Bacterial-like phosphoprotein phosphatase genes entered eukaryotes from archaea or with ancestral mitochondrial formation, their various protein types showing important differences in subcellular localization, and differing at specific sequence motifs that could serve as accessory protein-binding sites.
Abstract
Protein phosphorylation is a reversible regulatory process catalyzed by the opposing reactions of protein kinases and phosphatases, which are central to the proper functioning of the cell. Dysfunction of members in either the protein kinase or phosphatase family can have wide-ranging deleterious effects in both metazoans and plants alike. Previously, three bacterial-like phosphoprotein phosphatase classes were uncovered in eukaryotes and named according to the bacterial sequences with which they have the greatest similarity: Shewanella-like (SLP), Rhizobiales-like (RLPH), and ApaH-like (ALPH) phosphatases. Utilizing the wealth of data resulting from recently sequenced complete eukaryotic genomes, we conducted database searching by hidden Markov models, multiple sequence alignment, and phylogenetic tree inference with Bayesian and maximum likelihood methods to elucidate the pattern of evolution of eukaryotic bacterial-like phosphoprotein phosphatase sequences, which are predominantly distributed in photosynthetic eukaryotes. We uncovered a pattern of ancestral mitochondrial (SLP and RLPH) or archaeal (ALPH) gene entry into eukaryotes, supplemented by possible instances of lateral gene transfer between bacteria and eukaryotes. In addition to the previously known green algal and plant SLP1 and SLP2 protein forms, a more ancestral third form (SLP3) was found in green algae. Data from in silico subcellular localization predictions revealed class-specific differences in plants likely to result in distinct functions, and for SLP sequences, distinctive and possibly functionally significant differences between plants and nonphotosynthetic eukaryotes. Conserved carboxyl-terminal sequence motifs with class-specific patterns of residue substitutions, most prominent in photosynthetic organisms, raise the possibility of complex interactions with regulatory proteins.
Reversible protein phosphorylation is a posttranslational mechanism central to the proper function of living organisms (Brautigan, 2013). Governed by two large groups of enzymes, protein kinases and protein phosphatases, this mechanism has been suggested to regulate upwards of 70% of all eukaryotic proteins (Olsen et al., 2010). Protein phosphatases represent one-half of this dynamic regulatory system and have been shown to be highly regulated proteins themselves (Roy and Cyert, 2009; Shi, 2009; Uhrig et al., 2013). Classically, protein phosphatases have been placed into four families defined by a combination of their catalytic mechanisms, metal ion requirements, and phosphorylated amino acid targets (Kerk et al., 2008). These four families are the phosphoprotein phosphatases (PPPs), metallo-dependent protein phosphatases, protein Tyr phosphatases, and Asp-based phosphatases. The PPP protein phosphatases, best known to include PP1, PP2A, PP2B, and PP4 to PP7 (Kerk et al., 2008; Shi, 2009), have been found to regulate a diverse number of biological processes in plants ranging from cell signaling (Ahn et al., 2011; Di Rubbo et al., 2011; Tran et al., 2012) to metabolism (Heidari et al., 2011; Leivar et al., 2011) and hormone biosynthesis (Skottke et al., 2011). The classical PPP protein phosphatase family has been expanded to include three novel classes that show greatest similarity to PPP-like protein phosphatases of prokaryotic origin (Andreeva and Kutuzov, 2004; Uhrig and Moorhead, 2011a; Uhrig et al., 2013). These bacterial-like phosphatase classes were annotated as Shewanella-like (SLP) phosphatases, Rhizobiales-like (RLPH) phosphatases, and ApaH-like (ALPH) phosphatases based on their similarity to prokaryotic sequences from these respective sources (Andreeva and Kutuzov, 2004). Recent characterization of the SLP phosphatases from Arabidopsis (Arabidopsis thaliana) provided biochemical evidence of insensitivity to the classic PPP protein phosphatase inhibitors okadaic acid and microcystin in addition to revealing a lack of genetic redundancy across sequenced plant genomes (Uhrig and Moorhead, 2011a).
The characterization of eukaryotic protein evolution can provide insight into individual protein or protein class conservation across the domains of life for biotechnological applications in addition to furthering our understanding of how multicellular life evolved. In particular, investigation into the evolution of key signaling proteins, such as protein kinases and phosphatases from plants, can have wide-ranging agribiotechnological and medical potential. This can include the development of healthier, disease- or stress-resistant crops in addition to treatments for parasitic organisms such as Plasmodium spp. (malaria; Patzewitz et al., 2013) and other chromoalveolates (Kutuzov and Andreeva, 2008; Uhrig and Moorhead, 2011b) that are derived from photosynthetic eukaryotes and maintain a remnant chloroplast (apicoplast; Le Corguillé et al., 2009; Janouskovec et al., 2010; Kalanon and McFadden, 2010; Walker et al., 2011). The existence of proteins that are conserved across diverse eukaryotic phyla but absent in metazoa, such as the majority of bacterial-like PPP protein phosphatases described here, presents unique research opportunities.
Conventional understanding of the acquisition by eukaryotes of prokaryotic genes and proteins largely involves ancient endosymbiotic gene transfer events stemming from primary endosymbiosis of α-Proteobacteria and Cyanobacteria to form eukaryotic mitochondria and chloroplasts, respectively (Keeling and Palmer, 2008; Dorrell and Smith, 2011; Tirichine and Bowler, 2011). Over time, however, it has become apparent that alternative modes of eukaryotic gene and protein acquisition exist, such as independent horizontal or lateral gene transfer (LGT) events (Keeling and Palmer, 2008; Keeling, 2009). Targeted studies of protein evolution have seen a steady rise in documented LGT events across a wide variety of eukaryotic organisms, including photosynthetic eukaryotes (Derelle et al., 2006; Raymond and Kim, 2012; Schönknecht et al., 2013), nematodes (Mayer et al., 2011), arthropods (Acuña et al., 2012), fungi (Wenzl et al., 2005), amoebozoa (Clarke et al., 2013), and oomycetes (Belbahri et al., 2008). Each instance documents the integration of a bacterial gene(s) into a eukaryotic organism, seemingly resulting in an adaptive advantage(s) important to organism survival.
Utilizing a number of in silico bioinformatic techniques and available sequenced genomes, the molecular evolution of three bacterial-like PPP classes found in eukaryotes is revealed to involve ancient mitochondrial or archaeal origin plus additional possible LGT events. A third, more ancient group of SLP phosphatases (SLP3 phosphatases) is defined in green algae. Subcellular localization predictions reveal distinctive subsets of bacterial-like PPPs, which may correlate with altered functions. In addition, the large sequence collections compiled here have allowed the elucidation of two highly conserved C-terminal domain motifs, which are specific to each bacterial-like PPP class and whose differences are particularly pronounced in photosynthetic eukaryotes. Together, these findings substantially expand our knowledge of the molecular evolution of the bacterial-like PPPs and point the way toward attractive future research avenues.
RESULTS
Eukaryotic Bacterial-Like SLP, RLPH, and ALPH Protein Phosphatases Are PPP Phosphatases
Consistent with previous findings, the vast majority of the SLP, RLPH, and ALPH phosphatases identified here were found to maintain the key catalytic motifs indicative of being PPP protein phosphatases (Supplemental Figs. S1–S3; Andreeva and Kutuzov, 2004; Uhrig and Moorhead, 2011a). These motifs are represented by GDxHG, GDxVDRG, GNHE, and HGG (Shi, 2009) and in some instances can possess conservative substitutions. In a typical sequence, all four of these motifs can be clearly identified upon individual inspection of the amino acid sequence or as part of larger computer-assisted alignment (Supplemental Figs. S1–S3). In a few instances, sequences are clearly lacking fragments of the native N terminus and thus represent incomplete gene models (Supplemental Table S1). Of sequences that have an initial Met, a small proportion in each class nevertheless lack one or more of the conserved N-terminal motifs: about 4% of SLPs (seven of 163) and ALPHs (two of 49) and about 6% of RLPHs (three of 47). It is possible that these represent incomplete or incorrect gene models, but a genuine lack of one or more N-terminal motifs cannot be completely ruled out.
