Expression of a chloroplast-encoded splicing factor is regulated on the level of mRNA stability and translation, including a possible autoregulatory loop.
Abstract
The chloroplast genome of land plants contains only a single gene for a splicing factor, Maturase K (MatK). To better understand the regulation of matK gene expression, we quantitatively investigated the expression of matK across tobacco (Nicotiana tabacum) development at the transcriptional, posttranscriptional, and protein levels. We observed striking discrepancies of MatK protein and matK messenger RNA levels in young tissue, suggestive of translational regulation or altered protein stability. We furthermore found increased matK messenger RNA stability in mature tissue, while other chloroplast RNAs tested showed little changes. Finally, we quantitatively measured MatK-intron interactions and found selective changes in the interaction of MatK with specific introns during plant development. This is evidence for a direct role of MatK in the regulation of chloroplast gene expression via splicing. We furthermore modeled a simplified matK gene expression network mathematically. The model reflects our experimental data and suggests future experimental perturbations to pinpoint regulatory checkpoints.
Two classes of genes dominate the chloroplast genome of land plants: those coding for components of the photosynthetic apparatus and those encoding components of the chloroplast gene expression system. The latter class is represented by a larger number of genes for ribosomal proteins, several genes for subunits of the plastid-encoded RNA polymerase, and genes for tRNAs and ribosomal RNAs. In addition, there is a single gene for a protein involved in splicing, designated maturase K (matK). matK encodes a protein with homology to bacterial intron maturases. matK, like its bacterial relatives, is located within an intron, in this case within the tRNA-K (trnK) precursor RNA (Neuhaus and Link, 1987). This intron belongs to a class known as the group II introns, which are characterized by six secondary structure domains (Michel et al., 1989; Lambowitz and Zimmerly, 2004). Maturases are almost always found within domain IV of group II introns. The bacterial maturases are multifunctional proteins that are required for splicing as well as for making their intron mobile (Lambowitz and Zimmerly, 2011). They are with few exceptions specific for their own intron (Carignani et al., 1986; Anziano et al., 1990; Anziano and Butow, 1991; Lambowitz and Zimmerly, 2004) and are capable of down-regulating their own expression (Singh et al., 2002). In the case of MatK, however, genetic and phylogenetic evidence suggests that it aids in splicing multiple introns (Wolfe et al., 1992; Hess et al., 1994; Hübschmann et al., 1996; Vogel et al., 1999; Funk et al., 2007; Duffy et al., 2009; Gao et al., 2009; McNeal et al., 2009). Binding studies indicated an association with the trnK intron at least in vitro (Liere and Link, 1995). Recently, it was shown that MatK associates in vivo with seven group II introns, including the trnK intron (Zoschke et al., 2010).
Splicing in land plant chloroplasts is carried out by a multitude of proteins, many of which have been characterized over the last 15 years (Khrouchtchova et al., 2012). Among these splicing factors, MatK is distinguished because it is the only one expressed from the chloroplast genome. All other factors are nucleus encoded. The matK gene has an unbroken chloroplast heritage in land plants and, in fact, can also be found in green algae sister groups of embryophytes (Turmel et al., 2006). MatK is a standard molecular marker for phylogenetic studies and has been amplified from tens of thousands of plant species (CBOL Plant Working Group, 2009). It has been lost in parasitic species of the genus Cuscuta (Funk et al., 2007; McNeal et al., 2009) and a parasitic orchid (Delannoy et al., 2011). These plants have also lost the group II introns shown to be associated with the MatK protein, thus making the matK gene dispensable.
Little is known about the expression characteristics of matK or of its intron targets, information that is necessary to appreciate a potential regulatory role of this peculiar splicing factor. Therefore, we set out to perform a comprehensive expression analysis of MatK and its target RNAs in order to assess possible checkpoints within this small gene expression network. Furthermore, we developed a mathematical model that was fit to our experimental data. Theoretical predictions derived from simulations using this model indicate that MatK plays a critical role in the splicing of intron-containing chloroplast tRNA genes early in development. Moreover, the model implies an autoregulatory feedback mechanism required for the idiosyncratic accumulation kinetic of MatK. Testable predictions made by the model are discussed.
RESULTS
MatK Protein Accumulation Is Inversely Correlated with matK mRNA Levels during Early Tobacco Development
It was previously shown by western analysis of protein extracts prepared from tobacco (Nicotiana tabacum) plants of different ages that MatK displays a complex accumulation pattern across tobacco development (Zoschke et al., 2010).
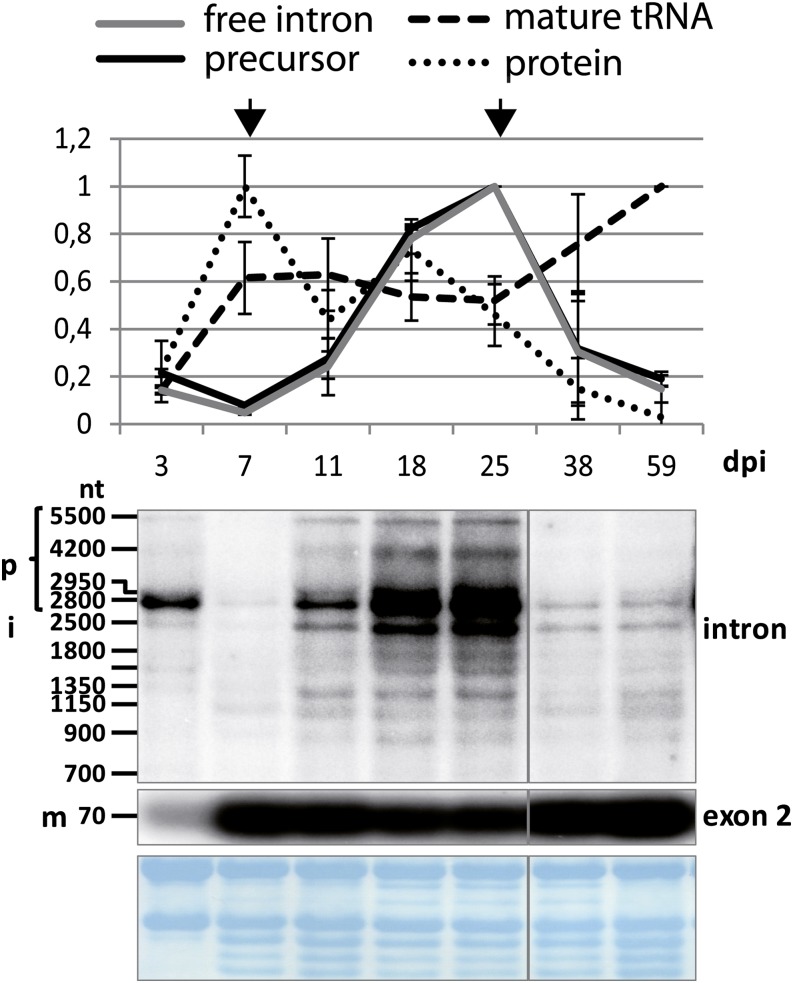
The main peak of MatK protein accumulation is found at day 7 after imbibition. Here, we extracted RNA from samples of the same age groups used in this previous study to analyze protein accumulation. For plant ages of 3 to 38 d, we used whole seedlings for RNA extraction, excluding roots. For 59-d-old plants, only leaf tissue from mature leaves was used. The samples are from plants that had been grown in parallel to those used for protein analyses published previously (Zoschke et al., 2010). With these samples, we performed RNA gel-blot hybridization using probes located within the matK reading frame and within exon 2 of tRNA-K(UUU) (Fig. 1). The signals obtained were scored based on their length and whether they are detected with intron- or exon-specific probes. Thus, we identified unspliced precursor RNAs (larger than 2.5 kb; the unspliced tRNA including the matK reading frame is approximately 2,600 nucleotides long), free intron RNA, and the mature tRNA. The autoradiographs of two biological replicates were quantified (Fig. 1). The quantification demonstrates that the levels of the mature, spliced tRNA-K(UUU) increase early to reach a plateau after only 7 d of development. By contrast, the precursor RNAs show a more complex accumulation pattern, with a main peak in 25-d-old seedlings and a minimum at 7 d. The band at 2.5 kb, presumably the free intron, shows the same pattern as the precursor, suggesting that there is no independent regulation of these two transcript types. Both tRNA precursor and free intron are putative templates for the translation of the matK reading frame. Therefore, we compared the accumulation of these trnK mRNAs with the accumulation of the MatK protein determined previously (Fig. 1, dotted graph). Initially, both mRNA and protein levels start out in parallel (0–3 d post imbibition [dpi]). At days 7 and 11, the two curves show an inverse correlation. In fact, peak MatK protein accumulation coincides with the lowest matK mRNA accumulation at 7 d of tobacco development. Later, mRNA levels increase until 25 d of development, while protein levels decline after day 18. This suggests that the translation or protein stability of MatK is independently and, at least during early development, inversely regulated relative to mRNA levels.
