A conserved chimeric genomic sequence found exclusively in apomictic Boechera species generates a putative long non-protein-coding transcript with secondary structure folding capability and is highly expressed in antherheads characterized by meiotically unreduced pollen formation.
Abstract
In apomictic Boechera spp., meiotic diplospory leads to the circumvention of meiosis and the suppression of recombination to produce unreduced male and female gametes (i.e. apomeiosis). Here, we have established an early flower developmental staging system and have performed microarray-based comparative gene expression analyses of the pollen mother cell stage in seven diploid sexual and seven diploid apomictic genotypes to identify candidate factors for unreduced pollen formation. We identified a transcript unique to apomictic Boechera spp. called UPGRADE2 (BspUPG2), which is highly up-regulated in their pollen mother cells. BspUPG2 is highly conserved among apomictic Boechera spp. genotypes but has no homolog in sexual Boechera spp. or in any other taxa. BspUPG2 undergoes posttranscriptional processing but lacks a prominent open reading frame. Together with the potential of stably forming microRNA-like secondary structures, we hypothesize that BspUPG2 functions as a long regulatory noncoding messenger RNA-like RNA. BspUPG2 has apparently arisen through a three-step process initiated by ancestral gene duplication of the original BspUPG1 locus, followed by sequential insertions of segmentally duplicated gene fragments, with final exonization of its sequence structure. Its genesis reflects the hybridization history that characterizes the genus Boechera.
Haploid gamete formation is an important step during the diplohaplontic plant life cycle, which marks the switch from sporophyte to gametophyte. During female and male sporogenesis, the diploid megaspore mother cell and pollen mother cell (PMC), respectively, undergo one reductional and equational mitotic division to form four haploid daughter cells. During subsequent male spore development (microgametogenesis), each of the four microspores undergoes two mitotic divisions to form a tricellular pollen grain containing two mature reduced gametes (generative nuclei) and one vegetative nucleus.
Despite a relatively simple structure, the male gametophyte is regulated by complex gamete-specific molecular programs that are characterized by (1) enrichment of transcripts responsible for DNA repair, cell cycle, and chromosome organization, (2) underrepresentation of the RNA-processing machinery (Pina et al., 2005; Borges et al., 2008), and (3) developmental stage-specific transcript sets whose activities vary temporally from the uninuclear microspore toward the mature pollen grain (Mascarenhas, 1989; Honys and Twell, 2004).
Loss-of-function studies of male and female meiosis have revealed cell cycle defects (Yang et al., 2003), effects on homologous chromosome recombination (Lu et al., 2008), and defects in meiotic cell fate (Sundaresan et al., 1995; Yang et al., 1999), all of which lead to reduced meiocyte fertility or death. Other mutations lead to unreduced (2C) gametes (Brownfield and Köhler, 2011).
Apomeiosis is a naturally occurring form of sensu stricto unreduced egg formation, which is the first step of gametophytic apomixis or asexual (clonal) seed formation in plants (Nogler, 1984; Grimanelli et al., 2001a). During apomixis, the apomeiotically derived egg cell undergoes parthenogenetic (i.e. without fertilization) development, while endosperm develops with (pseudogamous) or without (autonomous) fertilization (Nogler, 1984; Grimanelli et al., 2001a). Apomixis bears the potential to fix hybrid genotypes over generations and, therefore, is of high interest to agronomy (Grimanelli et al., 2001a; Grossniklaus, 2001).
Apomixis research can be divided into studies of de novo induction of apomixis-like processes in sexual plants and naturally occurring apomixis (Savidan et al., 2001). Mutant screening and attempts to introgress apomixis into sexual taxa have been hindered by its molecular genetic complexity (d’Erfurth et al., 2008; Hörandl and Temsch, 2009; Marimuthu et al., 2011). Comparative mapping strategies in natural apomicts, with an emphasis on aposporic species, have identified candidate factors, although proof of function in crop plants is still pending (Guerin et al., 2000; Albertini et al., 2004; Matzk et al., 2005; Schallau et al., 2010). Besides theories of apomixis inheritance through a single dominant locus (Mogie, 1988) or via a complex of physically linked coadapted genes (Van Dijk et al., 1999), the hybridization-derived floral asynchrony (HFA) theory proposes the deregulation of sexual genetic pathways via the asynchronous expression of duplicated gene sets as a mechanism for apomixis induction (Carman, 1997). Reproductive deregulation was recently supported by heterochronic expression patterns during ovule development in sexual and apomictic Boechera spp. (Sharbel et al., 2010), pointing to a network of epigenetic and posttranscriptional regulation during germline specification (Twell, 2011).
The approximately 110 species that constitute the North American genus Boechera (Brassicaceae) exist as diploid sexual or diploid and allopolyploid apomictic forms (Böcher, 1951; Al-Shehbaz and Windham, 2010), the latter of which are highly correlated with interspecific hybridization (Sharbel et al., 2009, 2010; Beck et al., 2012). Diploid apomixis is rare and facilitates comparisons between sex and apomixis without the added complexity of differing ploidy (Sharbel et al., 2009, 2010). Apomictic Boechera taxa are characterized by Taraxacum-type pseudogamous diplospory (Rollins, 1941; Böcher, 1951). Besides the formation of unreduced female gametes, apomictic Boechera spp. also produce unreduced pollen, as demonstrated by the fact that diploids and triploids produce seeds almost exclusively with hexaploid (6C = [4Cmaternal] + [2Cpaternal]) and nonaploid (9C = [6Cmaternal] + [3Cpaternal]) endosperm (Voigt et al., 2007; Aliyu et al., 2010; Voigt-Zielinski et al., 2012). Hence, strong selection pressure to maintain a balanced two maternal-to-one paternal genome ratio (i.e. endosperm balance number; Johnston et al., 1980) during endosperm formation apparently characterizes apomictic Boechera spp. (Johnston et al., 1980; Voigt et al., 2007; Aliyu et al., 2010). While we have thus far focused our transcriptome comparisons on sexual and apomictic ovules (Sharbel et al., 2009, 2010), strong selection for genetically balanced endosperm during apomictic seed formation has led us to study unreduced pollen formation.
Hence, we have undertaken a microarray-based transcriptome comparison of a single stage of antherhead development differentiating reduced and unreduced pollen formation between diploid sexual and diploid apomictic Boechera spp. Here, we describe the isolation and characterization of a differentially expressed candidate gene, BspUPG2 (for Boechera species UPGRADE2 [unreduced pollen grain development]), which is associated with unreduced pollen formation and is potentially part of a homology-dependent gene-silencing mechanism. BspUPG2 has apparently arisen through gene duplication and subsequent insertions of genic and nongenic fragments in the hybrid apomictic genome. Its putative molecular function in apomictic genomes of Boechera spp. is discussed.
RESULTS
Apomictic Boechera spp. Produce (First Division Restitution-Type) Unreduced Gametes
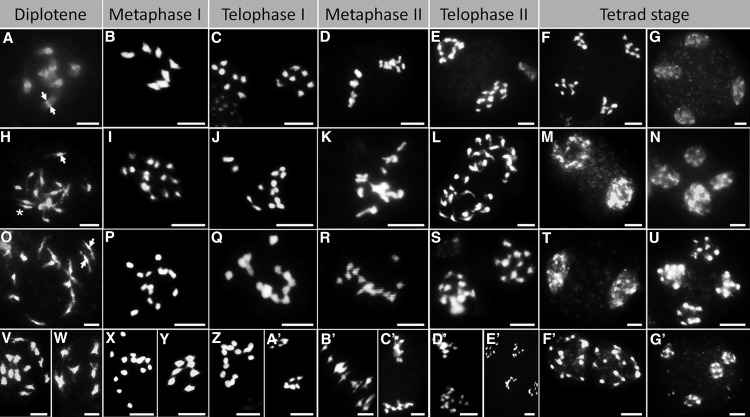
One major mechanism that leads to unreduced pollen formation is the meiotic nuclear restitution, which comprises a failure during the first or second meiotic division. Meiotic chromosome behavior (Ross et al., 1996) was examined to determine the exact mechanism by which unreduced pollen is formed in facultative and obligate apomictic Boechera spp. and showed that pollen formation differed between apomicts and sexuals (Fig. 1; Supplemental Table S1). The sexual Boechera stricta ES 612.1 showed expected homologous chromosome pairing, with the juncture of nonsister chromatids leading to seven bivalents (Fig. 1A). The subsequent two nuclear divisions lead to a tetrad with four haploid nuclei (Fig. 1, B–G). In contrast, the aneuploid obligate apomictic Boechera polyantha (ES 776.2; 2n = 2x = 15) and the euploid Boechera lignifera (ES 753; 2n = 2x = 14) exhibited complete or partial chromosomal asynapsis, resulting in univalents at metaphase I that do not (or only partially) segregate during meiosis I (Fig. 1, I, J, P, and Q). As a result, metaphase II plates were frequently fused (Fig. 1, K and R), in contrast to their perpendicular orientation in the sexual reference accession (Fig. 1D), leading to the generation of dyads with balanced and unreduced chromosome numbers (Fig. 1, L, M, S, and T). Low levels of chromosomal synapsis leading to tetrad formation were also observed in all examined diploid obligate apomicts (Fig. 1, N and U). Compared with obligate apomicts, the facultative apomictic Boechera divaricarpa ES 514 exhibited higher levels of nuclei with bivalents (Fig. 1, W and Y), which proceeded through both meiotic cell divisions (Fig. 1A′, C′, and E′), generating tetrads with four haploid cells (Fig. 1G′). Nonetheless, nuclei with univalents were frequently observed (Fig. 1, V, X, and Z) whose sister chromatids were equally separate at metaphase II (Fig. 1B′) to develop balanced dyads (Fig. 1, D′ and F′). Meiotic chromosome counts of all apomicts strongly support first division restitution without crossover. Univalents remain together during meiosis I and disjoin in meiosis II, where sister chromatids are equationally separated to opposite poles, leading to two balanced diploid chromosome sets in dyads.
Figure 1.
Meiotic chromosome behavior of male meiocytes. Chromosome spreads of a diploid obligate sexual (A–G), an aneuploid (H–N) and a euploid (O–U) obligate apomictic, and a high facultative apomictic (V–G′) Boechera spp. genotype are displayed. Arrowheads show synapsed homologs in sexual (A) and nonsynapsed homologs in apomictic (H and O) Boechera spp. In addition, homologs with close juxtaposition were occasionally observed in apomictic Boechera spp. (asterisk). Bars = 5 μm.
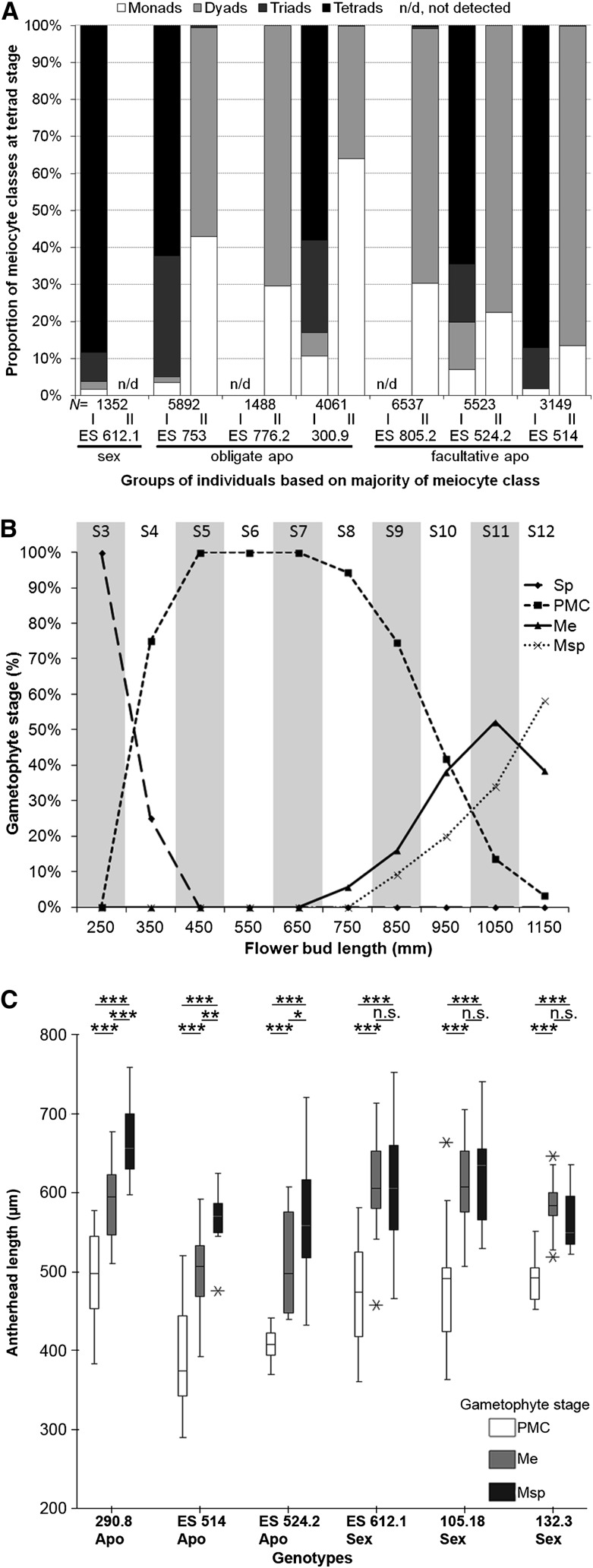
Approximately 28,000 meiocytes from single antherheads of six apomictic genotypes and one sexual reference genotype were counted to quantify meiotic product frequencies. The sexual reference genotype produced the expected high frequency of tetrads (88%), whereas the tetrad frequency varied among all tested apomicts (0%–87%) between individuals of one genotype and even between different flower buds from the same individual (Fig. 2A; Supplemental Table S2). Low levels of triads and tetrads were observed for apomictic genotypes ES 805.2 (0.55% and 0.35%, respectively) and ES 776.2 (0.00% and 0.00%, respectively), both of which showed high levels of monad and dyad formation. However, surprisingly high levels of triads and tetrads were found for other individuals of both obligate (300.9, 24.98% and 57.98%; ES 753, 32.78% and 62.16%) and facultative (ES 514, 11.22% and 87.00%; ES 524.2, 15.67% and 64.53%) apomicts (Fig. 2A). The formation of high levels of balanced dyads confirmed that unreduced pollen in apomictic Boechera spp. is produced by a defect in chromosome segregation during meiosis I. However, unequal sister chromatid segregation during meiosis II, resulting in the formation of extra nuclei, was low (in total, 0.19% meiocytes with micronuclei and 0.02% polyads) and mainly detected in the apomict ES 753 (0.88% meiocytes with micronuclei; Supplemental Fig. S1).