Distribution and Interrelationships of Bacterial-Like Protein Phosphatases
SLP Phosphatases
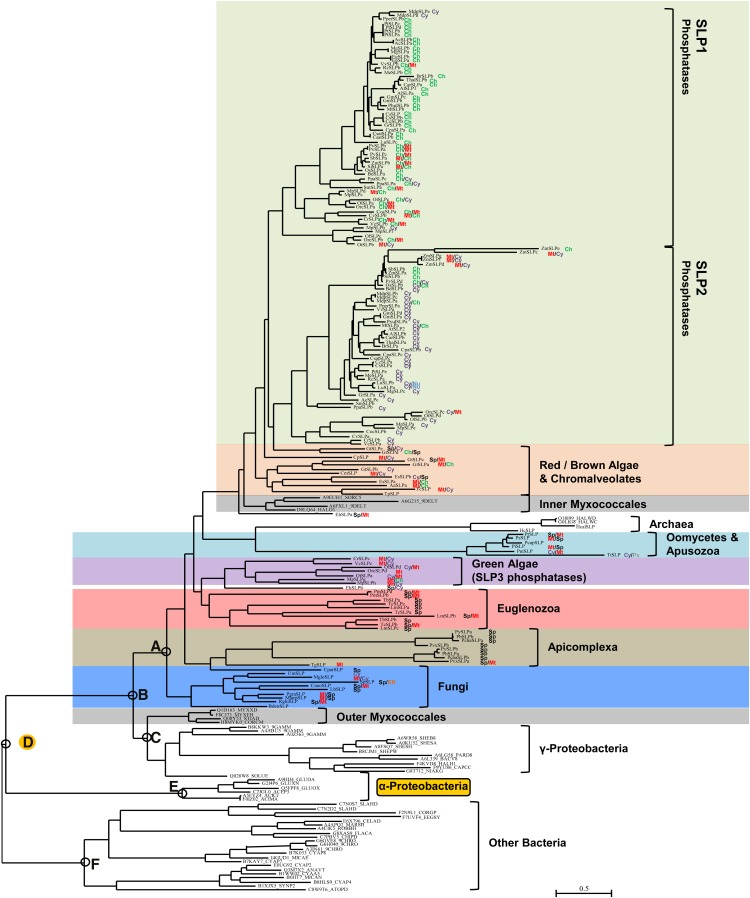
We searched protein databases compiled from the completely sequenced genomes of a large number of eukaryotes with a hidden Markov model (HMM) derived from SLP phosphatases. Additional sequences were derived by BLASTP searches (retrieving some sequences from organisms without complete genome sequencing) and some by TBLASTN searching of nucleotide sequence databases. The latter proved to be sequences that were unannotated in the protein sequence databases (for details, see “Materials and Methods”; individual sequence derivations are summarized in Supplemental Table S1). After multiple sequence alignment and phylogenetic tree inference using our candidate SLP sequence set, we obtained the data presented in Figure 1 (a radial view of this tree is presented as Supplemental Fig. S4, and the original sequence alignment is given in Supplemental Fig. S1). We found SLPs in representative species from four of the five major eukaryotic supergroups (Plantae, chromalveolates, excavates, and opisthokonts). It is clear from inspection of the sequence composition of this tree that organisms that are now photosynthetic (green algae [Chlorophyta], red algae [Rhodophyta], plants [Streptophyta], and diverse chromalveolates) or that are thought to be derived from photosynthetic ancestors (Apicomplexa, oomycetes, possibly Euglenozoa) predominate. Fungi are the only nonphotosynthetic group represented in strength. A single sequence was found in Apusozoa (an animal ally), and none was found in animals. Thorough TBLASTN searching failed to reveal any additional SLPs among previously unannotated sequences in animals or any other eukaryotic group.
Figure 1.
Phylogenetic orthogonal tree depicting SLP protein phosphatase distribution and interrelationships across eukaryotes, archaea, and bacteria. Phylogenetic tree inference was performed as outlined in “Materials and Methods.” The most crucial nodes are labeled. Branch support values with the four inference methods (PhyML [aBayes], RAxML [RBS], MrBayes [PP], and PhyloBayes_MPI [PP]) are as follows (for details, see “Materials and Methods”): node A, 0.998, 75, 0.86, 1.00; node B, 0.995, 36, 0.80, 0.52; node C, 0.755, 58, 0.86, 0.52; node D, 1.00, 80, 0.93, 1.00; node E, 1.00, 100, 1.00, 1.00; node F, 0.999, 88, 0.93, 1.00. Branch support values for all trees are summarized in Supplemental Table S3. Predicted in silico subcellular localizations are represented as follows: Ch, chloroplast; Cy, cytosol; ER, endoplasmic reticulum; Mt, mitochondria; Nu, nuclear; Px, peroxisome; SP, signal peptide. Sequences used in phylogenetic tree generation are listed in Supplemental Table S1, while compiled in silico subcellular localization data can be found in Supplemental Table S2. Plant SLP1 and SLP2 (green), red/brown/chromalveolate (orange), oomycetes/Apusozoa (aqua), SLP3 (purple), Euglenozoa (red), Apicomplexa (tan), fungi (blue), and outer and inner Myxococcales (gray) SLP phosphatases are shown along with archaea, γ-Proteobacteria, α-Proteobacteria, and other bacteria phosphatases. The root α-Proteobacteria group is outlined in yellow. [See online article for color version of this figure.]
The previously described SLP1 and SLP2 forms of plants and their associated green algae are seen here to represent the terminal, most derived aspect of a broad SLP radiation that spreads across eukaryotes (Fig. 1). The SLP1 and SLP2 sequences have presumably arisen by gene duplication and divergence from the deeper group of SLP sequences (which we here term “SLP3”), which are present as a distinct lineage in green algae. At the base of the SLP tree is a cluster of bacterial sequences from class δ-Proteobacteria, order Myxococcales (which we here term “outer Myxococcales” sequences), plus γ-Proteobacteria (including the genus Shewanella). Within the structure of the eukaryotic SLP radiation itself is a second cluster of δ-proteobacterial sequences from the order Myxococcales (which we here term “inner Myxococcales” sequences).
Intensive searching revealed four archaeal SLPs, all from organisms in the family Halobacteriaceae. In three of four phylogenetic tree inference methods, these clustered with eukaryotic SLPs from the oomycetes and Apusozoa. Finally, closely associated with the SLP sequence assemblage is a group of sequences from α-Proteobacteria (Fig. 1). In another, more basal cluster, are representative distantly related sequences from a diverse group of bacterial phyla, including Cyanobacteria, Bacteroidetes, and Actinobacteria.
We subjected eukaryotic sequences with intact N termini to a battery of subcellular localization prediction methods. Summary data organized by phosphatase class and organismal group are presented in Table I (detailed prediction data for each sequence are presented in Supplemental Table S2). These results are superimposed on the phylogenetic tree clustering data in Figure 1. SLPs have been shown previously to have differing subcellular localizations in plants, with Arabidopsis SLP1 (AtSLP1) being chloroplastic and AtSLP2 likely being cytosolic, when transiently expressed in Vicia faba epidermal leaf cells (Uhrig and Moorhead, 2011a). Our results using bioinformatic predictions of subcellular localization for the plant SLP1 and SLP2 group sequences are in agreement with these previous results; however, the potential for tissue-specific subcellular localization differences still exists. SLPs in the green algae associated with each of these two groups show predicted localizations in accord with their related plant sequences. This suggests that these differing protein isoform localizations may have been established early in evolution, before the advent of land plants. Furthermore, it is interesting that in the group of green algal sequences deeper in the tree (SLP3 phosphatases), the predominant predicted localization is mitochondrial (Fig. 1). This is also true of the sequences from other photosynthetic organisms in the deeper SLP radiation and suggests that protein retargeting may have occurred during SLP sequence evolution. A clear example of this is provided by the group of sequences from Apicomplexa. This group contains the only other SLP protein that has been characterized in detail biochemically, the SHLP1 protein of Plasmodium berghei (the causative agent of malaria in the mouse; Patzewitz et al., 2013). This protein (corresponding to our sequence PbSLPa) has been shown to be localized in the endoplasmic reticulum membrane, which is consistent with our prediction of a signal peptide. Our analysis indicates that this is a conserved feature of most of the SLP proteins from Apicomplexa. It is interesting in this regard that most of the SLP sequences in our data set from the parasitic Euglenozoa also manifest a predicted signal peptide. However, it should be noted that the nonpathogenic fungi Schizosaccharomyces pombe and Laccaria bicolor also possess SLP proteins with predicted signal peptides. Indeed, previous findings indicate that the S. pombe SLP phosphatase is also endoplasmic reticulum localized (Matsuyama et al., 2006).
Table I. Summary of subcellular localization predictions.
This table summarizes consensus subcellular localization predictions for sequences from each bacterial-like protein phosphatase class (SLP, RLPH, and ALPH) and major eukaryotic organismal group: Plantae, chromalveolates (photosynthetic and nonphotosynthetic), rhizaria, excavates, and opisthokonts. Subcellular localization predictions were generated as detailed in “Materials and Methods.” Consensus localizations are abbreviated as follows: Chloro, chloroplast; Cyto, cytoplasmic; Cyto or Nuc, cytoplasmic or nuclear; Mito, mitochondria; No prediction (sequence lacked native N terminus, so no prediction was possible); SP, signal peptide. Complete subcellular localization data are presented in Supplemental Table S2.