Figure 1.
RNA gel-blot analysis of matK across tobacco development. Total RNA (5 μg per lane) from seedlings of different ages (dpi) and from leaves of older plants was extracted, separated on an agarose gel, blotted to a nylon membrane, and hybridized with different radioactive probes. RNA gel-blot analyses with an exon and an intron probe were carried out. Bands that could be identified as unspliced precursor RNAs (p), free intron (i), and mature, spliced RNA (m) are indicated on the left. nt, Nucleotides. Probe positions for each autoradiogram are indicated on the right. The labeled bands were densitometrically quantified on a phosphorimager. Values were normalized to the top value of all seedling samples, which was arbitrarily set to 1. These values are displayed in the chart shown at top (black graph = precursor RNA; gray graph = free intron; dashed graph = mature tRNA). The experiment was replicated with independent RNA samples (data not shown), which allowed the determination of sd values shown here. As a loading control, blots were stained with methylene blue prior to hybridization; this is shown below the autoradiograms. Protein values (dotted line) are taken from western analyses published previously (Zoschke et al., 2010). Arrows indicate the largest discrepancies in the dynamics of protein and mRNA levels. [See online article for color version of this figure.]
RNA Stability of matK Increases in Mature Tobacco Seedlings
The changes identified in the accumulation of tRNA-K(UUU) precursors (i.e. matK mRNA) prompted us to investigate whether they are due to changes in transcription or to changes in RNA stability. We used run-on transcription assays to analyze transcription rates in 7- and 25-d-old plants, because these represent the lowest and highest points of matK mRNA accumulation in tobacco development (Fig. 1). Equal amounts of chloroplasts were used for each experiment. Two biological replicates (independent isolations of chloroplasts) were performed, and results from one of these replicates are shown in Figure 2A.
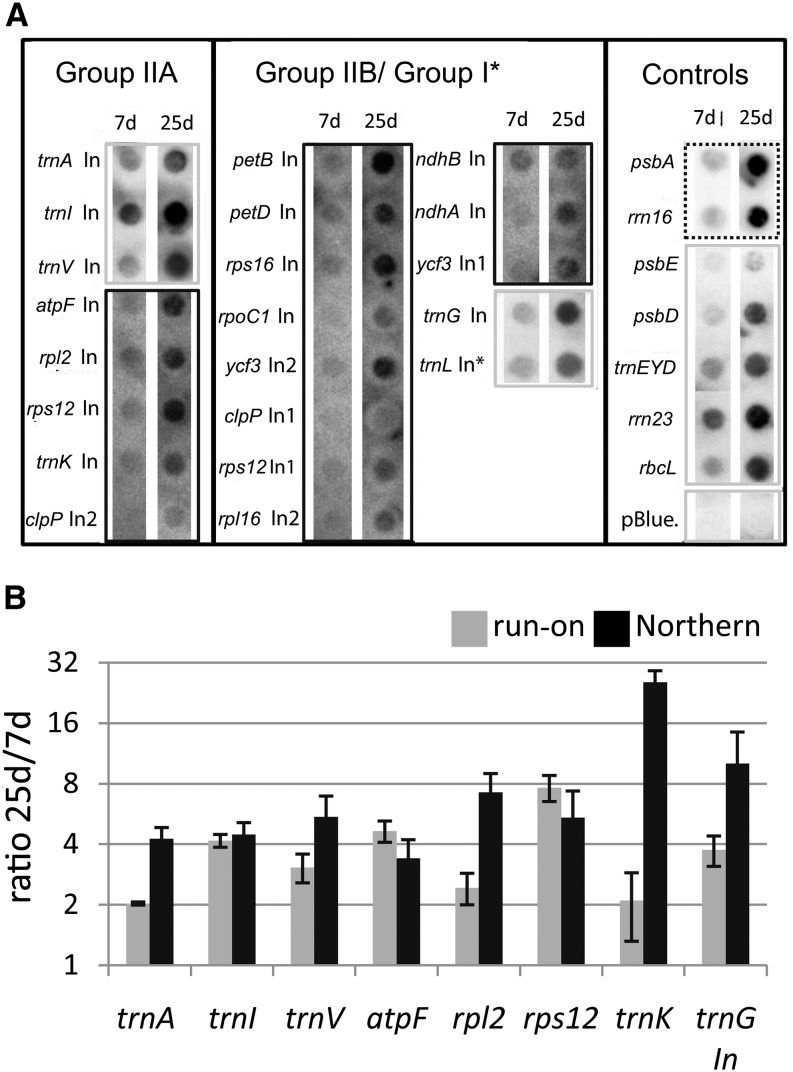
Figure 2.
Run-on transcription analysis of all intron-containing genes in the chloroplast and additional controls. A, RNA was extracted from run-on reactions, which were carried out with chloroplasts purified from 7- and 25-d-old plants and hybridized on macroarrays. The array comprised probes for all chloroplast introns (In) plus additional controls, including a negative control (pBlue., pBluescript II SK+). Probes were spotted as duplicates (only one spot shown). The signals shown here are examples of two biological replicates. The data were used to generate the charts in B. For better visualization, different exposure times are shown for different probes (dashed line = short; gray lines = intermediate; black lines = long). B, Comparison of transcription rates and RNA steady-state levels in 25- versus 7-d-old material. mRNA levels were assayed by northern blot and quantified from two independent experiments (gray bars). Transcription rates were assayed by run-on analysis and quantified from two independent assays (black bars; see A). Note the logarithmic scale of the y axis. Assayed genes are indicated below. sd values were derived from two biological replicates.
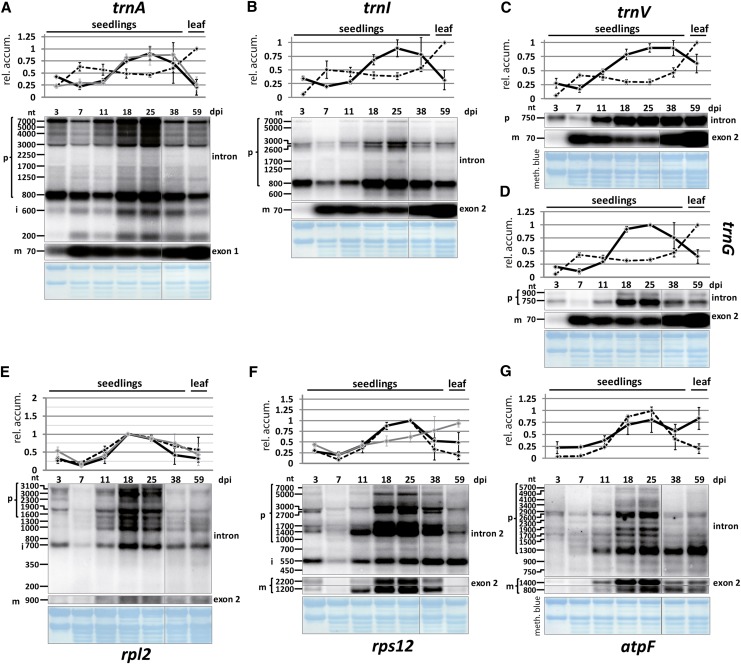
Radiolabeled run-on RNA was hybridized to probes representing all chloroplast introns as well as a number of intron-free chloroplast RNAs. Even inspection of the autoradiographs by eye reveals an overall increase in signal in the mature plant sample. This becomes even clearer when the results are quantified and the ratios of run-on transcription signals in 25-d-old plants versus 7-d-old plants are displayed for the MatK-dependent introns (Fig. 2B). Changes in transcriptional activity vary between 2-fold for trnK and trnA and almost 8-fold for rps12. In order to compare run-on data with changes in steady-state levels, we performed RNA gel-blot analyses of all seven known MatK target RNAs (trnK, trnA, trnI, trnV, ribosomal proteinL2 (rpl2), rps12, and atpF; Fig. 3, A–C and E–G) and one non-MatK target group II intron control (trnG; Fig. 3D). We used the same RNA samples as used to investigate the accumulation of trnK and matK (Fig. 1). As for matK, we identified precursor RNAs, spliced RNAs, and, where possible, free introns. The pattern for all investigated tRNAs is similar to tRNA-K (UUU) in that the precursors show a pronounced peak at day 25 of development, whereas the mature tRNAs reach a plateau early during development without much further change. The unspliced mRNA precursors of rpl2, rps12, and atpF also display maximal accumulation on days 18 and 25 of tobacco development. In contrast to tRNAs, spliced mRNAs closely follow the behavior of unspliced precursor RNAs across development. For rpl2, the free intron displays this behavior as well, while for rps12, the presumably free intron 2 shows an increase in accumulation until the end point of the measurements done here. The rps12 intron 2 is the only detected intron here that displays such unusual accumulation behavior, which suggests an active stabilization of this RNA species. Overall, the ratio of spliced to unspliced RNAs is constant for mRNAs across tobacco development but appears regulated for tRNAs.
