A GDSL lipase regulates ethylene-dependent pathogen resistance and ethylene-salicylic acid signaling interactions in plants.
Abstract
Ethylene is a key signal in the regulation of plant defense responses. It is required for the expression and function of GDSL LIPASE1 (GLIP1) in Arabidopsis (Arabidopsis thaliana), which plays an important role in plant immunity. Here, we explore molecular mechanisms underlying the relationship between GLIP1 and ethylene signaling by an epistatic analysis of ethylene response mutants and GLIP1-overexpressing (35S:GLIP1) plants. We show that GLIP1 expression is regulated by ethylene signaling components and, further, that GLIP1 expression or application of petiole exudates from 35S:GLIP1 plants affects ethylene signaling both positively and negatively, leading to ETHYLENE RESPONSE FACTOR1 activation and ETHYLENE INSENSITIVE3 (EIN3) down-regulation, respectively. Additionally, 35S:GLIP1 plants or their exudates increase the expression of the salicylic acid biosynthesis gene SALICYLIC ACID INDUCTION-DEFICIENT2, known to be inhibited by EIN3 and EIN3-LIKE1. These results suggest that GLIP1 regulates plant immunity through positive and negative feedback regulation of ethylene signaling, and this is mediated by its activity to accumulate a systemic signal(s) in the phloem. We propose a model explaining how GLIP1 regulates the fine-tuning of ethylene signaling and ethylene-salicylic acid cross talk.
The gaseous plant hormone ethylene plays important roles in growth and development, senescence, fruit ripening and abscission, and pathogen resistance (Abeles et al., 1992; van Loon et al., 2006; Cho and Yoo, 2009). The components of ethylene signaling have been identified through genetic screens for mutants in Arabidopsis (Arabidopsis thaliana) that exhibit defects in ethylene responses, the so-called triple response, exemplified by short, thick hypocotyl and root and exaggerated apical hook of ethylene-treated etiolated seedlings (Ecker, 1995; Chang, 1996; Woeste and Kieber, 1998). Ethylene response mutants include the constitutive triple response mutants constitutive triple response1 (ctr1), ethylene overproducer1 (eto1), eto2, and eto3 as well as the ethylene-insensitive mutants ethylene response1 (etr1), etr2, ethylene insensitive2 (ein2), ein3, ein4, ein5, ein6, and ein7 (Kieber et al., 1993; Guo and Ecker, 2004). In addition to these, mutants with an enhanced ethylene sensitivity have been isolated, including EIN3-binding F-box protein1 (ebf1), ebf2, enhanced ethylene response1 (eer1), eer3, eer4, eer5, and reversion-to-ethylene sensitivity1 (rte1; Larsen and Chang, 2001; Guo and Ecker, 2003; Resnick et al., 2006; Christians and Larsen, 2007; Robles et al., 2007; Christians et al., 2008). Epistasis and molecular analyses of mutants have revealed a linear ethylene signaling pathway and the core components involved in ethylene perception, signal cascading, and transcriptional regulation (Solano and Ecker, 1998; Chen et al., 2005).
Ethylene is recognized by a family of five endoplasmic reticulum-located integral membrane receptors in Arabidopsis, ETR1, ETR2, EIN4, ETHYLENE RESPONSE SENSOR1 (ERS1), and ERS2 (Chang et al., 1993; Hua et al., 1998; Sakai et al., 1998). In the absence of ethylene, ethylene receptors associate with the Raf-like Ser/Thr kinase CTR1 and repress downstream ethylene signaling (Kieber et al., 1993). When bound to ethylene, receptor signaling and CTR1 become inactivated, leading to derepression of the downstream positive regulators EIN2 and EIN3 (Chao et al., 1997). EIN2 has sequence similarity to mammalian NRAMP metal transporters and localizes at the endoplasmic reticulum membrane (Alonso et al., 1999; Bisson et al., 2009). Recent findings reveal that EIN2 undergoes proteolytic cleavage in response to ethylene, and the resultant C-terminal fragment of EIN2 moves into the nucleus and stabilizes EIN3 to activate ethylene responses (Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012). In the absence of ethylene, the F-box proteins EIN2 TARGETING PROTEIN1 (ETP1) and ETP2 lead to proteasomal degradation of EIN2 and thus negatively regulate ethylene signal transduction (Qiao et al., 2009). EIN3 and EIN3-LIKE1 (EIL1) are critical transcription factors downstream of EIN2 and also subjected to proteasomal degradation by EBF1 and EBF2 in the absence of ethylene (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). Ethylene stabilizes EIN3 and EIL1 by inducing the proteasomal degradation of EBF1 and EBF2 (An et al., 2010). EIN3 and EIL1 bind to a specific sequence in the target gene promoter and induce transcription (Chao et al., 1997; Solano et al., 1998). ETHYLENE RESPONSE FACTOR1 (ERF1), which contains the promoter sequence for EIN3 and EIL1, is thought to be the direct target gene of these proteins in Arabidopsis (Solano et al., 1998). ERF1 itself is a transcription factor belonging to the ethylene-responsive element-binding protein family, which can bind to the GCC box, and functions positively by activating ethylene responses (Fujimoto et al., 2000).
Ethylene, salicylic acid (SA), and jasmonic acid (JA) are key hormones regulating disease resistance, and they interact both synergistically and antagonistically in the signaling networks (Wang et al., 2002; Bostock, 2005; Beckers and Spoel, 2006; Broekaert et al., 2006). In Arabidopsis, each of these hormones is involved in different host-pathogen interactions. Whereas the SA-dependent pathway is generally implicated in resistance to biotrophic pathogens such as Pseudomonas syringae and Hyaloperonospora arabidopsidis, ethylene and JA pathways primarily confer resistance to necrotrophic pathogens such as Alternaria brassicicola and Botrytis cinerea (Penninckx et al., 1998; Pieterse and van Loon, 1999; Glazebrook, 2005; Spoel et al., 2007). There are many reports that SA antagonizes JA/ethylene signaling (Koornneef et al., 2008), but there is also evidence that positive interactions of these pathways lead to induced resistance (Penninckx et al., 1998; Thomma et al., 1998, 1999; Kwon et al., 2009). Systemic acquired resistance (SAR) is the best-studied SA-requiring induced immune response (Sticher et al., 1997), and other types of JA/ethylene-dependent induced resistance, including rhizobacteria-mediated induced systemic resistance (ISR), have also been demonstrated (Heil and Bostock, 2002; Kwon et al., 2009). Whereas SAR and ISR are differentially effective against some pathogens (e.g. Turnip crinkle virus and A. brassicicola), they also additively enhance resistance against others (e.g. P. syringae; van Wees et al., 2000; Ton et al., 2002). EIN3 and EIL1 transcription factors can both positively and negatively regulate pathogen-associated molecular pattern-triggered immunity at the transcriptional level, through up-regulation of FLAGELLIN-SENSING2 (FLS2), which is required for pathogen-associated molecular pattern flagellin binding, or down-regulation of SALICYLIC ACID INDUCTION-DEFICIENT2 (SID2), which is required for SA biosynthesis (Chen et al., 2009; Boutrot et al., 2010). These results suggest a mechanism of EIN3/EIL1-mediated cross talk between the ethylene and SA pathways.
Previously, we found that Arabidopsis GDSL LIPASE1 (GLIP1) is an ethylene-responsive secreted protein and regulates plant immunity (Oh et al., 2005; Kwon et al., 2009). Whereas GLIP1 is specifically involved in local resistance against necrotrophic pathogens, GLIP1 overexpression in plants induces resistance to a range of pathogens, including the necrotrophic pathogens A. brassicicola and Erwinia carotovora and the hemibiotrophic pathogen P. syringae pv tomato (Pst) DC3000 (Kwon et al., 2009). Local inoculation of GLIP1 proteins elicits systemic resistance, which is abolished in the ethylene-insensitive mutant etr1-1, suggesting that ethylene signaling plays a vital role in GLIP1 action. However, the mechanism of this interaction remains unknown. Here, we present data indicating that GLIP1 expression depends on ethylene signaling components. Furthermore, we show that activated GLIP1 or petiole exudates from GLIP1-overexpressing plants (35S:GLIP1) both positively and negatively modulate ethylene responses, thereby enhancing JA/ethylene- and SA-mediated pathogen resistance, respectively.
