Abstract
RNA interference (RNAi) has considerable potential as a therapeutic strategy, but the development of efficient in vivo RNA delivery methods remains challenging. To this end, we designed and synthesized chemically modified interfering nanoparticles (iNOPs) composed of functionalized poly-l-lysine dendrimers modified with reducible spacers to facilitate release of small interfering RNAs (siRNAs) in vivo. We show that the novel siRNA–iNOP complexes mediate efficient gene-specific RNAi in cultured cells and in mice, where they display enhanced tissue-targeting capabilities. At a clinically feasible dose of 1 mg kg–1, apolipoprotein B (apoB) siRNA–iNOP complexes achieved ∼40–45% reduction of liver apoB mRNA and plasma apoB protein levels within 48 h of administration to mice, without apparent toxicity. Collectively, these findings demonstrate that siRNA delivery by the modified reducible iNOPs can provide a clinically significant and potentially tissue-specific new approach for RNAi therapy.
Keywords: Nanoparticle, siRNA delivery, iNOP, reducible nanoparticles
Gene silencing by RNA interference (RNAi) is attracting increasing attention as a potential therapeutic strategy.1−3 During RNAi, small interfering RNA (siRNA) duplexes4,5 are introduced into the cell and assemble in the RNA-induced silencing complex (RISC),6 a multiprotein complex that cleaves the complementary targeted mRNA or blocks its translation. The sequence specificity of RNAi supports its potential to treat diseases caused by gene products that are inaccessible to traditional small molecule approaches and thus considered nondruggable. However, effective siRNA-based therapies require identification of target-specific siRNA sequences and development of efficient vehicles to deliver siRNA to the appropriate cells in vivo.7 Successful in vivo delivery of siRNA has recently been reported using cholesterol-functionalized siRNA and formation of stable nucleic acid–lipid siRNA particles (SNALPs).8−11 Moreover, polymeric nanoparticles for RNAi have been successfully administered to humans, providing encouragement for the development of RNAi-based therapies.12−14
We previously reported the design and synthesis of interfering nanoparticles (iNOPs) as new siRNA delivery agents.15 iNOPs can be conveniently assembled by mixing siRNA with functionalized poly-l-lysine dendrimers without the need for complex liposomal formulation procedures16,17 or harsh covalent reaction conditions.18 One formulation, iNOP-7, has been successfully used to deliver apolipoprotein B (apoB)-specific siRNA in mice at therapeutically feasible and affordable doses, and to deliver NOX2-specific siRNA to local arteries for prevention of restenosis in an atherosclerotic rat model.191
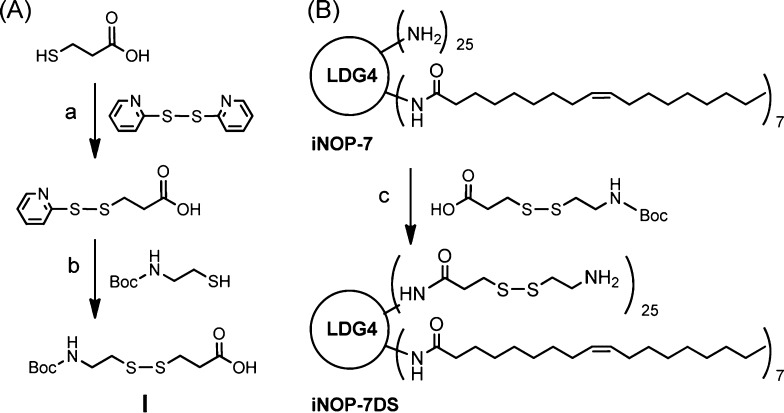
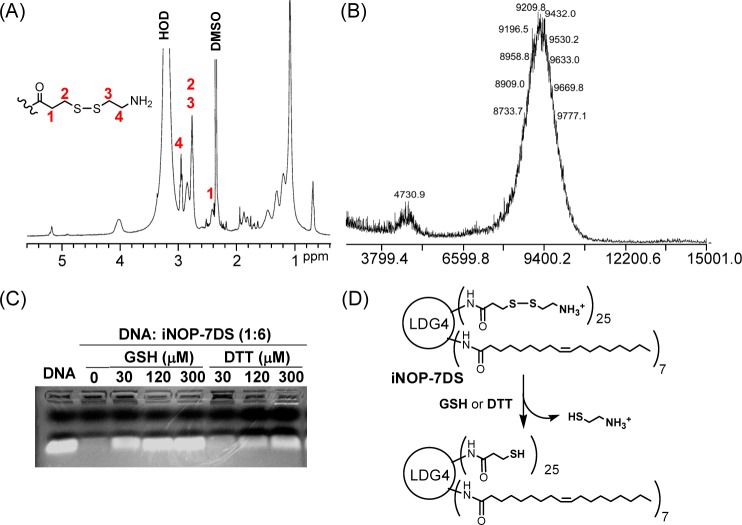
