergent behaviors of the system. Mechanism-based models help us conceptualize biological processes and uncover gaps in our understanding. These models can be used as simulations, which assist in experiential design by enabling researchers to make well-founded and testable predictions. Models, by nature, oversimplify the system we wish to study and should ideally be improved in an iterative loop, in association with experimental validation. The more quantitative data available for key components of a particular system, the greater the opportunity to construct meaningful models.
New mathematical and computational models probe the biological significance of identified interactions within increasingly complex GRNs. The BioModels Database serves as a reliable repository of computational models of biological processes (Li et al., 2010). The BioModels Database Web site (http://www.ebi.ac.uk/biomodels-main/) hosts a collection of models described in peer-reviewed scientific literature as well as models generated automatically from pathway resources (Path2Models). These freely available models can be used or adapted to address relevant biological questions. As information on root regulatory networks increases, so does the opportunity to implement mechanistic models for bottom-up studies (for review, see Middleton et al., 2012). Below, we discuss some examples of how these types of models have successfully addressed important biological questions related to root systems.
The molecular and biochemical reactions that underpin biological systems are often nonlinear, with feed-forward and feed-backward loops that contribute to the robustness of the system (Mosconi et al., 2008). For example, the phytohormone auxin regulates cell division, elongation, and differentiation in roots (Péret et al., 2009; Stahl and Simon, 2010; Sablowski, 2011). Auxin functions in part by mediating the degradation of auxin/indole-3-acetic acid (Aux/IAA) family proteins, which repress auxin response transcription factors (ARFs). ARFs regulate various downstream genes that define the auxin-responsive cellular processes described above, including Aux/IAAs. Thus, the pathway comprises multiple negative feedback loops. Middleton et al. (2010) developed a mathematical model of the auxin response network by employing a series of coupled ordinary differential equations. The model captures all key auxin response network components, including auxin, its receptor protein TIR1, ARFs, and interacting AUX/IAA repressor proteins, together with their known regulatory relationships. The quantitative data from molecular studies on protein turnover and transcription rates enabled Middleton et al. (2010) to build a model using realistic parameters. Simulations using this model revealed the importance of the turnover rates of Aux/IAA protein and mRNA for the dynamic behavior of the model.
Model predictions that are inconsistent with the experimental data can point to the existence of unknown interactions or components missing from the network model. For example, the model described above showed discrepancies between its behavior and the experimental data (Middleton et al., 2010). In experiments, the abundance of some mRNA species can change by up to 30-fold (Abel and Theologis, 1996). However, achieving this level of fold change in the original model developed by Middleton et al. (2010) requires an extremely large (and possibly unrealistic) increase in auxin levels. This points to the existence of additional regulatory mechanisms that might amplify the cellular response (as measured by Aux/IAA mRNA levels) to changes in auxin. A number of ARF-encoding genes respond to auxin (Okushima et al., 2005), which could represent a feed-forward loop that allows signal amplification.
The ARF family member MONOPTEROS (MP) up-regulates both its own expression and the expression of the gene encoding Aux/IAA family member BODENLOS (BDL; Lau et al., 2011). Thus, the network consists of an ARF-based positive feedback loop coupled to the Aux/IAA negative feedback. The presence of a positive feedback loop in a model can lead to bistability, in which the system can rest in either of two stable states. The two stable states allow for switch-like behavior that functions as a form of signal amplification. For example, increasing levels of exogenous auxin can switch the system from a low-MP/BDL concentration steady state to a high one (Lau et al., 2011). Bistable systems also exhibit hysteresis, where the behavior of a system depends not only on its current condition but also on its past environment. As auxin levels increase, MP expression increases and the system approaches a high-MP/BDL steady state. However, as auxin levels decrease, MP and BDL levels decrease much more slowly (i.e. the MP positive feedback loop is self-sustaining) and remain up-regulated for several hours (Lau et al., 2011). Moreover, the system can generate switch-like responses to changes in auxin concentration, so that an MP-dependent developmental program can only initiate over a certain threshold. Once the switch has been activated (and the system is in an MP up-regulated state), the network’s behavior will be robust to large changes in the level of auxin. Hence, modeling has revealed how auxin can induce all-or-nothing developmental outputs.
Another increasingly important aspect of root developmental biology is the study of cross regulation between two or more hormones. For example, the interplay between auxin and cytokinin controls the size of the root meristem in an antagonistic manner (Dello Ioio et al., 2008). Local accumulation of auxin promotes root cell division, whereas high cytokinin concentrations promote differentiation. To study the interplay between auxin and cytokinin signaling, Muraro et al. (2011) developed an ordinary differential equation-based mathematical model of known interactions. In addition to the auxin-mediated up-regulation of Aux/IAA genes (notably SHORT HYPOCOTYL2 [SHY2]), auxin also up-regulates the transcription of genes encoding ARR-A inhibitors of the cytokinin response. ARR-A genes are also up-regulated by ARR-Bs in response to cytokinin. In contrast, cytokinin signaling activates an ARR-B transcription factor, which in turn induces the transcription of SHY2. In silico perturbations to the model predict experimental changes in cell division observed in both gain-of-function and loss-of-function shy2 mutants, suggesting that the network had accurately captured the regulatory mechanisms controlling meristem size.
MIDDLE-OUT ROOT SYSTEMS BIOLOGY APPROACHES
The middle-out approach starts at the physical scale at which there is the most information and takes bottom-up and top-down approaches. Models constructed at the cell scale are particularly well suited to the middle-out approach because the cell comprises subcellular network systems (revealed using a top-down approach), and cell-cell interactions lead to the emergence of tissue-scale properties (revealed using a bottom-up approach). However, the middle-out approach can start at any scale with sufficient available information. In the following section, we review examples where root researchers have developed models focused at the subcellular, cell, tissue, organ, whole-plant, and plant-environment scales.
Subcellular-Scale Systems Models
At the subcellular scale, root models have been described that simulate gene regulatory, signaling, and metabolic networks. These typically employ Boolean, ordinary differential equations, or stochastic approaches (when low copy numbers are important; for review, see Band et al., 2012a).