RESULTS
GLIP1 Is Linked to the Ethylene Signaling Pathway
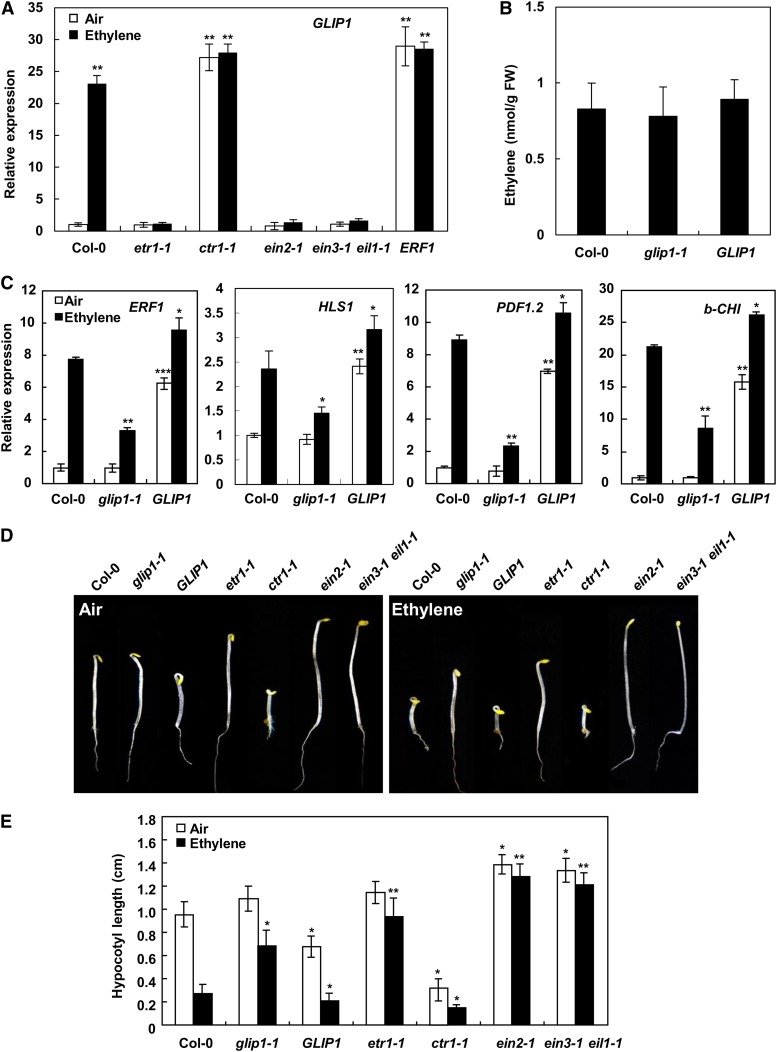
Our previous results demonstrated that GLIP1 is strongly induced by ethylene-releasing ethephon treatment and that it requires the ethylene pathway for the induction of pathogen resistance (Oh et al., 2005; Kwon et al., 2009). GLIP1 expression was additionally examined in wild-type plants treated with the ethylene precursor 1-aminocyclopropane-carboxylic acid (ACC) or ethylene (Supplemental Fig. S1A). All three treatments, ethephon, ACC, and ethylene, significantly induced GLIP1 expression. To gain insights into the relationship between ethylene and GLIP1, we evaluated GLIP1 expression in various ethylene mutants, including etr1-1, ctr1-1, ein2-1, and ein3-1 eil1-1 (Fig. 1A). Whereas GLIP1 expression was compromised in etr1-1, ein2-1, and ein3-1 eil1-1 mutant plants, strong and constitutive expression of GLIP1 was observed in the ctr1-1 mutant. In addition, GLIP1 expression was markedly higher in ERF1-overexpressing plants (Fig. 1A). These results demonstrate that GLIP1 expression is regulated by ethylene signaling components.
Figure 1.
GLIP1 is associated with ethylene signaling. A, Expression analysis of GLIP1 in ethylene mutants etr1-1, ctr1-1, ein2-1, and ein3-1 eil1-1 and in ERF1-overexpressing plants. The values represent means ± sd from three independent experiments. Asterisks indicate significant differences from the air-treated Col-0 (Student’s t test, **P < 0.01). B, Ethylene contents of Col-0, glip1-1, and 35S:GLIP1 plants. Ten-day-old seedlings were used for ethylene quantification. The values are means ± sd (n = 20). Experiments were carried out more than five times with similar results. FW, Fresh weight. C, Expression analysis of ethylene-responsive genes ERF1, HLS1, PDF1.2, and b-CHI in Col-0, glip1-1, and 35S:GLIP1 plants. Four-week-old plants were treated with air or 10 µL L−1 ethylene for 12 h. The values represent means ± sd from three independent experiments. Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001). D, Triple response of 4-d-old etiolated seedlings of Col-0, glip1-1, 35S:GLIP1, and ethylene mutants grown in air or 10 µL L−1 ethylene. E, Hypocotyl lengths of the plants in D. The values are means ± sd (n = 10). Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01).
We then evaluated whether the expression of ethylene-responsive genes is altered in glip1-1 mutant and 35S:GLIP1 plants (Fig. 1C; Supplemental Fig. S1, B–F). We analyzed the expression levels of ERF1, HOOKLESS1 (HLS1), required for apical hook curvature (Lehman et al., 1996; An et al., 2012), and two pathogenesis-related genes, PLANT DEFENSIN1.2 (PDF1.2) and BASIC CHITINASE (b-CHI), induced by ERF1 (Fig. 1C; Lorenzo et al., 2003). They were significantly up-regulated in 35S:GLIP1 plants to the levels in ethylene-treated wild-type plants, but not in the glip1-1 mutant, and were somewhat increased in glip1-1 and 35S:GLIP1 plants by ethylene treatment, although the increase was much smaller than that in the ethylene-treated wild type. We additionally analyzed the expression of other ethylene-responsive genes, such as the ethylene response factor gene ERF5, the ethylene receptor genes ETR2 and ERS1, ACO2 encoding an ACC oxidase, and EBP encoding an ethylene-responsive element-binding protein (Supplemental Fig. S1, B–F). GLIP1 expression did not much affect their ethylene-inducible gene expression, although 35S:GLIP1 plants showed increased basal expression of ERF5, ETR2, and ERS1. This suggests that their expression is largely affected by other factors in ethylene signaling. According to gene expression patterns, GLIP1 expression depends on the ethylene pathway and GLIP1 overexpression leads to the induction of ERF1 and the downstream effector genes, suggesting that GLIP1 may modulate ethylene signaling through a positive feedback mechanism. However, GLIP1 does not likely have a universal effect on the ethylene pathway but rather modulates a subset of ethylene responses.
To check whether GLIP1-induced gene expression was a result of enhanced ethylene production in 35S:GLIP1 plants, ethylene contents were measured in plants (Fig. 1B). Ethylene levels did not differ among wild-type, glip1-1, and 35S:GLIP1 plants, indicating that GLIP1 is related to ethylene signaling but not to ethylene biosynthesis.
GLIP1 Expression Triggers Ethylene Responses
Activation of ethylene response genes in 35S:GLIP1 plants prompted us to compare ethylene response phenotypes of glip1 and 35S:GLIP1 plants with those of wild-type and ethylene mutant plants. Etiolated 35S:GLIP1 seedlings constitutively exhibited features of the triple response (increased hook curvature and shorter, thicker hypocotyl and root), although not so dramatically as ctr1-1 (Fig. 1, D and E). In the presence of ethylene or ACC, 35S:GLIP1 seedlings displayed the enhanced triple response, but glip1 mutants were less sensitive than the wild type (Fig. 1, D and E; Supplemental Figs. S2 and S3). The increased and decreased ethylene sensitivity of 35S:GLIP1 and glip1 seedlings were similar to, but less marked than, those of the constitutive triple response mutant (i.e. ctr1-1) and ethylene-insensitive mutants (i.e. etr1-1, ein2-1, and ein3-1 eil1-1), respectively. Ethylene also accelerates leaf senescence (Chao et al., 1997; Yoo et al., 2008). When exposed to ACC, 35S:GLIP1 plants, like ctr1-1, had reduced chlorophyll content (Supplemental Fig. S3C). However, the leaf senescence of glip1-1 plants was little affected by ACC treatment, as observed in etr1-1, ein2-1, and ein3-1 eil1-1. These results indicate that GLIP1 plays a positive role in ethylene responses, consistent with the induction of ethylene response genes in 35S:GLIP1 plants (Fig. 1C).
To determine whether lipase activity is required for GLIP1-mediated ethylene responses, the triple response was evaluated in wild-type plants (35S:GLIP1TM [for GLIP1 with triple mutations]) and glip1 plants (35S:GLIP1TM glip1-1) overexpressing GLIP1TM, encoding an inactive GLIP1 in which residues of the catalytic triad (Ser, Asp, and His) were replaced with Ala (Supplemental Fig. S4A; Kwon et al., 2009). Transgenic lines of 35S:GLIP1TM and 35S:GLIP1TM glip1-1 showing GLIP1 expression similar to that of 35S:GLIP1 plants were used for the test (Supplemental Fig. S4B). Both 35S:GLIP1TM and 35S:GLIP1TM glip1-1 plants were less sensitive to ethylene than wild-type and 35S:GLIP1 plants, implying that lipase activity is important for the activation of ethylene responses by GLIP1. Ag2+ ions are known to inhibit the ethylene response by replacing Cu2+, an ethylene receptor cofactor (Beyer, 1976). Treatment with AgNO3 effectively eliminated the ACC-induced ethylene response of wild-type, glip1-1, and 35S:GLIP1 seedlings, but not the constitutive triple response of 35S:GLIP1 seedlings, suggesting that GLIP1 may trigger ethylene responses independently of ethylene receptors (Supplemental Fig. S4C).