Figure 3.
RNA gel-blot analysis of MatK target RNAs and trnG across tobacco development. Total RNA (5 μg per lane) from seedlings of different ages (dpi) and from leaves of older plants were extracted, separated on an agarose gel, blotted to a nylon membrane, and hybridized with different radioactive probes. Each panel includes two RNA gel-blot analyses with an exon and an intron probe. Bands that could be identified as unspliced precursor RNAs (p), free intron (i), and mature, spliced RNA (m) are indicated on the left. nt, Nucleotides. Probe positions for each autoradiogram are indicated on the right. The labeled bands were densitometrically quantified on a phosphorimager. Values were normalized to the top value of all seedling samples, which was arbitrarily set to 1. These values are displayed in the charts shown at the top of each panel (black graphs = precursor RNAs; gray graphs = free introns; dashed graphs = mature RNAs). The experiment was replicated with independent RNA samples (data not shown), which allowed the determination of sd values shown here. As a loading control, blots were stained with methylene blue prior to hybridization; this is shown below the autoradiograms. A to D, Probes for tRNAs. E to G, Probes for mRNAs. [See online article for color version of this figure.]
We next used data points for 7- and 25-d-old plants from RNA gel-blot hybridization experiments and calculated the 25-d/7-d ratios of the precursor RNA signals for each probe (Fig. 2B). Overall, RNA accumulation ratios resemble ratios from run-on transcription experiments. Both transcription and RNA accumulation rise in mature tissue for the genes investigated. For most genes, the increase in transcription activity and the increase in RNA accumulation are equal or differ maximally by a factor of 2. By contrast, RNA accumulation of the trnK precursor increases 29-fold in the mature tissue, while transcriptional activity only increases 2-fold (Fig. 2B). This suggests that trnK precursor RNA is stabilized in mature tissue. Alternatively, reduced splicing of the precursor in mature tissue increases precursor abundance. In any case, trnK is the most strongly regulated gene in the set investigated here.
Dynamic Association of MatK with Different Target RNAs across Tobacco Development
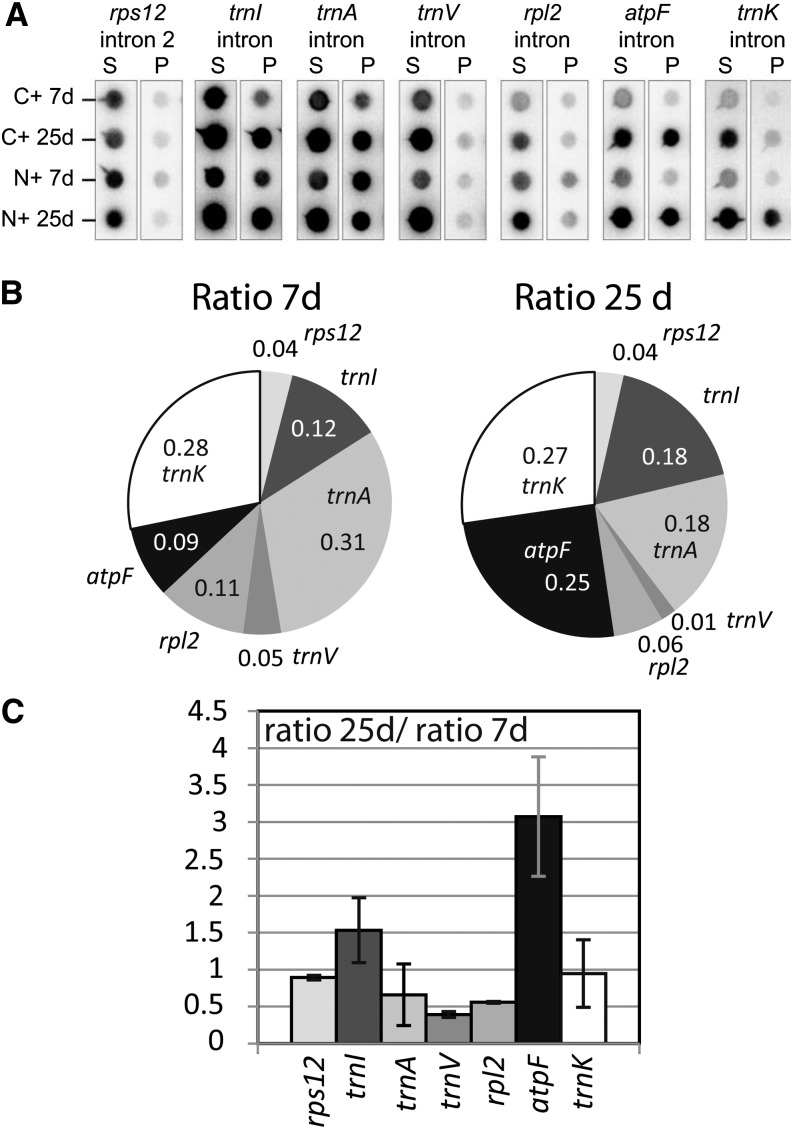
It has been shown that the RNA-binding activity of many RNA-binding proteins is regulated (Dreyfuss et al., 2002). This has also been demonstrated for several nuclear splicing factors (Manley and Tacke, 1996; Huang et al., 2007). In chloroplasts, the RNA affinity of certain RNA recognition proteins depends on posttranslational modifications in vitro (Lisitsky and Schuster, 1995). With our interest in the regulation of MatK, we sought to determine the association of MatK with its target introns in two developmental stages. We isolated chloroplasts from 7- and 25-d-old plants carrying an engineered version of MatK with a hemagglutinin (HA) epitope tag either at the N or C terminus (transplastomic plants described by Zoschke et al. [2010]). The tagged MatK protein was immunoprecipitated from stroma extracts with an anti-HA antibody (Supplemental Fig. S1). RNA was extracted from precipitated and supernatant fractions of the immunoprecipitation and dot blotted onto a nylon membrane. These membranes were hybridized with radiolabeled probes for the seven known target introns of MatK (Fig. 4A). The experiment was carried out starting from two independent extracts of each N-terminally and C-terminally tagged MatK:HA plants. Signals obtained after hybridization were quantified using a phosphorimager. While it is difficult to compare the absolute values between pellet signals within individual experiments due to differences in probe lengths and base composition, we can easily score relative enrichment within one developmental stage and also the relative signal changes in the pool of MatK-associated RNAs of 7- and 25-d-old plants. To do so, we calculated the mean pellet-to-supernatant ratio for the two replicate experiments for each probe. This ratio indicates the enrichment of a particular RNA in MatK immunoprecipitation experiments. The ratios are displayed for each of the two developmental stages in a pie chart as fractions of the sum of the ratios of all MatK targets (Fig. 4B). We call these the relative enrichment ratios. They allow scoring differences in immunoprecipitation results within each developmental stage. For example, at day 7, the most strongly enriched intron is the one in trnA, followed by the trnK intron. The least enriched intron is intron 2 in the rps12 mRNA precursor. The atpF intron is only number five in the list of enrichment ratios at day 7. This changes toward day 25, when enrichment of the atpF intron increases and is only topped by the trnK intron. To visualize and quantify such changes better, we plotted the ratio of relative enrichment ratios at 25 d over 7 d in Figure 4C. This shows that enrichment of the atpF mRNA increases 3-fold in the mature seedlings, which marks the largest change of all MatK-associated introns. Other RNAs show a reduced enrichment, like trnV and the rpl2 mRNA. These changes do not correlate with changes in transcript abundance between the two growth stages (Fig. 2B). In sum, this analysis shows that (1) MatK associates with different affinity to different introns and (2) affinities change over tobacco development.
Figure 4.
Association of MatK with its target RNAs changes across tobacco development. A, Dot-blot analysis of the RNA from pellet (P) and supernatant (S) fractions of MatK:HA immunoprecipitations from the stroma of 7- and 25-d-old tobacco seedlings. C+, Plants tagged at the C terminus of MatK; N+, plants tagged at the N terminus of MatK. B, The ratios of pellet and supernatant signals were calculated and summarized for experiments with N+ and C+ plants. Means of these experiments are represented in a pie chart as ratios of the sum of all MatK targets. C, The ratio to ratio (25 d/7 d) is indicative of the developmental dependency of RNA-MatK interactions. This chart was derived from the experiment shown in A with immunoprecipitations from C- and N-terminally tagged plants considered as replicate experiments (error bars show sd). RNAs with values below 1 show a decrease in the association with MatK:HA in 25-d-old plants when compared with 7-d-old plants; values above 1 are the opposite.