| Phosphatase | Organismal Group | No. | Consensus Subcellular Localization |
|---|---|---|---|
| Eukaryotic SLP phosphatases | Plantae (Chlorophyta, Streptophyta) | 107 | Chloro 51; Cyto 37; Mito 9; SP 1; No prediction 9 |
| Chromalveolates | |||
| Photosynthetic | 16 | Mito 6; SP 5; Cyto 3; Chloro 1; No prediction 1 | |
| Nonphotosynthetic | 17 | SP 12; Mito 2; Chloro 1; Cyto 1; No prediction 1 | |
| Excavates | 9 | SP 8; No prediction 1 | |
| Opisthokonts | 9 | SP 4; Mito 3; Cyto 1; No prediction 1 | |
| Eukaryotic RLPH phosphatases | Plantae (Chlorophyta, Streptophyta) | 40 | Cyto or Nuc 31; Cyto 4; Chloro 2; Mito 2; No prediction 1 |
| Rhizaria | 1 | Mito 1 | |
| Excavates | 1 | Cyto or Nuc 1 | |
| Eukaryotic ALPH phosphatases | Plantae (Chlorophyta only) | 6 | Mito 5; Cyto 1 |
| Chromalveolates | |||
| Photosynthetic | 12 | Mito 5; Cyto 3; Chloro 2; No prediction 2 | |
| Nonphotosynthetic | 5 | Cyto 4; Mito 1 | |
| Rhizaria | 2 | Mito 2 | |
| Excavates | 7 | Cyto 4; Chloro 1; Mito 1; No prediction 1 | |
| Opisthokonts | 19 | SP 8; Mito 6; Cyto 4; No prediction 1 | |
| Eukaryotic ApaH phosphatases | Plantae (Streptophyta only) | 7 | Cyto 3; No prediction 4 |
| Chromalveolates | |||
| Photosynthetic | 2 | Cyto 2 | |
| Opisthokonts | 14 | Cyto 9; SP 2; No prediction 3 | |
| Eukaryotic PA3087 phosphatases | Chromalveolates | ||
| Photosynthetic | 1 | Cyto 1 | |
| Nonphotosynthetic | 1 | No prediction 1 | |
| Rhizaria | 1 | Cyto 1 | |
| Opisthokonts | 1 | No prediction 1 |
RLPH Phosphatases
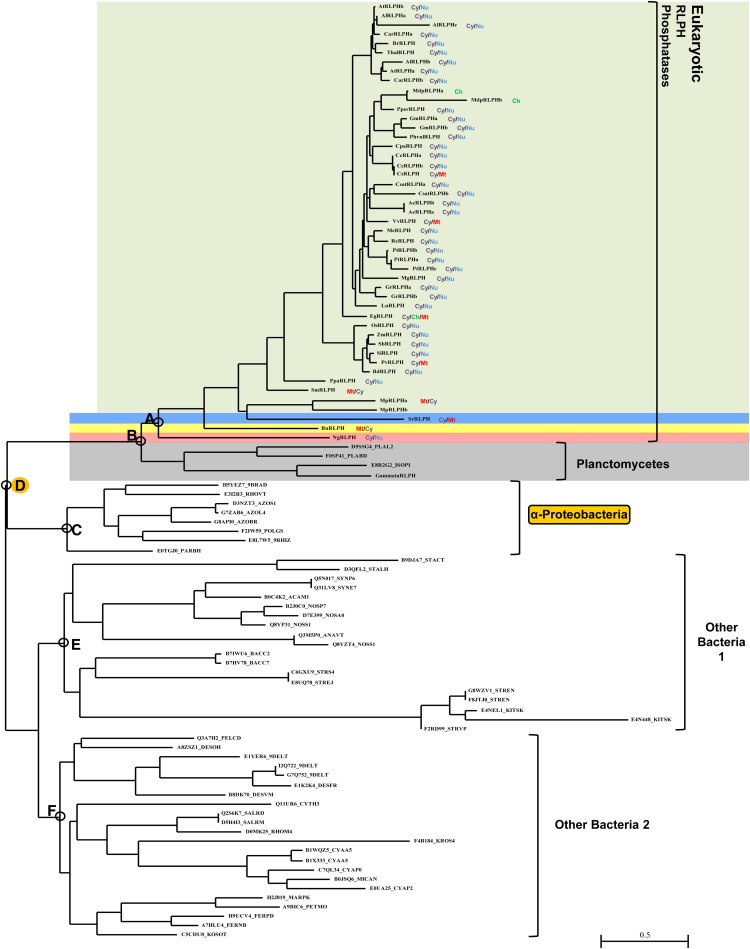
Our data on the distribution and interrelationships of the RLPHs are presented in Figure 2 (a radial view of this tree is presented as Supplemental Fig. S5, and the original sequence alignment is given in Supplemental Fig. S2). Once again, we see that the species representation is heavily weighted toward photosynthetic organisms, with the only exceptions being the heterolobosean Naegleria gruberi (an excavate) and the choanoflagellate Salpingoeca rosetta (an opisthokont). Among photosynthetic organisms, the RLPH distribution is dominated by land plants. Despite intensive searching, RLPH sequences could only be detected in two different strains of a single green algal species, Micromonas pusilla. None of the photosynthetic chromalveolates contained a RLPH sequence, with the sole remaining eukaryotic organism being the photosynthetic rhizarian Bigelowiella natans. Intensive searching by TBLASTN failed to reveal any additional RLPHs among previously unannotated sequences from other species of green algae or any other eukaryotic group. At the base of the RLPH distribution is a closely related set of sequences from planctomycete bacteria.
Figure 2.
Phylogenetic orthogonal tree depicting RLPH protein phosphatase distribution and interrelationships across both eukaryotes and bacteria. Phylogenetic tree inference was performed as outlined in “Materials and Methods.” The most crucial nodes are labeled. Branch support values with the four inference methods (PhyML [aBayes], RAxML [RBS], MrBayes [PP], and PhyloBayes_MPI [PP]) are as follows (for details, see “Materials and Methods”): node A, 0.999, 99, 0.98, 1.00; node B, 0.575, 80, 0.95, 0.86; node C, 0.999, 90, 0.93, 1.00; node D, 1.00, 100, 0.98, 1.00; node E, 0.999, 16, 0.83, 0.95; node F, 0.999, 12, 0.90, 0.91. Branch support values for all trees are summarized in Supplemental Table S3. Predicted in silico subcellular localizations are represented as follows: Cy, cytosol; Mt, mitochondria; Nu, Nuclear. Sequences used in tree generation are listed in Supplemental Table S1, and in silico subcellular localization data are listed in Supplemental Table S2. Plant RLPH2 (green), choanoflagellida (blue), rhizaria (yellow), heterolobosea (red), and Planctomycetes (gray) are shown along with α-Proteobacteria, other bacteria 1, and other bacteria 2. The root α-Proteobacteria group is outlined in yellow. [See online article for color version of this figure.]
Closely associated with the RLPH sequence distribution is a set of sequences from α-Proteobacteria. Other, more distantly related bacterial sequences include representatives from a variety of groups including Cyanobacteria, δ-Proteobacteria, Bacteroidetes, and Thermotogae. No RLPH sequences were detected from archaea by HMM searching of protein databases derived from completely sequenced archaeal genomes, BLASTP searching of archaeal protein databases, or TBLASTN searching among archaeal nucleotide databases.
The RLPH proteins have a distinctive predicted subcellular localization not shared by the SLP or ALPH proteins. Most sequences have a predicted cytoplasmic/nuclear localization. This is true not only of the land plants but also the N. gruberi sequence, the most deeply diverging in the tree, which suggests that a distinctive targeting of RLPH class sequences may have occurred early in eukaryotic evolution.
ALPH Phosphatases
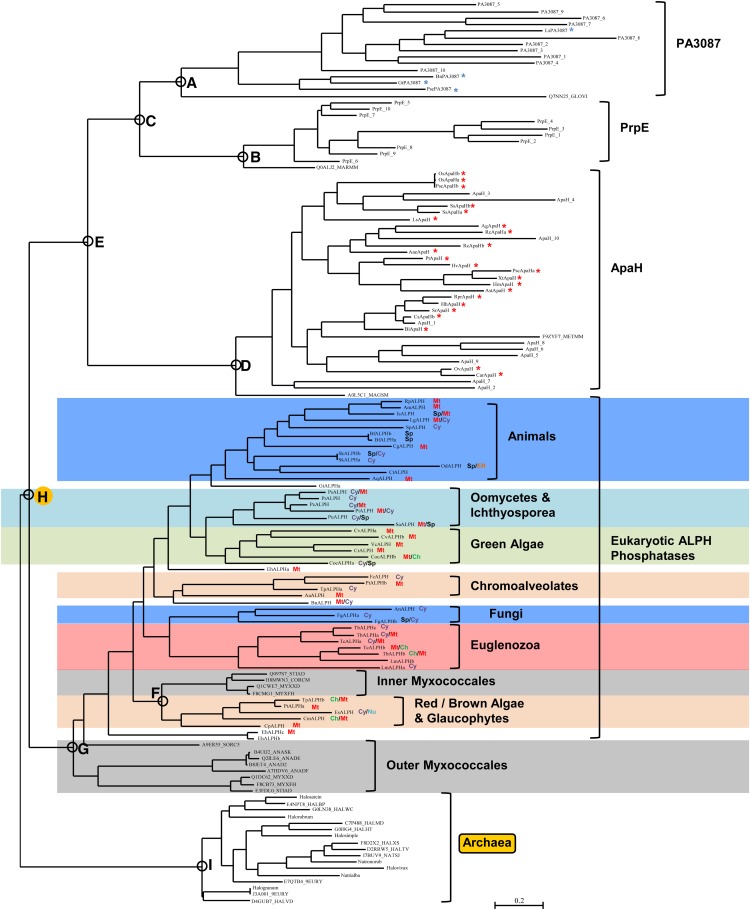
The original work of Andreeva and Kutuzov (2004) established the similarity of a class of eukaryotic protein phosphatase sequence (ALPHs) to the ApaH (diadenosine tetraphosphatase) sequences of bacteria. To lay the foundation for our characterization of ApaH-like protein phosphatase sequences in eukaryotes, we examined the “sequence neighborhood” of the ApaH class by a consideration of conserved domains documented in the National Center for Biotechnology Information (NCBI) Conserved Domain Database. According to their annotations, which we confirmed independently by our own preliminary sequence alignments and phylogenetic trees (data not shown), bacterial ApaHs (cd07422: MPP_ApaH [Escherichia coli ApaH and related proteins, metallophosphatase domain]) are related to bacterial PrpEs (cd07423: MPP_PrpE [Bacillus subtilis PrpE and related proteins, metallophosphatase domain]) and bacterial PA3087s (cd07413: MPP_PA3087 [Pseudomonas aeruginosa PA3087 and related proteins, metallophosphatase domain]). In practice, searches with our HMM derived from eukaryotic ALPH sequences, against protein databases from completely sequenced eukaryotic genomes, confirmed these relationships, as eukaryotic sequences were detected in two of these three sequence classes. The results of our multiple sequence alignment and phylogenetic tree analysis of candidate eukaryotic ALPHs and associated “accessory” group sequences are presented in Figure 3 (for a radial tree view, see Supplemental Fig. S6; for the multiple sequence alignment used, see Supplemental Fig. S3).