Epistasis of GLIP1 and Ethylene Pathway Genes
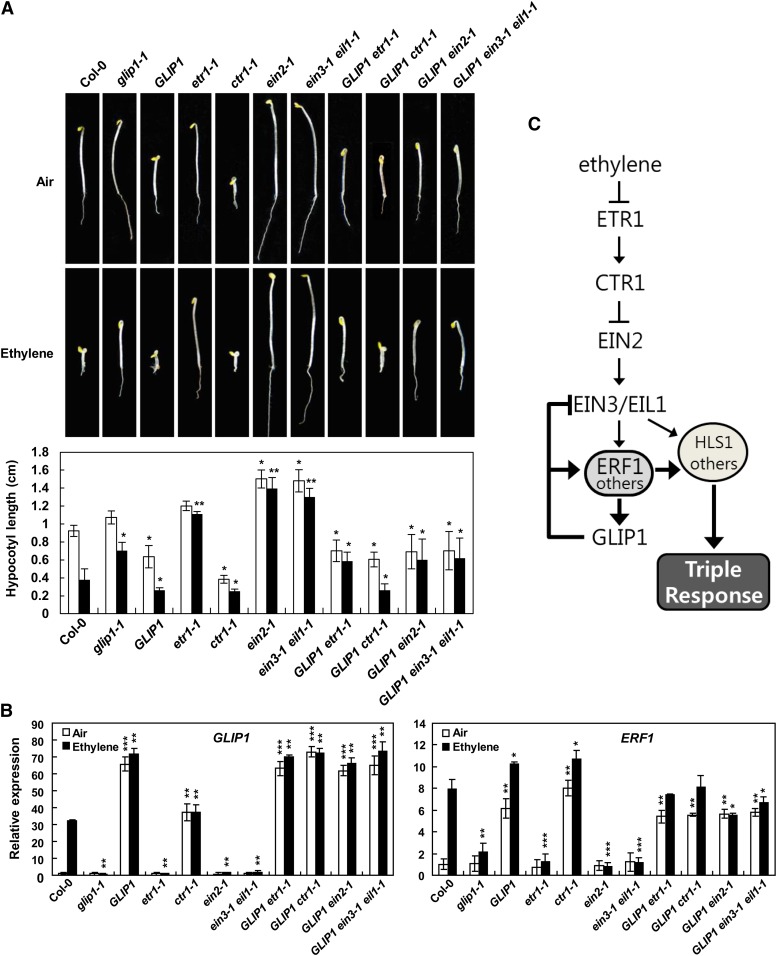
To further dissect how GLIP1 interacts with components of the ethylene signaling pathway, we performed an epistasis analysis by crossing 35S:GLIP1 plants with ethylene mutants (etr1-1, ctr1-1, ein2-1, and ein3-1 eil1-1; Supplemental Figs. S5 and S6). Homozygous crossed lines were obtained, and their growth phenotypes were observed. Ethylene has an inhibitory effect on cell elongation, leading to a reduction in leaf epidermal cell size (Guzman and Ecker., 1990; Kieber et al., 1993; Chao et al., 1997). As reported, ein2-1 and ein3-1 eil1-1 mutants had larger leaves, and the ctr1-1 mutant showed a great reduction in leaf size (Supplemental Fig. S5). However, crossed lines lost the leaf size changes associated with ethylene mutants and were similar in appearance to their 35S:GLIP1 counterpart.
The triple response was compared in 35S:GLIP1, ethylene mutants, and their crossed lines (Fig. 2A). GLIP1 overexpression in ethylene-insensitive mutants, etr1-1, ein2-1, and ein3-1 eil1-1, induced the constitutive triple response as shown in 35S:GLIP1 seedlings, but the triple response was not further enhanced by ethylene treatment. This indicates that GLIP1 acts positively and does not require the core components of ethylene signaling for activation of the ethylene response, but the increased ethylene sensitivity of 35S:GLIP1 seedlings depends on ethylene pathway components such as ETR1, EIN2, and EIN3/EIL1. On the other hand, the phenotype that resulted from crossing ctr1-1 with 35S:GLIP1 was quite unexpected: 35S:GLIP1 ctr1-1 seedlings lost the strong constitutive triple response of ctr1-1 but displayed the ethylene response phenotype of 35S:GLIP1 seedlings (Fig. 2A). Together with the observation that the leaf size change associated with the ctr1-1 mutant was abolished in 35S:GLIP1 ctr1-1 plants (Supplemental Fig. S5), this implies that GLIP1 regulates ethylene signaling in a dominant way and may play a dual role in ethylene signaling. In addition to activating ERF1 and ethylene-related genes, GLIP1 may eliminate the constitutive triple response effect of the ctr1-1 mutant by negatively affecting downstream components of CTR1, such as EIN2 and/or EIN3.
Figure 2.
Effect of GLIP1 on phenotypes and gene expression of ethylene mutants. A, Triple response phenotypes (top) and hypocotyl lengths (bottom) of 4-d-old etiolated seedlings of Col-0, glip1-1, 35S:GLIP1, ethylene mutants, and crossed lines grown in air or 10 µL L−1 ethylene. The values are means ± sd (n = 20). Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01). B, Expression analysis of GLIP1 and ERF1 in Col-0, glip1-1, 35S:GLIP1, ethylene mutants, and crossed lines. Four-week-old plants were treated with air or 10 µL L−1 ethylene for 12 h. The values represent means ± sd from three independent experiments. Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001). C, A model for the modulation of ethylene responses by GLIP1. GLIP1, which requires ethylene signaling for its expression, may form negative and positive feedback loops at EIN3 and ERF1, respectively, to regulate ethylene responses. In addition to ERF1, other ERFs are probably activated by GLIP1, leading to the expression of effector genes, such as HLS1, necessary for provoking ethylene responses.
Gene expression was then examined in ethylene mutants and crossed lines (Fig. 2B). Whereas the ethylene-responsive genes GLIP1 and ERF1 were ethylene inducible in wild-type plants, 35S:GLIP1, ctr1-1, and 35S:GLIP1-crossed ethylene mutants (35S:GLIP1 etr1-1, 35S:GLIP1 ctr1-1, 35S:GLIP1 ein2-1, and 35S:GLIP1 ein3-1 eil1-1) all showed marked constitutive expression of GLIP1 and ERF1. However, strong induction of GLIP1 and ERF1 was abolished in ethylene-insensitive etr1-1, ein2-1, and ein3-1 eil1-1 mutants. HLS1, which requires EIN3/EIL1 for ethylene-responsive expression, was also constitutively activated in 35S:GLIP1 and GLIP1 ein3-1 eil1-1 plants (Supplemental Fig. S7).
GLIP1 Plays a Negative Role in Ethylene Signaling by Down-Regulating EIN3
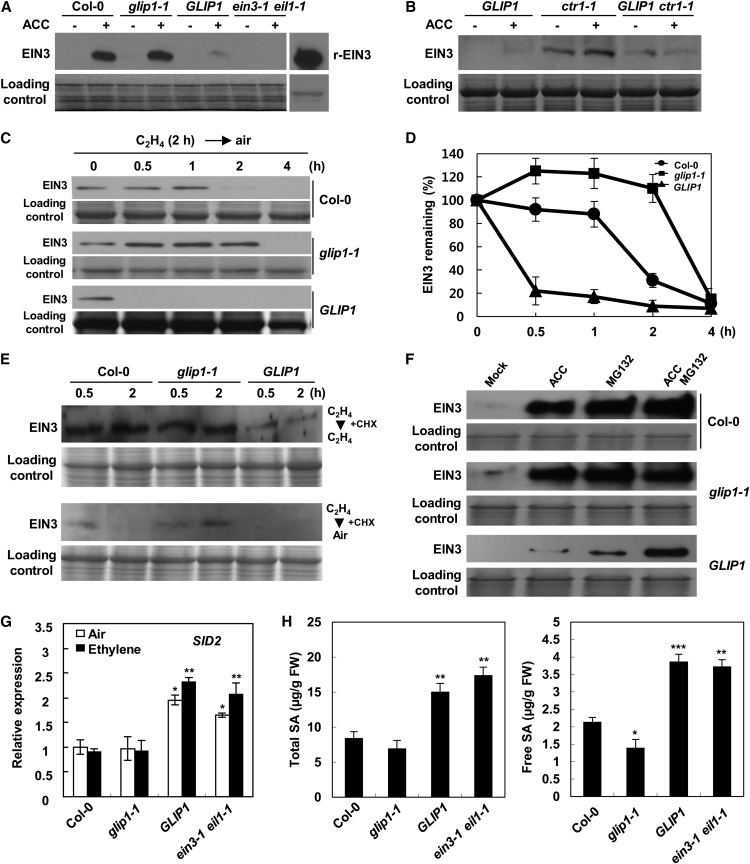
It was assessed whether GLIP1 negatively affects the expression of EIN2 and EIN3. Because EIN2 and EIN3 are modulated at the protein level (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004; Yoo et al., 2008; Qiao et al., 2009; Wang et al., 2009), we determined the expression of EIN2 and EIN3 in wild-type, glip1-1, and 35S:GLIP1 plants by western-blot analysis (Fig. 3A; Supplemental Fig. S8A). EIN2 proteins (C-terminal fragments) accumulated to comparable levels in ACC-treated wild-type, glip1-1, and 35S:GLIP1 plants, although the basal level of EIN2 was higher in glip1-1 plants than in the wild type and 35S:GLIP1 (Supplemental Fig. S8A). On the other hand, EIN3 expression was elevated by ACC treatment in wild-type and glip1-1 plants, but it was significantly reduced in 35S:GLIP1 plants (Fig. 3A). It was also observed that the protein level of EIN3 in 35S:GLIP1 ctr1-1 was much lower than that in ctr1-1 plants, suggesting that elimination of the constitutive triple response of ctr1-1 in 35S:GLIP1 ctr1-1 seedlings was due to EIN3 down-regulation by GLIP1 (Fig. 3B). We monitored EIN3 levels in plants treated with ethylene for 2 h and then moved back into air (Fig. 3, C and D). In the absence of ethylene, EIN3 proteins rapidly disappeared within 30 min in 35S:GLIP1 plants but persisted for 2 h in glip1-1 plants. We further checked whether this is related to EIN3 protein stability. Ethylene-exposed plants were treated with the protein synthesis inhibitor cycloheximide and kept in ethylene or moved into air (Fig. 3E). In the presence of ethylene, EIN3 proteins accumulated in the wild type and glip1-1, but EIN3 stability markedly decreased in 35S:GLIP1 plants. Treatment with the proteasome inhibitor MG132 enhanced EIN3 stability in 35S:GLIP1 plants as well as in wild-type and glip1-1 plants, indicating that GLIP1-induced EIN3 degradation is proteasome dependent (Fig. 3F). There were no differences in EIN2 and EIN3 transcript levels among wild-type, glip1-1, and 35S:GLIP1 plants, with the exception of a slight increase in EIN3 in untreated 35S:GLIP1 plants (Supplemental Fig. S8B). Expression analysis demonstrates that GLIP1 promotes the proteasome-mediated proteolysis of EIN3 proteins. Taken together, these results suggest that GLIP1 regulates ethylene signaling via both positive (i.e. ERF1 activation) and negative (i.e. EIN3 degradation) feedback mechanisms, and this is the molecular basis of how GLIP1 affects ethylene responses (Fig. 2C).