Here, we describe a novel form of iNOP designed to increase the efficiency of in vivo siRNA delivery. We considered that reducible nanoparticles may have advantages over nonhydrolyzable materials by allowing siRNA to be released within the cellular environment.19−21 To test this, we modified iNOP-7 by conjugating reducible spacers to the surface of the core lipid-functionalized poly-l-lysine dendrimer. We prepared a spacer (I) containing a disulfide bond (Scheme 1A) and coupled it to iNOP-7 to give the new reducible derivative, iNOP-7DS (Scheme 1B). Excess spacer I was included during the coupling reaction to ensure highly substituted dendrimers were obtained, which was confirmed by monitoring free amines in the product by Kaiser’s test (data not shown), as well as analysis by 1H nuclear magnetic resonance (NMR) and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF MS). As shown in Figure 1A, signals from the deprotected spacer I (2.42, 2.79, and 2.95 ppm), the oleoyl group (0.75, 1.12, ∼1.75–1.85, and 5.21 ppm), and lysine residues (∼1.20–1.54, 2.85, and 4.06 ppm) were observed in the 1H NMR spectrum. iNOP-7DS showed a broad distribution in the MALDI-TOF mass spectrum (Figure 1B). Major peaks were seen at approximately m/z 9432.0, which corresponds to a molecule with a degree of substitution 23, and minor peaks were present at m/z 4730.9, corresponding to [M + 2H]. These analyses confirmed we had successfully synthesized the highly substituted iNOP derivative.
Scheme 1. Synthesis of Disulfide Bond-Containing Spacer I (A) and Reducible iNOP-7DS (B).
Reagents and conditions: (a) acetic acid, EtOH, r.t., 4 h, 65%. (b) EtOH, r.t., 6 h, 73%. (c) BOP reagent, DIPEA, DMF, 0 °C to r.t., 24 h, 84% then TFA, r.t., 30 min.
Figure 1.
Characterization of iNOP-7DS. (A) 1H NMR spectrum showing signals from the disulfide-containing spacer, lipid chains, and lysine residues. (B) MALDI-TOF MS analysis of iNOP-7DS. (C) DNA complexed with iNOP-7DS at 1:6 weight ratio (N/P ratio 5:1) was treated with or without reducing agents at 37 °C for 10 min and then analyzed on a 2% agarose gel. (D) Schematic of iNOP-7DS reduction by glutathione (GSH) or dithiothreitol (DTT).
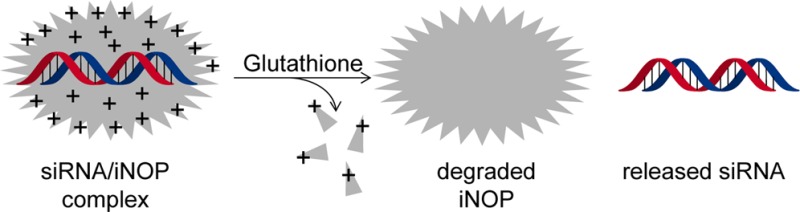
To test the ability of iNOP-7DS to release nucleic acid following cleavage of the spacer, we mixed a 21-nucleotide dsDNA with iNOP-7DS for 20 min at room temperature to allow electrostatic association of the components. The complex was then treated with reducing agents, and the products were analyzed by electrophoretic mobility shift assay. We found that the electrophoretic mobility of DNA was completely inhibited when complexed with iNOP-7DS at a weight ratio of 1:6 (N/P ratio 5:1) (Figure 1C). However, the DNA was released from the complex following a brief incubation (10 min at 37 °C) with glutathione or dithiothreitol, indicating that the disulfide bonds in iNOP-7DS are readily cleaved by reducing agents (Figure 1C). Taken together, these data showed that iNOP-7DS is reducible and, given the physiological abundance of glutathione (3–10 mM), the new iNOP derivative may be expected to facilitate siRNA delivery by promptly releasing siRNA upon entering the cell.