Figure 3.
Phylogenetic orthogonal tree depicting ALPH protein phosphatase distribution and interrelationships across eukaryotes, archaea, and bacteria. Phylogenetic tree inference was performed as outlined in “Materials and Methods.” The most crucial nodes are labeled. Branch support values with the four inference methods (PhyML [aBayes], RAxML [RBS], MrBayes [PP], and PhyloBayes_MPI [PP]) are as follows (for details, see “Materials and Methods”): node A, 0.892, 83, 0.81, 0.67; node B, 1,00, 91, 1.00, 1.00; node C, 0.999, 100, 1.00, 0.74; node D, 0.885, 100, 1.00, 0.87; node E, 1.00, 40, 0.96, 1.00; node F, 0.996, 54, 0.99, 0.98; node G, 0.994, 25, 0.85, 0.70; node H, 0.998, 59, 0.99, 0.97; node I, 1.00, 100, 0.99, 1.00. Branch support values for all trees are summarized in Supplemental Table S3. Predicted in silico subcellular localizations are represented as follows: Ch, chloroplast; Cy, cytosol; ER, endoplasmic reticulum; Mt, mitochondria; Nu, nuclear; SP, signal peptide. Sequences used in tree generation are listed in Supplemental Table S1, while compiled in silico subcellular localization data can be found in Supplemental Table S2. Eukaryotic ApaH (red) and eukaryotic PA3087 (blue) sequences are starred; the other sequences in these clusters are bacterial. Animal and fungi (blue), oomycetes/Ichthyosporea (aqua), green algae (green), red/brown/chromalveolate/glaucophyte (orange), Euglenozoa (red), and outer and inner Myxococcales (gray) SLP phosphatases are shown along with archaea, ApaH, PrpE, and PA3087 bacteria phosphatases. The root archaea group is outlined in yellow. [See online article for color version of this figure.]
Eukaryotic ALPHs comprise a large clade with representatives from every currently recognized eukaryotic supergroup (Plantae, rhizaria, chromalveolates, excavates, opisthokonts). It is notable, however, that while there are green algal representatives, land plants are missing (Fig. 3). It should also be noted that ALPH sequences were not found in land plant genomic sequences by TBLASTN searching. Intermixed, and closely associated with the base of the eukaryotic ALPH clade, are two sets of sequences from class δ-Proteobacteria, order Myxococcales (inner Myxococcales and outer Myxococcales). Furthermore, closely associated with the eukaryotic ALPH clade is a group made up of sequences from archaea (Fig. 3). These archaeal sequences are all from closely related genera in the family Halobacteriaceae.
Surprisingly, eukaryotic sequences were also detected that cluster with bacterial sequences within highly supported “accessory” sequence groups related to the eukaryotic ALPHs. A mixed ApaH cluster was composed of sequences from a number of bacterial groups (α-, β-, γ-, and ε-Proteobacteria) together with eukaryotic sequences from plants, nonphotosynthetic chromalveolates, and animals (opisthokonts). Most of the eukaryotic sequences are not annotated in current protein databases. A mixed PA3087 cluster was composed of sequences from a number of bacterial groups (Actinobacteria, α-Proteobacteria, Cyanobacteria, γ-Proteobacteria, Verrucomicrobia, Lentisphaerae, and Bacteroidetes) plus eukaryotic sequences from a photosynthetic rhizarian, photosynthetic and nonphotosynthetic chromalveolates, and animals. Similarly, two of these eukaryotic sequences are not annotated in current protein databases. Despite intensive searching by HMMs, BLASTP, and TBLASTN, no archaeal sequences were found that clustered with the ALPH accessory groups.
Subcellular localization predictions for ALPH sequences from photosynthetic eukaryotes, considered as a group, tend to be either mitochondrial or cytoplasmic, with the former more prominent (12 mitochondrial, four cytoplasmic, and two chloroplast; Table I; Supplemental Table S2). The preference for mitochondrial localization is more marked in glaucophyte and green algal ALPH sequences (six mitochondrial and one cytoplasmic), whereas cytoplasmic localization is more marked in land plant ApaHs (three cytoplasmic and no mitochondrial). In ALPH and ApaH sequences of nonphotosynthetic organisms, the clearly predominant characteristic is predicted cytoplasmic localization (23 sequences), followed by prediction of a signal peptide (10 sequences) or predicted mitochondrial localization (eight sequences). Predictions of a signal peptide are restricted to sequences from fungi and animals, suggesting that this may be an evolutionary innovation restricted to the opisthokonts.
Combined SLP, RLPH, and ALPH Phosphatase Set
As a final check on the validity of the individual phylogenetic trees presented here for the SLP, RLPH, and ALPH bacterial-like phosphatases, we combined these three sequence sets, produced a joint alignment, and inferred a combined sequence phylogenetic tree. The result is shown in radial form in Supplemental Figure S7. Inspection of this tree shows that all the major relationships of the individual trees are preserved.
Sequence Motif Identification
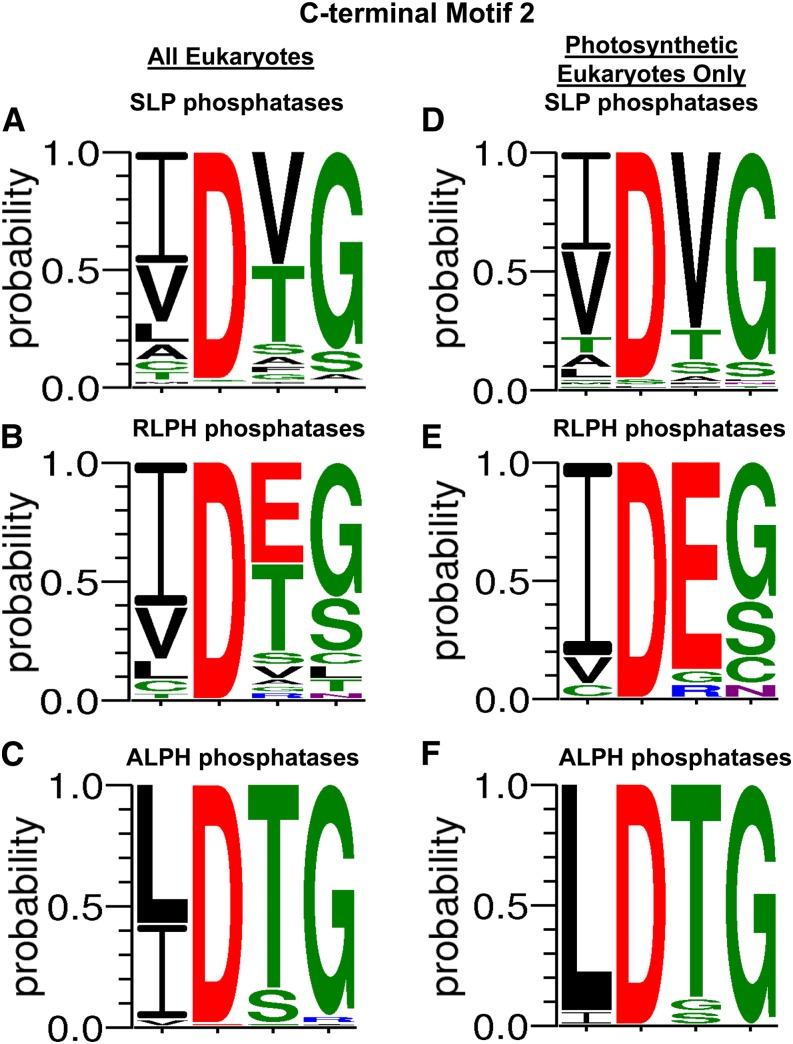
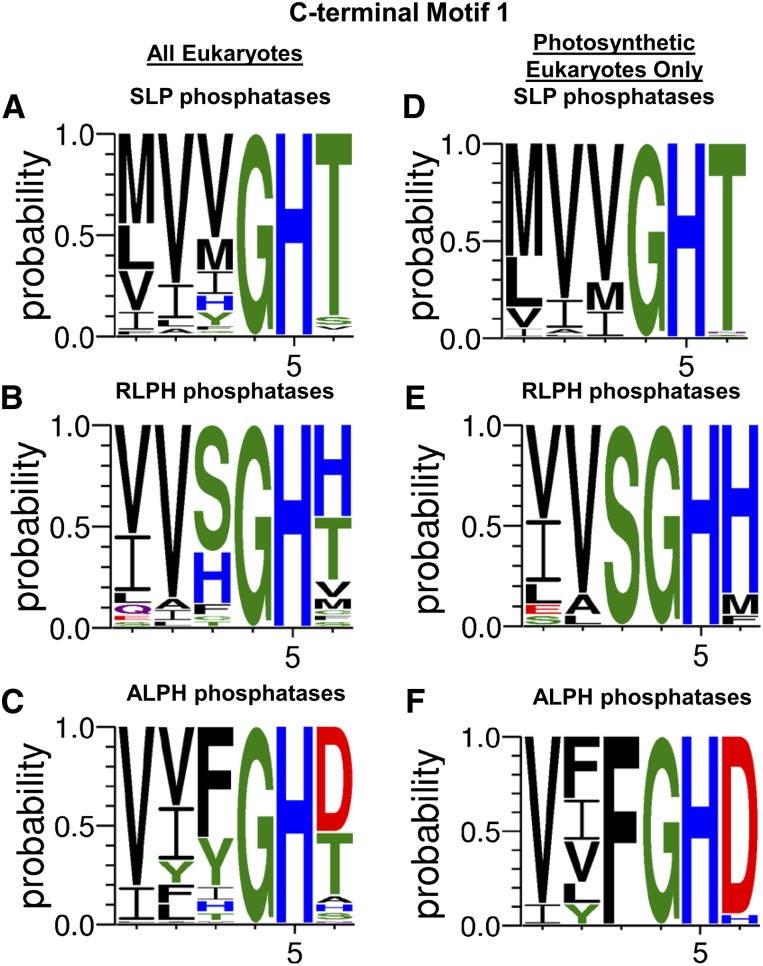
Upon the classification of SLP, RLPH, and ALPH phosphatases, a novel C-terminal sequence motif, I/L/V-D-S/T-G (labeled motif 2 here), was revealed (Andreeva and Kutuzov, 2004). Our data confirm the conservation of sequence motif 2 across all eukaryotic bacterial-like phosphatases (Fig. 4) in addition to revealing a second C-terminal motif (motif 1), (M/I//V)-(I/L/V)-(V/S/F)-G-H-(T/H/D), upstream of motif 2 (Fig. 5). Within both of these sequence motifs, each eukaryotic bacterial-like phosphatase class was found to maintain distinct diversity at specific motif positions that parallel their classification (Figs. 4 and 5). This was most pronounced when examining these motifs from photosynthetic eukaryotes (Figs. 4 and 5). Figure 6 summarizes the distinctive sequence features of the bacterial-like phosphatases in comparison with other representative members of the PPP family.