A Mathematical Model Accurately Reflects the Characteristics of the MatK Gene Expression Network
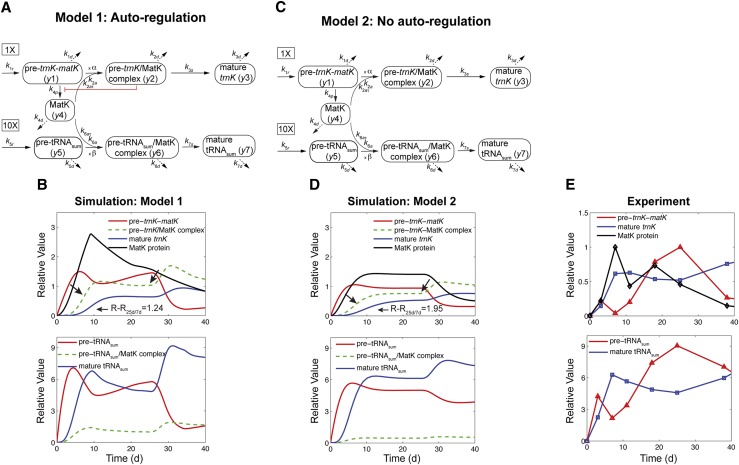
The experimental data have shown so far that MatK associates with different RNA precursors forming pre-RNA/MatK complexes, including pre-trnK/MatK complexes (Zoschke et al., 2010), MatK protein accumulation varies inversely with changes in the matK mRNA levels (Fig. 1), and there is no correlation between levels of the spliced trnK and the matK (trnK precursor) RNA (Fig. 1). What mechanisms cause these observed inverse correlations? We emphasize that such a question cannot be answered intuitively but requires mathematical models. Here, we developed two models of the regulation of matK expression (Fig. 5). In model 1 (Fig. 5A), we assumed that MatK down-regulates its own production through a negative feedback loop mediated by pre-trnK/MatK repression complexes, in light of the fact that direct autoregulation exists for bacterial maturases (Singh et al., 2002). In model 2 (Fig. 5C), we considered the case that there is no such feedback regulation of matK translation to analyze whether this model can still explain the observed expression dynamics. Both models were built with the intention to use a minimum number of variables and parameters that are nevertheless capable of explaining the dynamics within the matK system. Eventually, a model is aimed to identify important regulatory points. To set the stage for our theoretical investigation, we first state our assumptions, also using the simplified models as outlined in Figure 5, A and C.
Figure 5.
Model 1 with autoregulation of MatK reproduces the observed expression dynamics. Two possible models of matK gene expression were analyzed with respect to how well both models fit to our experimental data. A, Reaction scheme of the tRNA-MatK splicing network for model 1. The MatK transtargets trnA, trnV, and trnI are lumped into one tRNA species assigned as tRNAsum. The values of trnA accumulation (Fig. 3A) were used to describe the expression dynamics of tRNAsum, and tRNAsum is assumed to be 10-fold more abundant than trnK (for details, see “Results”). k1r and k5r are the transcription rates of the tRNA precursors pre-trnK-matK (y1) and pre-tRNAsum (y5), respectively. The protein MatK (y4) is encoded within the intron of the trnK-matK precursor gene and translated with the rate k4p. As the levels of the tRNA precursors y1 and y5 as well as the MatK protein y4 increase, they form pre-trnK/MatK (y2) and pre-tRNAsum/MatK complexes (y6), respectively. The pre-trnK/MatK repression complex (y2) inhibits the translation of matK via a negative feedback loop (red line with a blunt end). The subscript τ in k2aτ and k6aτ is the delay introduced in the models to signify the time taken for the formation of pre-trnK/MatK and pre-tRNAsum/MatK complexes at day 25 of tobacco development. Within the tRNA/protein complexes, MatK splices the introns with rates k3s and k7s, leading to spliced tRNAs: mature trnK (y3) and mature tRNAsum (y7). Competition of pre-trnK-matK (y1) and tRNAsum (y5) for MatK (y4) is assigned by the factors 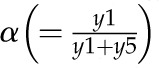 and
and 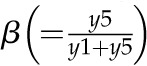 . To simplify the model, the involvement of nucleus-encoded splicing factors was not considered. Complex dissociation was assumed to be slow and, therefore, was neglected. Dashed arrows represent the degradation of RNAs, complexes, and the MatK protein with rates k1d to k7d. B, Accumulation of tRNA precursors, mature tRNAs, MatK protein, and tRNA/MatK complexes during tobacco development, derived from theoretical predictions of model 1 in comparison with northern-blot and western-blot analyses (E). Model 1 (top) fits certain experimental data displayed in E, such as the inverse correlation of MatK expression and matK mRNA (pre-matK-trnK) accumulation around day 7, the sharp decline of the MatK protein level after day 7, and the enrichment of pre-trnK/MatK complexes in mature seedlings. The model consistently reproduces the ratio-to-ratio value between pre-trnK/MatK complexes at days 25 and 7 (RR25d/7d = 1.24), indicated with black arrows. Model 1 (bottom) shows temporal expression profiles of tRNAsum (trnA, trnV, and trnI). The days of maximal accumulation of tRNAsum are the same as those for trnK and those experimentally observed. C, Model 2 shows the same reaction scheme depicted in A, but negative autoregulation is removed from the system. D, Accumulation of tRNA precursors, mature tRNAs, MatK protein, and tRNA/MatK complexes during tobacco development derived from theoretical predictions of model 2 (C) in comparison with northern-blot and western-blot analyses (E). Model 2 (without autoregulation) fails to capture the distinctive expression dynamics of matK mRNA (pre-trnK-matK) and MatK protein. E, Experimental data for the accumulation of tRNA precursors, mature tRNAs, and MatK protein during tobacco development derived from northern-blot and western-blot analyses. The accumulation of MatK is plotted relative to the maximum obtained 7 dpi, while the abundance of the RNA precursor (pre-trnK-matK) and mature trnK is plotted relative to the maximum 25 and 59 dpi, respectively (set to 1). Reference parameters of the reaction kinetics of model 1 and model 2 are given in “Materials and Methods.” Note the different scaling.
. To simplify the model, the involvement of nucleus-encoded splicing factors was not considered. Complex dissociation was assumed to be slow and, therefore, was neglected. Dashed arrows represent the degradation of RNAs, complexes, and the MatK protein with rates k1d to k7d. B, Accumulation of tRNA precursors, mature tRNAs, MatK protein, and tRNA/MatK complexes during tobacco development, derived from theoretical predictions of model 1 in comparison with northern-blot and western-blot analyses (E). Model 1 (top) fits certain experimental data displayed in E, such as the inverse correlation of MatK expression and matK mRNA (pre-matK-trnK) accumulation around day 7, the sharp decline of the MatK protein level after day 7, and the enrichment of pre-trnK/MatK complexes in mature seedlings. The model consistently reproduces the ratio-to-ratio value between pre-trnK/MatK complexes at days 25 and 7 (RR25d/7d = 1.24), indicated with black arrows. Model 1 (bottom) shows temporal expression profiles of tRNAsum (trnA, trnV, and trnI). The days of maximal accumulation of tRNAsum are the same as those for trnK and those experimentally observed. C, Model 2 shows the same reaction scheme depicted in A, but negative autoregulation is removed from the system. D, Accumulation of tRNA precursors, mature tRNAs, MatK protein, and tRNA/MatK complexes during tobacco development derived from theoretical predictions of model 2 (C) in comparison with northern-blot and western-blot analyses (E). Model 2 (without autoregulation) fails to capture the distinctive expression dynamics of matK mRNA (pre-trnK-matK) and MatK protein. E, Experimental data for the accumulation of tRNA precursors, mature tRNAs, and MatK protein during tobacco development derived from northern-blot and western-blot analyses. The accumulation of MatK is plotted relative to the maximum obtained 7 dpi, while the abundance of the RNA precursor (pre-trnK-matK) and mature trnK is plotted relative to the maximum 25 and 59 dpi, respectively (set to 1). Reference parameters of the reaction kinetics of model 1 and model 2 are given in “Materials and Methods.” Note the different scaling.