Figure 2.
Meiocyte constitution at the tetrad stage and correlation of flower bud size and antherhead length with different gametophyte stages. A, Squashes of single antherheads from several individuals per genotype demonstrate meiocyte constitution at the tetrad stage. Roman numbers denote groups of individuals per genotype primarily (greater than 50%) producing reduced gametes (I) versus groups of individuals per genotype primarily (greater than 50%) producing unreduced gametes (II). B, Relationship between flower bud stages S3 to S12 and proportion of anthers at a particular developmental stage of gametophytes in Boechera spp. C, Horizontal bars above the box plots demonstrate significant comparisons between gametophytic stages per antherhead length within each genotype (*P < 0.05, **P < 0.01, ***P < 0.001, n.s. = not significant) as conducted for a 95% binomial proportion confidence interval with a one-way ANOVA including Tukey’s HSD post hoc test. Despite observations of antherhead-related gametophyte stages for flower bud stages S3 to S12, the correlation analysis focuses only on S8 to S12. Me, Meiosis; Msp, microspore; Sp, sporocyte.
Considering the highly variable pattern of meiocyte frequencies between individuals and between flowers of single specific apomictic genotypes (Fig. 2A), only plants producing exclusively unreduced pollen were chosen for comparative gene expression analyses. Plant selection, therefore, was based upon measurements of the nuclear DNA content of pollen and seed nuclei for all individuals (Supplemental Fig. S1). Interestingly, flow cytometric seed screen data of all tested obligate apomictic individuals suggest successful fertilization with only unreduced pollen, although also some individuals produce high levels of reduced pollen (embryo:endosperm ratio of 2C [1Cmaternal + 1Cpaternal]:6C [4Cmaternal + 2Cpaternal]; Fig. 2A; Supplemental Fig. S1; Supplemental Table S3).
Staging of Meiosis in Boechera spp.
To optimize for comparative gene expression analyses between reduced (sex) and unreduced (apomeiosis) pollen, the morphological development of antherheads was quantified through staging (Sharbel et al., 2010). Analyses of early flower development from about 780 flower buds in six diploid Boechera spp. genotypes (Figs. 2, B and C, and 3; Table I; Supplemental Table S3) showed a linear relationship between bud length and flower organ size (e.g. for stages S4 to S12 antherheads, r2apo = 0.87, F = 1273.50, P < 0.001; r2sex = 0.95, F = 4237.23, P < 0.001). Minor variations among genotypes, but no overall differences between sex and apomixis, were detected [i.e. stages predict similar organ size classes; general linear model for regression line slopes (b) of stages S4 to S12 antherheads: H0, the effective null hypothesis which assumes that there is no relation between two phenomena, bapo = bsex; r2 = 0.95, F(44, 8) = 0.68, P = 0.804; Supplemental Fig. S2].
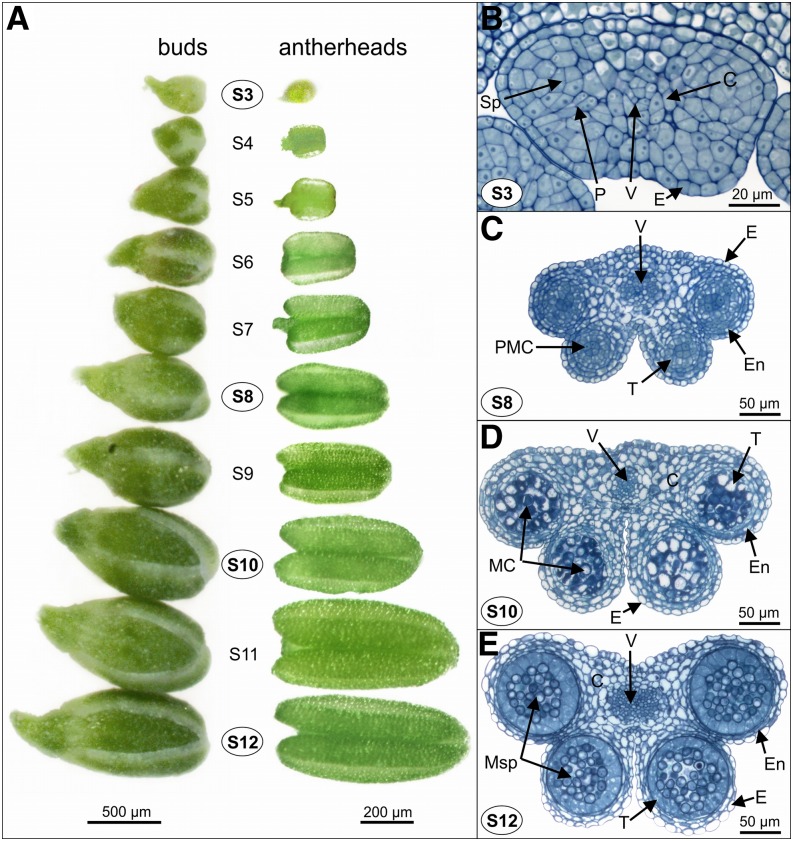
Figure 3.
Light microscopic analysis of Boechera spp. antherhead development and corresponding gametophyte stages during pollen formation. A, Flower buds and appropriate antherheads of different developmental stages S3 to S12 in a sexual genotype. B to E, Light microscopy images of semithin sections of resin-embedded antherheads after histological staining displaying the development of the sporocyte into a mature microspore, used for cytological validation of major gametophyte stages. C, Connective; E, endodermis; En, endothecium; MC, meiotic cell; Me, meiosis; Msp, microspores; P, parietal cells; Sp, sporocyte; T, tapetum; V, vascular region. [See online article for color version of this figure.]
Table I. Developmental markers of pollen formation corresponding to gametophyte and flower stages.
| Flower Stage | Size Range | Arabidopsis Flower Stagea | Pollen Stageb | Developmental Marker Identifier | Developmental Marker and Histological Eventsc |
|---|---|---|---|---|---|
| mm | |||||
| S1 | 0–0.1 | n/ad | n/a | n/a | n/a |
| S2 | 0.1–0.2 | n/a | n/a | n/a | n/a |
| S3 | 0.2–0.3 | 7–8 | n/a | Sp | Sporocytes, premeiotic, anther in differentiation stage, occurrence of 1° and 2° parietal cells, vascular region initiated |
| S4 | 0.3–0.4 | 9 | 3 | Pattern of anther defined, all four locules present | |
| S5 | 0.4–0.5 | Rapid lengthening of all flower organs, especially antherheads and filament | |||
| S6 | 0.5–0.6 | ||||
| S7 | 0.6–0.7 | ||||
| S8 | 0.7–0.8 | PMC | PMC enlarged and clearly separated from tapetum, prior to meiosis, no callose deposition | ||
| S9 | 0.8–0.9 | 4 | PMC enters pollen meiosis, crushed middle layer, vacuolated tapetum, initial callose deposition, binucleate tapetum | ||
| S10 | 0.9–1.0 | 4–6 | Me | Meiosis, tetrads of microspores, accumulated callose deposition | |
| S11 | 1.0–1.1 | 10 | 5–6 | Msp | Microspores in interphase, early-vacuolated, minorities of tetrads or binucleate microspores |
| S12 | 1.1–1.2 | 11 | 6–8 | Microspores vacuolated and undergo first mitotic division, callose degradation, binucleate microspores |
Arabidopsis flower developmental stages taken from Smyth et al. (1990). bArabidopsis pollen developmental stages after Regan and Moffatt (1990). cObserved histological events characterized according to Sanders et al. (1999). dn/a, Not applicable.
We focused on four major histodifferentiation steps of male gametophyte development that are characterized by their premeiotic, meiotic, and postmeiotic appearance (Fig. 1; Table I). Despite some marker-specific variations in correlation strength, flower bud length predicts the gametophytic stage of anthers with relatively high accuracy (n = 455; sporocyte, 100%, PMC, 100%, meiosis, 52.27%, microspores, 58.24%; Fig. 2B). Antherhead length predicts the gametophytic stage of anther development, except for meiotic and microspore stages in sexuals, which overlapped in antherhead size class (one-way ANOVA with Tukey’s honestly significant difference [HSD] post hoc test, between P < 10−2 and P < 10−9; Fig. 2C). The correlation between antherhead length and gametophyte stage led to the selection of 400- ± 30-µm antherheads, a stage that is significantly enriched for PMCs, being close to meiosis in both sexual and apomictic genotypes, for the collection of total RNA (P < 0.001, Fisher’s exact test; Supplemental Table S4; the decision to take the PMC stage is detailed in “Materials and Methods”).
Identification of the Apo-Specific UPGRADE Transcript
In search for candidate transcripts that are differentially expressed between the sexual and the apomictic microsporogenesis programs, we performed pairwise comparisons of RNA expression levels in microdissected antherheads from seven sexual and seven apomictic genotypes using a custom Agilent microarray (detailed in “Materials and Methods”). Using a 2-fold or greater change threshold and corrected P ≤ 0.05, while allowing one outlier sample, five microarray probes were identified to be highly up-regulated in all apomicts, except in genotype ES 753 (B. lignifera), exhibiting absolute fold change levels between 48.74 and 849.31 compared with sexuals (Supplemental Fig. S3; Supplemental Table S5).
Homologous complementary DNAs (cDNAs) corresponding to the five candidate array probes were identified from a flower-specific cDNA library (Supplemental Table S5; Sharbel et al., 2009). A BLASTN (Altschul et al., 1997) search of these cDNAs against the whole GenBank nucleotide collection (http://www.ncbi.nlm.nih.gov/genbank/) revealed hits for only two of the candidate probes; probe Sharb1199059 (GenBank/EMBL/DNA Data Bank of Japan [DDBJ] accession no. ERS317552) was homologous to an Arabidopsis (Arabidopsis thaliana) S-locus lectin protein kinase involved in pollen recognition, and probe Sharb0501554 (GenBank/EMBL/DDBJ accession no. ERS317556) matched two S-LOCUS RECEPTOR KINASE genes (S-12 and S-15 type) encoding a Brassica oleracea S-locus receptor kinase involved in pollen self-recognition specificity (Supplemental Table S5).
Real-time quantitative reverse transcription (qRT)-PCR confirmed that four of the five microarray probes had expression profiles corresponding to the microarray data (Supplemental Fig. S4; Supplemental Tables S6 and S7). The single probe that could not be qRT-PCR validated (Sharb1199059; lectin homolog) was furthermore characterized by a negative bacterial artificial chromosome (BAC) screen result (Supplemental Table S8) and was not considered for further analysis. With the exception of the apomictic outlier sample (ES 753), the four microarray probes were highly up-regulated in apomictic antherheads. In contrast to sexuals, in which the relative mRNA levels were close to the detection limit, ubiquitous expression of all probe sequences was detected in apomictic somatic and reproductive tissues. Moreover, for microarray probes Sharb0931225, Sharb0501554, and Sharb0690829, expression was substantially higher in antherheads compared with the levels in leaf tissue (one-way ANOVA with Tukey’s HSD post hoc test, P = 0.022, P = 0.008, and P = 0.027, respectively; Supplemental Fig. S4; Supplemental Table S7).
RACE was performed using gene-specific primers from microarray probe Sharb0690829 (Supplemental Table S5) to obtain full-length cDNA including the 3′ and 5′ cDNA ends. Although total RNA of antherheads and whole flower tissue from two sexuals and eight apomicts was used, 5′ and 3′ RACE generated similar DNA fragments exclusively from apomicts, except for the apomictic outlier ES 753 (Supplemental Fig. S5).
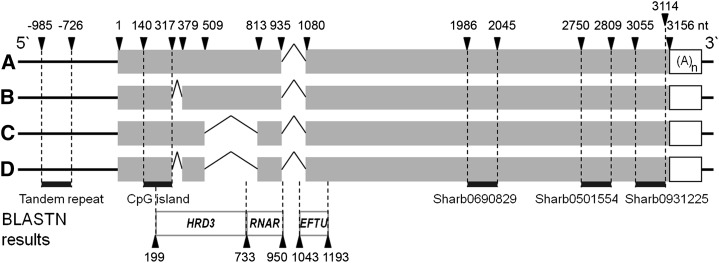
All 3′ and 5′ RACE fragments from five apomicts were cloned and sequenced and led to the identification of a single polyadenylated full-length transcript, BspUPG (GenBank/EMBL/DDBJ accession no. HF930769; Fig. 4) with length of about 2,648 nucleotides (nt; for B. divaricarpa ES 524), excluding the poly(A) sequence. The microarray probes Sharb0931225, Sharb0501554, and Sharb0690829 were found in the most 3′ exon of the BspUPG transcript (Fig. 4). A comparison of different BspUPG full-length cDNA variants with genomic DNA by isolation of 3′ RACE and the various 5′ RACE fragments in different Boechera spp. genotypes (Supplemental Fig. S5, white arrows) showed that the gene has an overall length of 3,156 nt (for B. divaricarpa ES 524) and contains two putative alternative splicing sites, 61 bp (intron 1) and 303 bp (intron 2), in addition to a 144-bp intron common to all apomicts (intron 3; Fig. 4; Supplemental Fig. S5). The U2-dependent classified introns (5′-GT/3′-AG splicing site; Simpson and Filipowicz, 1996) are located toward the 5′ end of the transcript.