Dupeux et al. (2011) described a stochastic model developed to study abscisic acid (ABA) signal perception within a cell. The key hormone ABA regulates root growth and responses to biotic and abiotic stresses. Within the cell, ABA binds to a family of intracellular receptors, termed PYRABACTIN RESISTANCE (PYR)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR (RCAR) proteins. ABA binding promotes the formation of complexes involving the PYR/PYL/RCAR receptors and several types of PROTEIN PHOSPHATASE TYPE 2C (PP2C) proteins that activate ABA responses. PYR/PYL/RCAR family members bind ABA as a monomer or dimer. Dupeux et al. (2011) developed a computational model capturing this relatively simple network of interactions to probe the response to ABA when both monomeric and dimeric PYR/PYL/RCAR receptors compete for ABA and PP2C molecules. Intriguingly, the study revealed that monomeric receptors have a competitive advantage for binding, particularly at lower ABA concentrations. Hence, tissue receptor composition and oligomerization likely impact ABA responsiveness.
Cruz-Ramírez et al. (2012) recently used a mathematical model of a small GRN to demonstrate how two nested feed-forward loops precisely control asymmetric cell divisions within the root stem cell niche (Fig. 2). The authors employed modeling to help understand how spatial restriction of these divisions requires physical binding of the stem cell regulator SCARECROW (SCR) by the RETINOBLASTOMA-RELATED (RBR) protein. They generated coupled ordinary differential equations to describe the wiring of the network. Initial models tested the importance of the CycD6 feedback loop on the phosphorylation of RBR and its interaction with SCR and subsequent feedback on SCR expression. Because asymmetric cell divisions occur at low or high SCR-RBR levels, the authors analyzed equilibrium levels of nuclear SHORT ROOT (SHR) and free SCR (unbound to RBR). SHR and auxin distribution and their effect on the precise positioning of CycD6 in the cortex endodermis determine radial and longitudinal positional information. Coupling of cell cycle progression to protein degradation resets the circuit, resulting in a “flip-flop” (i.e. a circuit with two stable states) that constrains asymmetric cell division to the stem cell region. RBR binds SCR and, together with the RBR regulator CYCD6;1, defines the position of asymmetric cell divisions. The resulting network creates a robust, bistable switch combining the auxin gradient along the longitudinal axis with the SHR distribution pattern along the radial axis (Cruz-Ramírez et al., 2012).
Figure 2.
Examples of hypothesis-driven bottom-up and middle-out approaches. Models were constructed for processes at different spatial and temporal scales. A, At the subcellular level, GRNs can be modeled to include the input of signals such as auxin and output in the form of cell-scale events, such as mitosis. B, Stem cell division and the bistable switch that regulates it are modeled using longitudinal gradients of auxin and radial gradients of the transcription factor SHR (Cruz-Ramírez et al., 2012). C, Root system scale models such as RootTyp (Pagès et al., 2004) can be linked to hydraulic models to explicitly simulate water flow in the soil-plant domain (Javaux et al., 2008).
Cell-Scale Systems Models
At the cell level, models capture interactions between neighboring root cells. For example, Savage et al. (2008) modeled the patterning of the Arabidopsis root epidermis, which forms alternating files of trichoblasts (bearing root hairs) and atrichoblasts (lacking root hairs). Epidermal cells overlying two cortical cell files form trichoblasts. The mobile activator proteins GALABRA3/ENHANCER OF GALABRA3 (GL3/EGL3) regulate this patterning and form an activator complex with TRANSPARENT TESTA GALABRA1, WEREWOLF (WER), and the mobile inhibitor protein CAPRICE (CPC). Savage et al. (2008) formulated a stochastic Boolean model, embedded into a circle of cells, with the levels of CPC and GL3/EGL3 depending on the rate of transcription in the neighboring cells. They modeled potential network structures, comparing WER self-activation and CPC inhibition via competition for GL3/EGL3, with constitutive WER transcription and CPC directly inhibiting WER. While both models replicated the patterning in wild-type roots, the latter model best explained the mutant phenotypes, in particular cpc.
Benítez et al. (2008) explored the same problem using a three-state deterministic model within a two-dimensional array of cells; they also found that WER autoregulation was not required for pattern formation in wild-type roots, but it proved necessary to explain some mutant phenotypes in the two-dimensional geometry. The two-dimensional model was also used to explore the effects of cell shape, with cell elongation acting to stabilize patterns. While the experiments of Savage et al. (2008) provided evidence that WER autoregulation is not important for the early patterning of epidermal cells, local self-activation does occur at later stages (Kwak and Schiefelbein, 2008; Kang et al., 2009), although its functional role remains unclear.
Tissue- to Organ-Scale Systems Models
Tissue- and organ-scale models describe growth, transport, and deformation on the macroscale, usually employing continuum (mainly partial differential) equations that do not explicitly subdivide a tissue into discrete cells. Péret et al. (2012) described a tissue-scale modeling approach to investigate how aquaporins regulate lateral root emergence. In Arabidopsis, lateral root primordia originate from pericycle cells located deep within the parental root and emerge through endodermal, cortical, and epidermal tissues. Recent studies have highlighted the importance of the localized auxin signal originating from the tip of the lateral root primordia to induce specific physiological responses in overlying tissues (Swarup et al., 2008; Péret et al., 2012). Biomechanical properties targeted by auxin include changes in aquaporin spatial expression patterns to modify water fluxes. Based on these data, Péret et al. (2012) included a mathematical model describing how aquaporin-dependent tissue hydraulics could affect the timing of lateral root development. In the model, water fluxes are coupled to primordium expansion, with the lateral root primordia and overlying tissue represented as two fluid compartments. The dividing primordium cells have a prescribed increasing osmotic potential. The model predicts the resulting water fluxes and pressure dynamics and shows how these drive primordium expansion. The model predicts that aquaporin repression by auxin promotes lateral root emergence and explains the experimentally observed delay in lateral root primordia emergence for both aquaporin loss-of-function and overexpression lines.
Plant-Scale Systems Models
Plant-scale models aim to integrate the behavior of individual plant organs, or even suborgan characteristics, at the whole-plant level, into functional structural plant models (Godin and Sinoquet, 2005). At this level, major dynamic processes come into play, including resource capture (carbon, water, nutrients), long-distance transport, resource allocation, long-distance signaling, and concurrent growth of many organs. This type of model implements most processes, such as the production and consumption of carbohydrate assimilates (Drouet and Pagès, 2007) or the development of the plant (growth, branching) at the organ or suborgan level. These processes are integrated at the plant level by connecting the different parts (up to several thousand) according to plant topology. These models often adopt a tree-like structure, well suited to representing plant networks.