The electrostatic interaction between iNOP-7DS and nucleic acids could yield a range of particle sizes. To examine this, we measured the size distribution profiles of siRNA-iNOP-7DS complexes by dynamic light scattering. At a weight ratio of 1:10 (N/P ratio 8:1), siRNA and iNOP-7DS formed nanoparticles ∼140 nm in diameter (Supplementary Figure 1), confirming that the siRNA-iNOP-7DS nanoparticles are an ideal size for application in drug or siRNA delivery.22
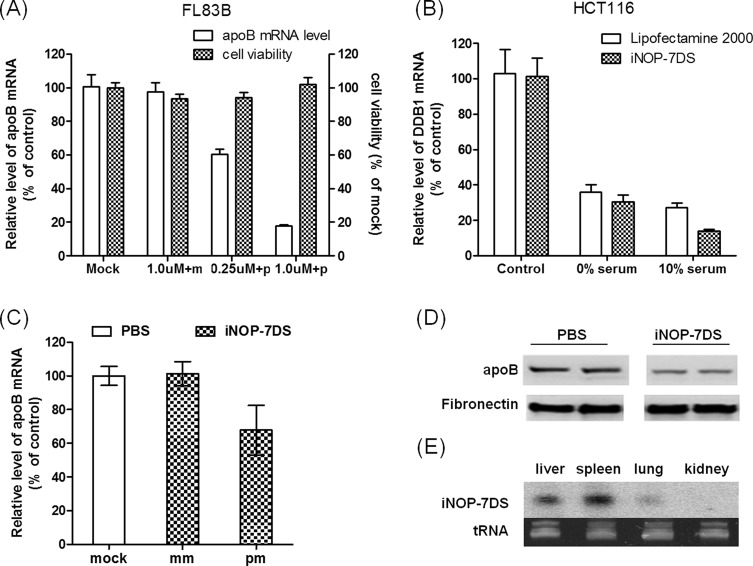
To determine whether iNOP-7DS could efficiently deliver complexed siRNA to target cells in vitro, we tested silencing of apoB transcription in FL83B mouse hepatocytes. ApoB is a critical ligand for the low-density lipoprotein receptor and plays an important role in cholesterol metabolism. For these experiments, the stability of the apoB siRNA was enhanced by chemical modification according to rules established in our previous studies.6,7,23 FL83B cells were treated for 3 h at 37 °C with iNOP-7DS complexed with apoB-specific or mismatched siRNA, and apoB mRNA levels were then measured by quantitative RT-PCR. As shown in Figure 2A, incubation of cells with apoB siRNA-iNOP-7DS complexes effectively silenced apoB expression (90% inhibition) compared with cells treated with the control siRNA-iNOP-7DS (Figure 2A). Cell viability was unchanged in the presence of iNOPs, indicating that inhibition of apoB transcription was not due to toxicity (Figure 2A). These results demonstrate that the reducible iNOP derivative is nontoxic and efficiently delivers siRNA into cells.
Figure 2.
Efficient in vitro and in vivo delivery of siRNA by iNOP-7DS. (A) ApoB siRNA-iNOP-7DS complexes specifically silence apoB expression in FL83B cells. Cells were treated for 3 h with iNOP-7DS with mismatched siRNA (m) or complementary siRNA (p). ApoB mRNA levels were measured by qRT-PCR, and viability was assessed with the MTS assay. Results are expressed as percent of control cells and are the mean ± SD of duplicate cultures from two representative experiments. (B) DDB1 siRNA-iNOP-7DS specifically silences DDB1 in HCT116 cells in the absence or presence of serum. Cells were treated for 3 h in medium containing 0% or 10% FBS with control or DDB1 siRNA (50 nM) delivered by iNOP-7DS (N/P ratio 6:1) or Lipofectamine (5 μL per well for 6-well plate). DDB1 mRNA levels were measured by qRT-PCR and are expressed as percent of control cells. (C) In vivo silencing of apoB by apoB siRNA-iNOP-7DS complexes. Mice were injected via the tail vein with PBS (mock) or with 2.0 mg kg–1 iNOP-7DS containing apoB-specific siRNA (pm) or mismatched siRNA (mm). Mice were sacrificed 48 h later and liver apoB mRNA levels were measured by qRT-PCR. (D) ApoB protein levels are reduced in plasma of mice injected with apoB siRNA-iNOP-7DS. Animals were treated as in panel B, and blood was drawn 24 h later. ApoB protein was analyzed in plasma samples by Western blotting. Fibronectin blotting was performed as a protein loading control. Samples from two mice are shown. (E) Biodistribution of apoB guide strand siRNA in mouse tissues after i.v. injection with iNOP-7DS complexes. Northern blots of total RNA (∼10 μg) isolated from the indicated tissues harvested 48 h after i.v. injection. Ethidium bromide staining of tRNA is shown as a loading control.