Figure 4.
Schematic representation of putative splicing forms of BspUPG2. All four splice forms could be extracted by end-to-end sequencing in five apomictic Boechera spp. Exons are represented by gray boxes, and numbers indicate intron-exon boundary positions. BLAST search results from the GenBank nucleotide collection and the TAIR10 gene database are given below.
UPGRADE Is a Chimeric Gene Encoding a Transcript without Translational Capacity
BspUPG fragments are homologous to sequences in Arabidopsis located on chromosomes 1, 4, and 5, which encode an elongation factor TU/EF1-A protein (EFTU/EF1-A; AT4G02930), an RNA recognition motif-containing protein (RNAR; AT5G19960), and the HRD3 protein (for 3-hydroxy-3-methyl glutaryl-CoA reductase degradation), which is homologous to components of the yeast HRD1 complex (AT1G18260), respectively (Fig. 4). From these candidates, AtHRD3, which plays a central role in endoplasmic reticulum-associated protein degradation, exhibits the highest similarity to BspUPG, with fragments of 145 nt (83% identity; P < 5E-31), 118 nt (87% identity; P < 6E-30), and 47 nt (89% identity; P < 0.0002).
The transcript carries a poly(A) tail with some variation in length and initiation position. In search for regulatory motifs on BspUPG, a putative near-upstream element (AATAAA) of a polyadenylation signal, which is common in plants (Hunt, 1994), was identified 31 nt upstream of the polyadenylation site. Furthermore, a single CpG island was detected at the 5′ end of the transcript locus (+140 to 312 nt, C + G > 60%; Fig. 4; Supplemental Fig. S6C). Sequences that are rich in the CpG/CpNpG patterns are predominantly nonmethylated and tend to be associated with genes that are frequently switched on (Deaton and Bird, 2011). A screen for putative regulatory features in the upstream sequences (between +47,809 and +48,921 nt on Assembly 2, PlantPAN database; Chang et al., 2008; http://plantpan.mbc.nctu.edu.tw/) detected a single tandem repeat (−726 to −985 nt, 82-nt consensus size, 3.2 copies; Fig. 4) and multiple transcription factor-binding sites (Supplemental Table S9).
Coding potential calculation of BspUPG (for ES 524; Kong et al., 2007), excluding the constitutive intron 3, revealed a single short open reading frame (ORF) for each strand. The sense strand gave an ORF with 196 nt (66 amino acids, +895 to +1,090 nt, coding potential score = −0.0298), and the reverse complement strand revealed a 202-nt ORF (68 amino acids, +1,958 to +2,159 nt, coding potential score = −1.180). The lack of an ORF length typical for a protein encoding mRNA suggested a long non-protein-coding mRNA-like RNA (lncRNA) product of BspUPG (ORF typically more than 100 amino acids; Kondo et al., 2007).
The genomic sequence information of the candidate locus was augmented by a screen of a B. divaricarpa BAC library. Colony PCR using target-specific primers from the chromosome-walking approach (detailed in “Materials and Methods”; Supplemental Table S10) led to the identification of 12 Boechera spp. BAC clones that hybridized with one or several probes simultaneously (Supplemental Table S8). None of the clones were positive for probe Sharb1199059, whereas three single hits for probe Sharb0425060 and nine triple hits for probes Sharb0931225, Sharb0501554, and Sharb0690829 were detected, which confirmed the results from the RACE experiment (Fig. 4). Restriction digests of the 12 BACs suggested partial overlap of their DNA inserts. Sanger sequencing was thus performed on BAC clones A4O22, E7K5, C8B11, and F8G11, each being positive for three of the five microarray probes (Sharb931225, Sharb501554, and Sharb690829). The C8B11 BAC sequence contig (Assembly 1; GenBank/EMBL/DDBJ accession no. HF954100), which could not be aligned together with the sequence contigs of the other three BAC clones, reached 57,458 bp in length with 33.5% average GC content, which increased to 40.1% in genic regions. BAC sequence contigs from clones A4O22, F8G11, and E7K5 overlapped and were assembled into the 58,769-bp Assembly 2 (GenBank/EMBL/DDBJ accession no. HF954101), with 32.9% average GC content increasing to 42.1% in genic regions.
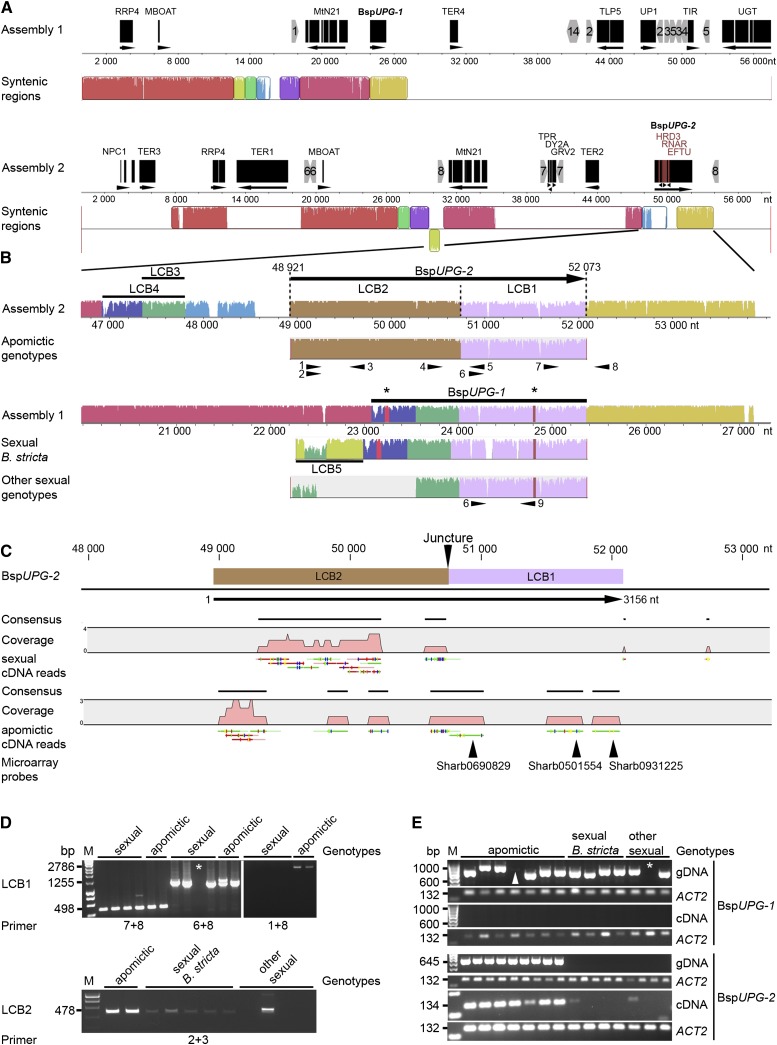
Annotation of Assembly 1 identified one transposon-related gene, five protein-encoding genes, and two fragments of protein-encoding genes, all of which (except for TER4) were homologous to genes located on Arabidopsis chromosome 1 (Fig. 5A; Supplemental Fig. S6, A and B; Supplemental Table S11). In contrast, Assembly 2 contained a higher level of transposon-related genes (three) and gene fragments (nine), whose homologs were found on all Arabidopsis chromosomes except for chromosome 3 (Fig. 5A; Supplemental Fig. S6, A and C; Supplemental Table S11). Comparison of both assemblies showed that both fragments share several highly similar sequences (e.g. for the two protein-encoding genes MtN21 and RRP4, for a fragment of the protein-encoding gene MBOAT, and for the 3′ end of BspUPG). Allowing for rearrangements, Assembly 1 and Assembly 2 aligned along orthologous regions covering 27.15 kb of Assembly 1 and 46.20 kb of Assembly 2. Synteny gaps in Assembly 2 contained transposon-related genes (e.g. TER1 and TER2) and insertions composed of short protein-encoding gene fragments flanked by inverted repeat sequences (e.g. IR7; Fig. 5A; Supplemental Fig. S6C; Supplemental Table S12), suggesting that parts of Assembly 2 arose via partial duplications of Assembly 1. Hence, considering the partial presence of the candidate gene at both loci, we subsequently labeled the original locus in Assembly 1 BspUPG1 and the duplicated variant on Assembly 2 BspUPG2 (Fig. 5A; Supplemental Fig. S6).
Figure 5.
Original and duplicated UPGRADE loci. A, Schematic representation of the gene annotation of Assembly 1 containing the original BspUPG1 locus and Assembly 2 containing the duplicated apo-specific BspUPG2 locus. Syntenic regions of the two assemblies are shown below. Inverted repeats are denoted as gray numbered arrowheads. Insertions of genic fragments are marked in red letters. RRP4, Exosome complex component RRP4; MBOAT, membrane-bound O-acyl transferase-like protein; MtN21, nodulin MtN21/EamA-like transporter protein; BspUPG1, original locus of candidate gene UPGRADE; TER1-4, transposable element-related protein; TLP5, Tubby-like F-box protein5; UP1, uncharacterized protein; TIR, Toll/interleukin1 receptor-nucleotide binding site class of disease resistance protein; UGT, sterol 3β-glucosyltransferase; NPC1, Niemann-Pick C1 protein; TPR, tetratricopeptide repeat domain-containing protein; DY2A, dynamin2A; GRV2, DNAJ heat-shock N-terminal domain-containing protein; HRD3, HRD3-like protein; BspUPG2; duplicated locus of UPGRADE. B, Mapping of genomic sequences of BspUPG1 in apomictic (e.g. B. divaricarpa, ES 514) and sexual (e.g. B. stricta, ES 612.1) genotypes onto both BAC clone assemblies identified by rearrangements of LCBs. Black asterisks denote indels for the sex-specific identity of LCB1 and LCB4. The LCB5 position on Assembly 1 and Assembly 2 is displayed in Supplemental Figure S5. Black arrowheads denote primers 1 (CON234X2L), 2 (CON234X14L), 3 (CON234X10R), 4 (PC1pol1L), 5 (PC1pol1R), 6 (GSP4), 7 (TSP33R), 8 (CON234X5R), and 9 (Indel9minus). C, CLC Genomics Workbench (version 4.5.1) output file showing different distributions of separate mapped cDNA reads from sexual and apomictic genotypes onto BspUPG2. Both sense-oriented (green) and antisense-oriented (red) cDNA reads are displayed below gray regions that demonstrate cDNA coverage (in pink). Colored vertical bars on cDNA reads show single-nucleotide polymorphisms in comparison with genomic DNA of BspUPG2. D, PCR with independent primer pairs, one located on LCB1 and one on the cDNA mapping positions of LCB2 in sexual and apomictic genomes illustrate that LCB1 is highly conserved in both sexuals and apomicts and that LCB2 is also present in sexual genotypes but in a different position in the genome. Primer locations are shown as numbered black arrows in B. The white asterisk denotes a missing band for genotype 105.18, which lacks the priming site for primer 6. E, PCR with primers (4 and 5) combining LCB1 and LCB2 shows the apo-specific presence of BspUPG2, whereas BspUPG1 is present in both sexual and apomictic genomes (primers 6 and 9). In contrast to BspUPG1, which is not transcribed in sexuals and apomicts (primers 6 and 9), BspUPG2 is solely transcribed from apomicts (primers CON234B2L and CON234B2R; Supplemental Table S6). The white arrowhead marks a faint band for genotype ES 753, and the white asterisk marks a missing band for genotype 105.18, which lacks the priming site for primer 6.
Both apomictic and sexual gene variants were aligned with Assembly 1 and Assembly 2 (i.e. BAC DNA sequences), allowing for rearrangements. The 3′ end of all gene variants from sexuals and apomicts, here named locally collinear block (LCB) 1 (LCB1 in violet; see definition in “Materials and Methods”), shares high similarity with both assemblies. The 5′ ends of apomictic gene copies (LCB2 in brown) were only identified on Assembly 2 (Fig. 5, A and B; Supplemental Fig. S6). Unlike apomictic gene variants (now BspUPG2; see nomenclature above), the 5′ ends (LCB3–LCB5) of variants from sexuals (now BspUPG1) share high sequence similarity with Assembly 1 but also exhibit a mosaic distribution of their LCBs relative to the duplicated locus of Assembly 2 (Supplemental Figs. S6 and S7).
Transcriptional Activity of BspUPG2 Is Restricted to Apomictic Boechera spp.
The transcriptional functionality of BspUPG1 and BspUPG2 was inferred by mapping sexual and apomictic cDNAs (Sharbel et al., 2009) independently onto both and revealed that only the apo-specific LCB2 is transcribed in both sexuals and apomicts (see sexual/apomictic cDNA reads; Fig. 5C). In contrast, apomictic cDNA maps to LCB1 and LCB2, including the reads with homology to the candidate microarray 60-mer oligonucleotide probes (Sharb931225, Sharb501554, and Sharb690829; Fig. 5C). No cDNA maps to any of the remaining LCBs. PCR on genomic DNA using specific primers for both loci demonstrates the presence of BspUPG1 in sexuals and apomicts but absence using cDNA as a PCR template. In contrast, genomic copies and transcripts of BspUPG2 were solely identified in apomicts (Fig. 5, D and E).
BspUPG1 is flanked by four genes centered in the centromeric region on the lower arm of Arabidopsis chromosome 1 (Fig. 5A; Supplemental Fig. S8; Supplemental Table S11; approximately 0.57-Mb distance from T3P8-sp6; Hosouchi et al., 2002). Interestingly, two genetic markers (Bst006701 and BstES0032) from the synthetic F2 linkage map of sexual B. stricta (Schranz et al., 2007), both of which are homologous to the Arabidopsis locus identifiers AT1G51310 and AT1G43245, flank BspUPG1. Both markers span the interval of genomic block C1, which localizes BspUPG1 on the BstLG1 linkage group of B. stricta, for which very low levels of recombination were detected (Schranz et al., 2007). The putative location of the duplicated BspUPG2 is unknown.