Figure 4.
Compiled canonical bacterial-like phosphatase motif 2 from SLP, RLPH, and ALPH protein phosphatases. A to C, Amino acid positional probability consensus within the bacterial-like motif 2 of SLP (A), RLPH (B), and ALPH (C) phosphatases from eukaryotic organisms outlined in each respective phylogenetic tree and listed in Supplemental Table S1. D to F, Amino acid positional probability consensus within bacterial-like motif 2 of photosynthetic eukaryote SLP (D), RLPH (E), and ALPH (F) phosphatases only. The greatest diversity was observed in motif position 3, where Thr (T), conserved among prokaryotic and eukaryotic SLP and RLPH phosphatases alike, was replaced with Val (V) and Glu (E) in photosynthetic eukaryote SLP and RLPH phosphatases, respectively. Amino acid colors represent polar (green), neutral (purple), basic (blue), acidic (red), and hydrophobic (black) amino acids. Each amino acid positional probability consensus was constructed using MAFFT-aligned sequences submitted to WebLogo 3 (http://weblogo.threeplusone.com/). [See online article for color version of this figure.]
Figure 5.
Compiled canonical bacterial-like phosphatase motif 1 from SLP, RLPH, and ALPH protein phosphatases. A to C, Amino acid positional probability consensus within the bacterial-like motif 1 of SLP (A), RLPH (B), and ALPH (C) phosphatases from eukaryotic organisms outlined in each respective phylogenetic tree and listed in Supplemental Table S1. D to F, Amino acid positional probability consensus within bacterial-like motif 1 of photosynthetic eukaryote SLP (D), RLPH (E), and ALPH (F) phosphatases only. Bacterial-like motif 1 exhibited greatest diversity in motif positions 1 through 3, where predominantly a mixed variety of hydrophobic amino acids were observed. Similar to position 3 of motif 2, position 6 of motif 1 also exhibited conserved bacterial-like class diversity, with photosynthetic eukaryote SLP, RLPH, and ALPH phosphatases predominantly maintaining Thr (T), His (H), and Asp (D) residues, respectively. Amino acid colors represent polar (green), neutral (purple), basic (blue), acidic (red), and hydrophobic (black) amino acids. Each amino acid positional probability consensus was constructed using MAFFT-aligned sequences submitted to WebLogo 3 (http://weblogo.threeplusone.com/). [See online article for color version of this figure.]
Figure 6.
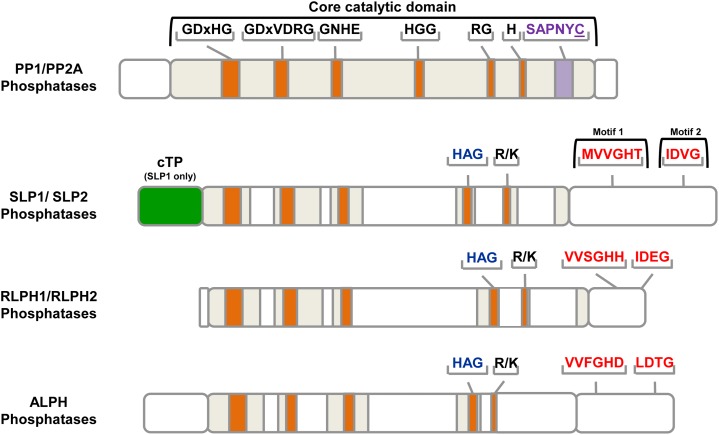
Unique motif features of the eukaryotic, bacterial-like PPP family SLP, RLPH, and ALPH phosphatases. The highly conserved core catalytic domains of representative PPP family phosphatases PP1 and PP2A are depicted in gray with signature motifs highlighted. Amino acids involved in metal ion coordination and phosphate binding are depicted by orange bars, while the microcystin inhibition docking motif SAPNYC is illustrated with a purple bar. The reactive Cys (C) to which microcystin covalently attaches is underlined. Unique motifs defining each bacterial phosphatase subfamily are depicted (red) and labeled as motif 1 and 2. A chloroplast transit peptide (cTP) is also denoted (green); however, this feature is only found in SLP1 phosphatases. Protein models depicted here were derived from At2g29400 (TOPP1; AtPP1), At1g69960 (AtPP2A-1), At1g07010 (AtSLP1), At1g18480 (AtSLP2), At3g09970 (AtRLPHa), At3g09970 (AtRLPHb), and VcALPH (Vocar20010015m). [See online article for color version of this figure.]
DISCUSSION
For two types of eukaryotic bacterial-like PPPs investigated here (SLPs and RLPHs), a well-supported group of sequences from α-Proteobacteria lies in close association in phylogenetic trees. The most straightforward interpretation of this observation is that these bacterial-like PPP genes entered eukaryotes very early in their history, with the advent of mitochondria. This is consistent also with the broad extant eukaryotic distribution of the SLP sequences. Current concepts of the origin of mitochondria and early eukaryotes differ somewhat in their details, with either endosymbiosis of an α-proteobacterium within an amitochondriate eukaryotic host or symbiogenesis combining an α-proteobacterium with another prokaryote (usually deemed to be an archaeon; Koonin, 2010). These concepts are embodied in competing and as yet unresolved models of early eukaryotic evolution (Embley and Martin, 2006; Poole and Penny, 2007). However, all are agreed that the advent of mitochondrial formation, with its attendant large-scale genetic transfer to the eukaryotic nucleus, together with intracellular retargeting of translated proteins, was a major driver of eukaryotic evolution. Classically, the donor α-proteobacterium was held to be an ancient Rickettsia-like organism (Gray, 1998; Lang et al., 1999). Our data fail to support this hypothesis. None of the deeply placed α-proteobacterial sequences we found in either of these bacterial-like PPP trees are from the order Rickettsiales. These findings are consistent with a recent review (Gray, 2012) that emphasized that the true identity of the ancestral α-proteobacterium has yet to be definitively established.