First, we deliberately based our model exclusively on chloroplast genetic information to test whether we still could arrive at an output that is congruent with our experimental data. We do not dispute that nucleus-encoded factors are essential for all steps of chloroplast gene expression, including splicing, and that these factors can be expected to also exert regulatory control over most individual gene expression steps. However, to date, there is little evidence for nuclear factors actively regulating chloroplast gene expression (Raynaud et al., 2007; Boulouis et al., 2011), and chloroplast autoregulation has been observed for several chloroplast genes (Choquet et al., 2001; Ramundo et al., 2013). Second, we disregarded the mRNA targets of MatK, such as atpF, rpl2, and rps12, and focused only on tRNA-MatK interactions (hereafter called the tRNA-MatK splicing network), because these mRNAs are expressed to a much lower level than the tRNAs. In particular, trnI and trnA are expressed as part of the ribosomal RNA operon, which has been shown to have the highest transcription levels of all genes in the plastid genome (Legen et al., 2002; Nakamura et al., 2003). By comparison, rps12 and rpl2 transcription and mRNA levels are much lower, close to the detection level (Legen et al., 2002). Third, we assumed that the formation of tRNA/MatK complexes is a prerequisite for splicing, since a role of MatK for splicing of these introns is very likely (Zoschke et al., 2010). Dissociation constants of other group II intron-binding factors are low, and complex recycling is inferred to occur slowly (Saldanha et al., 1999; Wank et al., 1999; Rambo and Doudna, 2004; Ostersetzer et al., 2005). Fourth, accordingly, we assumed the same kinetics for tRNA/MatK complexes and neglected any complex dissociation, as there are neither data on intron affinities (kD) for the group II intron-binding factor MatK available nor are turnover rates or functions of free introns known. Fifth, we assumed that the tRNAs trnA, trnV, trnI, and trnK compete for MatK. Sixth, since the levels of trnA, trnV, and trnI show similar dynamics, we lumped them into one tRNA species, tRNAsum, and chose the values of RNA accumulation of trnA (Fig. 3A) as a representative example that describes the dynamics of tRNAsum. Because of known differences in the expression levels between trnA, trnV, trnI, and trnK, we calculated tRNAsum as 10-fold more abundant than trnK in our model. Seventh, the processes that cause the observed increase of the matK mRNA levels until 25 d of development and their subsequent decline are unclear at present. However, our data on matK (trnK precursor) RNA and MatK protein accumulation point to the putative rate-limiting role of nucleus-encoded splicing factors at the later stage of development when MatK is no longer rate limiting (see “Discussion and Conclusion”). Correspondingly, we modeled this transition in the form of a delay in tRNA/MatK complex formation. This has the advantage of not having to specify all the processes explicitly and thus reduced the numbers of effective variables and parameters to be fixed.
In both models (Fig. 5, A and C), the variables y1 and y5 represent the concentration of the unspliced tRNAs: pre-trnK-matK and pre-tRNAsum, respectively. The variable y4 represents the concentration of the MatK protein. MatK (y4) binds to pre-trnK-matK (y1) and pre-tRNAsum (y5) transcripts, leading to pre-trnK/MatK (y2) and pre-tRNAsum/MatK complexes (y6), respectively. y3 describes the concentration of mature (spliced) trnK transcripts, and y7 describes the amount of mature tRNAsum transcripts.
For model 1, the dynamics of these variables is described by the following system of differential equations:
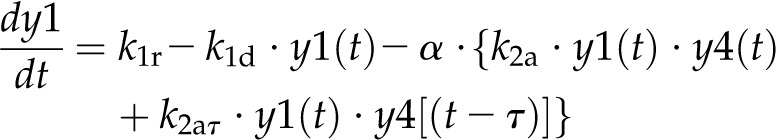 |
(1) |
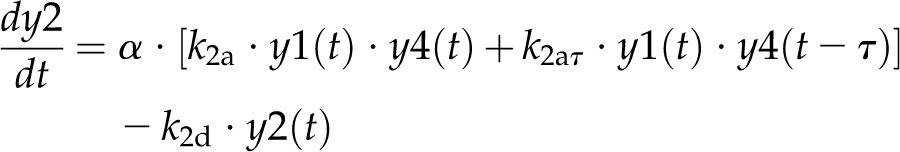 |
(2) |
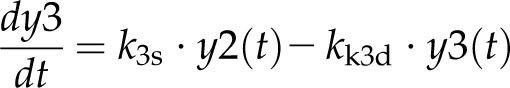 |
(3) |
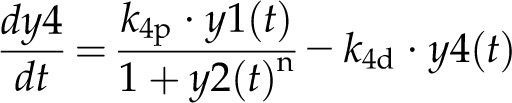 |
(4) |
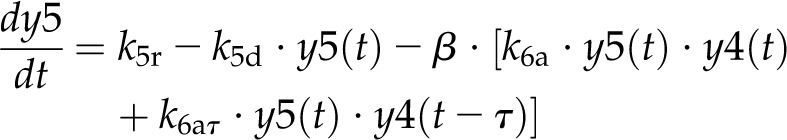 |
(5) |
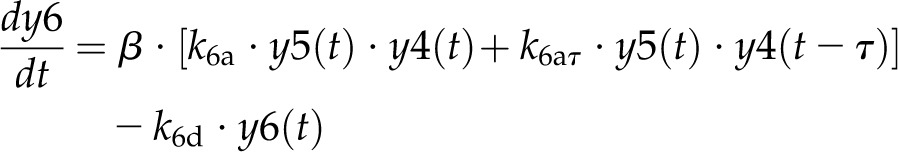 |
(6) |
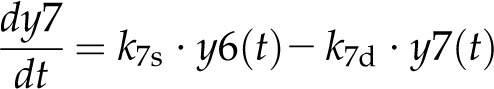 |
(7) |
with
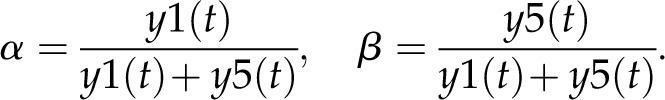 |
(8) |
Here, the various rate constants parameterize transcription (k1r, k5r), degradation (k1d–k7d), translation (k4p), complex formation (k2a/k2aτ, k6a/k6aτ), and splicing (k3s, k7s). The subscript τ in k2aτ and k6aτ is the delay introduced in the model to signify the time taken for the formation of pre-trnK/MatK and pre-tRNA/MatK complexes at the later stage of development. To simulate the competition of tRNA precursors for MatK, we include the factors α and β (Eq. 8; see also Eqs. 1, 2, 5, and 6). A Hill function with the coefficient n was used to describe the translational repression term of the MatK protein (Eq. 4).
In model 2 (Fig. 5C), Equations 1 to 3 and 5 to 8 are the same, but we took autoregulation out of Equation 4 to analyze whether this model can still explain the observed expression dynamics. Equation 4 changes as follows:
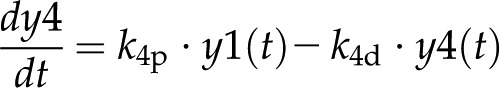 |
(9) |
We determined the parameters of both models using an optimization procedure that fit model 1 and model 2 to our experimental data, including values for RNA accumulation of the trnK-matK precursor, the spliced trnK tRNA taken from northern-blot analyses shown in Figure 1, and MatK protein values taken from previous western-blot analyses (Zoschke et al., 2010). The optimal parameters we obtained for both models are listed in “Materials and Methods.”
With autoregulation included (model 1), the dynamic behavior of the accumulation of unspliced and spliced transcripts observed later than 7 dpi (Fig. 1) was only achieved when a restart of complex formation at a later stage (25 dpi) of tobacco development was assumed (Fig. 5B). We modeled this delayed complex formation explicitly as delay differential equations (Eqs. 1, 2, 5, and 6). A model of delay differential equations enables us to group unknown biological processes together to account only for the time required for these processes to occur. Here, the unknown processes are those that cause the expression dynamics of tRNA precursor and mature tRNAs around 25 dpi. As shown in Figure 5, B and E, model 1 produces time courses quantitatively similar to the experimental ones (Figs. 1 and 3). Moreover, model 1 predicts the likely time course of the formation of pre-trnK/MatK repression complexes and pre-tRNAsum/MatK complexes. Notably, model 1 is also able to simulate the time point of maximum production of the MatK protein (Fig. 5, B and E), although we did not use any direct constraints to arrive at the characteristic shape of its accumulation. We also calculated the ratio to ratio (25 d/7 d) of the pre-trnK/MatK repression complex (R-R25d/7d = 1.2) and again found agreement with the experiments (Fig. 4C). Thus, we demonstrate that this minimal regulatory tRNA-MatK network is enough to explain our data.
The model without autoregulation (model 2) reflects the expression dynamics of mature tRNAs but fails to reproduce the sharp decline of the MatK protein levels around day 25 (Fig. 5, D and E). The observed inverse correlation with the maximum MatK protein accumulation coinciding with minimum matK RNA accumulation at 7 d of tobacco development is lost as well. trnK (matK) precursor RNA and MatK protein show instead a parallel expression dynamics. Their levels increase early in development, reach a plateau, and decline after 25 d. Furthermore, model 2 incorrectly predicts a 2-fold enrichment of pre-trnK/MatK complexes in the mature seedlings. Thus, the model without autofeedback regulation fails to fit the main characteristics of the MatK gene expression network.