BspUPG2 Has High Secondary Structure Folding Potential
Apart from other lncRNA types in plants, some lncRNAs act as precursors for small non-protein-coding RNAs (npcRNAs), such as microRNAs (miRNAs) or endogenous trans-acting small interfering RNAs, and may regulate translation in cis or trans (Hirsch et al., 2006; Pikaard et al., 2008). A search for miRNA-binding sites of known Arabidopsis (www.mirbase.org; default parameter, one mismatch) and Boechera spp. miRNAs (Amiteye et al., 2011, 2013) revealed no mature miRNAs matching the candidate transcript. Considering this, in addition to the obvious lack of a long ORF on BspUPG2, a bioinformatics approach was used to examine whether thermodynamically stabile npcRNA structural elements could be predicted from its genomic sequence (detailed in Supplemental Protocol S1).
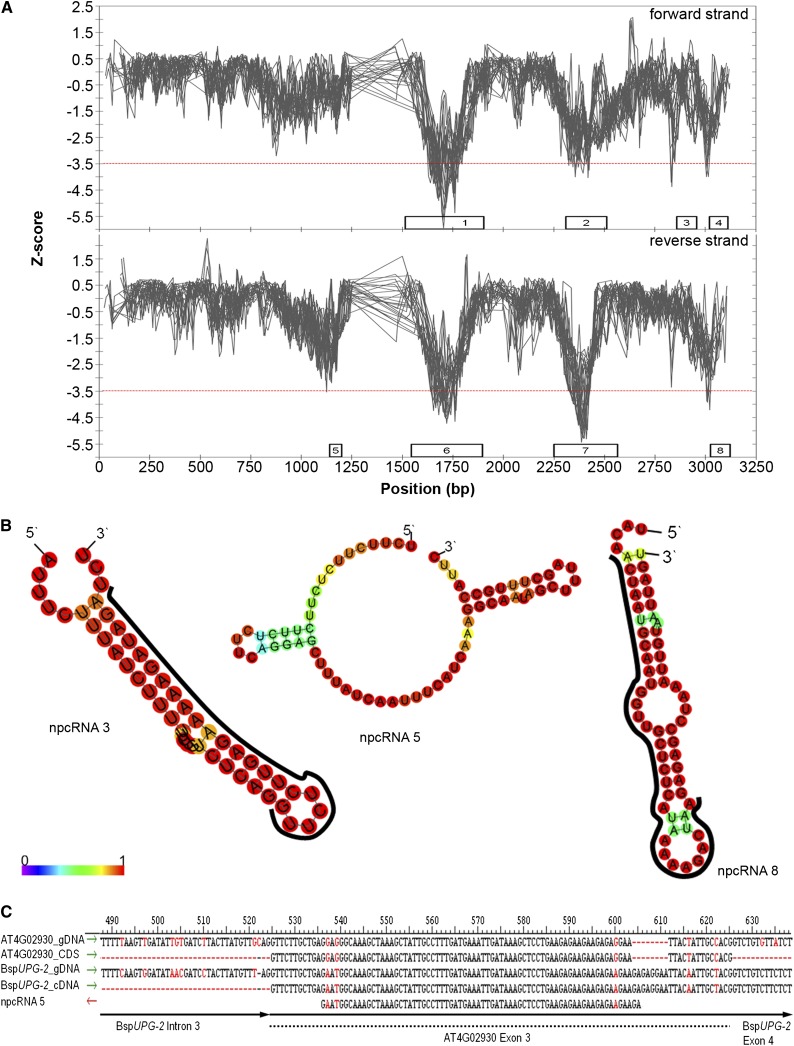
As shown in Figure 6A, the BspUPG forward and reverse strands contain eight regions (i.e. overlapping windows are combined to determine the total length of a candidate region; Supplemental Table S13) characterized by significant Z-score values, suggesting that parts of BspUPG are able to form stable secondary structures (Fig. 6A; Supplemental Fig. S9). These candidate regions span only exons, comprising together 25% and 27% of the total length of each strand of BspUPG, whereby npcRNAs 1/6, 2/7, and 4/8 cover similar regions on both strands of the genomic sequence. The predicted secondary structures were classified based upon their (1) minimal folding free energy, (2) adjusted minimal folding free energy, (3) minimal folding free energy index, and (4) A + U content (Seffens and Digby, 1999; Bonnet et al., 2004; Zhang et al., 2006). Interestingly, all detected secondary structures met most of the criteria for miRNA precursors (Fig. 6B; Supplemental Fig. S9; Supplemental Table S14). Comparable with known primary miRNAs (pri-miRNAs) in Boechera spp. (Amiteye et al., 2011, 2013) and other plant species (Zhang et al., 2006), npcRNAs 1 to 8 demonstrate a lower minimal folding free energy (ranging from −18.27 to −73.72 kcal mol−1), have an elevated A + U level (ranging from 56.70% to 70.00%), and, with the exception of npcRNA 5 (0.70), have a minimal folding free energy index greater than 1.07, which is much higher compared with tRNA (0.64), ribosomal RNA (0.59), or random mRNA (0.65).
Figure 6.
Secondary structure prediction for BspUPG2. A, The Z-score for the sliding window with length between 50 and 300 nt (step size = 10) is plotted versus the position on BspUPG2. Any window producing a significant Z-score during the scanning process was considered a candidate region for a structural npcRNA. If multiple, overlapping windows of several lengths produced significant Z-scores, the region encompassed by all the overlapping windows defined the candidate region (represented as boxes). Numbers in the boxes are putative npcRNAs having the most negative Z-scores (Supplemental Fig. S8; Supplemental Tables S13 and S14). B, Proposed minimum free energy structures of putative npcRNAs with significant BLASTN hits in the GenBank and TAIR10 genes databases. The structures above are colored by base-pairing probabilities from 0% (violet; see color scale) to 100% (red). For unpaired regions, the color denotes the probability of being unpaired. C, Overlap of npcRNA 5 with exon 3 of EFTU/EF-1A (AT4G02930). Red letters indicate dissimilarities between sequences.
A BLASTN search of the GenBank nucleotide and The Arabidopsis Information Resource 10 (TAIR10) genes databases showed that three npcRNAs are significantly similar to known protein-coding genes (i.e. potential regulatory targets). Two of these potential targets, npcRNAs 5 and 8, encode for proteins with ribosomal functions. The 70-nt npcRNA 5 (position +1,090 nt on the forward strand and +1,160 nt on the reverse strand of BspUPG) covers 75% of the third exon of the EFTU/EF-1A protein homolog in Arabidopsis, which binds tRNAs in a GTP-dependent reaction to the acceptor site of ribosomes (AT1G18260; 92.86% similarity; P = 7.00E-24; Fig. 6, B and C; Supplemental Table S14). A fragment of npcRNA 8 maps to a homolog in Arabidopsis lyrata and Arabidopsis that is associated with a U3 small nucleolar RNA involved in the processing of pre-ribosomal RNA (AT3G06530; P = 0.008). The third, npcRNA 3, maps to exon 25 of the myosin XI B protein-coding gene (AT1G04160; P = 0.01), which belongs to a class of myosins potentially involved in the process of spindle/phragmoplast alignment during cytokinesis (Hepler et al., 2002) and whose mutation led to abnormal pollen development in rice (Oryza sativa), suggesting a crucial role of myosin XI B in pollen formation (Jiang et al., 2007).
BspUPG2 Is Highly Conserved in Apomictic Boechera spp.
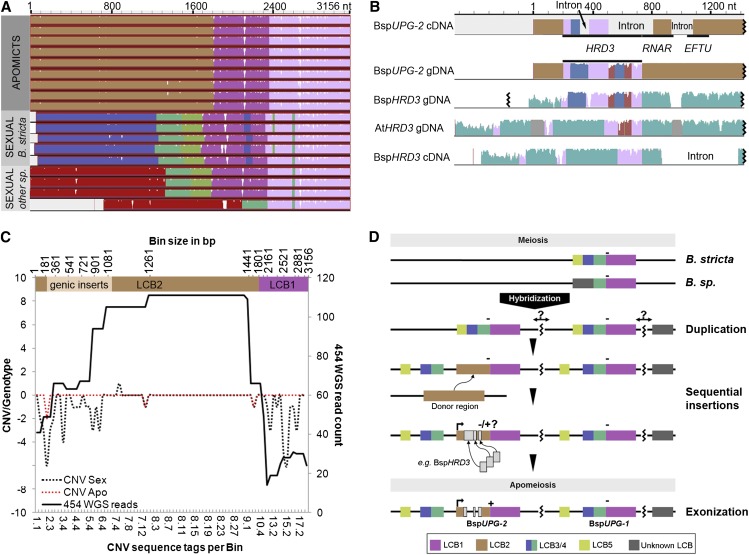
Within-individual (i.e. allelic) polymorphisms were not identified for BspUPG2 among nine tested apomicts. Furthermore, both the original locus (BspUPG1) and its duplicated variant (BspUPG2) are highly conserved at the 3′ end between all tested genotypes, which decreases markedly toward the 5′ end (Fig. 7A; Supplemental Table S15). Sequence conservation is high between all apomicts (i.e. BspUPG2), including the transcriptional outlier ES 753, with the highest nucleotide divergence between genotypes ES 753 and ES 514 (99.33% ± 0.13% similarity), while BspUPG2 in all other apomicts shares complete sequence identity (Fig. 7A). In contrast, sexual sequences can be split into two subgroups based on low sequence conservation at the 5′ end of BspUPG1 (39.95% ± 0.02% similarity), one representing B. stricta genotypes (99.69% ± 0.05% similarity between B. stricta genotypes) and the second representing the remaining sexual genotypes (98.87% ± 0.30% similarity between non-B. stricta genotypes; Fig. 7A; Supplemental Table S15).
Figure 7.
Genesis of BspUPG2 via duplication from the original locus BspUPG1 and sequential insertion of genic fragments. A and B, Structural overview of full-length BspUPG2 in apomictic and sexual Boechera spp. (Table I), adapted and modified from the progressive Mauve algorithm, exhibits LCBs between the candidate genes in various genotypes (A) and between BspUPG2 and the source genes from which BspUPG2 hosts fragments (B). Identical color displays highly similar sequences; black zigzag lines denote truncated sequences due technical reasons. gDNA, Genomic DNA. C, Distribution of 454 whole-genome sequencing (WGS) reads mapping across sliding windows (bins) of BspUPG2 versus CNV in 10 sexual versus 10 apomictic genotypes (Aliyu et al., 2013; for details, see “Materials and Methods”; note that “genic inserts” refers to positions +199 to +1,191 nt in B and that whole-genome sequencing reads did not overlap between LCB1 and LCB2). D, Schematic representation of parental and chimeric BspUPG genes. Colored boxes illustrate different LCBs. +, Transcription activity; −, no transcription activity.
Among all insertions/deletions (indels) that were detected between BspUPG1 and BspUPG2 (less than 50 nucleotides; Albers et al., 2011; Supplemental Table S16), a single 27-nt sequence was exclusively present in BspUPG1 but not in BspUPG2 (Fig. 7A, indel 9). Interestingly, Chellappan et al. (2010) identified a new 27-nt small RNA species that is associated with AGO4 to regulate gene expression at the transcriptional level by directing DNA methylation to some of their target loci in trans. Hence, indel 9 was tested as a small RNA-binding site by northern-blot screening of small RNAs from sexual and apomictic pooled flower buds using specific sense and antisense probes (detailed in Supplemental Protocol S2; Supplemental Fig. S10). Expression of the highly conserved plant miRNA167, which was used as a positive control, was observed in all flower tissues, whereas no signal was detected for any indel 9-specific probe in both apomictic and sexual flower tissues (Supplemental Fig. S10).
Together, the lack of intraindividual allelic variation and the absence of BspUPG2 at any other locus in both sexuals and apomicts (Fig. 5) point to a homozygous or hemizygous state for BspUPG2 in apomicts.
BspUPG2 Arose via Genome Rearrangements
Two lines of evidence were used to validate the proposed fusion of the 5′ (LCB1) and 3′ (LCB2) ends of the transcriptionally active BspUPG2. A BLASTN search between BspUPG2 and the complete genomic sequence-read archive of a sexual B. stricta identified a gap at the juncture between all reads mapping to LCB1 and LCB2, while all reads per LCB overlapped, suggesting their separate genomic origins in the sexual genotype (position +1,820 nt; Fig. 7C). We confirmed this result by amplifying a 478-bp fragment in sexual genotypes using primers corresponding to a fragment of the BspUPG2-specific LCB2. Additionally, this fragment is mapped in sexual and apomictic Boechera spp. by cDNA belonging to the Boechera spp. homolog of AtHRD3 (BspHRD3; Figs. 4 and 7B).
BspHRD3 was used to test whether the 5′ end of BspUPG2 derived directly from parental genes or from putative duplicated variants. Although BspHRD3 cDNA is conserved with Arabidopsis HRD3 (93.0%), variation (i.e. insertions) between the transcript and the detected genomic copy, which are not caused by RNA splicing, was identified (Fig. 7B). This would point to two different genomic variants of BspHRD3: (1) one from which the transcript could be detected but not its genomic copy and that lacks the insertions, and (2) a second variant from which the genomic copy could be sequenced but not the transcript and that includes the inserts (Fig. 7B). Interestingly, comparing the genomic sequence of BspHRD3 with BspUPG2, a 550-bp fragment, identified it as being homologous to BspUPG2, including these insertions (93.6% similarity; Fig. 7B, blue boxes denote insertions), thus pointing to the integration of a fragment of one of the two genomic variants of BspHRD3 into BspUPG2.