The analysis of carbon transport and allocation within the root system has been described using the MassFlowDyn model (Bidel et al., 2000), which explicitly simulates the phloem transport of sugars following simple source/sink rules in relation to the growth and development of complex root system. The authors demonstrated the importance of the initial diameter of individual root meristems as a determinant of sink strength. The formation of a hierarchy of root meristems (primary > lateral) leads to herringbone-type systems (akin to the arrangement of bones in a fish), while similar meristem sizes lead to fibrous root systems (composed of many roots of approximately equivalent lengths).
Plant-Environment-Scale Systems Models
A second category of functional structural plant models explicitly takes into account the bidirectional flow of material or energy and the biological, physical, and chemical interactions between the plant and the soil compartment. Compared with the previous models, which generally exclude environmental effects or consider them uniform, these models explicitly include the environment, with a focus on the processes of interest. As for plant-scale models, the different processes, in the plant, in the environment, or at the plant-environment interface, are implemented at the local scale and integrated at the whole-plant scale. The plant structure still follows a tree-like structure, but the environment, especially the soil, may require a three-dimensional graph, as all neighboring soil voxels are interconnected. The integration of the plant tree and the environment graph requires a complex modeling framework in which the positioning of the different structures can be matched in time and space. Indeed, since most of the processes are computed locally, the exact position of every plant and environment element is needed.
Soil-root models developed in the last two decades address mostly water and nutrient dynamics. SimRoot (Lynch et al., 1997; Postma and Lynch, 2011a) and ROOTMAP (Diggle, 1988; Dunbabin et al., 2002) link root growth and root system architecture with nutrient acquisition (for review, see Dunbabin et al., 2013). SimRoot, for example, was used to demonstrate the influence of root-type gravitropism on phosphorus acquisition in bean (Phaseolus vulgaris; Ge et al., 2000). Other models have been developed to analyze the dynamics of water flow in the soil-root system, taking into account the long-distance movement of water in the soil (Doussan et al., 1998; Javaux et al., 2008; Schröder et al., 2008; Lobet and Draye, 2013; Fig. 2C). The three-dimensional model R-SWMS, for example, simulates the uptake of water by individual root segments as a function of the root hydraulic architecture and soil properties (Doussan et al., 2006; Draye et al., 2010) and will help our understanding of the contribution of long-distance regulatory signals (e.g. ABA) to whole-plant hydraulics and water use.
The recent development of methods for imaging live roots in real soil at the microscopic scale (Mooney et al., 2012) enables a step change in the modeling of nutrient uptake. The ability to uncover the three-dimensional interactions between the root surface and soil porosity has allowed the development of a model of phosphate uptake by root hairs based directly on the real geometry of hairs and associated soil pores (Keyes et al., 2013). The new model questioned the currently accepted relative contribution of the root surface versus root hairs during the uptake of phosphorus.
The interface between the root and its environment also has many biological interactions between the root system and its surrounding communities. The German plant physiologist Lorenz Hiltner in 1904 coined the term “rhizosphere” to describe the region inhabited by a unique population of microorganisms, which he postulated were influenced by the chemicals released from plant roots (Hartmann et al., 2008). Root exudation represents a significant carbon cost to the plant (Lynch and Whipps, 1990). In addition, the presence of exudates and microbes profoundly affects the physical properties of the soil-root interface, such as its hydraulic conductivity (Carminati and Vetterlein, 2013). Therefore, understanding plant behavior in the field requires a fundamental understanding of the rhizosphere. The functional structural plant models framework has all the required components to address the spatiotemporal dynamics of the rhizosphere, yet such work remains to be initiated.
Crop-Scale Systems Models
At the crop scale, the description of plants can be further simplified, mainly to reduce computational cost as the total number of plants (and, subsequently, of individual plant elements) increases. Compared with the plant-environment scale, modeling at the crop scale still requires the space and time localization of the plants, but to a lesser extent; for example, the position of every root is not required anymore. Such models mainly aim to predict the construction of final crop yield under different environmental conditions, in different agronomic management practices, or in different genetic backgrounds.
Despite the fact that plants are conceptualized at a higher level than in lower scale models, some crop models include physiological and genetic modules influencing crop development. For example, SUNFLO (Casadebaig et al., 2011) models sunflower (Helianthus annuus) adaptation to environmental stimuli (water and nitrogen stress) and aims to assess genotypes for their success in these changing environments, as evaluated by their yield. The SUNFLO model successfully predicted genotype when tested against validation data from over 50 field experiments. This could be due to the incorporation into the model of genotype via the gene relationship to dynamic processes such as nutrient uptake and oil content rather than to the model’s inherent predictive power. These results also highlight the need to incorporate information about rooting phenotypes to enhance the model’s applicability in water stress situations.
In development for more than 15 years, the crop-level model Agricultural Production Systems Simulator (APSIM) aims to integrate genetic information, management practices, and environmental conditions to predict the yields of economically important crop plants (Keating et al., 2003). APSIM has been used to test the effect of genetic variation either at the root (Hammer et al., 2010; Manschadi et al., 2010) or the shoot (Chenu et al., 2008) level. Using APSIM, Chenu et al. (2009) modeled the effect of organ-level quantitative trait loci (QTLs) on maize (Zea mays) yield. Using the model, they simulated the effect of QTLs of two selected developmental processes (leaf and silk elongation) by generating hundreds of hypothetical recombinant inbred lines and testing them across several patterns of drought seasons. The simulations predicted the QTL-by-environment interactions observed under field conditions. In particular, yield increase depended on higher leaf elongation rate in well-watered conditions but depended on decreased anthesis to silking interval in all environments. Additionally, the pleiotropic effects of the different QTLs limited possible trait combinations. Using APSIM, Manschadi et al. (2010) evaluated the effect on final yield of specific root traits. By reducing the insertion angle of primary roots in wheat (Triticum aestivum), the authors demonstrated the capacity of the crop as a whole to extract more water and increase yield. Hence, the integration of local processes (root emergence, leaf growth rate) at the crop scale can be useful to help understand the crop system as a whole.
Crop models initially aimed to simulate yield construction over the season based on resource capture and environmental conditions. They do not obviously lend themselves to organ-based representation of the plant, because the elementary unit is the crop itself. Rather, parameters such as mass, leaf area index, and root depth represent aspects of plant architecture. These variables are essential to compute light interception or soil resource capture. Although crop models are much less mechanistic than the previous examples discussed, reasons to include them into the root systems biology approach include the following. (1) Crop models represent one further step in upscaling, and, in essence, that step resembles the step from cell to organ or from organ to plant. Each step requires the identification of the most relevant source of variation between elementary units. (2) These models generate unique paths in time that integrate environmental, management, and genetic factors that influence the future of the crop. They have been used to simulate the emergence of genotype-environment interactions, which remain one of the major challenges to moving from the laboratory to the field. Therefore, they provide very useful insights to orient research at lower scales. (3) They offer the only simulation framework that allows testing (systematically) of many (virtual) genotypes in many seasons, which has become a major step in plant breeding, and provide a means to address the agronomic potential of gene targets identified in basic research.