Figure 3.
GLIP1 overexpression down-regulates EIN3 but up-regulates SID2 and SA levels. A, EIN3 levels in Col-0, glip1-1, 35S:GLIP1, and ein3-1 eil1-1 plants. r-EIN3, Recombinant EIN3 (full length). B, EIN3 levels in 35S:GLIP1, ctr1-1, and 35S:GLIP1 ctr1-1 plants. C, Time course of EIN3 degradation in the absence of ethylene in Col-0, glip1-1, and 35S:GLIP1 plants. Four-week-old plants were treated with 10 µL L−1 ethylene for 2 h, moved back into air, and incubated for the indicated times. For 35S:GLIP1, about 3-fold more proteins were loaded into lanes for a fair comparison of EIN3 degradation in different plants. D, Quantitative analysis of the data in C. EIN3 levels were assessed by densitometric measurement and calculated as the amounts of EIN3 remaining. The values represent means ± sd from five independent experiments. E, EIN3 levels in Col-0, glip1-1, and 35S:GLIP1 plants treated with the protein synthesis inhibitor cycloheximide (CHX). Three-day-old seedlings grown in the presence of 10 µL L−1 ethylene were treated with 100 μm cycloheximide for 2 h, moved into air or ethylene, and incubated for the indicated times. F, EIN3 levels in Col-0, glip1-1, and 35S:GLIP1 plants treated with the proteasome inhibitor MG132. Three-day-old seedlings were treated with either 10 μm ACC or 50 μm MG132, or both together, for 2 h. G, Expression analysis of SID2 in Col-0, glip1-1, 35S:GLIP1, and ein3-1 eil1-1 plants. Four-week-old plants were treated with air or 10 µL L−1 ethylene for 12 h. The values represent means ± sd from three independent experiments. Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01). H, Quantification of total and free SA in Col-0, glip1-1, 35S:GLIP1, and ein3-1 eil1-1 plants. The values are means ± sd (n = 20). Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001). The experiment was repeated three times with similar results. FW, Fresh weight. For A and B, 10-d-old seedlings were either untreated or treated with 10 μm ACC for 4 h. For A to F, protein samples were separated by SDS-gel electrophoresis and subjected to Coomassie staining (bottom) and western-blot analysis with anti-EIN3 antibody (top).
GLIP1 Leads to Increased SID2 Expression and SA Production
We previously showed that 35S:GLIP1 plants are more resistant to Pst DC3000 than wild-type plants (Kwon et al., 2009). Moreover, it was previously reported that EIN3 and EIL1 negatively regulate plant innate immunity by repressing SID2 expression; as a result, ein3-1 eil1-1 mutant plants accumulate SA and exhibit enhanced resistance to Pst DC3000 (Chen et al., 2009). Demonstrating the negative effect of GLIP1 on EIN3 stability, we tested glip1-1 and 35S:GLIP1 plants for SID2 expression (Fig. 3G). 35S:GLIP1 plants showed a marked increase in SID2 expression as in the ein3-1 eil1-1 mutant. SID2 expression was correlated with SA accumulation in these plants (Fig. 3H). These results suggest that GLIP1 induces SA production and, thus, SA-dependent pathogen resistance through negative regulation of EIN3.
GLIP1-Mediated Regulation of Ethylene Signaling Is Associated with Its Activity to Induce Systemic Resistance
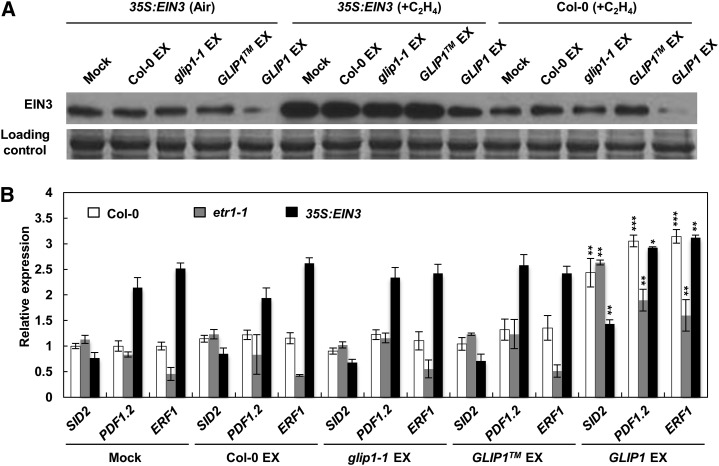
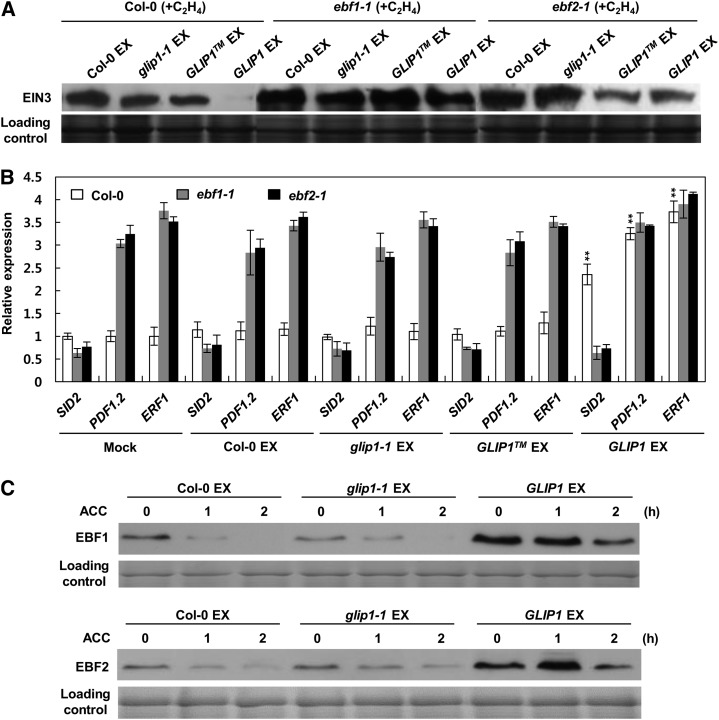
The up-regulation of ERF1 and SID2 in 35S:GLIP1 plants suggests that the feedback regulation of ethylene signaling by GLIP1 may be a mechanism by which GLIP1 regulates immune responses. 35S:GLIP1 plants accumulate PDF1.2-inducing activity in petiole exudates, suggesting that GLIP1 elicits systemic resistance by mediating the production of a systemic signal (Kwon et al., 2009). We thus evaluated whether positive (ERF1 activation) and negative (EIN3 degradation and SID2 activation) regulation by GLIP1 are associated with GLIP1 activity to generate and accumulate a systemic signal(s). Petiole exudates were collected from wild-type, glip1-1, 35S:GLIP1TM, and 35S:GLIP1 plants (Supplemental Fig. S9) and inoculated into wild-type, etr1-1, and 35S:EIN3 plants to assess their effect on the stability of EIN3 proteins and the expression of ERF1 and SID2 (Fig. 4). Upon inoculation with collected petiole exudates, EIN3 expression was significantly decreased in ethylene-treated wild-type and 35S:EIN3 plants in response to 35S:GLIP1 exudates, although a smaller reduction in EIN3 levels was observed in ethylene-treated 35S:EIN3 plants than in untreated 35S:EIN3 plants (Fig. 4A). We further examined whether the expression of ERF1 and SID2 was also altered by 35S:GLIP1 exudate treatment (Fig. 4B). Significant induction of PDF1.2, ERF1, and SID2 was detected in wild-type, etr1-1, and 35S:EIN3 plants when they were inoculated with 35S:GLIP1 exudates but not with exudates of other plants. These results suggest that feedback regulation of GLIP1 occurs through the production of a systemic signal(s) that is independent of upstream ethylene signaling involving ETR1 and that it is related to GLIP1-elicited induced resistance against pathogens.
Figure 4.
Petiole exudates of 35S:GLIP1 plants decrease EIN3 but induce the expression of SID2 and ERF1. A, Immunoblot analysis of EIN3 in Col-0 and 35S:EIN3 plants in response to petiole exudates (EX). Four-week-old plants were infiltrated with 10 μL of exudates (0.3 μg μL−1) and kept for 12 h in air or 10 µL L−1 ethylene. Protein samples were separated by SDS-gel electrophoresis and subjected to Coomassie staining (bottom) and western-blot analysis with anti-EIN3 antibody (top). B, Expression analysis of SID2, ERF1, and PDF1.2 in Col-0, etr1-1, and 35S:EIN3 plants in response to petiole exudates. Total RNAs were extracted from 4-week-old plants infiltrated with 10 μL of exudates (0.3 μg μL−1) for 24 h and used for quantitative real-time PCR analysis. The values represent means ± sd from three independent experiments. Asterisks indicate significant differences from the mock treatments (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001).
GLIP1-Induced EIN3 Degradation Is EBF1/EBF2 Dependent
Since EIN3 degradation in 35S:GLIP1 plants was proteasome dependent (Fig. 3F), it led us to investigate whether GLIP1-induced EIN3 degradation occurs through the action of the F-box proteins EBF1 and EBF2. We first checked for EIN3 levels in ebf1-1 and ebf2-1 mutant plants (Fig. 5A). Whereas EIN3 proteins underwent rapid turnover in ethylene-treated wild-type plants upon inoculation with 35S:GLIP1 exudates, EIN3 stability was remarkably increased in ebf1-1 and ebf2-1 mutants. In line with increased EIN3 protein levels, SID2 expression was not induced in ebf1-1 and ebf2-1 mutants in response to 35S:GLIP1 exudates (Fig. 5B). Expression levels of ERF1 and PDF1.2 were constitutively high in ebf1-1 and ebf2-1 plants, reflecting the increased EIN3 accumulation (Guo and Ecker, 2004).