As gene duplication is strongly correlated with insertions and pseudogenization (Ohno, 1970; Kaessmann, 2010), we tested for duplication by mapping a set of sequence tags from an array-based comparative genome hybridization experiment in sexual and apomictic Boechera spp. (Aliyu et al., 2013) against BspUPG2 (Fig. 7C). In total, 79 array probes from the comparative genome hybridization experiment mapped to BspUPG2. Whereas most probes mapping onto BspUPG2 show no copy number variations (CNVs) in apomicts (96%, with the remaining 4% showing depletion), 68% of the tags were depleted in at least one sexual genotype. Approximately half of the depleted sequence tags in sexual genotypes were situated at the 3′ end of BspUPG2, representing the start of LCB1. The remaining depleted sequence tags were distributed toward the 5′ end of BspUPG2, between +0 and +1,081 nt. To summarize, both LCB1 and the extreme 5′ end of BspUPG2 are present in fewer copies in sexual compared with apomictic genomes, whereas the middle part of BspUPG2 shows no variation in copy number in either reproductive mode. The duplication of LCB1 in apomictic genomes is explained by its presence in both assemblies. The duplication of the 5′ end in apomictic genomes is evidenced by the presence of duplicated source gene insertions covering almost exactly the region represented by depleted sequence tags in sexuals (HRD3, +199 to +731 nt; RNAR, +732 to +948 nt; EFTU, +1,041 to +1,191 nt; Figs. 5 and 7C).
Since transposable elements (TEs) are considered to drive genome rearrangements, including duplications, after interspecific hybridization, Assembly 1 and Assembly 2 were screened for Viridiplantae transposable elements. In total, 100 and 113 TEs mapped onto Assembly 1 and Assembly 2, respectively, many of which are simple repeats with low sequence complexity. Excluding all simple repeats left 68 and 73 TEs that mapped onto Assembly 1 and Assembly 2, respectively, with about half of them identified in Arabidopsis (27 and 35, respectively; Supplemental Fig. S11). The major repeat families are copia-like (18 and 16) and gypsy-like (8 and 9) long terminal repeat (LTR) retrotransposons, followed by DNA transposons En-Spm (9 and 12), MuDR (11 and 7), hAT (7 and 7), and Helitrons (6 and 9). Proportions of the TE superfamilies do not vary significantly between Assembly 1 and Assembly 2 but do when compared with their distribution across the whole Arabidopsis genome. Copia-like LTR retrotransposons represent the largest proportion of Arabidopsis TEs (57.70%) and those on the Boechera spp. BAC clone assemblies (26.47% and 21.92%, respectively). Nonetheless, copia-like LTR retrotransposons are more prominent across the whole Arabidopsis genome compared with both Boechera spp. assemblies [one-tailed Fisher’s exact test; LTR/Copia: P(Assembly1) = 1.83E-07, P(Assembly2) = 5.23E-10], in contrast to DNA transposons of the En-Spm and hAT class and non-LTR retrotransposons of the LINE/L1 class, which are more prominent on both Boechera spp. assemblies [one-tailed Fisher’s exact test; DNA/En-Spm: P(Assembly1) = 4.21E-10, P(Assembly2) = 3.83E-14; DNA/hAT: P(Assembly1) = 2.00E-06, P(Assembly2) = 3.24E-06; LINE/L1: P(Assembly1) = 0.001, P(Assembly2) = 0.008; Supplemental Fig. S11; Supplemental Tables S17 and S18].
Together, these results (Figs. 5 and 7) indicate a duplication of BspUPG1 fragments from the original locus on Assembly 1 to form the basis of BspUPG2, which subsequently underwent sequential insertions of genome fragments derived from at least two unlinked genomic regions. Thereby, LCB1 was inserted between LCB2 and LCB4. The newly formed locus was then the insertion target for the duplicated variant of at least one functional gene (e.g. BspHRD3), with subsequent exonization leading to a gain of BspUPG2 transcriptional activity (Fig. 7D).
DISCUSSION
High Quantitative Variation for Unreduced Pollen Formation in Apomictic Boechera spp.
High variability in pollen morphology and unreduced pollen formation, in addition to tolerance to deviations from the sexual endosperm balance number, have been described for some Boechera taxa (Böcher, 1951, 1954; Voigt et al., 2007; Aliyu et al., 2010; Voigt-Zielinski et al., 2012). Despite this variability, castration experiments (Böcher, 1951) and extensive flow cytometric analyses of seeds (Aliyu et al., 2010) strongly support selection pressure for the maintenance of unreduced pollen development to fulfill endosperm balance requirements in diploid apomicts.
Here, we have performed detailed quantitative analyses of anther growth and microsporogenesis to identify the optimal developmental stage for comparative expression profiling of reduced (sexual) and unreduced (apomictic) pollen formation in 14 genotypes. A strong correlation between microsporogenesis and anther growth was evident at the premeiotic and postmeiotic stages, whereas it was difficult to identify antherhead lengths corresponding to the meiotic stage (Fig. 2B). Considering the spatial and temporal variability of expression profiles in reproductive tissues (Mascarenhas, 1989; Honys and Twell, 2004), the identification of a specific stage of antherhead development and length characterized by PMCs at the onset of meiosis enabled targeted expression profiling of the meiotic stage, which differentiates reduced (sexual) and unreduced (apomeiotic) pollen formation (Supplemental Table S4). Variable levels of apomeiosis frequencies have been reported for Boechera spp. (Aliyu et al., 2010), and here, both obligate and highly facultative apomicts were analyzed. No evidence for sex- or apomixis-specific flower morphological variation was found, although meiosis and the appearance of microspores are developmentally uncoupled in apomicts relative to sexuals (Fig. 2A; Supplemental Fig. S1). This latter observation supports the HFA theory, which describes the temporal shift (heterochrony) between both reproductive modes (Carman, 1997), which has also been identified on the transcriptomic level (Sharbel et al., 2010).
The meiotic chromosome behavior of apomicts producing unreduced pollen (the same used for expression profiling) was primarily asynaptic (Fig. 1), as has previously been shown (asyndetic; Böcher, 1951). Although ultimately producing higher levels of dyads compared with sexuals, the same apomicts demonstrated variability in terms of monad, dyad, triad, and tetrad formation (Fig. 2A), mirroring reported genotype-specific variability for meiotic chromosome synapsis potential in both diploid and polyploid Boechera spp. (Böcher, 1951; Naumova, 2001; Kantama et al., 2007). Interestingly, variation between individuals of the same clonal lineage for dyad and tetrad formation was also apparent in the majority of tested apomictic genotypes (Fig. 2A); hence, phenotypic variability for pollen formation exists despite genetically clonal reproduction. In this light, the observed sexual allopolyploid progeny from both diploid and triploid obligate apomicts (e.g. ES 753, B. divaricarpa; Schranz et al., 2005) could be explained by fertilization with reduced self-pollen rather than with pollen from another plant (Aliyu et al., 2010), since the genus Boechera is a highly selfing system (Roy, 1995). On a broader scale, our data indicate that fixation of the genotype via apomixis does not necessarily lead to phenotypic stability of male meiocytes.
While these data are consistent with previous work (Kantama et al., 2007; Voigt et al., 2007), the lack of correlation with apomeiosis expression (e.g. egg cell formation as measured by flow cytometric seed screen; Supplemental Fig. S1; Supplemental Table S1) suggests separate mechanisms leading to unreduced male versus female gametes in Boechera spp. and supports independent genetic control for at least some of the developmental steps required to form apomictic seeds (Van Dijk et al., 1999; Noyes and Rieseberg, 2000; Matzk et al., 2005).
The analyses of seed production, anther development, meiosis, and pollen formation have shown that (1) unreduced pollen formation occurs via first division restitution in apomictic Boechera spp., (2) this mechanism is not fully penetrant, and (3) selection pressure for a balanced endosperm in apomictic Boechera spp. leads to the almost exclusive contribution of unreduced pollen to endosperm formation.
BspUPG2, Unreduced Pollen, and Stable Apomixis in Boechera spp.
The detailed phenotypic analyses and selection of a specific antherhead stage that differentiated reduced versus unreduced PMCs, in conjunction with a microarray-based analysis of diploid sexual versus diploid apomictic Boechera spp. genotypes, led to the identification of BspUPG2. Assuming all other developmental aspects of sexual and apomictic antherheads to be the same, the transcriptional activity of this locus is directly correlated with meiotic nonreduction during male sporogenesis and, furthermore, was qRT-PCR validated in 14 biological replicates (i.e. seven sexual and seven apomictic genotypes) and three technical replicates (i.e. microarray probes Sharb0931225, Sharb0501554, and Sharb0690829 mapping to different regions of the same locus) to show strong apomeiosis-specific up-regulation of BspUPG2 in reproductive tissues of apomictic Boechera spp. (Supplemental Fig. S4; Supplemental Table S7). It is unclear why genotype ES 753 was an outlier (Supplemental Fig. S5), despite sharing a highly similar BspUPG2 sequence with other apomictic genotypes. We suspect that genotype-specific shifts in anther development (as observed in other samples) may have led to the sampling of pollen formation outside of the developmental window that characterized other genotypes.
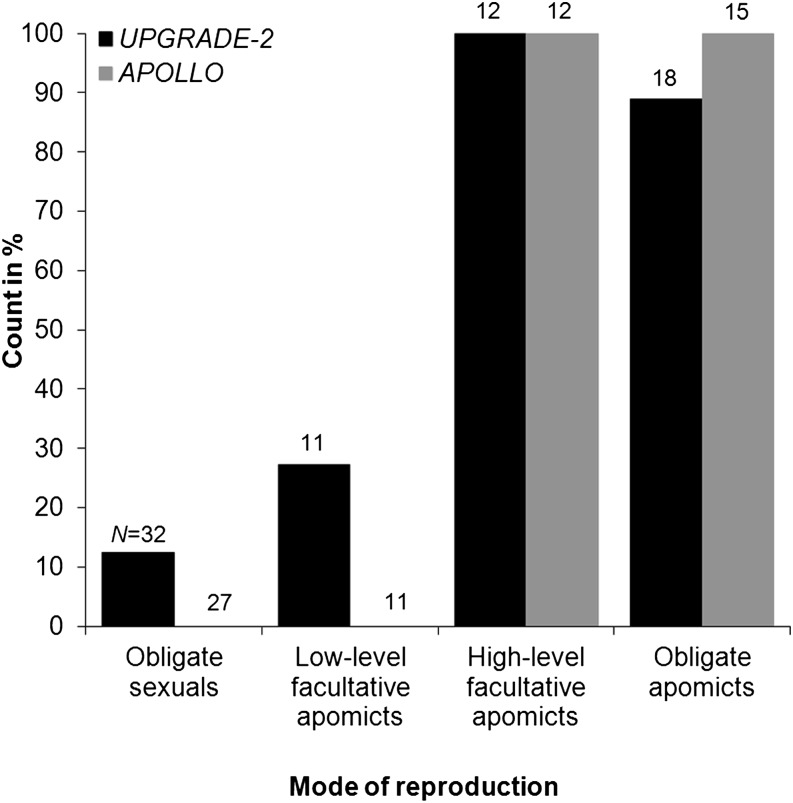
Flow cytometric analyses of over 20,000 single seeds have demonstrated that virtually all apomictic Boechera spp. are characterized by balanced endosperm (6C = [4Cmaternal] + [2Cpaternal] in diploids and 9C = [6Cmaternal] + [3Cpaternal] in triploids), leading to the hypothesis that both meiotically unreduced egg and pollen are required for stable apomictic seed production (Aliyu et al., 2010). Similar to the analyses here that led to the identification of BspUPG2, a second approach identified APOLLO, a similarly conserved factor that is correlated with unreduced egg cell formation in apomictic Boechera spp. (Corral et al., 2013). We tested the hypothesis that both factors should be required to form stable apomictic seed by analyzing for the presence of both factors in the same 73 genotypes examined by Aliyu et al. (2010). Both factors are highly cocorrelated (BspUPG2, 93.33%; APOLLO, 100%; Corral et al., 2013; Fig. 8) with highly facultative and obligate apomictic genotypes (Aliyu et al., 2010), thus supporting our hypothesis.
Figure 8.
Correlation between BspUPG2 and APOLLO with apomictic Boechera spp. The high correlation of BspUPG2 (primers 4 and 5 as denoted in Fig. 5B) with diploid and triploid high-level and obligate apomicts (see “Appendix” in Aliyu et al., 2010) is also mirrored by similar results for the female meiosis marker APOLLO (Corral et al., 2013). Numbers above the columns denote counts of tested genotypes per mode of reproduction as determined by Aliyu et al. (2010).
BspUPG2 Is a Long pri-miRNA with Potential Regulatory Functions in Trans
Although the apo specificity of BspUPG2 attests to its novelty, the lack of homologs in other species and a missing ORF typical of a protein-coding gene leave the question open whether BspUPG2 acts in a complementary fashion to increase the penetrance of an existing predisposition for environmentally induced unreduced gamete formation in sexual genotypes (Aliyu et al., 2010) or whether it is part of a completely novel pathway.
It is unclear whether BspUPG2 is a key genetic factor or one of several tightly linked factors that control different aspects of unreduced pollen formation (i.e. apomeiosis). Furthermore, modifier genes (Bicknell et al., 2000), genetic background, and/or environmental conditions (Nogler, 1984) could explain variation in the level of apomictic trait expression (Figs. 2A and 8). The detection of the apo-specific indel 9 is one example of a potential coregulatory target site in BspUPG2, although no homologous small RNAs were detected (Fig. 7A; Supplemental Fig. S10). Alternatively, the four detected mRNA isoforms (i.e. potentially caused by alternative splicing; Fig. 4; Supplemental Fig. S5) could regulate transcript abundance (as is predicted for at least 25% of all alternative exons; Stamm et al., 2005), leading to the observed variation in terms of reduced and unreduced pollen formation in both facultative and obligate apomicts (Fig. 2A).