Superimposed on a basic pattern of α-proteobacterial ancestry in the SLP and RLPH trees is a more complex picture of bacterial-like PPP protein phosphatase origins. In each of these classes, there is a group of bacterial sequences that very closely clusters with the radiation of each sequence type in eukaryotes. In the case of the SLPs, these are from the order Myxococcales of the class δ-Proteobacteria, while in the case of the RLPHs, these are from the Planctomycetes. As befits their positioning in the phylogenetic trees, these sequences are much more closely related to their respective eukaryotic sequence group than are those of the presumably ancestral α-Proteobacteria. This is reflected, for example, in much higher scores with their respective eukaryotic sequence-derived HMM type. One possible interpretation of these results is horizontal gene transfer or LGT. One of the hallmarks of this process is a “discordant” clustering of sequences from distant organismal sources in the same gene tree (Keeling and Palmer, 2008; Boto, 2010). Alternatively, given the likelihood of the α-proteobacterial ancestry detailed above, a more attractive possibility is that, in each case, a particular bacterial-like PPP sequence radiation in eukaryotes (e.g. the SLPs) would be viewed as the “sister group” of the closely related bacterial sequence cluster (e.g. the outer Myxococcales sequences). Both would derive from the same α-proteobacterial source. This interpretation is given as inset diagrams in the figures for each of the radial phylogenetic tree representations (Supplemental Figs. S4 and S5).
The above mechanisms are sufficient to explain the structure of the RLPH tree (Supplemental Fig. S5), which is the simpler of the two. In the case of the SLP tree, there is a further complication in that there is a second group of bacterial sequences (inner Myxococcales) sequestered within the overall eukaryotic radiation (Supplemental Fig. S4). This can be explained by a second application of the sister group argument above, where this time a more basal eukaryotic SLP sequence ancestor gave rise to both a further, more derived eukaryotic SLP radiation and also a second side cluster of Myxococcales sequences. However, given that the origin of the sequences would be eukaryotic and the destination bacterial, this would qualify as a possible instance of LGT.
Other hallmarks of LGT besides phylogenetically discordant clustering patterns are so-called “patchy distributions,” where there is nonuniform sequence representation among a broad organismal phylogenetic group (Snel et al., 2002). It is important to emphasize that it is generally possible to model such unusual sequence-clustering patterns as either LGT or as differential gene amplification, vertical transmission, and survival in descendant organismal lineages (Snel et al., 2002; Kurland et al., 2003). Instances must be judged as individual situations, and sometimes it still remains difficult or impossible to establish an unambiguous mechanism. In the case of the inner Myxococcales sequences within the eukaryotic SLP sequence distribution, we favor LGT, as it would be difficult to conceptualize this as a case of differential gene transmission and loss.
Another possible instance of LGT in the SLP tree is the clustering of the four archaeal SLP sequences with the deep eukaryotic SLP cluster from oomycetes and Apusozoa. However, caution must be exercised here. It has been recognized previously that rapid rates of sequence evolution may bias the branching patterns within phylogenetic trees, giving an artifactual appearance of LGT (Kurland et al., 2003; Keeling and Palmer, 2008). The branches for both the oomycete and archaeal SLP sequence clusters are the longest in the tree, indicating rapid sequence evolution. This is consistent with these sequences being the most divergent in the sequence alignment (Supplemental Fig. S2). It is possible, therefore, that this clustering may be an instance of the “long branch attraction” artifact well known in phylogenetic tree inference work (Brinkmann et al., 2005; Embley and Martin, 2006; Koonin, 2010).
The ALPH sequence-derived phylogenetic tree presents one fundamental difference from the SLP and RLPH trees considered above. In this tree, rather than an α-proteobacterial sequence cluster in association with the eukaryotic ALPHs, there is a cluster from archaea. This strongly suggests that the origin of the eukaryotic ALPH sequences was in an ancient archaeal ancestor. The proposed ancient archaeal root of this tree is indicated in the radial representation depicted in Supplemental Figure S6. This hypothesis is consistent with the strong case recently presented (Koonin, 2010) for the formation of the eukaryotic cell from an archaeal ancestor via symbiogenesis. In this scenario, the observed archaeal sequences would be persisting in the living descendants of the original archaeal ancestor population. This model is depicted in the radial tree presented in Supplemental Figure S6. Since the present ALPH sequences are restricted to the family Halobacteriaceae, this might suggest that the archaeal ancestor of eukaryotes was a halophile. While this is conceivable, Koonin (2010) proposes that eukaryotes received several critical cellular systems from a more complex, basal archaeal ancestor and that current archaea represent the products of selective genomic loss and streamlining. If this is so, the current ALPH-containing archaea may not accurately reflect either the lifestyle or the genomic complexity of the eukaryotic ancestor population.
Another feature of the ALPH tree is reminiscent of the SLP tree. There are two clusters of sequences from Myxococcales very tightly associated with the eukaryotic ALPH sequences. The outer Myxococcales and inner Myxococcales sequences can be explained by serial applications of the sister group argument presented above. However, unlike the SLP tree, in the ALPH tree each of these steps would involve an interdomain transfer and might thus be considered possible examples of LGT. This model is depicted in the radial tree presented in Supplemental Figure S6. Once again, it would be difficult to explain these results by postulating differential ALPH gene transmission and loss within both bacteria and eukaryotes.
Our ALPH tree also confirms evidence derived from the study of conserved domains (NCBI Conserved Domains Database), that the bacterial PrpE and PA3087 classes are related to the bacterial ApaH class. Since no archaeal sequences were found to cluster in the accessory groups portion of the ALPH tree, it appears that these sequence types are not of archaeal origin.
It is also very interesting that the ApaH and PA3087 clusters each contain a mixture of sequences from bacteria, photosynthetic eukaryotes, and animals. The eukaryotic sequences were often unannotated, discovered by searching organismal nucleotide sequence databases. Once again, LGT appears to be a possible explanation. These newly documented ApaH and PA3087 sequences of photosynthetic eukaryotes deserve further characterization to determine if they are expressed and functional in their host species.
It is intriguing that the two bacterial groups whose sequences are most closely related to the eukaryotic bacterial-like PPPs (Myxococcales and Planctomycetes) have been noted as being “eukaryote like” in terms of possessing features unusual for bacteria: in the former case, a complex life cycle and social behavior heavily dependent on intercellular signaling (Goldman et al., 2006; Pérez et al., 2008); in the latter case, intracellular compartmentation (Fuerst and Sagulenko, 2011). This might suggest that further research into the role of the ALPH and SLP proteins in Myxococcales and RLPHs in Planctomycetes is warranted.
It would appear that the ALPH gene lineage has become extinct in land plants, although it is widely represented in green algae and, therefore, was presumably present in the land plant ancestor. This suggests that the function(s) of the ALPH protein either became unnecessary in a terrestrial organism or became redundant due to the acquisition of this function by another gene lineage. In contrast, the SLPs underwent gene expansion in green algae, with an ancestral form (SLP3) giving rise to the SLP1 and SLP2 forms that were later inherited by land plants. This suggests that each related gene product might serve distinct cellular functions (Kutuzov and Andreeva, 2012). This inference is supported by the distinct localizations shown by Arabidopsis SLP1 (chloroplast) and SLP2 (cytoplasm; Uhrig and Moorhead, 2011a), whose generality among land plants is indicated by our in silico subcellular localization prediction data. Finally, the RLPH gene lineage has become nearly extinct in living green algae while it is ubiquitous in land plants. This might suggest again a cooption of gene function in algae, as discussed above. It is noteworthy that the RLPHs of land plants show a predicted cytoplasmic/nuclear localization that is unique in all the eukaryotic bacterial-like PPPs. This suggests a marked change in cellular function, which deserves further research exploration.
It is ironic that, at present, the most is known about the function of an SLP protein from a nonphotosynthetic organism. In P. berghei (the causative organism of malaria in mice), the SHLP1 protein has been shown to be necessary for a critical life cycle stage transition and for the development of ultrastructural features important for host cell infection (Patzewitz et al., 2013). It is well established that Plasmodium spp. (like all alveolates) were ancestrally photosynthetic, retaining an altered chloroplast remnant, the apicoplast (Kalanon and McFadden, 2010). This indicates that, in this organism, the SHLP1 gene, freed from possible previous functional constraints in a photosynthetic ancestor, evolved a novel role important to the pathogenic lifestyle. Since both the mouse and human hosts of malaria parasites lack any evidence of SLP genes and proteins, SHLP1 represents an attractive target for therapeutic drug development.
It is interesting that in our SLP phylogenetic tree (especially notable in the radial representation in Supplemental Fig. S4) there are several groups of sequences in the deep eukaryotic portion of the tree that have long branches (indicating probable rapid sequence evolution) and that are encoded by parasitic organisms. Most sequences in the Apicomplexa group (including several species of Plasmodium) have a predicted signal peptide, confirming previous findings (Kutuzov and Andreeva, 2008). This correlates with the discovery that the SHLP1 protein discussed above is localized to the endoplasmic reticulum membrane. There are two sequences from the genus Perkinsus (a marine shellfish pathogen), a group from the Euglenozoa (including the genera Leishmania and Trypanosoma), and a group including oomycete plant pathogens from the genera Phytophthora and Pythium. In the case of the Euglenozoa and genus Perkinsus, there is also a marked tendency toward the possession of a signal peptide. The predicted localizations are more mixed for the oomycete sequences; this may be because they are the most divergent SLP sequences in our data set, and the N termini may have been misannotated. In contrast, among SLP sequences from photosynthetic organisms, predicted signal peptides were rare. Taken together, these observations suggest that the SLP sequences of pathogens of both plants and animals may have taken alternative evolutionary trajectories from those in currently photosynthetic organisms. These genes and proteins may thus represent attractive targets of further research efforts.