MULTISCALE ROOT SYSTEMS BIOLOGY APPROACHES
Multiscale models consider behaviors on two or more scales, ranging from the subcellular up to whole-organism scales and beyond (Fig. 3). They must also incorporate different temporal scales, ranging from minutes for transcription to weeks for developmental adaptation. Relatively few examples of multiscale models have been described to date for roots (or other plant organs). We selected the following examples to illustrate the potential for developing multiscale models to probe the mechanisms underlying complex, nonlinear biological processes in roots. One key to predicting the relationship between genotype and phenotype is linking the spatiotemporal concentrations of regulators, such as auxin, with cell growth and division. We currently lack a mechanistic understanding of the effect of auxin on growth, but several existing examples employ multiscale approaches, incorporating phenomenological models to describe this missing step.
Figure 3.
Root biological models can be multiscale. Band et al. (2012b) demonstrated the power of multiscale modeling for understanding GA dynamics during root elongation. The elongation zone is modeled as a collection of cells (organ scale; A) and used to motivate the modeling of cell expansion and dilution in the vacuole (cell scale; B). This naturally leads to the link between cell partitioning of GA and subcellular processes, revealing the influence of the dilution of GA concentrations on DELLA expression and growth. MZ, Meristematic zone.
Grieneisen et al. (2007) proposed that a gradient of auxin controls the specification of the root’s developmental zones, supposing that cell division occurs at high hormone concentrations and cell elongation at lower concentrations. Due to the predicted auxin distribution, the model simulates cell division and growth dynamics in the meristem and elongation zone, capturing the gradual expansion of the meristem over the first 8 d post germination and the reduction in meristem size after root excision. Chavarría-Krauser et al. (2005) modeled the growth dynamics at the root tip, supposing that the ratio between auxin and cytokinin governs the production and degradation of a remodeling enzyme, which in turn regulates cell growth and division. For root developmental responses, Lucas et al. (2008), in predicting lateral root initiation, considered a pool of auxin at the root tip that is consumed by root gravitropic bending and/or emergence. Using such a phenomenological model between auxin and root development, the authors accurately predicted experimentally observed perturbations in lateral root initiation for the aux1 root gravitropic and lax3 lateral root emergence mutants.
The hormone GA controls root growth by, for example, controlling cell growth within the elongation zone (Ubeda-Tomás et al., 2008). Band et al. (2012b) recently developed a multiscale model of GA dynamics in the Arabidopsis root elongation zone, prescribing cell growth (using experimental measurements) and simulating GA dilution, the diffusion of GA between cellular compartments, and the response network through which GA degrades the growth-repressing DELLA proteins (describing the dynamics with a system of ordinary differential equations). The model (Fig. 3) revealed that, as cells pass through the elongation zone, dilution (rather than degradation) creates a decline in GA concentration, leading to spatial gradients in the levels of downstream DELLA mRNAs and proteins. This model prediction was confirmed experimentally with knockout mutants lacking every root-expressed GA2ox degradation enzyme. The study also considered the dynamics in plants treated with paclobutrazol (an inhibitor of GA biosynthesis) and plants with mutations in the GA biosynthesis and signaling pathways; these cases revealed that the growth rates appear to reflect the fold change in DELLA as cells traverse the elongation zone. Furthermore, the model provided new insights into the normal phenotype exhibited in the ga1-3/gai-t6/rga-24 triple mutant. The model demonstrated that the effect of the ga1-3 mutation in reducing GA biosynthesis (leading to higher functional DELLA levels) counteracts the effect of the gai-t6/rga-24 mutation in reducing the translation of functional DELLA; if these two processes are suitably balanced, the levels of functional DELLA are similar to that in the wild type, explaining why the triple mutant exhibits normal cell elongation. In summary, by assimilating a range of data and knowledge, the model revealed the dominant effect of GA dilution (rather than degradation) on the emergent DELLA distribution, providing new insights into GA’s growth regulation.
Jeuffroy et al. (2012) also used multiscale modeling to identify genotypes with desirable characteristics for breeding programs. The pea (Pisum sativum) crop model AFISOL for yield characteristics (incorporating water and nitrogen flow) was linked to SISOL modeling soil parameters (soil moisture content, soil biota activity effects, impacts of machinery, and natural soil weathering) and OTELO for farm practices (fertilizer, pesticides, and soil amendments). These models all include dynamic parameters, and this work demonstrates the capability to link several models of different scale, revealing the applicability of crop models in the systems biology approach across multiple plant physical and temporal scales. Additionally, the model assessment included real and virtual genotypes, which shows the value of isolating desirable characteristics that could then be harnessed through more molecular techniques.
FUTURE CHALLENGES FOR ROOT SYSTEMS BIOLOGY
Major advances have recently been made employing top-down systems approaches to identify the key molecular players (genes, RNAs, proteins, etc.) and to elucidate several GRNs that control root growth and development (Benfey et al., 2010; Ruffel et al., 2010; Bassel et al., 2012). However, regulatory networks not only operate at the molecular scale but must also integrate information from cell to organ to rhizophere scales, to probe the mechanisms controlling root development. Mathematical and computational models provide invaluable tools to bridge these distinct spatial and temporal scales and generate new mechanistic insights about the regulation of root growth and development (Band et al., 2012a). Nevertheless, we are only at the beginning of this new research area, and several challenging issues remain to be addressed for this field to move forward.
Integrating Models
In general, the models highlighted in this review have been designed independently of one another, raising important issues relating to interoperability. To proceed further, models must start to be assembled into unified frameworks. A number of conceptual and technological factors make model integration challenging. First, models may operate at different spatial resolutions or dimensions. For example, coupling a two-dimensional mechanical model of root tissues at a subcellular resolution with a two-dimensional model of auxin transport designed at a cellular resolution would require homogenizing the spatial resolution such that the mechanical model updates cell geometries over time. Second, models may also operate at different temporal resolutions. For example, mechanical processes are often assumed to be much faster than biochemical processes. Hence, cell size and shape can be considered constant in computing chemical simulations. Third, combining different types of mathematical models (e.g. Boolean, stochastic, ordinary differential equations, and partial differential equations) can often prove challenging. Nevertheless, models integrating mechanistic and stochastic models have been successfully designed at the organism level for aerial tissues (Costes et al., 2008).