The stability of siRNA-containing nanoparticles may be affected by serum components. To assess this, we incubated human colon carcinoma (HCT116) and hepatocellular carcinoma (HepG2) cell lines in 0% or 10% serum-containing medium. Control or DDB1 siRNA was added to cells either complexed with iNOP-7DS or in the presence of a standard transfection reagent Lipofectamine 2000. Treatment of cells with siDDB1-iNOP-7DS not only knocked down mRNA by ∼90% in HT116 cells (Figure 2B) but also efficiently delivered DDB1 siRNA to HepG2 cells, which are notoriously difficult to transfect (Supplementary Figure 2).
Moreover, iNOP-7DS was as efficient as Lipofectamine 2000 in delivering siRNA into the cells (Figure 2B and Supplementary Figure 2). Significantly, serum did not affect DDB1 silencing in either cell type, indicating that iNOP-7DS is stable in serum-containing medium.
We next evaluated the efficiency of siRNA delivery by iNOP-7DS in vivo. For this, iNOP-7DS complexes were prepared with chemically modified control or mouse apoB siRNA (N/P ratio 8:1) and injected at 1–2 mg kg–1 into wild-type C57BL/6 mice. Mice were bled after 24 h and sacrificed after 48 h. Analysis of plasma and tissues from animals treated with apoB siRNA complexes showed ∼40% decrease in liver apoB mRNA levels (Figure 2C) and ∼45% decrease in plasma apoB protein (Figure 2D). Moreover, Northern blot analysis of total RNA isolated from the major organs of these mice showed the apoB siRNA guide strand was present mainly in liver, spleen, and lung at 48 h after injection (Figure 2E). These results showed that iNOP-7DS complexed to chemically modified siRNA was able to silence apoB expression efficiently in vivo. Remarkably, apoB silencing with the new iNOP derivative required only 2.0 mg kg–1 siRNA, which is a therapeutically feasible dose when scaled to humans. Of note, iNOP-7DS did not affect liver enzymes and did not activate an immune response in mice (data not shown).
Collectively, these findings demonstrate that siRNA delivery by the novel reducible iNOP derivative may provide a clinically significant new approach for RNAi therapy. Furthermore, the derivatization strategy could be applied to create iNOPs for tissue-specific RNAi therapies.
Acknowledgments
We thank Craig Mello, Greg Hannon, Michael Czech, and members of the Rana lab for helpful discussions and critical reading of the manuscript.
Supporting Information Available
Complete experimental details and supporting Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
∥ Boston Biomedical, Inc., 333 Providence Highway, Norwood, Massachusetts 02062, United States.
Author Present Address
⊥ Children’s Cancer Institute Australia Randwick, NSW 2031, Australia.
This work was supported in part by grants from the NIH to T.M.R.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Fire A.; Xu S.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Castanotto D.; Rossi J. J. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457, 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi M.; Tesz G. J.; Nicoloro S. M.; Wang M.; Chouinard M.; Soto E.; Ostroff G. R.; Czech M. P. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature 2009, 458, 1180–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen N. J.; Parrish S.; Imani F.; Fire A.; Morgan R. A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S. M.; Harborth J.; Lendeckel W.; Yalcin A.; Weber K.; Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Rana T. M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [DOI] [PubMed] [Google Scholar]
- Chiu Y. L.; Rana T. M. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell 2002, 10, 549–561. [DOI] [PubMed] [Google Scholar]
- Palliser D.; Chowdhury D.; Wang Q. Y.; Lee S. J.; Bronson R. T.; Knipe D. M.; Lieberman J. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 2006, 439, 89–94. [DOI] [PubMed] [Google Scholar]
- Santel A.; Aleku M.; Keil O.; Endruschat J.; Esche V.; Fisch G.; Dames S.; Loffler K.; Fechtner M.; Arnold W.; Giese K.; Klippel A.; Kaufmann J. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006, 13, 1222–1234. [DOI] [PubMed] [Google Scholar]