Modeling Suggests That the Disruption of Autoregulation Leads to an Overaccumulation of MatK
The analysis of the two models suggests that an autoregulation like that in model 1 is able to explain the distinctive pattern of matK mRNA (trnK precursor RNA) and MatK protein accumulation. We next exploited this model as a predictor for what happens if we perturb the tRNA-MatK splicing system. We were most interested in the putative impact of autoregulation on the overall expression dynamics predicted by model 1. To analyze this, we removed the autoregulation from model 1 (see “Materials and Methods”). Please note that unlike for model 2 (Fig. 5, C and D), this simulation does not involve data fitting and optimization but instead predicts the expression dynamics of the tRNA-MatK network when negative autoregulation is absent/disrupted.
Experimentally, this prediction could be tested with transplastomic tobacco plants in which matK segments are ectopically overexpressed. If MatK indeed regulates its own translation, it would preferably interact with these overexpressed sequence elements. As a result, fewer MatK molecules would associate with the endogenous repression site in the trnK (matK) precursor RNA, causing a disturbance of the negative autoregulation.
With disrupted autoregulation, model 1 predicts a striking overaccumulation of MatK paralleled by a decline in trnK (matK) precursor RNA (Supplemental Fig. S2A). Both the peak of MatK protein levels at 7 d as well as the double peak of matK mRNA levels are lost. Moreover, unspliced tRNAs are sharply decreasing while mature tRNAs are increasing, which could be explained by increased splicing of tRNAs fostered by MatK and pre-trnK/MatK complexes. Most importantly, the simulation suggests that the autoregulatory feedback seems to prevent an overproduction of MatK.
Development-Dependent Complex Formation Could Ensure the Correct Timing of Intron Splicing
We also asked how increased MatK target tRNA levels (tRNAsum) would influence the expression dynamics of the tRNA-MatK splicing network. Constitutive overexpression was simulated through increasing the transcription rate of the pre-tRNAsum gene 10- and 20-fold (k5r in Eq. 5) while keeping the degradation rates (k1d in Eq. 1 and k5d in Eq. 5) of the tRNA precursors constant. Model 1 predicts an increase in the levels of tRNAsum/MatK complexes and mature tRNAsum (Supplemental Fig. S2B). Furthermore, elevated levels of tRNAsum transcripts cause increased accumulation of the MatK protein. An explanation for this finding is that MatK preferentially binds to the tRNAsum transcripts due to the excess of tRNA targets. Consequently, the MatK protein synthesis is no longer inhibited by pre-trnK/MatK repression complex formation. Interestingly, a delay in the maturation of trnK transcripts at the early stage of development is predicted for the tRNAsum overexpressors relative to the simulated wild type (Supplemental Fig. S2B). Because of this delay, we suppose that early pre-trnK/MatK repression complexes are essential to adjust the splicing of RNAs required for chloroplast translation.
In a next step, we investigated what effect the absence of early pre-trnK/MatK complexes would have on the splicing pattern of other MatK target tRNAs. As expected, the pre-trnK-matK level as well as the MatK protein abundance strongly increase, because much less pre-trnK (matK) RNAs are spliced (Supplemental Fig. S2C). The RNA is thus free for the translation of matK. More MatK protein is now available to bind to alternative MatK targets, here tRNAsum. As predicted by model 1, this results in a sharp increase of tRNAsum/MatK complexes such that spliced tRNA products already start to saturate at an early stage of plant development. Almost all pre-tRNAsum transcripts are used up. When they reach their minimum, MatK binds preferentially to the pre-trnK-matK precursors. As a result, the level of mature trnK tRNAs increases rapidly and greatly exceeds the simulated wild-type amount. Interestingly, this simulation strongly indicates that insufficient quantities of pre-trnK/MatK repression complexes at an early stage of development lead to the overproduction of MatK, which is potentially harmful for chloroplast development (see “Discussion and Conclusion”).
The absence of early tRNAsum/MatK complexes, as simulated in Supplemental Figure S2D, leads to excess quantities of unspliced tRNAsum precursors. They are predicted to form complexes with MatK at a later stage of development in order to finally be spliced very fast. By contrast, the pre-trnK-matK precursors are processed more and earlier in this simulated mutant. Later on, trnK maturation is again balanced due to enhanced maturation of the MatK trans-tRNA targets. Interestingly, earlier buildup of MatK/trnK repression complexes leads to reduced levels of MatK.
DISCUSSION AND CONCLUSION
MatK Gene Expression Is Regulated at the Level of Protein and RNA Accumulation
The unique position of MatK as the only chloroplast-encoded splicing factor conserved in land plant evolution prompted us to investigate its expression dynamics in plant development to get insights into how MatK might be regulated. We included the expression of six additional plastid genes in our analysis, because these genes are targets of the splicing factor MatK (Zoschke et al., 2010). Monitoring transcription, RNA accumulation, RNA processing, and protein accumulation is necessary to uncover potential regulatory steps in gene expression (i.e. steps where the different processes are not only determined by input of the proceeding process). In this way, we identified a striking discrepancy between the maximum amounts of MatK protein accumulation early in plant development (day 7) and the minimal matK mRNA levels at the same time. Most parsimoniously, our data suggest that the translation of MatK is induced actively at this stage, although increased protein stability would lead to the same results. Given that it is not easy to detect even steady-state amounts of MatK, it will be a challenge to differentiate between these two options in the future. Noteworthy, 7-d-old plants consist almost entirely of cotelydons, while at later stages, primary leaves emerge. As we find comparatively low RNA amounts for all transcripts tested at this stage (Figs. 1 and 3), a cotyledon-specific reduction in RNA accumulation has to be considered here. In any case, a strong accumulation of the MatK protein at early stages in plant development seems sensible, as this is a time when chloroplast biogenesis is in full gear. Since this requires plastid translation, MatK as an essential splicing factor for a set of tRNAs and two ribosomal protein-coding mRNAs is needed as well.
Besides effects on the protein level, we also found differences between the changes in matK transcriptional activity and changes in matK RNA accumulation over development. This effect stood out over the much more mild changes found for the other genes tested, suggesting that matK mRNA is specifically stabilized in mature tissue (Fig. 2B). PENTATRICOPEPTIDE REPEAT proteins are known to stabilize selected transcripts in the chloroplasts (Pfalz et al., 2009), although no information is available that this would be regulated in plant development. For the trnK/matK transcript, a 54-kD endonuclease has been shown to bind to the 3′ untranslated region and to mediate its processing (Nickelsen and Link, 1993). This or other proteins might contribute to the differential stabilization of the matK mRNA.
Age-Dependent Binding of MatK to Target Introns
To date, there is no evidence that chloroplast RNA splicing would be a rate-limiting step in gene expression in land plants (e.g. no correlation between splicing rates and the amount of the final gene product has been established for chloroplast RNAs). In a few cases, tissue-specific changes in the abundance of spliced to unspliced mRNAs have been found (Barkan, 1989; McCullough et al., 1992), but whether this is caused by altered splicing rates or via differential stability of spliced RNA versus precursor RNA was not shown. For tRNAs with introns, changes in splicing would directly impact the amount of the final gene product. Tissue-dependent changes in the ratio of spliced to unspliced transcripts have been described for the mustard (Sinapis alba) trnG intron (Liere and Link, 1994). Here, we show that precursor mRNAs and mature mRNAs display similar curves of accumulation during development, while tRNA precursors and mature tRNAs show striking differences (Figs. 1A and 3, A–D). All five mature tRNAs analyzed reach a plateau early during development, while precursor tRNA levels peak late, at 25 dpi. Whether this means that splicing rates decrease later in tobacco development or whether the precursors are stabilized remains to be determined. The observations of potential tissue-specific splicing described above imply changes in the activity of splicing factors. Previously, the amounts of the splicing factor CHLOROPLAST RNA SPLICING1 (CRS1) have been demonstrated to correlate with the splicing of its target intron in the atpF mRNA, incidentally also a target of MatK (Till et al., 2001). MatK levels are high in young tissues when atpF splicing rates are high as well (Fig. 1). Later in development, MatK levels have decreased dramatically, while atpF splicing rates are invariantly high. On first sight, this suggests that MatK might be responsible for rate limiting the splicing of atpF only at early stages of development, while later, nuclear factors like CRS1 might limit it. However, we have gone beyond a simple correlation of splicing factor/target introns and asked whether affinities of MatK for atpF and its other targets change during development. Intriguingly, the affinity for atpF increases 3-fold in mature tissue, while the association with most other introns remains stable or declines slightly (Fig. 4). This suggests that the binding of MatK to the atpF intron is a regulated process and that decreased MatK levels could be compensated by changes in RNA affinity. In the absence of indications for MatK protein modifications, it will be challenging to understand how such affinity changes could be induced. Again, nuclear factors might be responsible. Alternatively, modifications to MatK change affinities for RNAs and/or other splicing factors.