Nevertheless, its tissue-specific expression pattern in addition to conservation at the nucleotide level strongly support a developmental role for the lncRNA BspUPG2. Given its chimeric structure, the novel lncRNA BspUPG2 could have attained neofunctionalization in the context of apomeiosis for pollen development through a number of mechanisms (Kaessmann, 2010), similar to the lncRNAs BcMF11 from Brassica campestris (Song et al., 2007, 2013) and Zm401 in maize (Zea mays; Ma et al., 2008). BspUPG2 could conceivably have a regulatory function via a homology-dependent gene-silencing (HDGS) mechanism (for review, see Meyer and Saedler, 1996), such as posttranscriptional gene silencing of related genes in trans (Eamens et al., 2008), which has been found for chimeric Helitron and Pack-MULE RNAs in maize (Jiang et al., 2004; Morgante et al., 2005), or via the modulation of DNA methylation patterns, such as reported for lncRNA-like loci that are associated with polycomb components and histone modifications (De Lucia and Dean, 2011).
Consistent with this are the sequence fragments homologous to three different functional parental genes across the 5′ end of BspUPG2, which putatively could serve as targets for HDGS (BspHRD3, RNAR, and EFTU/EF-1A; Figs. 4, 5A, and 7B). Thereby, BspUPG2 could belong to a novel class of miRNA-containing lncRNAs that serve as a matrix (i.e. as long pri-miRNA) for several unidentified precursor miRNAs (premiRNAs; Altuvia et al., 2005) or endogenous trans-acting short interfering RNAs. Such regulatory elements are processed from the excised intronic region of a spliced transcript, or from the full-length unspliced transcript, and have been detected in vegetative developmental pathways (Peragine et al., 2004; Vazquez et al., 2004; Hirsch et al., 2006) as well as in generative tissues (e.g. in mature pollen; Grant-Downton et al., 2009b).
Computational analysis of RNA folding detected several sections of BspUPG2 that were predicted to form with high probability nonrandom and stable secondary structures (Fig. 6, A and B; Supplemental Fig. S8; Supplemental Table S13) and that fulfill most criteria for premiRNAs (e.g. elevated A + U content; Supplemental Table S14). Interestingly, these potential premiRNAs were not detected in previous screens (Amiteye et al., 2011, 2013), which may reflect (1) its tissue-specific and short-term up-regulation at the onset of male meiosis or (2) limitations of homology-based analyses considering that BspUPG2 is unique to apomictic Boechera spp. The detection of a stable secondary structure in a region that is homologous to a known protein-coding gene with translation elongation activities during polypeptide synthesis at the ribosome and activities in signal transduction (i.e. npcRNA 5 similar to GTP-binding EFTU/EF-1A family protein; E = 7.00E-24; GO:0003746, AT4G02930) could be a first indication for a HDGS function of BspUPG2. For example, a homologous factor to EFTU/EF-1A has been implicated with meiosis progression in Xenopus laevis (Bellé et al., 1990; Peters et al., 1995), while one of the other two sequence fragments of the 5′ end of BspUPG2 is also homologous to a gene with nucleotide-binding function (RNAR; GO:0003723).
High Sequence Conservation and Hemizygous or Homozygous Status Imply a Selective Advantage of BspUPG2
Importantly, BspUPG2 is a chimera of nongenic (e.g. LCB1) and genic (e.g. HDR3, RNAR, EFTU) regions in which the genic regions were derived from partial duplicated variants of their parental genes (e.g. BspHRD3, homologous to AT1G18260; i.e. a pseudogene derived through duplication; Ohno, 1972; Fig. 7, C and D). Although the long-term survival of chimeric genes is described to be rare (Bennetzen, 2005), BspUPG2 exhibited an unexpectedly high degree of conservation in DNA sequence (Fig. 7A; Supplemental Table S15) and expression (Supplemental Fig. S4) between apomictic Boechera spp. representing different taxa and geographic origins. In contrast, higher sequence variation was identified for BspUPG1 in sexuals (e.g. 5′ end; Fig. 7A; Supplemental Table S15). Thus, BspUPG2 appears to be under selective maintenance (Casillas et al., 2007), which would be consistent with its assumed role in unreduced pollen formation for balanced endosperm in apomicts (Aliyu et al., 2010).
The hemizygous or homozygous status of BspUPG2 reflects dominant inheritance as a proposed characteristic for an apomeiosis-controlling locus (Grossniklaus et al., 2001; Table I), as has been found, for example, in Pennisetum squamulatum (Ozias-Akins et al., 1998). Hemizygosity would reflect an extended period of low to no recombination in its genomic location and would be consistent with the distribution of TEs at both the original and duplicated loci (Supplemental Fig. S11). The similar abundance of TEs from gypsy-type and En-Spm superfamilies, which insert preferentially into gene-poor regions (i.e. heterochromatic regions; Fiston-Lavier et al., 2012) and which are enriched in both assemblies compared with their abundance within the total number of Arabidopsis TEs (Supplemental Fig. S11), points to a colocalization of the original and duplicated loci of BspUPG in the same pericentromeric region (Supplemental Fig. S9) or to regions with similar characteristics (i.e. interstitial heterochromatic regions comparable to hk4S in Arabidopsis; Fransz et al., 2000).
We hypothesize that the genesis of BspUPG2 is associated with the recurrent interspecific hybridization that is highly correlated with the origins of apomictic Boechera spp. (Schranz et al., 2005; Beck et al., 2012). Assuming colocalization of BspUPG2 with BspUPG1 and the formation of highly heterochromatic regions in apomicts as protected zones (e.g. Het and Del chromosomes; Kantama et al., 2007), the genome-wide effects of hybridization (Comai, 2000) could have provided the mechanism through which BspUPG2 originated and gained regulatory function.
To summarize, despite variability for unreduced pollen formation in apomictic Boechera spp., a single novel transcription unit (BspUPG2) is consistently up-regulated in apomictic flower tissues at the PMC stage, and its chimeric sequence structure might reflect the interspecific hybridization history of this genus. We hypothesize that BspUPG2 arose via sequential (segmental) duplication-insertion events involving at least five loci, from which three originated from transcribed genes (Fig. 7D). In this regard, it is interesting that only the apomixis-specific duplicated locus (BspUPG2) shows transcriptional activity, while the original locus (BspUPG1), which is present in both sexuals and apomicts, does not (Fig. 5E). Whereas many studies have focused on mutation accumulation and deregulation with respect to the origins of apomixis elements (Tucker et al., 2003; d’Erfurth et al., 2008), the emergence of novel genes in apomicts has not been appreciated, although various identified apomixis-associated loci suggest species-specific apomixis factors (Grossniklaus et al., 2001) for which gains in function are hypothesized (Vielle-Calzada et al., 1996).
CONCLUSION
The identification of the novel apo-specific BspUPG2 supports the HFA theory, which proposes that apomeiosis, and in a broader perspective apomixis, originates from hybrid-specific “genome collisions” and associated induction of gene duplication and TE activation (Carman, 1997). BspUPG2 could have a potential application in stabilizing the endosperm balance in the course of the implementation of apomixis into crop plants. How BspUPG2 has undergone neofunctionalization to develop a trans-regulatory function remains to be clarified.
MATERIALS AND METHODS
Plant Material and Cultivation Conditions
Several sets of the same diploids, 10 apomictic and 12 sexual Boechera spp., were used for all experiments (Supplemental Table S1; note the recent taxonomic information; Koch et al., 2010). Ten seeds per genotype were cultured on moist filter paper in sealed petri dishes and vernalized at 4°C in the dark for 2 weeks until germination. Seedlings were transplanted to plastic pots (11 × 11 × 13 cm) containing autoclaved substrate and grown in a phytotron without insecticides and herbicides under long-day conditions (16 h of light and 8 h of dark, 21°C).
Measurement of Relative Nuclear DNA Content and Reproductive Mode
Relative nuclear DNA content (referred to as ploidy) in leaf, seed (Matzk et al., 2000), and pollen (De Storme et al., 2013) was quantified using leaf tissue from a diploid sexual Boechera stricta (ES 558.2; Supplemental Table S1) as an external control. Leaf and seed material (20 seeds per individual plant) were chopped with a razor blade in a drop of Galbraith’s buffer containing 4 µg mL−1 4′,6-diamino-phenylindole (Galbraith et al., 1983). Tissue-specific ploidy measurements were performed on a FACSAria II (BD Biosciences) equipped with a 375-nm near-UV laser. Data were measured using the FACSDiva Software (version 6.1; BD Biosciences) and analyzed using WinMDI version 2.9 (Scripps Research Institute; http://facs.scripps.edu/software.html); “C” refers to DNA content of a haploid anaphase cell, and “x” is the basic chromosome number.
Microscopy Analysis of Gametophyte Stages
Pollen developmental stages were defined (Fig. 3; Regan and Moffatt, 1990) from each of 12 flower bud size stages (S1–S12) that differed in 100-µm length increments (Smyth et al., 1990; Sanders et al., 1999). For dissection of flower buds at stage S3 and antherheads at flower developmental stages S8 to S12 using a Zeiss Discovery V20 (Carl Zeiss), stereomicroscope sterile glass needles bent to an angle of approximately 100° to 120° and tip size of 50 to 100 µm using a Narishige PC-10 puller (Narishige Group) were used. Sample preparation of size-staged buds and antherheads for histology and light microscopy was carried out according to Grasser et al. (2009).
The decision to take the PMC stage for comparative gene expression profiling is based on two criteria: (1) apomeiosis candidate genes should affect early meiotic stages (Grimanelli et al., 2001b), and (2) in previous analyses, the spike of gene expression change between sexual and apomictic ovules was detected in the microspore mother cell (Sharbel et al., 2010), a stage that corresponds to the PMC in antherheads.
Cytochemical Analysis of Fixated Boechera spp. Anthers
Cytological observations of meiosis and PMCs were made from sexuals and apomicts at flower bud stages S9 to S10. Meiotic chromosome spreads of anthers were prepared according to Ross et al. (1996) with minor changes and observed with a Zeiss Axioplan 2 imaging microscope (Carl Zeiss). Photographs were taken with an AxioCam HRc Rev. 2 camera under 100-fold magnification using a 49 DAPI BP reflector block. Meiocytes at the tetrad stage were examined by separate squashes of all six antherheads per flower bud, from one sexual and six apomictic genotypes, to determine the number of monads, dyads, triads, and tetrads in each antherhead. Anthers were squashed according to Peterson et al. (2010), stained (Alexander’s stain; Alexander, 1969), and examined with a Zeiss Axioplan 2 imaging microscope (Carl Zeiss). All available meiocytes per anther were counted, and statistical analyses of meiocyte behavior and anther size correlations of gametophyte stages were evaluated with SPSS version 11.5 (LEAD Technologies).
RNA Isolation, Microarray Hybridization, and qRT-PCR of Antherhead Tissue
Microdissections and total RNA extractions were prepared using tools and a dissection area cleaned with ethanol, treated with RNAseZap (Ambion), and washed with diethyl phosphorocyanidate-treated distilled water. Approximately 30 antherheads of each genotype, corresponding to the PMC stage, were live microdissected from fresh whole flower buds using a Zeiss Discovery V20 stereomicroscope (Carl Zeiss) using sterile glass needles (see above) and collected in 500 µL of RNAlater (Qiagen). Eighteen microliters of RNase-free DNase I (Qiagen)-digested total RNA extracts (RNeasy Micro kit; Qiagen) was eluted through RNeasy MinElute Spin columns (Qiagen) and quantified on a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The RNA quality of all samples was assessed with the RNA 6000 Nano LabChip Kit II (Agilent Technologies) on the Agilent 2100 Bioanalyzer. Purified total RNA was stored at −80°C.
To ensure optimal copy RNA yield after labeling, 200 ng of total RNA with absorbance readings of A260/A280 > 1.8 and A260/A230 > 1.8 (recommended by Agilent) was used for the labeling procedure. Approximately 1.5 μg of the generated Cy3-labeled copy RNA per sample (One-color Quick Amp Labeling kit; Agilent Technologies) was hybridized for 18 h at 65°C to the custom Boechera spp. whole flower-105k-Agilent microarrays, which were scanned at 5-μm double-pass resolution with an Agilent G2565BA Microarray Scanner. The One-color RNA Spike-in kit (Agilent Technologies) was used to ensure optimal microarray processing. Microarray hybridization quality was assessed with Feature Extraction 10.1 software (Agilent Technologies), whereas quantile normalization with baseline-to-median transformation and gene expression analysis were performed with GeneSpring GX 10 software (Agilent Technologies). Differentially expressed microarray probes were considered validated with P ≤ 0.05 as assessed by an unpaired Student’s t test with a mean difference of 2-fold or greater. P values were corrected for the family-wise error rate as a control for false positives using the Bonferroni method.
Real-time PCR, using primers that were designed on the cDNA reads homologous to the candidate microarray probes Sharb0690829 (GenBank/EMBL/DDBJ accession no. ERS317557), Sharb0501554 (GenBank/EMBL/DDBJ accession no. ERS317556), and Sharb0931225 (GenBank/EMBL/DDBJ accession no. ERS317555; Primer3 version 0.4.0; http://frodo.wi.mit.edu/primer3/), was performed according to Sharbel et al. (2010). Seven biological and four technical replicates were run for each probe and tissue on a 384-well plate together with two endogenous control genes tested on Boechera spp. anther material (ACTIN2 and ELONGATION FACTOR α1; Pellino et al., 2011), negative template, and reverse transcriptase controls. PCR efficiencies and normalized cycle threshold values of each set of four technical replicates were processed with the Real-time PCR Miner version 2.0 software (Zhao and Fernald, 2005). Relative quantification and normalized cycle threshold values of the amplified targets were calculated separately with reference to the expression levels of each of the two housekeeping genes employing the ∆∆Ct method (Pfaffl, 2001) using a calibrator sample (ES 910.2; Supplemental Table S7). The corresponding mean relative expression ratio for each genotype was calculated with SPSS version 11.5 (LEAD Technologies), and significant differences between samples were evaluated using a one-way ANOVA (α = 0.05) with Tukey’s HSD post hoc test for differences between multiple pairs of means.