Figure 5.
Petiole exudates of 35S:GLIP1 plants trigger EIN3 degradation via EBF1/EBF2. A, Immunoblot analysis of EIN3 in Col-0, ebf1-1, and ebf2-1 plants in response to petiole exudates (EX). Four-day-old seedlings were treated with 10 μL of exudates (0.3 μg μL−1) and kept for 12 h in 10 µL L−1 ethylene. B, Expression analysis of SID2, ERF1, and PDF1.2 in Col-0, ebf1-1, and ebf2-1 plants in response to petiole exudates. Total RNAs were extracted from 4-week-old plants infiltrated with 10 μL of exudates (0.3 μg μL−1) for 24 h and used for quantitative real-time PCR analysis. The values represent means ± sd from three independent experiments. Asterisks indicate significant differences from the mock treatments (Student’s t test, **P < 0.01). C, Immunoblot analysis of EBF1/EBF2 in 35S:EBF1-TAP and 35S:EBF2-TAP plants in response to petiole exudates. Four-day-old seedlings were pretreated with 10 μL of exudates (0.3 μg μL−1) for 4 h and then treated with 10 μm ACC for the indicated times. For A and C, protein samples were separated by SDS-gel electrophoresis and subjected to western-blot analysis with anti-EIN3 (A) or anti-MYC (C) antibody.
We next examined whether GLIP1-induced EIN3 degradation is related to changes in the levels of EBF1/EBF2 proteins and/or EBF1/EBF2 transcripts (Fig. 5C; Supplemental Fig. S10). EBF1/EBF2 protein levels were checked in 35S:EBF1-TAP and 35S:EBF2-TAP plants, as previously (An et al., 2010). ACC treatment promoted EBF1/EBF2 protein degradation, and this was substantially suppressed by pretreatment of 35S:EBF1-TAP and 35S:EBF2-TAP seedlings with 35S:GLIP1 exudates (Fig. 5C). EBF1/EBF2 expression was not significantly different among wild-type, glip1-1, and 35S:GLIP1 plants, although EBF1/EBF2 transcript levels were slightly higher in untreated 35S:GLIP1 plants (Supplemental Fig. S10). These results suggest that GLIP1 triggers EIN3 degradation via the EBF1/EBF2-dependent proteasome pathway.
GLIP1 Interacts with Ethylene Signaling to Control Disease Resistance
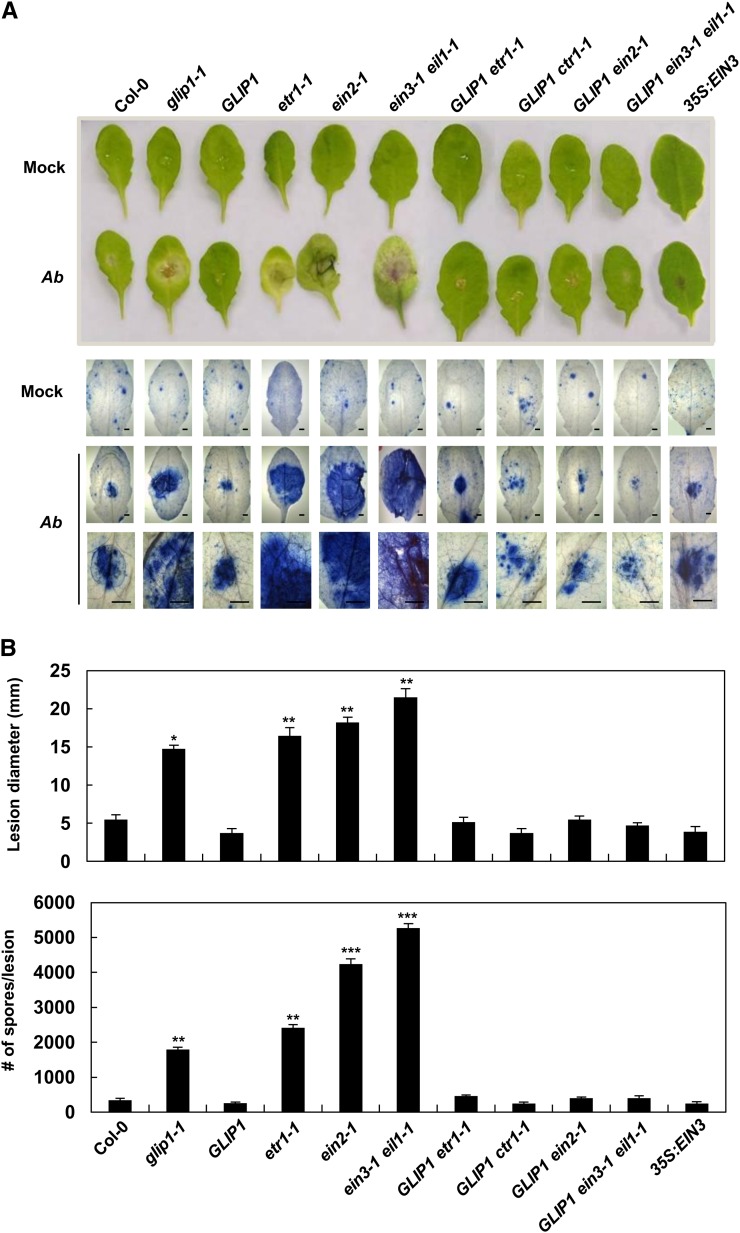
We then monitored how ethylene mutants and their 35S:GLIP1-crossed lines respond to pathogens. Because of tiny leaf size, ctr1-1 plants were excluded from this test. 35S:GLIP1 ctr1-1 plants, however, had normal-sized leaves and were included. First, plants were inoculated with the JA/ethylene-associated necrotrophic fungus A. brassicicola and assessed for disease development (Fig. 6). Whereas the glip1 mutant was highly susceptible to A. brassicicola infection, 35S:GLIP1 plants, like wild-type ecotype Columbia (Col-0), formed hypersensitive response-like small necrotic lesions (Kwon et al., 2009). A. brassicicola-inoculated 35S:GLIP1 ctr1-1 leaves did not differ from 35S:GLIP1 in resistance phenotype. Supposing that ctr1-1 plants with high expression of ERF1 and GLIP1 would be resistant to A. brassicicola, pathogen response phenotypes of 35S:GLIP1 ctr1-1 plants were consistent with ethylene response and gene expression data. The ethylene-insensitive mutants ein2-1 and ein3-1 eil1-1 had susceptible phenotypes, whereas overexpression of GLIP1 in 35S:GLIP1-crossed ein2-1 and ein3-1 eil1-1 plants restored resistance, as in wild-type and 35S:GLIP1 plants. 35S:EIN3 plants also showed resistance phenotypes, consistent with high expression of PDF1.2 and ERF1 (Fig. 4B, mock treatment).
Figure 6.
Functions of GLIP1 and ethylene components in resistance to A. brassicicola. A, Phenotypes (top) and necrotic lesions (bottom) of leaves inoculated with 10 µL of water (mock) or drops of A. brassicicola spore suspension (5 × 105 spores mL−1). Necrotic lesions of leaves were stained with lactophenol-aniline blue. Ab, A. brassicicola. Bars = 100 μm. B, Measurement of lesion diameter (top) and number of newly formed spores (bottom) in leaves from A. The values are means ± sd (n = 10). Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001). The experiment was repeated three times with similar results.