A Model of a Simplified MatK Gene Expression Network Reflects the Experimental Data with Great Precision
We present here a detailed analysis of the kinetics of transcription rates, RNA accumulation, RNA processing, product accumulation, and RNA-protein interactions. Identifying key steps inside such a diverse data set is difficult. Therefore, we used a mathematical approach to gain an improved understanding of the system. To the best of our knowledge, this is the first theoretical investigation applied to problems in organellar gene expression. We investigated two models. In model 1, we assumed that the MatK protein autoregulates its own production via pre-trnK/MatK repression complexes. In model 2, we assumed that there is no such autoregulation. Both models were fit to our experimental data. We found that only model 1 could capture the expression dynamics of MatK as well as the MatK targets, and even our simulated pre-trnK/MatK-complex ratio (25 d/7 d) was in perfect agreement with our experimental observations (Fig. 5). A particularly beneficial outcome from simulating of this system is that our mathematical model could produce predictions of the dynamic process of RNA-protein complex formation. Thus, our model predicts a first increase in the amount of RNA/protein complexes at the early stage of development and a second one at day 25. The second temporal peak formation of complexes was explicitly modeled as delay differential equations. Generally, observed dynamics that result from unknown biological processes can be lumped together into a form of time delay (Epstein, 1990). Here, the two different time points of maximum formation of tRNA/MatK complexes may account for a transition in the roles of MatK and nucleus-encoded splicing factors. Indeed, splicing in chloroplast was demonstrated to rely on a multitude of different factors. MatK is only one of them (Khrouchtchova et al., 2012). A simple interpretation is that MatK within the chloroplast “spliceosome” limits the splicing rate at the early stage of development, whereas nucleus-encoded splicing factors become rate-limiting later, when MatK protein production is steadily suppressed by pre-trnK/MatK complexes. The explicit delay could reflect the time lag of changes in the composition of RNA/protein complexes, with nucleus-encoded splicing factors becoming rate limiting for the splicing process in mature tissue and possibly replacing MatK as the limiting factor for the splicing process. This is a prediction that we propose to test by measuring the temporal variation of composition of the chloroplast splicing machinery on MatK-dependent introns. As more data become available, it will be possible to determine how to include nucleus-encoded splicing factors with intron target ranges overlapping that of MatK (e.g. RNC1, WHAT'S THIS FACTOR1, CRS1, WHIRLY1, and THYLAKOID ASSEMBLY8) into the network model.
Testable Predictions from the Mathematical Model
An intriguing feature of our analysis was that the fit of the model to our experimental data was dramatically improved by the assumption of MatK autoregulation. The importance of autoregulation is amenable to straightforward experimental testing, most directly by transferring the gene to the nucleus. Another interesting test of autoregulation would be in situ overexpression of partial matK sequences by stable chloroplast transformation. Such an overexpression should lead to a depletion of MatK repression complexes and, thus, would free matK mRNA for translation. One contact site of MatK has been shown to be located in the 5′ untranslated region of the matK reading frame (Zoschke et al., 2010), a potential site for autoregulation of intron maturases in general (Singh et al., 2002). More detailed studies on this binding site of MatK would help to choose the right fragment for overexpression. In bacteria, where a maturase in Lactobacillus lactis shows autoregulation, it has been speculated that overexpression of any maturase could be deleterious, because their reverse transcriptase and endonuclease activities could lead to nonspecific harmful effects in the bacterial transcriptome (Singh et al., 2002). However, domains for such activities have been lost in MatK. Nevertheless, MatK could interfere with the splicing of the other nontarget group II introns if overexpressed. Chloroplast introns are known to be degenerated and to deviate strongly from the group II consensus sequence, thus potentially allowing for promiscuous binding of splicing factors.
A surprising side effect of the simulated overexpression of pre-tRNAsum genes was that the maturation of trnK transcripts was slightly delayed, particularly at the early stage of development (Supplemental Fig. S2B, mature trnK). In contrast, missing quantities of early pre-tRNAsum/MatK complexes could counteract this delay and cause splicing to occur slightly earlier (Supplemental Fig. S2D, mature trnK). Most importantly, even insufficient amounts of early pre-trnK/MatK repression complexes gave rise to potentially harmful overproduction of MatK (Supplemental Fig. S2C). Therefore, we reason that early pre-trnK/MatK repression complexes appear to (1) ensure the accumulation of spliced tRNAs in time before they are needed for bulk photosystem translation and (2) prevent an excess of MatK proteins. The predicted delay in maturation indicates a developmentally persistent molecular defect in plants with reduced pre-trnK/MatK complexes within the first days of chloroplast development. Such mutants are supposed to be delayed in their development from germination onward. One potential additional role of MatK besides autoregulation, therefore, could be to ensure tight translation regulation in the chloroplast, starting from the very early developmental stage of the tobacco plant. We chose a minimal set of parameters that regulate chloroplast tRNA splicing via MatK. Additional components of the chloroplast MatK splicing mechanism almost certainly remain to be considered, but we believe that our minimal mathematical model is a significant step forward in understanding the role of MatK in the chloroplast splicing system. It aids experimental design and allows the identification of sensitive nodes in the network or the analysis of perturbation effects on the system. The detailed kinetics of RNA/protein complex formation has not been measured. Consequently, our model cannot be regarded as a precise quantitative model of the tRNA-MatK splicing network. However, our model can be extended stepwise by different kinetic mechanisms in order to gain deeper insights into the various underlying processes of MatK-dependent tRNA splicing in chloroplasts.
MATERIALS AND METHODS
Plant Material
Previously described transplastomic tobacco (Nicotiana tabacum ‘Petit Havana’) plants with in vivo HA epitope-labeled MatK and control plants with unlabeled MatK were grown in a growth chamber with a long-day regime (16 h of light, 300 μE, 27°C; 8 h of dark, 27°C; Zoschke et al., 2010). Plant material was harvested 3, 7, 11, 18, 25, 38, and 59 dpi (in each case, 1 h after the end of the dark period). Additionally dry seeds were harvested as preimbibition material.
RNA Gel-Blot Analyses and Run-On Transcription
Five micrograms of total RNA was used for RNA gel-blot analyses as described previously (Beick et al., 2008). PCR probes were radioactive body labeled with [α-32P]dCTP using the DecaLabel DNA labeling kit (Fermentas). Oligonucleotide probes were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas). The sequences of oligonucleotide probes and the primers used for the amplification of PCR probes are given in Supplemental Table S1. Chloroplasts were isolated from seedlings/plants 7/25 d after imbibition as described previously (Zoschke et al., 2010). A total of 5 × 107 chloroplasts were used for a 10-min in vivo run-on labeling of transcribed transcripts with [α-32P]UTP at 25°C following a prior published protocol (Zoschke et al., 2007). As described previously, labeled transcripts were hybridized to a custom macroarray with spotted PCR probes for all plastid introns and several control genes (Beick et al., 2008). Signals were detected and quantified using a phosphorimaging system and its software (Bio-Rad). Primers used for the amplification of PCR probes are shown in Supplemental Table S1.
RNA Coimmunoprecipitation and Dot-Blot Analyses
Isolated chloroplasts of 7- and 25-d-old seedlings and plants were lysed, and stroma fractions were separated by centrifugation as described previously (Zoschke et al., 2010). A volume of stroma fractions corresponding to a total protein content of 5 mg and 5 μL of anti-HA antiserum (H3663; Sigma-Aldrich) was used for each immunoprecipitation of HA epitope-labeled MatK with Dynabeads protein G (Invitrogen) following the manufacturer’s instructions. Dot-blot analyses of coprecipitated RNA of stroma from 7-/25-d old transplastomic seedlings/plants were carried out as previously described slot-blot analyses (Zoschke et al., 2010). Signals were quantified using a phosphorimager system and its software (Bio-Rad). Enrichment ratios of pellet signals over supernatant signals were calculated for two biological replicates (transplastomic tobacco lines with N- and C-terminal HA epitope-labeled MatK) and compared between experiments with 7-d-old seedlings and 25-d-old plants. Primers used for the amplification of PCR probes are shown in Supplemental Table S1.
Protein Protocols
The isolation of proteins from plant tissues was carried out as described (Barkan, 1998). The protein amount of HA epitope-labeled MatK was analyzed by SDS-PAGE and immunoblotting (anti-HA antiserum; Sigma-Aldrich). Quantifications of immunological analyses were carried out as described (Zoschke et al., 2010).
Computational Methods
Optimization Process
Model 1 and model 2 were implemented using Matlab (R2011b; Mathworks) with the dde23 solver. In order to investigate whether the models sufficiently explain the data, we fit these models to the measured data points (see “Results”) using a least-squares method. The cost function to be optimized is the sum of the least squares
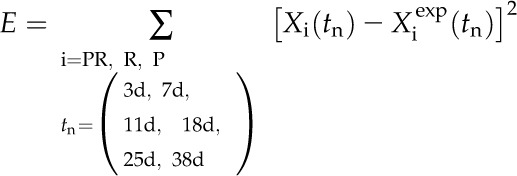 |
(10) |
where PR = precursor RNA, R = mature RNA, and P = protein.