Creating combinations of models often represents a large amount of theoretical and experimental effort. Hence, it is important that these models are freely available to the community as shared data sets, open-source model formats such as SBML (Hucka et al., 2003) or CellML (Cuellar et al., 2003), or common modeling software platforms such as VV (Prusinkiewicz, 2004), OpenAlea (Pradal et al., 2008), or MorphoGraphX (Kierzkowski et al., 2012). The move toward model integration and flexibility in the crop-modeling community should be translatable to the root systems biology ethos. For example, RECORD (for Renovation and Coordination of Agroecosystem Modeling) represents a concerted effort by French researchers at INRA to develop an “integrated modeling platform” that could eventually include Geographic Information System data, open source statistical resources, and data-handling resources (Bergez et al., 2012). Additionally, planned as a repository for crop models, the RECORD platform includes many principles of root systems biology, including multiple time scales (seconds to months), spatial variation, and environmental dynamics. RECORD aimed to use as many existing models as possible. However, to foster the incorporation of sub-crop-scale models as advocated in root systems biology, RECORD would need to look beyond crop-soil interactions to the root system itself.
Toward Digital Plants and Populations
Developing a mechanistic model of a whole plant represents a logical next step. Indeed, given the exchange of water, nutrients, and signals between root and shoot organs, developing a virtual root or shoot model independent of each other could be considered naive in the longer term. To date, models of diverse root system subprocesses have been developed at different scales. Compared with initial approaches in systems biology, most of these models make explicit use of spatial information. Such spatial information represents different aspects of realistic root structures and can take the form of a continuous medium, a branching structure of connected elements (e.g. root meristems), a multicellular population, or a set of interacting subcellular compartments. By progressively integrating more functional aspects into these realistic representations, researchers have created new models (Sievänen et al., 2000; Godin and Sinoquet, 2005). Functional structural plant models provide a promising platform with which to create a digital plant model.
Compared with many previous models developed on aerial parts (Vos et al., 2010) and on root systems (Bidel et al., 2000; Pagès et al., 2004), recent functional structural plant models also integrate gene regulation and signaling as a new dimension in the analysis of development (Prusinkiewicz et al., 2007; Han et al., 2010). Through the combined modeling of genetic networks, physiological processes, and spatial interaction between components, development of a new generation of functional structural plant models opens the way to building digital versions of real plants (Coen et al., 2004; Cui et al., 2010) and testing biological hypotheses in silico (Stoma et al., 2008; Perrine-Walker et al., 2010).
Ultimately, breeders measure plant performance at a population scale rather than an individual scale. Hence, mechanistic multiscale models need to be developed that bridge the remaining physical scale between the plant and the field to relate genotype to phenotype and enable the engineering of crop traits. Relatively few examples exist where modeling results have been used to direct the selection of new crop varieties. One promising example is the optimization of bean root systems for phosphorus and water acquisition. Using the model SimRoot, Lynch and coworkers investigated the impact of root gravitropism (Ge et al., 2000), root hair production (Ma et al., 2001), and aerenchyma production (Postma and Lynch, 2011a, 2011b) on plant performance under various phosphorus and/or water conditions. These results defined root system ideotypes (Lynch and Brown, 2001; Lynch, 2011, 2013) and were validated by several experimental trials (Bates and Lynch, 2001; Rubio et al., 2003). This identified QTLs for basal root gravitropism (Liao et al., 2004) and root hairs (Yan et al., 2004) and led to the selection of new bean varieties, now used by breeding programs in South America and Africa. This example illustrates how modeling at the plant level can drive research, untangle processes at the molecular level, and inform crop breeding.
Integrating Environmental Information
Phenotype represents the output of the interaction between genotype and environment. In this review, almost all of the models described study intrinsic root regulatory processes. This provides a solid foundation for describing basic developmental mechanisms, but to account for root plasticity, models must also integrate environmental information. Developing mixed genetic-ecophysiological models that bridge the gap between genetic and environmental regulation represents an important goal (Roose and Schnepf, 2008; Draye et al., 2010). These environmental factors include soil physical properties; water, nutrient, and macroelement/microelement availability and distribution; mycorrhization and nodulation; and competition and interaction with other root systems. Obtaining such spatial information remains very challenging. Researchers traditionally assess soil structure by imaging and then physically removing successive layers of soil and examine root structure (of soil-grown roots) by root washing. As both are destructive, the construction of combined root and soil data sets requires the integration of measurements obtained from different samples. High-resolution synchrotron and x-ray micro-computed tomography imaging has the potential to provide rich data sets from single samples, with concurrent measurements of root and soil (Mooney et al., 2012; Keyes et al., 2013).
Root systems biology also urgently requires new methods to assay rhizosphere parameters in addition to root and soil. For example, having tools to dynamically monitor quantitative changes in root biology (hormones, water status, nutrients, etc.) and the root environment (pH, nutrient content) would address a major challenge. The development of novel biosensors based on new understanding of the pathways responsible for the perception of small signaling molecules and advances in imaging technologies and mathematical modeling will enable a truly quantitative analysis of biological processes (Band et al., 2012b; for review, see Wells et al., 2013). Combined with new dynamic sensors for environmental parameters like optodes (Elberling et al., 2011), new biosensors promise to provide a deeper understanding of how plant systems interact with their environment. Moreover, these sensors could result in innovative technologies to monitor biotic or abiotic stresses in the field (Chaerle et al., 2009). Biosensors could help optimize the use of inputs such as water, fertilizers, or pesticides and, therefore, achieve more environment-friendly and sustainable agricultural practices. Such tools and the information generated will also greatly aid the development of more realistic root-rhizosphere models and, ultimately, help optimize crop root architectures for soil types and nutrient regimens.
Acknowledgments
We thank Rodrigo Gutierrez and Miriam Gifford for sharing information prior to publication.