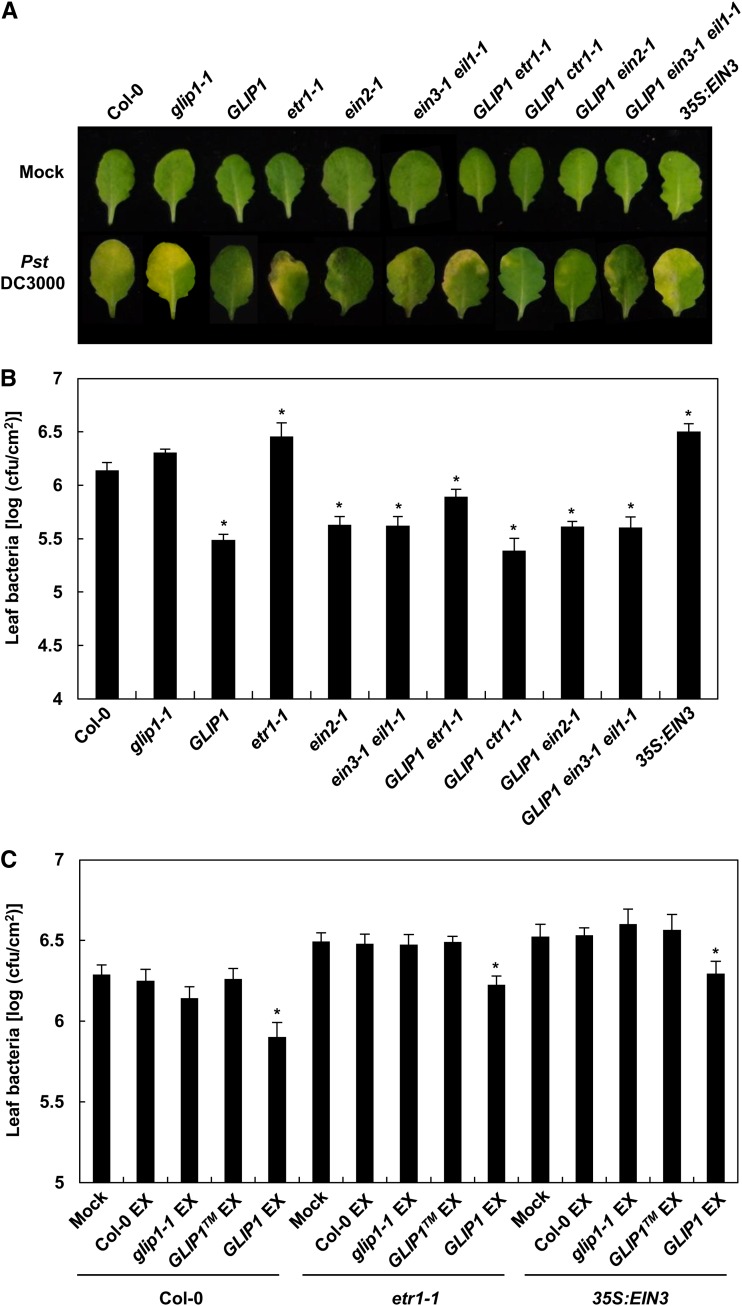
Plants were then challenged with the SA-associated hemibiotrophic bacterial pathogen Pst DC3000 (Fig. 7, A and B). Compared with the wild type, bacterial growth was significantly suppressed in 35S:GLIP1 plants but little altered in glip1 (Kwon et al., 2009). 35S:GLIP1 ctr1-1 plants exhibited the bacterial resistance phenotype of 35S:GLIP1, again correlating with triple response and gene expression patterns. It was previously reported that ein2-1 and ein3-1 eil1-1 plants have enhanced resistance to Pst DC3000 (Chen et al., 2009). This was supported by our observations that ein2-1 and ein3-1 eil1-1 plants experienced significantly lower levels of bacterial growth. Furthermore, this was maintained in their 35S:GLIP1-crossed lines. Consistently, 35S:EIN3 plants were more susceptible to Pst DC3000. As observed in our previous work (Kwon et al., 2009), etr1-1 plants had slightly increased levels of bacterial growth compared with the wild type, contrasting with the enhanced bacterial resistance in other ethylene-insensitive ein2-1 and ein3-1 eil1-1 mutants. We speculate that another EIN2/EIN3-independent ethylene pathway may exist downstream of ETR1 and cross talk with the SA pathway.
Figure 7.
Functions of GLIP1 and ethylene components in resistance to Pst DC3000. A, Phenotypes of leaves inoculated with 10 µL of 10 mm MgCl2 (mock) or aliquots of Pst DC3000 (106 colony-forming units [cfu] mL−1). B, Bacterial growth in leaves inoculated with 10-µL aliquots of Pst DC3000 (105 cfu mL−1). The values are means ± sd (n = 5). Asterisks indicate significant differences from Col-0 (Student’s t test, *P < 0.05). The experiment was repeated five times with similar results. C, Effect of petiole exudates on bacterial growth. Plant leaves were pretreated with petiole exudates (EX) from Col-0, glip1-1, 35S:GLIP1, and 35S:GLIP1 plants for 24 h and then infiltrated with 10-μL aliquots of Pst DC3000 (105 cfu mL−1). The values are means ± sd (n = 5). Asterisks indicate significant differences from the mock treatments (Student’s t test, *P < 0.05). The experiment was repeated three times with similar results.
Since 35S:GLIP1 exudates induced EIN3 down-regulation and SID2 activation in wild-type, etr1-1, and 35S:EIN3 plants (Fig. 4), we further tested whether 35S:GLIP1 exudates can lead to the suppression of bacterial growth (Fig. 7C). The growth of Pst DC3000 was significantly suppressed in wild-type, etr1-1, and 35S:EIN3 plants when they were pretreated with 35S:GLIP1 exudates but not with other exudates. This demonstrates that the bacterial resistance-inducing activity of 35S:GLIP1 exudates correlates with their EIN3-down-regulating and SID2-activating activities.
DISCUSSION
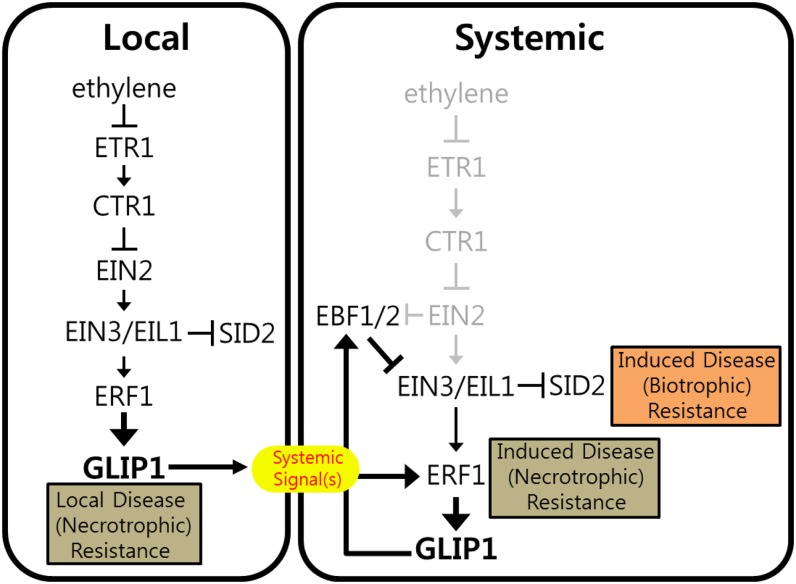
In this work, we present evidence that GLIP1 mechanistically interacts with ethylene signaling components to control plant immunity. GLIP1 expression depends on ethylene signaling and is up-regulated by ERF1. Activated GLIP1 increases ERF1 expression via positive feedback. Additionally, GLIP1 forms a negative feedback loop in ethylene signaling through EBF1/EBF2-dependent proteasomal degradation of EIN3 proteins. We propose that GLIP1-mediated positive and negative feedback regulation of the ethylene signaling pathway is the mechanism underlying GLIP1-induced systemic resistance (Fig. 8).
Figure 8.
A model for GLIP1 function in the regulation of ethylene signaling and immunity. GLIP1 expression depends on ethylene signaling components and positively regulates both local and systemic pathogen resistance. GLIP1 constitutes feedback regulation loops and modulates ethylene signaling in two ways: by inducing ERF1 (positive) and by suppressing EIN3 (negative) via EBF1/EBF2. ERF1 induction increases JA/ethylene-regulated pathogen resistance, and EIN3 down-regulation elevates SID2 expression and SA production, resulting in the enhanced SA-regulated pathogen resistance. We propose that GLIP1-mediated feedback regulation of ethylene signaling is the underlying mechanism of GLIP1 function in induced systemic resistance to pathogens and that it operates through GLIP1-mediated production of a systemic signal(s).
Reciprocal Activation of GLIP1 and Ethylene Signaling
We previously showed that GLIP1 is induced and functions in an ethylene-dependent manner (Oh et al., 2005; Kwon et al., 2009). In this study, ethylene induction of GLIP1 was significantly reduced in etr1-1, ein2-1, and ein3-1 eil1-1 mutants, but GLIP1 was highly expressed in the ctr1-1 mutant and ERF1-overexpressing plants. This indicates that GLIP1 is downstream of ERF1 and is regulated by ethylene signaling components. Noticeably, GLIP1 overexpression enhanced the triple response of ACC/ethylene-treated etiolated seedlings, which correlated with elevated expression of ethylene response genes, including ERF1 and HLS1. In fact, ERF1 expression was largely dependent on GLIP1, as ERF1 induction was much less in the glip1-1 mutant. Therefore, ethylene-regulated GLIP1 appears to affect ethylene signaling via a positive feedback mechanism. 35S:GLIP1 and glip1-1 seedlings showed increased and decreased ethylene sensitivity, respectively, but to a lesser extent than ethylene mutants. Whereas expression levels of GLIP1 and ERF1 in 35S:GLIP1 plants were similar to those in ctr1-1, the triple response was weakly constitutive but largely inducible, unlike in ctr1-1 plants. These data suggest that GLIP1 and ERF1 expression are important, but not sufficient, for ethylene responses. Other factors and/or additional posttranscriptional modifications may be necessary for and act in concert to facilitate a full ethylene response. It was previously shown that ERF1 overexpression rescued the mutant phenotypes of ein3 but only a subset of ethylene responses (Solano et al., 1998).
Epistatic Interaction of GLIP1 and Ethylene Signaling Genes
Supporting that GLIP1 positively acts downstream of ERF1 and ethylene signaling components, GLIP1 overexpression in etr1-1, ein2-1, and ein3-1 eil1-1 mutants rescued the loss of ethylene responses. Intriguingly, 35S:GLIP1 ctr1-1 displayed ethylene-response phenotypes of 35S:GLIP1 plants. Loss of the constitutive triple response of ctr1-1 in 35S:GLIP1 ctr1-1 seedlings led us to propose that GLIP1 constitutes a negative feedback loop downstream of CTR1 and upstream of ERF1, probably at EIN2 and/or EIN3. Western-blot analysis showed that the protein level of EIN3 is negatively modulated by GLIP1 expression, suggesting that GLIP1 contributes to the negative regulation of EIN3 in ethylene signaling. As shown by treatments with petiole exudates, GLIP1-mediated down-regulation of EIN3 depended on EBF1 and EBF2, which induce the degradation of EIN3/EIL1 (Guo and Ecker, 2003). It is known that EIN3 and EIL1 play a role in integrating other signals into the ethylene signaling pathway. Glc accelerated EIN3 degradation through the Glc sensor hexokinase and thus antagonized ethylene signaling (Zhou et al., 1998; Yanagisawa et al., 2003). Interaction between light and ethylene signaling has also been reported. EIN3/EIL1 activated PIF3 expression by directly binding to the promoter of PIF3, leading to ethylene-induced hypocotyl elongation in light (Zhong et al., 2012). In addition, EIN3/EIL1 protein stability increased in light-grown seedlings (Lee et al., 2006). Apical hook development is coordinately regulated by ethylene and GAs. In a recent study, DELLA proteins, key repressors of the GA pathway, were shown to associate with the DNA-binding domains of EIN3/EIL1 and to inhibit EIN3/EIL1-induced HLS1 expression, and GAs enhanced the hook curvature by derepressing EIN3/EIL1 (An et al., 2012). Moreover, as demonstrated both by previous work (Chen et al., 2009) and our current results, EIN3/EIL1 negatively controlled SID2 expression and thus the SA signaling pathway in plant immunity. EIN3 and EIL1, as key modulators of ethylene signaling, may serve as a molecular link connecting distinct signaling pathways to the ethylene pathway in order to coordinate plant growth and development and environmental responses.