We repeated the parameter search from three different initial conditions. For each tested model, three parameter sets were determined. An optimal parameter set was chosen from these three based on how a model fits best to the data points. The parameters of the optimal sets are given in Table I.
Table I. Parameters of the optimal sets.
| Parameters | Values |
Description | |
|---|---|---|---|
| Autoregulation Model (Model 1) | No Autoregulation Model (Model 2) | ||
| k1r | 0.35 d−1 | 0.34 d−1 | Transcription rate of matK-trnK precursor |
| k1d | 0.02 d−1 | 0.13 d−1 | Degradation rate of pre-trnK-matK RNA |
| k2a | 0.62 d−1 | 0.97 d−1 | Complex formation rate of pre-trnK/MatK |
| k2aτ | 3.79 d−1 | 8.02 d−1 | (Time-delayed) complex formation rate of pre-trnK/MatK |
| k2d | 0.29 d−1 | 0.29 d−1 | Decay rate of pre-trnK/MatK complex |
| k3s | 0.19 d−1 | 0.19 d−1 | Splicing rate for mature trnK |
| k3d | 0.30 d−1 | 0.26 d−1 | Degradation rate of mature trnK |
| k4p | 0.33 d−1 | 0.47 d−1 | Translation rate of MatK |
| k4d | 0.04 d−1 | 0.32 d−1 | Degradation rate of MatK protein |
| n | 90 | – | Hill coefficient of the inhibition of MatK translation |
| k5r | 3.51 d−1 | 2.17 d−1 | Transcription rate of MatK trans-tRNA target (tRNAsum precursor) |
| k5d | 0.29 d−1 | 0.21 d−1 | Degradation rate of tRNAsum precursor |
| k6a | 0.24 d−1 | 0.19 d−1 | Complex formation rate of tRNAsum/MatK |
| k6aτ | 0.93 d−1 | 0.20 d−1 | (Time-delayed) complex formation rate of tRNAsum/MatK |
| k7s | 10.0 d−1 | 10.0 d−1 | Splicing rate for mature tRNAsum |
| k7d | 2.08 d−1 | 0.74 d−1 | Degradation rate of tRNAsum |
| τ | 25 d | 25 d | Explicit delay of pre-trnK/MatK and pre-tRNA/MatK complex formation |
Simulation of the tRNA-MatK Gene Expression Network with Disrupted MatK Autoregulation
We took the autoregulation out of Equation 4. Equation 4 then changes to Equation 9 (Supplemental Fig. S2A).
Simulation of the tRNA-MatK Gene Expression Network with MatK Transtarget Genes Overexpressed
To simulate an overproduction of tRNAsum precursor transcript, we constitutively increased the rate k5r 10-fold and 20-fold: for 10×[pre-tRNAsum], k5r = 35.106 d−1; for 20×[pre-tRNAsum], k5r = 70.212 d−1 (Supplemental Fig. S2B).
Simulation of the tRNA-MatK Gene Expression Network in the Absence of Early tRNA/MatK Complexes
To simulate the absence of early pre-trnK/MatK complexes and tRNAsum/MatK complexes, the formation rates of pre-trnK/MatK complexes (k2a) in Equation 2 and pre-tRNAsum/MatK complexes (k6a) in Equation 6 were set to zero (Supplemental Fig. S2, C and D).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Immunological analysis of the immunoprecipitation of MatK:HA from stroma fractions of epitope-tagged tobacco plants (C+, N+).
Supplemental Figure S2. Predictions of model 1.
Supplemental Table S1. Sequences of oligonucleotide probes and the primers used for the amplification of PCR probes.
Supplementary Material
Acknowledgments
We thank Thomas Börner and Kenny Watkins for critical reading of the manuscript.
Glossary
- dpi
days post imbibition
- HA
hemagglutinin
References
- Anziano PQ, Butow RA. (1991) Splicing-defective mutants of the yeast mitochondrial COXI gene can be corrected by transformation with a hybrid maturase gene. Proc Natl Acad Sci USA 88: 5592–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anziano PQ, Moran JV, Gerber D, Perlman PS. (1990) Novel hybrid maturases in unstable pseudorevertants of maturaseless mutants of yeast mitochondrial DNA. Nucleic Acids Res 18: 3233–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1989) Tissue-dependent plastid RNA splicing in maize: transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell 1: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulouis A, Raynaud C, Bujaldon S, Aznar A, Wollman FA, Choquet Y. (2011) The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts. Plant Cell 23: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carignani G, Netter P, Bergantino E, Robineau S. (1986) Expression of the mitochondrial split gene coding for cytochrome oxidase subunit I in S. cerevisiae: RNA splicing pathway. Curr Genet 11: 55–63 [DOI] [PubMed] [Google Scholar]
- CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106: 12794–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman FA. (2001) Assembly-controlled regulation of chloroplast gene translation. Biochem Soc Trans 29: 421–426 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Duffy AM, Kelchner SA, Wolf PG. (2009) Conservation of selection on matK following an ancient loss of its flanking intron. Gene 438: 17–25 [DOI] [PubMed] [Google Scholar]
- Epstein IR. (1990) Differential delay equations in chemical kinetics: some simple linear model systems. J Chem Phys 92: 1702–1712 [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K. (2007) Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Yi X, Yang YX, Su YJ, Wang T. (2009) Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol 9: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Hoch B, Zeltz P, Hübschmann T, Kössel H, Börner T. (1994) Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tang Z, Lin RJ, Tucker PW. (2007) Phosphorylation by SR kinases regulates the binding of PTB-associated splicing factor (PSF) to the pre-mRNA polypyrimidine tract. FEBS Lett 581: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann T, Hess WR, Börner T. (1996) Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol Biol 30: 109–123 [DOI] [PubMed] [Google Scholar]
- Khrouchtchova A, Monde RA, Barkan A. (2012) A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. (2004) Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. (2011) Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM. (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Liere K, Link G. (1994) Structure and expression characteristics of the chloroplast DNA region containing the split gene for tRNA(Gly) (UCC) from mustard (Sinapis alba L.). Curr Genet 26: 557–563 [DOI] [PubMed] [Google Scholar]
- Liere K, Link G. (1995) RNA-binding activity of the matK protein encoded by the chloroplast trnK intron from mustard (Sinapis alba L.). Nucleic Acids Res 23: 917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I, Schuster G. (1995) Phosphorylation of a chloroplast RNA-binding protein changes its affinity to RNA. Nucleic Acids Res 23: 2506–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. (1996) SR proteins and splicing control. Genes Dev 10: 1569–1579 [DOI] [PubMed] [Google Scholar]
- McCullough AJ, Kangasjarvi J, Gengenbach BG, Jones RJ. (1992) Plastid DNA in developing maize endosperm: genome structure, methylation, and transcript accumulation patterns. Plant Physiol 100: 958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, Leebens-Mack J, dePamphilis CW. (2009) Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS ONE 4: e5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. (1989) Comparative and functional anatomy of group II catalytic introns: a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Furuhashi Y, Hasegawa K, Hashimoto H, Watanabe K, Obokata J, Sugita M, Sugiura M. (2003) Array-based analysis on tobacco plastid transcripts: preparation of a genomic microarray containing all genes and all intergenic regions. Plant Cell Physiol 44: 861–867 [DOI] [PubMed] [Google Scholar]
- Neuhaus H, Link G. (1987) The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet 11: 251–257 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Link G. (1993) The 54 kDa RNA-binding protein from mustard chloroplasts mediates endonucleolytic transcript 3′ end formation in vitro. Plant J 3: 537–544 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. (2005) CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17: 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Doudna JA. (2004) Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43: 6486–6497 [DOI] [PubMed] [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix JD. (2013) Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Loiselay C, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA, Choquet Y. (2007) Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc Natl Acad Sci USA 104: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM. (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38: 9069–9083 [DOI] [PubMed] [Google Scholar]
- Singh RN, Saldanha RJ, D’Souza LM, Lambowitz AM. (2002) Binding of a group II intron-encoded reverse transcriptase/maturase to its high affinity intron RNA binding site involves sequence-specific recognition and autoregulates translation. J Mol Biol 318: 287–303 [DOI] [PubMed] [Google Scholar]
- Till B, Schmitz-Linneweber C, Williams-Carrier R, Barkan A. (2001) CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. (2006) The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol 23: 1324–1338 [DOI] [PubMed] [Google Scholar]
- Vogel J, Börner T, Hess WR. (1999) Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res 27: 3866–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H, SanFilippo J, Singh RN, Matsuura M, Lambowitz AM. (1999) A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell 4: 239–250 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T. (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710–722 [DOI] [PubMed] [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C. (2010) An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.