Glossary
- GRN
gene regulatory network
- TF
transcription factor
- miRNA
microRNA
- ChIP
chromatin immunoprecipitation
- ARF
auxin response transcription factor
- Aux/IAA
auxin/indole-3-acetic acid
- ABA
abscisic acid
- APSIM
Agricultural Production Systems Simulator
- QTL
quantitative trait locus
- RECORD
Renovation and Coordination of Agroecosystem Modeling
- RNA-seq
RNA analysis through complementary DNA sequencing at a massive scale
References
- Abel S, Theologis A. (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais P, Moon SM, He K, Leitao R, Dreher K, Walk T, Sucaet Y, Barkan L, Wohlgemuth G, Roth MR, et al. (2010) PlantMetabolomics.org: a Web portal for plant metabolomics experiments. Plant Physiol 152: 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Fozard JA, Godin C, Jensen OE, Pridmore T, Bennett MJ, King JR. (2012a) Multiscale systems analysis of root growth and development: modeling beyond the network and cellular scales. Plant Cell 24: 3892–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, Sun J, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Péret B, et al. (2012b) Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci USA 109: 4668–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. (2007) How to infer gene networks from expression profiles. Mol Syst Biol 3: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Gaudinier A, Brady SM, Hennig L, Rhee SY, De Smet I. (2012) Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T, Lynch J. (2001) Root hairs confer a competitive advantage under low phosphorus availability. Plant Soil 236: 243–250 [Google Scholar]
- Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. (2007) Purdue Ionomics Information Management System: an integrated functional genomics platform. Plant Physiol 143: 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Bennett M, Schiefelbein J. (2010) Getting to the root of plant biology: impact of the Arabidopsis genome sequence on root research. Plant J 61: 992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez M, Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER. (2008) Interlinked nonlinear subnetworks underlie the formation of robust cellular patterns in Arabidopsis epidermis: a dynamic spatial model. BMC Syst Biol 2: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergez JE, Chabrier P, Gary C, Jeuffroy M, Makowski D, Quesnel G, Ramat E, Raynal H, Rousse N, Wallach D. (2012) An open platform to build, evaluate and simulate integrated models of farming and agro-ecosystems. Environ Model Softw 39: 39–49 [Google Scholar]
- Bidel LP, Pagès L, Rivière LM, Pelloux G, Lorendeau JY. (2000) MassFlowDyn I: a carbon transport and partitioning model for root system architecture. Ann Bot (Lond) 85: 869–886 [Google Scholar]
- Bielach A, Podlesáková K, Marhavy P, Duclercq J, Cuesta C, Müller B, Grunewald W, Tarkowski P, Benková E. (2012) Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. Plant Cell 24: 3967–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN. (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2: 615–619 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. (2011) A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV. (2007) The nature of systems biology. Trends Microbiol 15: 45–50 [DOI] [PubMed] [Google Scholar]
- Carminati A, Vetterlein D. (2013) Plasticity of rhizosphere hydraulic properties as a key for efficient utilization of scarce resources. Ann Bot (Lond) 112: 277–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadebaig P, Guilioni L, Lecoeur J, Christophe A, Champolivier L, Debaeke P. (2011) SUNFLO, a model to simulate genotype-specific performance of the sunflower crop in contrasting environments. Agric For Meteorol 151: 163–178 [Google Scholar]
- Chaerle L, Lenk S, Leinonen I, Jones HG, Van Der Straeten D, Buschmann C. (2009) Multi-sensor plant imaging: towards the development of a stress-catalogue. Biotechnol J 4: 1152–1167 [DOI] [PubMed] [Google Scholar]
- Chavarría-Krauser A, Jäger W, Schurr U. (2005) Primary root growth: a biophysical model of auxin-related control. Funct Plant Biol 32: 849–862 [DOI] [PubMed] [Google Scholar]
- Chenu K, Chapman SC, Hammer GL, McLean G, Salah HBH, Tardieu F. (2008) Short-term responses of leaf growth rate to water deficit scale up to whole-plant and crop levels: an integrated modelling approach in maize. Plant Cell Environ 31: 378–391 [DOI] [PubMed] [Google Scholar]
- Chenu K, Chapman SC, Tardieu F, McLean G, Welcker C, Hammer GL. (2009) Simulating the yield impacts of organ-level quantitative trait loci associated with drought response in maize: a “gene-to-phenotype” modeling approach. Genetics 183: 1507–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E, Rolland-Lagan AG, Matthews M, Bangham JA, Prusinkiewicz P. (2004) The genetics of geometry. Proc Natl Acad Sci USA 101: 4728–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes E, Smith C, Renton M, Guédon Y, Prusinkiewicz P, Godin C. (2008) MAppleT: simulation of apple tree development using mixed stochastic and biomechanical models. Funct Plant Biol 35: 936–950 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, et al. (2012) A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar AA, Lloyd CM, Nielsen PF, Bullivant DP, Nickerson DP, Hunter PJ. (2003) An overview of CellML 1.1, a biological model description language. Simulation 79: 740–747 [Google Scholar]
- Cui ML, Copsey L, Green AA, Bangham JA, Coen E. (2010) Quantitative control of organ shape by combinatorial gene activity. PLoS Biol 8: e1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- Diggle A. (1988) ROOTMAP: a model in three-dimensional coordinates of the growth and structure of fibrous root systems. Plant Soil 105: 169–178 [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Doussan C, Pagès L, Vercambre G. (1998) Modelling of the hydraulic architecture of root systems: an integrated approach to water absorption-model description. Ann Bot (Lond) 81: 213–223 [Google Scholar]
- Doussan C, Pierret A, Garrigues E, Pagès L. (2006) Water uptake by plant roots. II. Modelling of water transfer in the soil root-system with explicit account of flow within the root system: comparison with experiments. Plant Soil 283: 99–117 [Google Scholar]
- Draye X, Kim Y, Lobet G, Javaux M. (2010) Model-assisted integration of physiological and environmental constraints affecting the dynamic and spatial patterns of root water uptake from soils. J Exp Bot 61: 2145–2155 [DOI] [PubMed] [Google Scholar]
- Drouet JL, Pagès L. (2007) GRAAL-CN: a model of growth, architecture and allocation for carbon and nitrogen dynamics within whole plants formalised at the organ level. Ecol Modell 206: 231–249 [Google Scholar]
- Dunbabin V, Postma J, Schnepf A, Pagès L, Javaux M, Wu L, Leitner D, Chen Y, Rengel Z, Diggle A. (2013) Modelling root-soil interactions using three–dimensional models of root growth, architecture and function. Plant Soil 372: 93–124 [Google Scholar]
- Dunbabin VM, Diggle AJ, Rengel Z, van Hugten R. (2002) Modelling the interactions between water and nutrient uptake and root growth. Plant Soil 239: 19–38 [Google Scholar]