GLIP1 and Ethylene Signaling in the Control of Pathogen Resistance
Here, we tested ethylene mutants and their 35S:GLIP1-crossed lines for resistance responses to both necrotrophic A. brassicicola and hemibiotrophic Pst DC3000. Consistent with the epistatic interactions of GLIP1 and ethylene signaling genes, GLIP1 overexpression restored resistance to A. brassicicola in the ethylene mutants etr1-1, ein2-1, and ein3-1 eil1-1. 35S:GLIP1 plants displayed strong induction of ERF1, which was previously shown to be a key factor for the regulation of defense response genes (Lorenzo et al., 2003). Constitutive expression of ERF1 in Arabidopsis conferred resistance to several fungal pathogens, such as B. cinerea, Plectosphaerella cucumerina, and Fusarium oxysporum (Berrocal-Lobo et al., 2002). This indicates that ERF1 expression may be necessary for GLIP1-induced resistance to necrotrophic pathogens, meaning that GLIP1-mediated feedback regulation of ERF1 accumulation is critical for disease resistance responses in plants. On the other hand, enhanced resistance of ein2-1 and ein3-1 eil1-1 to Pst DC3000 was previously shown to be related to SID2 induction and SA accumulation (Chen et al., 2009), and this was further confirmed in our work here. We showed that the EIN3 protein level was decreased and that SID2 expression, repressed by EIN3 and EIL1, was increased in 35S:GLIP1 plants. These results suggest that GLIP1-induced resistance to Pst DC3000 may be regulated by GLIP1-mediated feedback suppression of EIN3. We propose that positive (i.e. ERF1 induction) and negative (i.e. EIN3 destabilization) feedback regulation of ethylene signaling by GLIP1 is an underlying mechanism for GLIP1 functions in plant immunity, specifically in induced systemic resistance (Fig. 8).
Inoculation of plants with petiole exudates from 35S:GLIP1 plants led to significant reduction of EIN3 and induction of SID2 and ERF1 and also suppressed the growth of Pst DC3000 in wild-type, etr1-1, and 35S:EIN3 plants. These results suggest that ETR1, and its ethylene binding, are required for GLIP1 expression, but that once it is activated, GLIP1 elicits induced resistance through the feedback regulation of ethylene signaling. This further supports our proposed model for GLIP1, in which GLIP1-mediated feedback regulation operates through a systemic signal(s), probably generated by catalytic processes involving GLIP1. However, our previous results demonstrated that, unlike 35S:GLIP1 exudates, GLIP1 proteins failed to induce systemic resistance upon inoculation into etr1-1 (Kwon et al., 2009). These differing effects of 35S:GLIP1 exudates and GLIP1 proteins suggest that GLIP1 proteins, unlike the systemic signals in petiole exudates, may not be sufficient for signal amplification and propagation in the etr1-1 mutant background.
There have been numerous reports about both antagonistic and synergistic interactions between ethylene/JA and SA pathways (Kunkel and Brooks, 2002; Glazebrook et al., 2003; Broekaert et al., 2006). The activation of local SA- and JA/ethylene-dependent resistance appears to be mutually exclusive. Upon attack by necrotrophic pathogens, plants may suppress the SA pathway via EIN3/EIL1-mediated SID2 suppression; this antagonistic effect of ethylene signaling allows plants to prioritize the ethylene/JA signaling pathway. On the other hand, two types of induced resistance, SAR and ISR, which require SA and JA/ethylene, respectively, both depend on NPR1 and seem to act additively (Pieterse et al., 1998; Ryu et al., 2004). In fact, positive interactions between ethylene, JA, and SA have previously been observed for induced resistance (Kwon et al., 2009). In the case of induced resistance, hormone pathways may act in concert to evoke resistance against multiple types of pathogens that plants often encounter in the natural environment. For this, ethylene signaling can positively affect the SA pathway through GLIP1-mediated EIN3 down-regulation. Further studies will hopefully allow us to elucidate how cross talk between hormone pathways is fine-tuned in the complex signaling networks that control plant immune responses.
MATERIALS AND METHODS
Plant Materials
Wild-type, mutant, and transgenic Arabidopsis (Arabidopsis thaliana Col-0) plants were grown at 23°C under long-day conditions in a 16-h-light/8-h-dark cycle. The following plants were used in this study: glip1-1 (Oh et al., 2005), 35S:GLIP1(3-2) (Kwon et al., 2009), etr1-1 (Hua et al., 1998), ctr1-1 (Kieber et al., 1993), ein2-1 (Roman et al., 1995), ein3-1 eil1-1 (Alonso et al., 2003), ebf1-1 (Guo and Ecker, 2003), ebf2-1 (Guo and Ecker, 2003), 35S:EBF1-TAP (An et al., 2010), 35S:EBF2-TAP (An et al., 2010), 35S:EIN3 (Chao et al., 1997), and 35S:ERF1 (Lorenzo et al., 2003). 35S:ERF1 (CS6142) seeds were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/). Mutation and insertion sites were verified by sequencing, and homozygous lines were selected. 35S:GLIP1 was crossed to etr1-1, ctr1-1, ein2-1, and ein3-1 eil1-1, and homozygous lines were confirmed by PCR and sequence analysis using gene-specific primers (Supplemental Table S1) for ethylene mutants and by segregation analysis and PCR using 35S primers for 35S:GLIP1.
Preparation of Petiole Exudates
Petiole exudates were prepared as described previously (Kwon et al., 2009). Petioles of wild-type, glip1-1, 35S:GLIP1TM, and 35S:GLIP1 plants were cut above the stem. The cut surface was briefly sterilized in a solution containing 50% (v/v) ethanol and 0.0006% (w/v) sodium hypochlorite and then rinsed in sterile distilled water. Exudates were collected in distilled water for 2 d. Prior to use, exudates were syringe filtered and tested on medium plates for bacterial contamination.
Chemical Treatments
Sterilized seeds were plated on Murashige and Skoog (MS)-Suc (2% [w/v]) agar medium alone or supplemented with ACC (0.5–10 μm) or AgNO3 (100 μm). For the triple response, plates were wrapped in foil and kept for 4 d at 23°C, as described (Yoo et al., 2008). For ethylene treatment, seedlings grown on MS plates in a 500-mL container for 2 weeks were supplemented with 10 µL L−1 ethylene in hydrocarbon-free air, wrapped in foil, and kept for 4 d (triple response) or 12 h (gene expression) at 23°C (Kieber et al., 1993). For ethephon treatment, 4-week-old plants were sprayed with water (mock) or with ethephon (1.5 mm) dissolved in water. For ACC treatment, 4-week-old plants were sprayed with water (mock) or with ACC (10 μm) dissolved in water. The treated plants were maintained at 100% humidity for the indicated times and then harvested. For inhibitor treatments, 3-d-old seedlings germinated on Whatman 3MM filter paper placed on MS plates were treated with 0.1% (v/v) dimethyl sulfoxide (mock), MG132 (50 μm), or cycloheximide (100 μm) for 2 h.
Pathogen Infection
For pathogen infection, 4-week-old plants grown under short-day conditions in an 8-h-light/16-h-dark cycle were used. Treatment with Alternaria brassicicola was performed by applying 10 μL of water or drops of spore suspension (5 × 105 spores mL−1) to each plant leaf, as described (Oh et al., 2005). The number of spores was counted, and the lesions and fungal hyphae were visualized by staining the infected leaves with lactophenol-aniline blue, as described (Oh et al., 2005). Treatment with Pseudomonas syringae was performed as described (Oh et al., 2005). Three other leaves of each plant were infiltrated at one site with 10 µL of MgCl2 (10 mm) or aliquots of P. syringae pv tomato DC3000 (105–106 colony-forming units mL−1). Inoculated plants were kept in a growth chamber at 100% humidity for 4 d as indicated. This experiment was designed as a randomized complete block with five replications and one plant per replication and was repeated at least three times.
Immunoblot Analysis
The coding regions corresponding to residues 800 to 1,294 of EIN2 and full-length EIN3 were cloned into the pDEST17 Gateway vector (Invitrogen) and expressed in Escherichia coli as described (Guo and Ecker, 2003). His-tagged recombinant EIN2 and EIN3 proteins were purified using nickel-nitrilotriacetic acid agarose columns (Qiagen) according to the manufacturer’s instructions and used to produce polyclonal antibodies in rats and rabbits (Komabiotech). Western-blot analysis was performed as described (Oh et al., 2005). Total proteins were extracted and separated by SDS-gel electrophoresis for immunoblotting. The blots were probed with anti-EIN2 and anti-EIN3 antisera for EIN2 and EIN3 immunoblots or with anti-MYC antibody (AbChem) for EBF1/EBF2-TAP immunoblots. Antibody-bound proteins were detected following incubation with secondary antibody conjugated to horseradish peroxidase using the ECL system (Amersham Biosciences).