The catalytic subunits of eukaryotic PPPs such as PP1 and PP2A are well known to combine with a variety of regulatory subunits to form holoenzymes, which provides for substrate specificity, subcellular localization, and enzymatic regulation (Virshup and Shenolikar, 2009). In PP1, for example, these interactions are mediated by small canonical motifs such as the RVxF and S/GILK motifs (Templeton et al., 2011). It has been recently suggested that SLP phosphatases might also interact with a diverse set of regulatory proteins (Uhrig and Moorhead, 2011b; Kutuzov and Andreeva, 2012). The data on C-terminal motifs presented here demonstrate that they maintain conserved class-specific alterations, which are most pronounced in photosynthetic eukaryotes (Figs. 4 and 5). Amino acid substitutions in positions 6 and 3 of motifs 1 and 2, respectively, would be expected to alter motif charge, polarity, and hydrophobicity. This could alter protein-binding specificity without an overall change in phosphatase conformation, suggesting that a regulatory protein-binding strategy might be a general feature of the bacterial-like PPP phosphatases. Exploration of this possibility represents an attractive option for future research.
MATERIALS AND METHODS
Multiple Sequence Alignments
Protein sequences were aligned using MAFFT, version 7 (Katoh et al., 2002; http://mafft.cbrc.jp/alignment/server/), with the BLOSUM45 scoring matrix, using the E-INS-i option (very slow, multiple domains with long inserts). Alignments were visualized and hand edited in GeneDoc (Nicholas et al., 1997; http://www.nrbsc.org/gfx/genedoc/).
Candidate Sequence Search, Retrieval, and Validation
Initial eukaryotic sequences of the ALPH, SLP, and RLPH phosphatases were obtained from the literature (Andreeva and Kutuzov, 2004; Uhrig and Moorhead, 2011a) and through database searching at NCBI with BLASTP and PSI-BLAST (Altschul et al., 1997; http://blast.ncbi.nlm.nih.gov/). These were then used to generate initial multiple sequence alignments, as described above. Edited multiple sequence alignments were converted into Stockholm format and used to generate HMMs by the HMMER (version 3.0) software suite (Eddy, 1998; http://hmmer.janelia.org/). Databases of protein sequences from completely sequenced eukaryotic and prokaryotic species were compiled locally. Eukaryotic sequences were obtained from the Joint Genome Institute (http://www.jgi.doe.gov/), Phytozome (http://www.phytozome.net/), Metazome (http://www.metazome.net/), or individual genome project Web sites. Prokaryotic sequences were obtained from UniProt (http://www.uniprot.org/). Databases were searched using HMMs, and candidate sequences were extracted and then placed into further multiple sequence alignments as described above. Potential candidate sequences were evaluated from the full range of HMM hits for each sequence class, from the strongest (lowest E value) to the weakest (the statistical inclusion threshold [E ∼ 0.01]). In some instances, further iteration was performed with BLASTP and HHblits (Remmert et al., 2012; http://toolkit.tuebingen.mpg.de/hhblits) searching of the UniProt database comprising all bacterial sequences, HAMAP (http://hamap.expasy.org/; Lima et al., 2009).
The rationale was to supplement previously identified candidate sequences from completely sequenced genomes with closely related homologs (approximately E < 1e-50) from nonsequenced genomes.
Candidate searching for each sequence class was supplemented by using two validated query sequences per class for NCBI TBLASTN searches against various databases: nucleotide collection, NCBI genomes (chromosome), high-throughput genomic sequences, and whole-genome shotgun contigs. Only candidate sequences with full-length hits to the query and credible matches to conserved C-terminal conserved sequence motifs (see below) were considered for further evaluation. The rationale was to search for previously unannotated sequences relevant to each target sequence class. Candidate sequence identity was confirmed through phylogenetic tree inference. All sequences found are given in Supplemental Table S1.
Phylogenetic Tree Inference
ProtTest, version 2.4 (Abascal et al., 2005; http://darwin.uvigo.es/software/prottest2_server.html) was used with completed multiple sequence alignments to assess the optimal amino acid substitution model to use for subsequent work. In all instances, the Le and Gascuel (LG) model (Le and Gascuel, 2008) with four γ-categories was optimal. Multiple sequence alignments were subjected to phylogenetic tree inference at both the CIPRES Science Gateway (Miller et al., 2010; http://www.phylo.org/index.php/portal/) and locally. Maximum likelihood analysis (RAxML version 7.4.2; Stamatakis et al., 2008; http://www.exelixis-lab.org/) was run at CIPRES under the LG amino acid substitution model, using a maximum of 1,000 rapid bootstraps or until automatic convergence was reached. Bayesian analysis (MrBayes version 3.1.2; Ronquist et al., 2012; http://mrbayes.sourceforge.net/) was performed at CIPRES, using four independent chains, under the mixed amino acid substitution model (the LG model is not available with this implementation), with four discrete γ-categories, running to a maximum of 7.5 million tree generations or until automatic convergence (average sd of split frequencies < 0.010) was achieved. Bayesian analysis (PhyloBayes_MPI version 1.3b; Lartillot et al., 2009; http://megasun.bch.umontreal.ca/People/lartillot/www/downloadmpi.html) was run on the WestGrid system of Compute Canada (https://computecanada.ca/index.php/en/), using two independent chains, under the LG amino acid substitution model, with four discrete γ-categories. Maximum likelihood analysis (PhyML-aBayes version 3.0.1beta; Anisimova et al., 2011; http://www.atgc-montpellier.fr/phyml/versions.php) was run locally, under the LG model, with four discrete γ-categories, with all other parameters at defaults, through 25 random starts, employing an initial parsimony input tree, and subtree pruning and regrafting moves. For each analyzed protein sequence class, trees were obtained by all the utilized inference methods that had concordant topologies at all major nodes. Within the body of this report, tree figures are presented that represent a typical topology (Figs. 1–3), with branch support given for each method at the most critical nodes. The branch support for all the trees is summarized in Supplemental Table S3. For Bayesian methods (MrBayes and PhyloBayes_MPI), branch support represents the posterior probability (PP; maximum value = 1.00). For the PhyML maximum likelihood method, branch support represents a Bayesian-like transformation of the approximate likelihood ratio test value (aBayes; Anisimova et al., 2011; [maximum value = 1.00]). For the RAxML maximum likelihood method, branch support represents rapid bootstrap support (RBS; [maximum value = 100]).
Subcellular Localization Prediction
A battery of methods (10 in total for plant or algal sequences with chloroplast potential, nine for sequences from nonplant species) was used to infer the probable subcellular localization of the eukaryotic proteins described in this study. These were TargetP (Emanuelsson et al., 2000; http://www.cbs.dtu.dk/services/TargetP/), WoLF PSORT (Horton et al., 2007; http://wolfpsort.org/), PREDOTAR (Small et al., 2004; http://urgi.versailles.inra.fr/predotar/predotar.html), Protein Prowler (Bodén and Hawkins, 2005; http://bioinf.scmb.uq.edu.au/pprowler_webapp_1-2/), PredSL (Petsalaki et al., 2006; http://hannibal.biol.uoa.gr/PredSL/input.html), SLP-Local (Matsuda et al., 2005; http://sunflower.kuicr.kyoto-u.ac.jp/~smatsuda/slplocal.html), iPSORT (Bannai et al., 2002; http://ipsort.hgc.jp/), PCLR (Schein et al., 2001; http://www.andrewschein.com/pclr/), MITOPROT (Claros and Vincens, 1996; http://ihg.gsf.de/ihg/mitoprot.html), and ChloroP (Emanuelsson et al., 1999; http://www.cbs.dtu.dk/services/ChloroP/). The top two in silico-predicted subcellular localizations for each protein sequence are displayed on each respective phylogenetic tree branch (Figs. 1–3). A single subcellular localization is given for those protein sequences where the prediction methods provided a clear preponderance of that location (80% of methods used). Protein sequences where no subcellular prediction is given are those that are fragments lacking a native N terminus (N-terminal Met) and therefore could not be properly assessed. The majority of in silico techniques applied here also have their own internal thresholds for compartment predictions and automatically convert the sequence score into a compartment prediction. A complete output from these prediction algorithms is found in Supplemental Table S2.
Analysis of Sequence Motifs
ALPH, SLP, and RLPH sequences were identified by HMM, BLASTP, and TBLASTN analyses as detailed above, aligned using MAFFT, with each alignment visualized and hand edited in GeneDoc. Highly conserved C-terminal regions were manually identified, and an amino acid positional probability consensus was generated using WebLogo 3 (Crooks et al., 2004; http://weblogo.threeplusone.com/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the phosphatase domain of SLP protein phosphatases from both prokaryotes and eukaryotes.
Supplemental Figure S2. Alignment of the phosphatase domain of RLPH protein phosphatases from both prokaryotes and eukaryotes.
Supplemental Figure S3. Alignment of the phosphatase domain of ALPH protein phosphatases from both prokaryotes and eukaryotes.
Supplemental Figure S4. Phylogenetic radial tree depicting SLP protein phosphatase distribution and interrelationships across eukaryotes, archaea, and bacteria.
Supplemental Figure S5. Phylogenetic radial tree depicting RLPH protein phosphatase distribution and interrelationships across both eukaryotes and bacteria.