- Dupeux F, Santiago J, Betz K, Twycross J, Park SY, Rodriguez L, Gonzalez-Guzman M, Jensen MR, Krasnogor N, Blackledge M, et al. (2011) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30: 4171–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AN, Schlueter J, Spooner DM. (2012) Applications of next-generation sequencing in plant biology. Am J Bot 99: 175–185 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elberling B, Askaer L, Jørgensen CJ, Joensen HP, Kühl M, Glud RN, Lauritsen FR. (2011) Linking soil O2, CO2, and CH4 concentrations in a wetland soil: implications for CO2 and CH4 fluxes. Environ Sci Technol 45: 3393–3399 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe’er D. (2000) Using Bayesian networks to analyze expression data. J Comput Biol 7: 601–620 [DOI] [PubMed] [Google Scholar]
- Ge Z, Rubio G, Lynch JP. (2000) The importance of root gravitropism for inter-root competition and phosphorus acquisition efficiency: results from a geometric simulation model. Plant Soil 218: 159–171 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. (2007) A predicted interactome for Arabidopsis. Plant Physiol 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin C, Sinoquet H. (2005) Functional-structural plant modelling. New Phytol 166: 705–708 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hamilton JP, Buell CR. (2012) Advances in plant genome sequencing. Plant J 70: 177–190 [DOI] [PubMed] [Google Scholar]
- Hammer GL, van Oosterom E, McLean G, Chapman SC, Broad I, Harland P, Muchow RC. (2010) Adapting APSIM to model the physiology and genetics of complex adaptive traits in field crops. J Exp Bot 61: 2185–2202 [DOI] [PubMed] [Google Scholar]
- Han L, Hanan J, Gresshoff PM. (2010) Computational complementation: a modelling approach to study signalling mechanisms during legume autoregulation of nodulation. PLoS Comput Biol 6: e1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A, Rothballer M, Schmid M. (2008) Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312: 7–14 [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. (2011) Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 21: 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javaux M, Schröder T, Vanderborght J, Vereecken H. (2008) Use of a three-dimensional detailed modeling approach for predicting root water uptake. Vadose Zone J 7: 1079–1088 [Google Scholar]
- Jeuffroy MH, Vocanson A, Roger-Estrade J, Meynard JM. (2012) The use of models at field and farm levels for the ex ante assessment of new pea genotypes. Eur J Agron 42: 68–78 [Google Scholar]
- Jiao Y, Tausta SL, Gandotra N, Sun N, Liu T, Clay NK, Ceserani T, Chen M, Ma L, Holford M, et al. (2009) A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet 41: 258–263 [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N. (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J. (2009) The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating BA, Carberry P, Hammer G, Probert ME, Robertson M, Holzworth D, Huth N, Hargreaves J, Meinke H, Hochman Z. (2003) An overview of APSIM, a model designed for farming systems simulation. Eur J Agron 18: 267–288 [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. (2003) Laser capture microdissection of cells from plant tissues. Plant Physiol 132: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes SD, Boardman RP, Marchant A, Roose T, Sinclair I. (2013) A robust approach for determination of the macro-porous volume fraction of soils with x-ray computed tomography and an image processing protocol. Eur J Soil Sci 64: 298–307 [Google Scholar]
- Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, Weber A, Bayer E, Schorderet M, Reinhardt D, Kuhlemeier C, Smith RS. (2012) Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, Carpita NC, Carr T, Chen JG, Cooke TR, et al. (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 7: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. (2010) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J. (2008) A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr Biol 18: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Kwasniewski M, Nowakowska U, Szumera J, Chwialkowska K, Szarejko I. (2013) iRootHair: a comprehensive root hair genomics database. Plant Physiol 161: 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Sero A, Pratelli R, Pilot G, Chen J, Sardi MI, Parsa SA, Kim DY, Acharya BR, Stein EV, et al. (2010) A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front Physiol 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jürgens G. (2011) Auxin triggers a genetic switch. Nat Cell Biol 13: 611–615 [DOI] [PubMed] [Google Scholar]
- Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan MI, et al. (2010) BioModels database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst Biol 4: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Yan X, Rubio G, Beebe SE, Blair MW, Lynch JP. (2004) Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Funct Plant Biol 31: 959–970 [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, Bilgin DD, Radwan O, Neece DJ, Clough SJ, May GD, et al (2010) Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol 152: 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WD, Liao YY, Yang TJ, Pan CY, Buckhout TJ, Schmidt W. (2011) Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol 155: 1383–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G, Draye X. (2013) Novel scanning procedure enabling the vectorization of entire rhizotron-grown root systems. Plant Methods 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L. (2008) Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot 59: 55–66 [DOI] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J, Brown K. (2001) Topsoil foraging: an architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237 [Google Scholar]
- Lynch JM, Whipps JM. (1990) Substrate flow in the rhizosphere. Plant Soil 129: 1–10 [Google Scholar]
- Lynch JP. (2007) Roots of the second Green Revolution. J Bot 55: 493–512 [Google Scholar]
- Lynch JP. (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol 156: 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann Bot (Lond) 112: 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP, Nielsen KL, Davis RD, Jablokow AG. (1997) SimRoot: modelling and visualization of root systems. Plant Soil 188: 139–151 [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. (2001) Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 24: 459–467 [Google Scholar]
- Manschadi A, Christopher J, Hammer G, Devoil P. (2010) Experimental and modelling studies of drought-adaptive root architectural traits in wheat (Triticum aestivum L.). Plant Biosyst 144: 458–462 [Google Scholar]
- Mardis ER. (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24: 133–141 [DOI] [PubMed] [Google Scholar]
- Middleton AM, Farcot E, Owen MR, Vernoux T. (2012) Modeling regulatory networks to understand plant development: small is beautiful. Plant Cell 24: 3876–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton AM, King JR, Bennett MJ, Owen MR. (2010) Mathematical modelling of the Aux/IAA negative feedback loop. Bull Math Biol 72: 1383–1407 [DOI] [PubMed] [Google Scholar]
- Mooney SJ, Pridmore TP, Helliwell J, Bennett MJ. (2012) Developing x-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 352: 1–22 [Google Scholar]