RACE
The SMARTer RACE method (Clontech) was employed to obtain 5′ end and poly(A) site information from BspUPG2 and BspHRD3, an Arabidopsis (Arabidopsis thaliana) homolog of HRD3 in Boechera spp. Primers (Primer3 version 0.4.0) for BspUPG2 RACE were derived from microarray probe homologous cDNA read ERS317557 (5′ end primer GSP3, 5′-TCTTCGCCATCGTTCATGTTTACTTCCG-3′; 3′ end primer GSP4, 5′-TCATCATGTCTTCTTCGCCATCGTTCA-3′), and those for BspHRD3 RACE were derived from the LCB2 of BspUPG2 (5′ end primer GSP11, 5′-TAATGCCCACTGGGGTCGTCATTGT-3′; 3′ end primer CON234X14_L, 5′-ACTGGAATTGGGTACTTGTATGTCA-3′). PCR was performed with the Advantage 2 PCR Kit (Clontech), and PCR fragments were cloned (pCR4-TOPO TA; Life Technologies) and Sanger sequenced (see above).
Computational Analysis of RNA-Folding Probabilities of BspUPG
Structural RNAs are usually characterized by an unusual thermodynamic stability and a conserved secondary structure. The minimum folding energy as a measure of thermodynamic stability for a sequence (i.e. negative values indicate that a sequence is more stable) was calculated using the RNAfold and RNAz version 1.0 software for Windows of the Vienna RNA package (Hofacker et al., 1994; http://www.tbi.univie.ac.at/ivo/RNA). The presence of statistically significant secondary structures of BspUPG2 was monitored using Z-score values as described by Crespi et al. (1994) and Bonnet et al. (2004) using similar thresholds to Kavanaugh and Dietrich (2009). All significant secondary structures of BspUPG2 were classified according to Zhang et al. (2006). The “npcRNA [number]” names were chosen to follow the naming convention established by previous investigators (Hirsch et al., 2006).
BAC Probe Preparation and Screening of a Boechera spp. BAC Library
Candidate 60-mer microarray probes were mapped against Boechera spp. 454 FLX cDNA libraries (Sharbel et al., 2009) using CLC Genomics Workbench version 4.5.1 (CLC Bio; standard parameters). Flanking regions to the mapped microarray probes were used to design specific primers (Primer3 version 0.4.0) for chromosome walking in sexuals and apomicts using the DNA Walking SpeedUp Premix Kit I from Seegene (Supplemental Table S10). Resultant products were transformed into Escherichia coli TOP10 cells using the TOPO TA Cloning Kit for Sequencing (Invitrogen), and individual clones were amplified using the TempliPhi DNA Sequencing Template Amplification Kit (Reagin et al., 2003) and Sanger sequenced on an ABI 3730 XL sequencing system. Sequence analysis and assembly were carried out with Lasergene 8 (DNAStar). The E. coli TOP10 subcloned, PCR-amplified, and finally gel-purified DNA walking products (NucleoSpin Extract II kit; Macherey-Nagel) were hybridized against a gridded BAC library (48 × 384 spotted wells onto a 22- × 22-cm filter membrane; binary vector pCLD04541; Bancroft, 1997). Individually radiolabeled and pooled probes were hybridized as a group using the overgo hybridization method (Ross et al., 1999).
Vector inserts of 500 ng of pure BAC clone DNA extracts were restriction digested to completion with HindIII, BglII, and BamHI (Amersham Pharmacia Biotech) at 37°C (HindIII and BglII) and 30°C (BamHI) for 8 h and examined on a 1% agarose gel. Based on their partial overlapping restriction patterns, DNA was isolated from four BAC clones (A4O22, E7K5, C8B11, and F8G11) using Nucleobond Xtra Midi Kits (Macherey-Nagel). BAC DNA was randomly sheared (Hydroshear; Digilab) and size fractionated by agarose gel electrophoresis in approximately 1-kb and approximately 4- to 5-kb size classes. These fragments were end repaired, blunt-end ligated into pUC19 (Life Technologies), transformed into E. coli ELECTROMAX DH5α-E electro-competent cells (Invitrogen), and sequenced on an ABI 3730 XL automatic DNA sequencer (PE Applied Biosystems). Vector clipping, quality trimming, and sequence assembly using stringent conditions (e.g. 95% sequence identity cutoff, 25-bp overlap) were done using Lasergene 8 (DNASTAR) and Staden (http://staden.sourceforge.net/). Assembly of the complete BAC clone sequences was performed using Seqman and the Gap4 algorithm implemented in Staden. Remaining gaps in the contiguous BAC sequences were manually inspected and closed with primer walking or PCR products crossing the gaps from adjacent contigs. The resulting sequences were assembled using Lasergene 8 (DNASTAR) set to an overlap minimum of 20 bp with 95% identity. All BAC assemblies were annotated using BLASTN and BLASTX searches against the nonredundant GenBank nucleotide and protein databases, respectively (http://www.ncbi.nlm.nih.gov/genbank/).
Processing of DNA Sequences and Sequence Analysis
The allelic constitution of BspUPG2 in nine each sexual and apomictic genotypes was determined by sequencing proofreading polymerase-amplified (Phusion high-fidelity polymerase; Thermo Scientific), gel-purified (NucleoSpin Extract II kit; Macherey-Nagel), and multiply cloned (CloneJet PCR cloning kit; Fermentas) PCR fragments using specific primers (5′ end primer in apomictic genotypes, CON234X5L, 5′-TCCGACCTAAATCCTACCAAACTGA-3′; in sexual B. stricta, CON234X11L, 5′-CAAAAATAAAAGATTTGATGTAGATTGC-3′, and in other sexual genotypes, FLTsexX2L, 5′-GAAGAAAGAGCTACGGCGTGAT-3′; 3′ end primer CON234X5R, 5′-TGCTCAATTTTGAACATCTTATTTGC-3′). Lasergene 8 (DNASTAR) was used for assembly and similarity analysis of Phred 20 quality-trimmed sequences. Coding potential was calculated from all six frames of all BspUPG2 splicing forms using the Coding Potential Calculator software (Kong et al., 2007).
Pairwise ClustalW comparisons in the presence of rearrangements were performed with Mauve (version 2.3.1; progressiveMauve, default parameters; Darling et al., 2004) to detect LCBs (conserved sequence segments, which are internally free from genome rearrangements) in BspUPG1 and BspUPG2 between different genotypes.
Local BLASTN search of the complete genomic sequence read archive (SRP007750; http://www.ncbi.nlm.nih.gov/sra/; 454 GS FLX) and sequence extraction were conducted with CLC Genomics Workbench (costs = match 1, mismatch 3, existence 5, extension 2; E value = 10; word size = 11, filter complexity = yes). For mapping of both 454 whole-genome sequencing and CNV sequence tags (Aliyu et al., 2013), the genomic sequence of BspUPG2 was divided into 18 200-bp bins with 20-bp overlap. All existing CNVs for each single sequence tag are represented, and relative CNV frequencies are considered independently for sexual and apomictic genotypes.
BAC sequence assemblies were annotated for transposable elements by screening the green plant section (Viridiplantae) of the Repbase repetitive element database (Jurka, 1998; http://www.girinst.org/server/Maps/AT/index.html) using CENSOR (Kohany et al., 2006). The programs einverted and EMBOSS (Rice et al., 2000) were employed to identify inverted repeats with 80% or more matches, and the Pipmaker software was used to identify simple repeats and CpG islands (Schwartz et al., 2003). LTR analyses of Assembly 1 and Assembly 2 were performed with LTR FINDER software (Xu and Wang, 2007).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ERS317552, ERS317553, ERS317554, ERS317555, ERS317556, ERS317557, ERS317558, HF930769, HF954100, HF954101, HG007837 and HG007838.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Representative meiocytes at the tetrad stage and flow cytometric ploidy confirmation.
Supplemental Figure S2. Correlation of flower organs and flower bud length for sexual and apomictic Boechera spp. genotypes.
Supplemental Figure S3. Constantly differentially regulated microarray probes in apomictic compared with sexual Boechera spp. genotypes.
Supplemental Figure S4. Validation of differentially expressed microarray probes by qRT-PCR.
Supplemental Figure S5. Rapid amplification of cDNA 3′ and 5′ ends of the candidate gene UPGRADE.
Supplemental Figure S6. Percentage identity plots showing the original UPGRADE locus and its duplicated variant.
Supplemental Figure S7. Distribution of locally collinear sequence blocks along the BAC clone Assembly 1 and Assembly 2.
Supplemental Figure S8. Distribution of Arabidopsis homologous loci corresponding to Assembly 1 and Assembly 2 in Boechera spp. along Arabidopsis chromosomes.
Supplemental Figure S9. Thermodynamically stable nonrandom secondary structures of BspUPG2.
Supplemental Figure S10. Northern-blot analysis for putative small RNAs homologous to indel 9 at BspUPG2.
Supplemental Figure S11. Distribution of the TE superfamilies along the original and duplicated UPGRADE locus.
Supplemental Table S1. List of Boechera spp. genotypes with population information used for histological examination of microsporogenesis and candidate gene analyses.
Supplemental Table S2. Analysis of meiocyte constitution at the tetrad stage in diploid sexual and high facultative and obligate apomictic Boechera spp. genotypes.
Supplemental Table S3. High-level 1C pollen individuals with corresponding flow cytometric seed screen data.
Supplemental Table S4. Frequencies of microspores, meiotic cells, and PMCs relative to antherhead length.
Supplemental Table S5. Microarray probes demonstrating significant absolute fold change up-regulation in apomictic genotypes.
Supplemental Table S6. qRT-PCR primers used for the validation of microarray candidate probes.
Supplemental Table S7. Relative mRNA expression values for four microarray probes in somatic and reproductive tissues.
Supplemental Table S8. Overview of BAC clones carrying one to three candidate microarray probes.
Supplemental Table S9. Transcription factor-binding sites upstream of BspUPG2.
Supplemental Table S10. Primers used for chromosome walking on microarray candidate probes and the source gene BspHRD3.
Supplemental Table S11. Gene annotation of Boechera spp. BAC clone Assembly 1 and Assembly 2.
Supplemental Table S12. Distribution of inverted repeats on Assembly 1 and Assembly 2.
Supplemental Table S13. Detection of candidate regions for stable non-protein-coding RNA secondary structures of BspUPG2.
Supplemental Table S14. Characteristics of thermodynamically stable secondary non-protein-coding RNA structures with highest Z-score from BspUPG2 in apomictic Boechera spp. genotypes.
Supplemental Table S15. Sequence divergence of BspUPG1 and BspUPG2.
Supplemental Table S16. Indels on BspUPG2 isolates of sexual and apomictic genotypes using a ClustalW multiple sequence alignment.
Supplemental Table S17. Green plant (Viridiplantae) homologous repetitive DNA element sequence mapping on Assembly 1.
Supplemental Table S18. Green plant (Viridiplantae) homologous repetitive DNA element sequence mapping on Assembly 2.
Supplemental Protocol S1. Computational analysis of RNA-folding probabilities of BspUPG2.
Supplemental Protocol S2. Small RNA northern blot.
Supplementary Material
Acknowledgments
The authors thank Heidi Block, Vera Katzorke, Monika Wiesner and Ines Walde for technical support, Ingo Schubert for constructive comments on the manuscript, and Thomas Mitchell-Olds and Eric Schranz for seed material.