Supplemental Figure S6. Phylogenetic radial tree depicting ALPH protein phosphatase distribution and interrelationships across eukaryotes, archaea, and bacteria.
Supplemental Figure S7. Phylogenetic radial tree depicting SLP, RLPH, and ALPH protein phosphatase distribution and interrelationships across eukaryotes, archaea, and bacteria.
Supplemental Table S1. Complete list of sequence gene identifiers used in HMM analysis, phylogenetic tree construction, and in silico subcellular localization analysis (Supplemental Table S2).
Supplemental Table S2. Complete spreadsheet of consensus in silico subcellular localization findings.
Supplemental Table S3. Phylogenetic tree branch support values.
Supplementary Material
Acknowledgments
This research has been enabled by the use of computing resources provided by WestGrid and Compute/Calcul Canada. D.K. thanks Justin Kerk for writing programming scripts.
Glossary
- PPP
phosphoprotein phosphatase
- LGT
lateral gene transfer
- SLP
Shewanella-like
- RLPH
Rhizobiale-like
- ALPH
ApaH-like
- HMM
hidden Markov model
- NCBI
National Center for Biotechnology Information
- RBS
rapid bootstrap support
- PP
posterior probability
- LG
Le and Gascuel
References
- Abascal F, Zardoya R, Posada D. (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Acuña R, Padilla BE, Flórez-Ramos CP, Rubio JD, Herrera JC, Benavides P, Lee SJ, Yeats TH, Egan AN, Doyle JJ, et al (2012) Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc Natl Acad Sci USA 109: 4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. (2011) The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA. (2004) Widespread presence of “bacterial-like” PPP phosphatases in eukaryotes. BMC Evol Biol 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60: 685–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Belbahri L, Calmin G, Mauch F, Andersson JO. (2008) Evolution of the cutinase gene family: evidence for lateral gene transfer of a candidate Phytophthora virulence factor. Gene 408: 1–8 [DOI] [PubMed] [Google Scholar]
- Bodén M, Hawkins J. (2005) Prediction of subcellular localization using sequence-biased recurrent networks. Bioinformatics 21: 2279–2286 [DOI] [PubMed] [Google Scholar]
- Boto L. (2010) Horizontal gene transfer in evolution: facts and challenges. Proc Biol Sci 277: 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigan DL. (2013) Protein Ser/Thr phosphatases: the ugly ducklings of cell signalling. FEBS J 280: 324–345 [DOI] [PubMed] [Google Scholar]
- Brinkmann H, van der Giezen M, Zhou Y, Poncelin de Raucourt G, Philippe H. (2005) An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol 54: 743–757 [DOI] [PubMed] [Google Scholar]
- Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Bürglin TR, et al. (2013) Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol 14: R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241: 779–786 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, et al. (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103: 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Russinova E. (2011) PP2A phosphatases: the “on-off” regulatory switches of brassinosteroid signaling. Sci Signal 4: pe25. [DOI] [PubMed] [Google Scholar]
- Dorrell RG, Smith AG. (2011) Do red and green make brown? Perspectives on plastid acquisitions within chromalveolates. Eukaryot Cell 10: 856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. (2006) Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Fuerst JA, Sagulenko E. (2011) Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9: 403–413 [DOI] [PubMed] [Google Scholar]
- Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, et al. (2006) Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci USA 103: 15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. (1998) Rickettsia, typhus and the mitochondrial connection. Nature 396: 109–110 [DOI] [PubMed] [Google Scholar]
- Gray MW. (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4: a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari B, Matre P, Nemie-Feyissa D, Meyer C, Rognli OA, Møller SG, Lillo C. (2011) Protein phosphatase 2A B55 and A regulatory subunits interact with nitrate reductase and are essential for nitrate reductase activation. Plant Physiol 156: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Horák A, Oborník M, Lukes J, Keeling PJ. (2010) A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA 107: 10949–10954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanon M, McFadden GI. (2010) Malaria, Plasmodium falciparum and its apicoplast. Biochem Soc Trans 38: 775–782 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. (2009) Role of horizontal gene transfer in the evolution of photosynthetic eukaryotes and their plastids. Methods Mol Biol 532: 501–515 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9: 605–618 [DOI] [PubMed] [Google Scholar]
- Kerk D, Templeton G, Moorhead GB. (2008) Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol 146: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. (2010) The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Canback B, Berg OG. (2003) Horizontal gene transfer: a critical view. Proc Natl Acad Sci USA 100: 9658–9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutuzov MA, Andreeva AV. (2008) Protein Ser/Thr phosphatases of parasitic protozoa. Mol Biochem Parasitol 161: 81–90 [DOI] [PubMed] [Google Scholar]
- Kutuzov MA, Andreeva AV. (2012) Prediction of biological functions of Shewanella-like protein phosphatases (Shelphs) across different domains of life. Funct Integr Genomics 12: 11–23 [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. (1999) Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet 33: 351–397 [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. (2008) LG: An improved, general amino-acid replacement matrix. Mol Biol Evol 25: 1307–1320 [DOI] [PubMed] [Google Scholar]
- Le Corguillé G, Pearson G, Valente M, Viegas C, Gschloessl B, Corre E, Bailly X, Peters AF, Jubin C, Vacherie B, et al (2009) Plastid genomes of two brown algae, Ectocarpus siliculosus and Fucus vesiculosus: further insights on the evolution of red-algal derived plastids. BMC Evol Biol 9: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Antolín-Llovera M, Ferrero S, Closa M, Arró M, Ferrer A, Boronat A, Campos N. (2011) Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 23: 1494–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. (2009) HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res 37: D471–D478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Vert JP, Saigo H, Ueda N, Toh H, Akutsu T. (2005) A novel representation of protein sequences for prediction of subcellular location using support vector machines. Protein Sci 14: 2804–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24: 841–847 [DOI] [PubMed] [Google Scholar]
- Mayer WE, Schuster LN, Bartelmes G, Dieterich C, Sommer RJ. (2011) Horizontal gene transfer of microbial cellulases into nematode genomes is associated with functional assimilation and gene turnover. BMC Evol Biol 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE). IEEE, New Orleans, pp 1–8 [Google Scholar]
- Nicholas KB, Nicholas HBJ, Deerfield DWI. (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW.news 4: 1–4 [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3. [DOI] [PubMed] [Google Scholar]
- Patzewitz EM, Guttery DS, Poulin B, Ramakrishnan C, Ferguson DJ, Wall RJ, Brady D, Holder AA, Szöőr B, Tewari R. (2013) An ancient protein phosphatase, SHLP1, is critical to microneme development in Plasmodium ookinetes and parasite transmission. Cell Rep 3: 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez J, Castañeda-García A, Jenke-Kodama H, Müller R, Muñoz-Dorado J. (2008) Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci USA 105: 15950–15955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ. (2006) PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics 4: 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AM, Penny D. (2007) Evaluating hypotheses for the origin of eukaryotes. Bioessays 29: 74–84 [DOI] [PubMed] [Google Scholar]
- Raymond JA, Kim HJ. (2012) Possible role of horizontal gene transfer in the colonization of sea ice by algae. PLoS ONE 7: e35968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Söding J. (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9: 173–175 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J, Cyert MS. (2009) Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signal 2: re9. [DOI] [PubMed] [Google Scholar]
- Schein AI, Kissinger JC, Ungar LH. (2001) Chloroplast transit peptide prediction: a peek inside the black box. Nucleic Acids Res 29: E82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, et al. (2013) Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339: 1207–1210 [DOI] [PubMed] [Google Scholar]
- Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A. (2011) Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet 7: e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. (2002) Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res 12: 17–25 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Templeton GW, Nimick M, Morrice N, Campbell D, Goudreault M, Gingras AC, Takemiya A, Shimazaki K, Moorhead GB. (2011) Identification and characterization of AtI-2, an Arabidopsis homologue of an ancient protein phosphatase 1 (PP1) regulatory subunit. Biochem J 435: 73–83 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Bowler C. (2011) Decoding algal genomes: tracing back the history of photosynthetic life on Earth. Plant J 66: 45–57 [DOI] [PubMed] [Google Scholar]
- Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A, Moorhead GB. (2012) Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J 71: 263–272 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, Labandera AM, Moorhead GB. (2013) Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci 18: 505–513 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, Moorhead GB. (2011a) Two ancient bacterial-like PPP family phosphatases from Arabidopsis are highly conserved plant proteins that possess unique properties. Plant Physiol 157: 1778–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Moorhead GB. (2011b) Okadaic acid and microcystin insensitive PPP-family phosphatases may represent novel biotechnology targets. Plant Signal Behav 6: 2057–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. (2009) From promiscuity to precision: protein phosphatases get a makeover. Mol Cell 33: 537–545 [DOI] [PubMed] [Google Scholar]
- Walker G, Dorrell RG, Schlacht A, Dacks JB. (2011) Eukaryotic systematics: a user’s guide for cell biologists and parasitologists. Parasitology 138: 1638–1663 [DOI] [PubMed] [Google Scholar]
- Wenzl P, Wong L, Kwang-won K, Jefferson RA. (2005) A functional screen identifies lateral transfer of beta-glucuronidase (gus) from bacteria to fungi. Mol Biol Evol 22: 308–316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.