- Mosconi F, Julou T, Desprat N, Sinha DK, Allemand JF, Croquette V, Bensimon D. (2008) Some nonlinear challenges in biology. Nonlinearity 21: T131 [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, Malitsky S, Toal TW, Belcher H, Yativ M, Brady SM, Benfey PN, Aharoni A. (2013) High-resolution metabolic mapping of cell types in plant roots. Proc Natl Acad Sci USA 110: E1232–E1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D, Byrne H, King J, Voss U, Kieber J, Bennett M. (2011) The influence of cytokinin-auxin cross-regulation on cell-fate determination in Arabidopsis thaliana root development. J Theor Biol 283: 152–167 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Noble D. (2008) Claude Bernard, the first systems biologist, and the future of physiology. Exp Physiol 93: 16–26 [DOI] [PubMed] [Google Scholar]
- Noor A, Serpedin E, Nounou M, Nounou H, Mohamed N, Chouchane L. (2013) An overview of the statistical methods used for inferring gene regulatory networks and protein-protein interaction networks. Adv Bioinformatics 2013: 953814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès L, Vercambre G, Drouet JL, Lecompte F, Collet C, Le Bot J. (2004) Root Typ: a generic model to depict and analyse the root system architecture. Plant Soil 258: 103–119 [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K. (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Parizot B, De Rybel B, Beeckman T. (2010) VisuaLRTC: a new view on lateral root initiation by combining specific transcriptome data sets. Plant Physiol 153: 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Larrieu A, Bennett MJ. (2009) Lateral root emergence: a difficult birth. J Exp Bot 60: 3637–3643 [DOI] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Perrin BE, Ralaivola L, Mazurie A, Bottani S, Mallet J, d’Alché-Buc F. (2003) Gene networks inference using dynamic Bayesian networks. Bioinformatics (Suppl 2) 19: ii138–ii148 [DOI] [PubMed] [Google Scholar]
- Perrine-Walker F, Doumas P, Lucas M, Vaissayre V, Beauchemin NJ, Band LR, Chopard J, Crabos A, Conejero G, Péret B, et al. (2010) Auxin carriers localization drives auxin accumulation in plant cells infected by Frankia in Casuarina glauca actinorhizal nodules. Plant Physiol 154: 1372–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, et al (2012) The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci USA 109: 6811–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma JA, Lynch JP. (2011a) Root cortical aerenchyma enhances the growth of maize on soils with suboptimal availability of nitrogen, phosphorus, and potassium. Plant Physiol 156: 1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma JA, Lynch JP. (2011b) Theoretical evidence for the functional benefit of root cortical aerenchyma in soils with low phosphorus availability. Ann Bot (Lond) 107: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poultney CS, Gutiérrez RA, Katari MS, Gifford ML, Paley WB, Coruzzi GM, Shasha DE. (2007) Sungear: interactive visualization and functional analysis of genomic datasets. Bioinformatics 23: 259–261 [DOI] [PubMed] [Google Scholar]
- Pradal C, Dufour-Kowalski S, Boudon F, Fournier C, Godin C. (2008) OpenAlea: a visual programming and component-based software platform for plant modelling. Funct Plant Biol 35: 751–760 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P (2004) Art and science for life: designing and growing virtual plants with L-systems. Acta Hortic 630: 15–28 [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. (2007) Evolution and development of inflorescence architectures. Science 316: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Roose T, Schnepf A. (2008) Mathematical models of plant-soil interaction. Philos Trans A Math Phys Eng Sci 366: 4597–4611 [DOI] [PubMed] [Google Scholar]
- Rubio G, Zhu J, Lynch JP. (2003) A critical test of the two prevailing theories of plant response to nutrient availability. Am J Bot 90: 143–152 [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Coruzzi GM. (2010) A systems view of responses to nutritional cues in Arabidopsis: toward a paradigm shift for predictive network modeling. Plant Physiol 152: 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. (2011) Plant stem cell niches: from signalling to execution. Curr Opin Plant Biol 14: 4–9 [DOI] [PubMed] [Google Scholar]
- Savage NS, Walker T, Wieckowski Y, Schiefelbein J, Dolan L, Monk NA. (2008) A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol 6: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder T, Javaux M, Vanderborght J, Körfgen B, Vereecken H. (2008) Effect of local soil hydraulic conductivity drop using a three-dimensional root water uptake model. Vadose Zone J 7: 1089–1098 [Google Scholar]
- Sievänen R, Nikinmaa E, Nygren P, Ozier-Lafontaine H, Perttunen J, Hakula H. (2000) Components of functional-structural tree models. Ann For Sci 57: 399–412 [Google Scholar]
- Stahl Y, Simon R. (2010) Plant primary meristems: shared functions and regulatory mechanisms. Curr Opin Plant Biol 13: 53–58 [DOI] [PubMed] [Google Scholar]
- Stoma S, Lucas M, Chopard J, Schaedel M, Traas J, Godin C. (2008) Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLoS Comput Biol 4: e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benková E, Swarup R, Casimiro I, Péret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. (2008) The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA. (2013) SUBA3: a database for integrating experimentation and prediction to define the subcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GT, Hedden P, Bhalerao R, Bennett MJ. (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10: 625–628 [DOI] [PubMed] [Google Scholar]
- Van Landeghem S, De Bodt S, Drebert ZJ, Inzé D, Van de Peer Y. (2013) The potential of text mining in data integration and network biology for plant research: a case study on Arabidopsis. Plant Cell 25: 794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA. (2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 110: 12840–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Tamayo KP, Gutierrez RA. (2010) Gene networks for nitrogen sensing, signaling, and response in Arabidopsis thaliana. Wiley Interdiscip Rev Syst Biol Med 2: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J, Evers JB, Buck-Sorlin GH, Andrieu B, Chelle M, de Visser PH. (2010) Functional-structural plant modelling: a new versatile tool in crop science. J Exp Bot 61: 2101–2115 [DOI] [PubMed] [Google Scholar]
- Wells DM, Laplaze L, Bennett MJ, Vernoux T. (2013) Biosensors for phytohormone quantification: challenges, solutions, and opportunities. Trends Plant Sci 18: 244–249 [DOI] [PubMed] [Google Scholar]
- Willis RC, Hogue CW (2006) Searching, viewing, and visualizing data in the Biomolecular Interaction Network Database (BIND). Curr Protoc Bioinformatics Chapter 8: Unit 8-9 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won SK, Lee YJ, Lee HY, Heo YK, Cho M, Cho HT. (2009) Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol 150: 1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Liao H, Beebe S, Blair M, Lynch J. (2004) QTL mapping of root hair and acid exudation traits and their relationship to phosphorus uptake in common bean. Plant Soil 265: 17–29 [Google Scholar]
- Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. (2004) Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics 20: 3594–3603 [DOI] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]