Glossary
- PMC
pollen mother cell
- HFA
hybridization-derived floral asynchrony
- HSD
honestly significant difference
- cDNA
complementary DNA
- DDBJ
DNA Data Bank of Japan
- qRT
quantitative reverse transcription
- BAC
bacterial artificial chromosome
- nt
nucleotides
- ORF
open reading frame
- lncRNA
long non-protein-coding mRNA-like RNA
- LCB
locally collinear block
- miRNA
microRNA
- npcRNA
small non-protein-coding RNA
- pri-miRNA
primary miRNA
- TAIR10
The Arabidopsis Information Resource 10
- indel
insertion/deletion
- CNV
copy number variation
- TE
transposable element
- LTR
long terminal repeat
- HDGS
homology-dependent gene-silencing
- premiRNAs
precursor miRNAs
References
- Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R. (2011) Dindel: accurate indel calls from short-read data. Genome Res 21: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini E, Marconi G, Barcaccia G, Raggi L, Falcinelli M. (2004) Isolation of candidate genes for apomixis in Poa pratensis L. Plant Mol Biol 56: 879–894 [DOI] [PubMed] [Google Scholar]
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Aliyu OM, Schranz ME, Sharbel TF. (2010) Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am J Bot 97: 1719–1731 [DOI] [PubMed] [Google Scholar]
- Aliyu OM, Seifert M, Corral JM, Fuchs J, Sharbel TF. (2013) Copy number variation in transcriptionally active regions of sexual and apomictic Boechera demonstrates independently derived apomictic lineages. Plant Cell http://dx.doi.org/10.1105/tpc.113.113860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA, Windham M. (2010) Brassicaceae In Flora of North America Editorial Committee, eds, Flora of North America North of Mexico, Vol 7. Oxford University Press, New York and Oxford, pp 224–746 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33: 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiteye S, Corral JM, Vogel H, Kuhlmann M, Mette MF, Sharbel TF. (2013) Novel microRNAs and microsatellite-like small RNAs in sexual and apomictic Boechera species. MicroRNA 2: 46–63 [PubMed] [Google Scholar]
- Amiteye S, Corral JM, Vogel H, Sharbel TF. (2011) Analysis of conserved microRNAs in floral tissues of sexual and apomictic Boechera species. BMC Genomics 12: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I. (1997) A strategy involving the use of high redundancy YAC subclone libraries facilitates the contiguous representation in cosmid and BAC clones of 1.7 Mb of the genome of the plant Arabidopsis thaliana. Weeds World 4: 1–9 [Google Scholar]
- Beck JB, Alexander PJ, Allphin L, Al-Shehbaz IA, Rushworth C, Bailey CD, Windham MD. (2012) Does hybridization drive the transition to asexuality in diploid Boechera? Evolution 66: 985–995 [DOI] [PubMed] [Google Scholar]
- Bellé R, Cormier P, Poulhe R, Morales J, Huchon D, Mulner-Lorillon O. (1990) Protein phosphorylation during meiotic maturation of Xenopus oocytes: cdc2 protein kinase targets. Int J Dev Biol 34: 111–115 [PubMed] [Google Scholar]
- Bennetzen JL. (2005) Transposable elements, gene creation and genome rearrangement in flowering plants. Curr Opin Genet Dev 15: 621–627 [DOI] [PubMed] [Google Scholar]
- Bicknell RA, Borst NK, Koltunow AM. (2000) Monogenic inheritance of apomixis in two Hieracium species with distinct developmental mechanisms. Heredity (Edinb) 84: 228–237 [DOI] [PubMed] [Google Scholar]
- Böcher TW. (1951) Cytological and embryologal studies in the amphiapomictic Arabis holboellii complex. Biologiske Skrifter/Kongelige Danske Videnskabernes Selskab 6: 1–59 [Google Scholar]
- Böcher TW. (1954) Experimental taxonomical studies in the Arabis holboellii complex. Sven Bot Tidskr 48: 31–44 [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. (2004) Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20: 2911–2917 [DOI] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L, Köhler C. (2011) Unreduced gamete formation in plants: mechanisms and prospects. J Exp Bot 62: 1659–1668 [DOI] [PubMed] [Google Scholar]
- Carman JG. (1997) Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Linn Soc Lond 61: 51–94 [Google Scholar]
- Casillas S, Barbadilla A, Bergman CM. (2007) Purifying selection maintains highly conserved noncoding sequences in Drosophila. Mol Biol Evol 24: 2222–2234 [DOI] [PubMed] [Google Scholar]
- Chang WC, Lee TY, Huang HD, Huang HY, Pan RL. (2008) PlantPAN: Plant Promoter Analysis Navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene group. BMC Genomics 9: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. (2010) siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2000) Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol 43: 387–399 [DOI] [PubMed] [Google Scholar]
- Corral JM, Vogel H, Aliyu OM, Hensel G, Thiel T, Kumlehn J, Sharbel TF. (2013) A conserved apomixis-specific polymorphism is correlated with exclusive DEDDh exonuclease expression in premeiotic ovules of apomictic Boechera Species. Plant Physiol 163: 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14: 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. (2011) CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Dean C. (2011) Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol 14: 168–173 [DOI] [PubMed] [Google Scholar]
- d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Simon M, Jenczewski E, Mercier R. (2008) Mutations in AtPS1 (Arabidopsis thaliana parallel spindle 1) lead to the production of diploid pollen grains. PLoS Genet 4: e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Zamariola L, Mau M, Sharbel TF, Geelen D. (2013) Volume-based pollen size analysis: an advanced method to assess somatic and gametophytic ploidy in flowering plants. Plant Reprod 26: 65–81 [DOI] [PubMed] [Google Scholar]
- Eamens A, Wang MB, Smith NA, Waterhouse PM. (2008) RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol 147: 456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Vejnar CE, Quesneville H. (2012) Transposable sequence evolution is driven by gene context. Cornell University Library, http://arxiv.org/abs/1209.0176 [Google Scholar]
- Fransz PF, Armstrong SJ, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH. (2000) Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell 100: 367–376 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H. (2009b) MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, Grasser KD. (2009) Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J Mol Biol 386: 598–611 [DOI] [PubMed] [Google Scholar]
- Grimanelli D, Leblanc O, Perotti E, Grossniklaus U. (2001a) Developmental genetics of gametophytic apomixis. Trends Genet 17: 597–604 [DOI] [PubMed] [Google Scholar]
- Grimanelli D, Tohme J, González-De-León D. (2001b) Applications of molecular genetics in apomixis research. In Y Savidan, JG Carman, T Dresselhaus, eds, The Flowering of Apomixis: From Mechanisms to Genetic Engineering. Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT), Institut de Recherche pour le Developpernent (IRD), European Commission DG VI (FAIR), Mexico, D.F., pp 83–94 [Google Scholar]
- Grossniklaus U. (2001a) From sexuality to apomixis: molecular and genetic approaches. In Y Savidan, JG Carman, T Dresselhaus, eds, The Flowering of Apomixis: From Mechanisms to Genetic Engineering. Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT), Institut de Recherche pour le Developpernent (IRD), European Commission DG VI (FAIR), Mexico, D.F., pp 168–211 [Google Scholar]
- Grossniklaus U, Nogler GA, van Dijk PJ. (2001) How to avoid sex: the genetic control of gametophytic apomixis. Plant Cell 13: 1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin J, Rossel JB, Robert S, Tsuchiya T, Koltunow A. (2000) A DEFICIENS homologue is down-regulated during apomictic initiation in ovules of Hieracium. Planta 210: 914–920 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Valster A, Molchan T, Vos JW. (2002) Roles for kinesin and myosin during cytokinesis. Philos Trans R Soc Lond B Biol Sci 357: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J, Lefort V, Vankersschaver M, Boualem A, Lucas A, Thermes C, d’Aubenton-Carafa Y, Crespi M. (2006) Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol 140: 1192–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer SL, Tacker M, Schuster P. (1994) Fast folding and comparison of RNA secondary structures. Chemical Monthly 125: 167–188 [Google Scholar]
- Honys D, Twell D. (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörandl E, Temsch EM. (2009) Introgression of apomixis into sexual species is inhibited by mentor effects and ploidy barriers in the Ranunculus auricomus complex. Ann Bot (Lond) 104: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosouchi T, Kumekawa N, Tsuruoka H, Kotani H. (2002) Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res 9: 117–121 [DOI] [PubMed] [Google Scholar]
- Hunt AG. (1994) Messenger RNA 3′ end formation in plants. Annu Rev Plant Physiol Plant Mol Biol 45: 47–60 [Google Scholar]
- Jiang N, Bao Z, Zhang X, Eddy SR, Sr, Wessler SR. (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Jiang SY, Cai M, Ramachandran S. (2007) ORYZA SATIVA MYOSIN XI B controls pollen development by photoperiod-sensitive protein localizations. Dev Biol 304: 579–592 [DOI] [PubMed] [Google Scholar]
- Johnston SA, den Nijs TPM, Peloquin SJ, Hanneman RE., Jr (1980) The significance of genic balance to endosperm development in interspecific crosses. Theor Appl Genet 57: 5–9 [DOI] [PubMed] [Google Scholar]
- Jurka J. (1998) Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol 8: 333–337 [DOI] [PubMed] [Google Scholar]
- Kaessmann H. (2010) Origins, evolution, and phenotypic impact of new genes. Genome Res 20: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantama L, Sharbel TF, Schranz ME, Mitchell-Olds T, de Vries S, de Jong H. (2007) Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc Natl Acad Sci USA 104: 14026–14031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh LA, Dietrich FS. (2009) Non-coding RNA prediction and verification in Saccharomyces cerevisiae. PLoS Genet 5: e1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Karl R, Kiefer C, Al-Shehbaz IA. (2010) Colonizing the American continent: systematics of the genus Arabis in North America (Brassicaceae). Am J Bot 97: 1040–1057 [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. (2006) Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. (2007) Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9: 660–665 [DOI] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 36: 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Liu X, An L, Zhang W, Sun J, Pei H, Meng H, Fan Y, Zhang C. (2008) The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis. Cell Res 18: 589–599 [DOI] [PubMed] [Google Scholar]
- Ma J, Yan B, Qu Y, Qin F, Yang Y, Hao X, Yu J, Zhao Q, Zhu D, Ao G. (2008) Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J Cell Biochem 105: 136–146 [DOI] [PubMed] [Google Scholar]
- Marimuthu MPA, Jolivet S, Ravi M, Pereira L, Davda JN, Cromer L, Wang L, Nogué F, Chan SWL, Siddiqi I, et al. (2011) Synthetic clonal reproduction through seeds. Science 331: 876. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. (1989) The male gametophyte of flowering plants. Plant Cell 1: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzk F, Meister A, Schubert I. (2000) An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J 21: 97–108 [DOI] [PubMed] [Google Scholar]
- Matzk F, Prodanovic S, Bäumlein H, Schubert I. (2005) The inheritance of apomixis in Poa pratensis confirms a five locus model with differences in gene expressivity and penetrance. Plant Cell 17: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saedler H. (1996) Homology-dependent gene silencing in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 23–48 [DOI] [PubMed] [Google Scholar]
- Mogie M. (1988) A model for the evolution and control of generative apomixis. Biol J Linn Soc Lond 35: 127–153 [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. (2005) Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet 37: 997–1002 [DOI] [PubMed] [Google Scholar]
- Naumova TN. (2001) Reproductive development in apomictic populations of Arabis holboellii (Brassicaceae). Sex Plant Reprod 14: 195–200 [DOI] [PubMed] [Google Scholar]
- Nogler GA (1984) Gametophytic apomixis. In BM Johri, ed, Embryology of Angiosperms. Springer-Verlag, Berlin, pp 475–518 [Google Scholar]
- Noyes RD, Rieseberg LH. (2000) Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 155: 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Springer-Verlag, Berlin [Google Scholar]
- Ohno S. (1972) So much “junk” DNA in our genome. Brookhaven Symp Biol 23: 366–370 [PubMed] [Google Scholar]
- Ozias-Akins P, Roche D, Hanna WW. (1998) Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc Natl Acad Sci USA 95: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellino M, Sharbel TF, Mau M, Amiteye S, Corral JM. (2011) Selection of reference genes for quantitative real-time PCR expression studies of microdissected reproductive tissues in apomictic and sexual Boechera. BMC Res Notes 4: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters HI, Chang YWE, Traugh JA. (1995) Phosphorylation of elongation factor 1 (EF-1) by protein kinase C stimulates GDP/GTP-exchange activity. Eur J Biochem 234: 550–556 [DOI] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C. (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. International Journal of Plant Biology 1: 66–69 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Haag JR, Ream T, Wierzbicki AT. (2008) Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13: 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagin MJ, Giesler TL, Merla AL, Resetar-Gerke JM, Kapolka KM, Mamone JA. (2003) TempliPhi: a sequencing template preparation procedure that eliminates overnight cultures and DNA purification. J Biomol Tech 14: 143–148 [PMC free article] [PubMed] [Google Scholar]
- Regan SM, Moffatt BA. (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rollins RC. (1941) Monographic study of Arabis in western North America. Rhodora 43: 289–483 [Google Scholar]
- Ross KJ, Fransz P, Jones GH. (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4: 507–516 [DOI] [PubMed] [Google Scholar]
- Ross M, LaBrie T, McPherson S, Stanton VPS (1999) Screening large-insert libraries by hybridization. In Current Protocols in Human Genetics. John Wiley & Sons, New York, unit 5.6 [Google Scholar]
- Roy BA. (1995) The breeding systems of six species of Arabis (Brassicaceae). Am J Bot 82: 869–877 [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Savidan Y. (2001) Transfer of apomixis through wide crosses. In Y Savidan, JG Carman, T Dresselhaus, eds, The Flowering of Apomixis: From Mechanisms to Genetic Engineering. Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT), Institut de Recherche pour le Developpernent (IRD), European Commission DG VI (FAIR), Mexico, D.F., pp 153–167 [Google Scholar]
- Schallau A, Arzenton F, Johnston AJ, Hähnel U, Koszegi D, Blattner FR, Altschmied L, Haberer G, Barcaccia G, Bäumlein H. (2010) Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J 62: 773–784 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Dobeš C, Koch MA, Mitchell-Olds T. (2005) Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae). Am J Bot 92: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Windsor AJ, Song BH, Lawton-Rauh A, Mitchell-Olds T. (2007) Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiol 144: 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Green ED, Hardison RC, Miller W. (2003) MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res 31: 3518–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seffens W, Digby D. (1999) mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res 27: 1578–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, Galla G, Kumlehn J, Klukas C, Schreiber F, Vogel H, Rotter B. (2010) Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22: 655–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel TF, Voigt ML, Corral JM, Thiel T, Varshney A, Kumlehn J, Vogel H, Rotter B. (2009) Molecular signatures of apomictic and sexual ovules in the Boechera holboellii complex. Plant J 58: 870–882 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Filipowicz W. (1996) Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol 32: 1–41 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JH, Cao JS, Wang CG. (2013) BcMF11, a novel non-coding RNA gene from Brassica campestris, is required for pollen development and male fertility. Plant Cell Rep 32: 21–30 [DOI] [PubMed] [Google Scholar]
- Song JH, Cao JS, Yu XL, Xiang X. (2007) BcMF11, a putative pollen-specific non-coding RNA from Brassica campestris ssp. chinensis. J Plant Physiol 164: 1097–1100 [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. (2005) Function of alternative splicing. Gene 344: 1–20 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Araujo AC, Paech NA, Hecht V, Schmidt ED, Rossell JB, De Vries SC, Koltunow AM. (2003) Sexual and apomictic reproduction in Hieracium subgenus pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D. (2011) Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod 24: 149–160 [DOI] [PubMed] [Google Scholar]
- Van Dijk PJ, Tas IC, Falque M, Bakx-Schotman T. (1999) Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity (Edinb) 83: 715–721 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Crane CF, Stelly DM. (1996) Apomixis: the asexual revolution. Science 274: 1322–1323 [Google Scholar]
- Voigt ML, Melzer M, Rutten T, Mitchell-Olds T, Sharbel TF. (2007) Gametogenesis in the apomictic Boechera holboellii complex: the male perspective. In E Hörandl, U Grossniklaus, PJ van Dijk, TF Sharbel, eds, Apomixis: Evolution, Mechanisms and Perspectives. A.R.G. Gantner Verlag, Ruggell, pp 235–257 [Google Scholar]
- Voigt-Zielinski ML, Piwczyński M, Sharbel TF. (2012) Differential effects of polyploidy and diploidy on fitness of apomictic Boechera. Sex Plant Reprod 25: 97–109 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H. (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35: 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Makaroff CA, Ma H. (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. (2006) Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci 63: 246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12: 1047–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.