Ethylene Quantification
Ethylene production in plants was measured by gas chromatography (GC 7890; Agilent) as described (Tamaoki et al., 2008). Seedlings (100 mg) grown on MS-Suc (2% [w/v]) agar medium for 10 d under long-day conditions were enclosed in 60-mL vials and incubated in light for 6 h. Ethylene standards with different concentrations were made by diluting ethylene in 1-L Tedlar sample bags. Gas-phase samples (5 mL) from the vials were injected into an HP-PLOT Q column (30 m × 0.53 mm, 40 µm; Agilent) with column and detector temperatures of 75°C and 250°C, respectively. The amount of ethylene from the plants was determined from ethylene peak area based on comparison with ethylene standards.
SA Quantification
Total and free SA were quantified as described (Segarra et al., 2006; Garcion et al., 2008). Leaf tissues (250 mg), together with an internal standard (1 µg of o-anisic acid), were ground in liquid nitrogen and extracted twice with 90% methanol. After methanol was evaporated from extracts, total SA was further extracted by acid hydrolysis with 4 n HCl at 95°C for 1 h. Liquid chromatography analysis was performed on an Optima Pak C18 column (250 × 4.6 mm, 5 µm; RS Tech). The injection volume was 50 µL, and elution was performed with a binary solvent system consisting of 0.05% (v/v) HOAc in water (solvent A) and 0.05% (v/v) HOAc in acetonitrile (solvent B) at a constant flow rate of 1 mL min−1. A linear gradient profile with the following proportions (v/v) of solvent B was applied (time [min], percentage B): (0, 15), (20, 50), (30, 100), (32, 100), (33, 15), and (45, 15) with 5 min for reequilibration. Fluorescence was recorded with excitation/emission wavelengths of 305/365 nm and 305/407 nm for o-anisic acid and SA, respectively.
Gene Expression Analysis
Quantitative real-time reverse transcription-PCR was performed using KAPA SYBR FAST qPCR master mix in a LightCycler 480 system (Roche). PCR was performed with gene-specific primers (Supplemental Table S1) according to the manufacturer’s protocol. The expression levels of the tested genes were standardized to the constitutive expression level of ACTIN1 and calculated using the geNorm program (Vandesompele et al., 2002). The experiments were repeated at least three times with biologically independent samples.
Determination of Chlorophyll Content
To measure chlorophyll content, leaves were submerged in 95% ethanol and incubated for 20 min at 80°C (Kwon et al., 2007). Absorbance was monitored at 648 nm (A648) and 665 nm (A664), and chlorophyll content was calculated according to the following formula: μg chlorophyll = [(13.36A664) – (5.19A648)] + [(27.43A648) – (8.12A664)] (Lichtenthaler, 1987).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_104918 (ACO2), M38240 (b-CHI), NM_180429 (CTR1), NM_112550 (EBP), NM_128106 (EBF1), NM_122444 (EBF2), NM_120406 (EIL1), NM_112968 (EIN3), NM_113225 (ERF1), NM_124094 (ERF5), NM_129658 (ERS1), NM_105305 (ETR1), NM_113216 (ETR2), NM_123464 (GLIP1), NM_119922 (HLS1), NM_123809 (PDF1.2), and NM_106129 (SID2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of ethylene-responsive genes in Col-0, glip1-1, and 35S:GLIP1 plants.
Supplemental Figure S2. Triple response of glip1 mutants and homozygous T3 lines of 35S:GLIP1.
Supplemental Figure S3. Positive regulation of ethylene responses by GLIP1.
Supplemental Figure S4. Effects of the catalytic mutation of GLIP1 and Ag2+ on the triple response of glip1-1 and 35S:GLIP1 seedlings.
Supplemental Figure S5. Genetic crosses between 35S:GLIP1 and ethylene mutants.
Supplemental Figure S6. Genomic DNA analysis of crossed lines.
Supplemental Figure S7. Expression analysis of HLS1 in Col-0, glip1-1, 35S:GLIP1, ein3-1 eil1-1, and 35S:GLIP1 ein3-1 eil1-1 plants.
Supplemental Figure S8. Expression analysis of EIN2 proteins and EIN2 and EIN3 transcripts in Col-0, glip1-1, and 35S:GLIP1 plants.
Supplemental Figure S9. Proteins isolated from petiole exudates of Col-0, glip1-1, 35S:GLIP1TM, and 35S:GLIP1 plants.
Supplemental Figure S10. Expression analysis of EBF1 and EBF2 in Col-0, glip1-1, and 35S:GLIP1 plants.
Supplemental Table S1. List of primers used for PCR and quantitative reverse transcription-PCR.
Supplementary Material
Acknowledgments
We thank Sang-Dong Yoo and Younghee Cho for etr1-1, ctr1-1, and ein2-1 and Jian-Min Zhou for ein3-1 eil1-1 and 35S:EIN3 seeds.
Glossary
- SA
salicylic acid
- JA
jasmonic acid
- SAR
systemic acquired resistance
- ISR
induced systemic resistance
- Pst
Pseudomonas syringae pv tomato
- ACC
1-aminocyclopropane-carboxylic acid
- Col-0
ecotype Columbia
- MS
Murashige and Skoog
References
- Abeles FB, Morgan PW, Saltveit ME (1992) Ethylene in Plant Biology, Ed 2. Academic Press, San Diego [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR. (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers GJ, Spoel SH. (2006) Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol (Stuttg) 8: 1–10 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Beyer EM. (1976) A potent inhibitor of ethylene action in plants. Plant Physiol 58: 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G. (2009) EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6 [DOI] [PubMed] [Google Scholar]
- Bostock RM. (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43: 545–580 [DOI] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delauré SL, De Bolle MF, Cammue BP. (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Chang C. (1996) The ethylene signal transduction pathway in Arabidopsis: an emerging paradigm? Trends Biochem Sci 21: 129–133 [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Etheridge N, Schaller GE. (2005) Ethylene signal transduction. Ann Bot (Lond) 95: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD. (2009) Emerging complexity of ethylene signal transduction. J Plant Biol 52: 283–288 [Google Scholar]
- Christians MJ, Larsen PB. (2007) Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J Exp Bot 58: 2237–2248 [DOI] [PubMed] [Google Scholar]
- Christians MJ, Robles LM, Zeller SM, Larsen PB. (2008) The eer5 mutation, which affects a novel proteasome-related subunit, indicates a prominent role for the COP9 signalosome in resetting the ethylene-signaling pathway in Arabidopsis. Plant J 55: 467–477 [DOI] [PubMed] [Google Scholar]
- Ecker JR. (1995) The ethylene signal transduction pathway in plants. Science 268: 667–675 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP. (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 147: 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Bostock RM. (2002) Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot (Lond) 89: 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CM. (2008) Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol 147: 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Kwon SJ, Jin HC, Lee S, Nam MH, Chung JH, Kwon SI, Ryu CM, Park OK. (2009) GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J 58: 235–245 [DOI] [PubMed] [Google Scholar]
- Kwon SJ, Kwon SI, Bae MS, Cho EJ, Park OK. (2007) Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell Physiol 48: 1713–1723 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Chang C. (2001) The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol 125: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Deng XW, Kim WT. (2006) Possible role of light in the maintenance of EIN3/EIL1 stability in Arabidopsis seedlings. Biochem Biophys Res Commun 350: 484–491 [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. (1996) HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophyll fluorescence signatures of leaves during the autumnal chlorophyll breakdown. J Plant Physiol 131: 101–110 [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK. (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17: 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, van Loon LC. (1999) Salicylic acid-independent plant defence pathways. Trends Plant Sci 4: 52–58 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SS, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen CK, Shockey JA, Chang C. (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103: 7917–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles LM, Wampole JS, Christians MJ, Larsen PB. (2007) Arabidopsis enhanced ethylene response 4 encodes an EIN3-interacting TFIID transcription factor required for proper ethylene response, including ERF1 induction. J Exp Bot 58: 2627–2639 [DOI] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. (1995) Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Murphy JF, Mysore KS, Kloepper JW. (2004) Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J 39: 381–392 [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang CR, Medrano LJ, Bleecker AB, Meyerowitz EM. (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra G, Jáuregui O, Casanova E, Trillas I. (2006) Simultaneous quantitative LC-ESI-MS/MS analyses of salicylic acid and jasmonic acid in crude extracts of Cucumis sativus under biotic stress. Phytochemistry 67: 395–401 [DOI] [PubMed] [Google Scholar]
- Solano R, Ecker JR. (1998) Ethylene gas: perception, signaling and response. Curr Opin Plant Biol 1: 393–398 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP. (1997) Systemic acquired resistance. Annu Rev Phytopathol 35: 235–270 [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Freeman JL, Pilon-Smits EA. (2008) Cooperative ethylene and jasmonic acid signaling regulates selenite resistance in Arabidopsis. Plant Physiol 146: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF. (1999) Requirement of functional Ethylene-Insensitive2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121: 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Van Pelt JA, Van Loon LC, Pieterse CM. (2002) Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol Plant Microbe Interact 15: 27–34 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: H0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC, Geraats BPJ, Linthorst HJM. (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- van Wees SC, de Swart EA, van Pelt JA, van Loon LC, Pieterse CM. (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 8711–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR. (2002) Ethylene biosynthesis and signaling networks. Plant Cell (Suppl) 14: S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kong H, Ma H. (2009) F-box proteins regulate ethylene signaling and more. Genes Dev 23: 391–396 [DOI] [PubMed] [Google Scholar]
- Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22: 1613–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste K, Kieber JJ. (1998) The molecular basis of ethylene signalling in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 353: 1431–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J. (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J. (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H. (2012) A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol 22: 1530–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.