- Zimmermann T. S.; Lee A. C.; Akinc A.; Bramlage B.; Bumcrot D.; Fedoruk M. N.; Harborth J.; Heyes J. A.; Jeffs L. B.; John M.; Judge A. D.; Lam K.; McClintock K.; Nechev L. V.; Palmer L. R.; Racie T.; Rohl I.; Seiffert S.; Shanmugam S.; Sood V.; Soutschek J.; Toudjarska I.; Wheat A. J.; Yaworski E.; Zedalis W.; Koteliansky V.; Manoharan M.; Vornlocher H. P.; MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature 2006, 441, 111–114. [DOI] [PubMed] [Google Scholar]
- Soutschek J.; Akinc A.; Bramlage B.; Charisse K.; Constien R.; Donoghue M.; Elbashir S.; Geick A.; Hadwiger P.; Harborth J.; John M.; Kesavan V.; Lavine G.; Pandey R. K.; Racie T.; Rajeev K. G.; Rohl I.; Toudjarska I.; Wang G.; Wuschko S.; Bumcrot D.; Koteliansky V.; Limmer S.; Manoharan M.; Vornlocher H. P. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. [DOI] [PubMed] [Google Scholar]
- Love K. T.; Mahon K. P.; Levins C. G.; Whitehead K. A.; Querbes W.; Dorkin J. R.; Qin J.; Cantley W.; Qin L. L.; Racie T.; Frank-Kamenetsky M.; Yip K. N.; Alvarez R.; Sah D. W.; de Fougerolles A.; Fitzgerald K.; Koteliansky V.; Akinc A.; Langer R.; Anderson D. G. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple S. C.; Akinc A.; Chen J.; Sandhu A. P.; Mui B. L.; Cho C. K.; Sah D. W.; Stebbing D.; Crosley E. J.; Yaworski E.; Hafez I. M.; Dorkin J. R.; Qin J.; Lam K.; Rajeev K. G.; Wong K. F.; Jeffs L. B.; Nechev L.; Eisenhardt M. L.; Jayaraman M.; Kazem M.; Maier M. A.; Srinivasulu M.; Weinstein M. J.; Chen Q.; Alvarez R.; Barros S. A.; De S.; Klimuk S. K.; Borland T.; Kosovrasti V.; Cantley W. L.; Tam Y. K.; Manoharan M.; Ciufolini M. A.; Tracy M. A.; de Fougerolles A.; MacLachlan I.; Cullis P. R.; Madden T. D.; Hope M. J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [DOI] [PubMed] [Google Scholar]
- Davis M. E.; Zuckerman J. E.; Choi C. H.; Seligson D.; Tolcher A.; Alabi C. A.; Yen Y.; Heidel J. D.; Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigude H.; McCarroll J.; Yang C. S.; Swain P. M.; Rana T. M. Design and creation of new nanomaterials for therapeutic RNAi. ACS Chem. Biol. 2007, 2, 237–241. [DOI] [PubMed] [Google Scholar]
- Heyes J.; Palmer L.; Bremner K.; MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Controlled Release 2005, 107, 276–287. [DOI] [PubMed] [Google Scholar]
- Jeffs L. B.; Palmer L. R.; Ambegia E. G.; Giesbrecht C.; Ewanick S.; MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm. Res. 2005, 22, 362–372. [DOI] [PubMed] [Google Scholar]
- Rozema D. B.; Lewis D. L.; Wakefield D. H.; Wong S. C.; Klein J. J.; Roesch P. L.; Bertin S. L.; Reppen T. W.; Chu Q.; Blokhin A. V.; Hagstrom J. E.; Wolff J. A. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12982–12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M.; Newburger P. E.; Gounis M. J.; Dargon P.; Zhang X.; Messina L. M. Local arterial nanoparticle delivery of siRNA for NOX2 knockdown to prevent restenosis in an atherosclerotic rat model. Gene Ther. 2010, 17101279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H.; Jeong J. H.; Kim T. I.; Kim S. W.; Bull D. A. VEGF siRNA delivery system using arginine-grafted bioreducible poly(disulfide amine). Mol. Pharmaceutics 2009, 6, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H.; Ou M.; Bull D. A.; Kim S. W. Reductive degradation behavior of bioreducible poly(disulfide amine) for enhancing SiRNA efficiency. Macromol. Biosci. 2010, 10, 898–905. [DOI] [PubMed] [Google Scholar]
- Dunn S. S.; Tian S.; Blake S.; Wang J.; Galloway A. L.; Murphy A.; Pohlhaus P. D.; Rolland J. P.; Napier M. E.; Desimone J. M. Reductively responsive siRNA-conjugated hydrogel nanoparticles for gene silencing. J. Am. Chem. Soc. 2012, 134, 7423–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaumet M.; Vargas A.; Gurny R.; Delie F. Nanoparticles for drug delivery: the need for precision in reporting particle size parameters. Eur. J. Pharm. Biopharm. 2008, 69, 1–9. [DOI] [PubMed] [Google Scholar]
- Chiu Y. L.; Rana T. M. siRNA function in RNAi: a chemical modification analysis. Rna 2003, 9, 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.