Abstract
Objective
Copy number variation is a common polymorphic phenomenon within the human genome. While the majority of these events are non-deleterious they can also be highly pathogenic. Herein we characterize five families with parkinsonism that have been identified to harbor multiplication of the chromosomal 4q21 locus containing the α-synuclein gene (SNCA).
Methods
A methodological approach employing fluorescent in situ hybridization (FISH) and Affymetrix 250K SNP microarrays (CHIPs) was used to characterize the multiplication in each family and identify the genes encoded within the region. The telomeric and centromeric breakpoints of each family were further narrowed using semi-quantitative PCR with microsatellite markers and then screened for transposable repeat elements.
Results
The severity of clinical presentation is correlated with SNCA dosage and does not appear to be overtly effected by the presence of other genes in the multiplicated region. With the exception of the Lister kindred, in each family the multiplication event appears de novo. The type and position of Alu/LINE repeats are also different at each breakpoint. Microsatellite analysis demonstrates two genomic mechanisms are responsible for chromosome 4q21 multiplications, including both SNCA duplication and triplication.
Interpretation
SNCA dosage is responsible for parkinsonism, autonomic dysfunction and dementia observed within each family. We hypothesize dysregulated expression of wild-type α-synuclein results in parkinsonism and may explain the recent association of common SNCA variants in sporadic Parkinson’s disease. SNCA genomic duplication results from intra-allelic (segmental duplication) or inter-allelic recombination with unequal crossing-over, whereas both mechanisms appear to be required for genomic SNCA triplication.
Keywords: Parkinsonism, SNCA, Genomic multiplication, Alu repeat, Parkinson’s disease
Introduction
The human genome displays a considerable level of inter-individual variability from simple single nucleotide polymorphisms (SNPs) and short repeats to large-scale deletions, multiplications and rearrangements. Recent studies have demonstrated that large gene copy number variations (CNVs) occur frequently in the general population with no determinable disadvantage to carriers. However this phenomenon can also be pathogenic and result in severe disease phenotypes 1–3.
In 2003, Singleton and colleagues reported a triplication on one allele of the chromosomal locus (4q21) containing the α-synuclein gene (SNCA) in affected members of a family with parkinsonism known as the Iowan kindred 4. Although a relatively rare event, several families have since been described that carry multiplications of this region including both triplications and duplications which segregate with disease 5–11. The severity of the clinical phenotype of SNCA duplication and triplication families appears to be associated with gene dosage and mRNA/protein expression levels in brain 6. The SNCA duplication families are generally reminiscent of typical, late-onset Parkinson’s disease (PD) 5, 7, 9, while the two families (Iowan & Swedish-American) with monoallelic triplication of SNCA present with a severe form of early-onset parkinsonism with autonomic dysfunction and subsequent dementia 6, 12.
In the Iowan kindred, the region triplicated is reported to contain seventeen gene transcripts (1.6–2.1Mb) whereas in both French and Japanese patients much smaller genomic intervals are duplicated (~0.5Mb) 4, 5, 7, 9. Although SNCA multiplication appears necessary for parkinsonism, whether increased dosage of adjacent genes contributes to the phenotype is unclear. The mechanism underlying chromosome 4q21 genomic multiplication has also to be elucidated. The region appears to be evolutionarily fragile given the spontaneous deletion of SNCA within an inbred strain of C57BL/6J (OlaHsd) mice, albeit with no apparent deleterious effects 13, 14.
Herein we compare the phenotypes of SNCA multiplication families and present data on the genomic copy number, size and breakpoints for each 4q21 multiplication mutation, using a combination of fluorescent in situ hybridization (FISH) and Affymetrix 250k SNP microarrays (CHIPS). Within each interval/family, we detail the genes with aberrant copy number and expression. We characterize the transposable repeat elements at each breakpoint and provide a mechanistic hypothesis for the genomic instability, rearrangement and multiplication of this locus.
Subjects and Methods
Frequency, Clinical manifestations and Neuropathology
The frequency of SNCA multiplication is low and appears to be a relatively rare event 15. World-wide seven families have been identified that harbor SNCA multiplication; one triplication (Iowa, US) 4, 6, 12, five duplication kindreds (2 French, 2 Japanese and 1 Italian) 5, 7, 9, 11, and one kindred with individuals with either duplication or triplication mutations (Swedish-US, now recognized as a branch of the ‘Lister family complex’) 6, 11. The clinical presentation and available pathological findings for five of these families are summarized in Table 1. In the preparation of this manuscript Ahn and colleagues reported the first SNCA duplication patients in Korea16. Intriguingly, of the three PD patients identified only one is described with a family history of parkinsonism. This familial SNCA duplication patient presented with symptoms at age 40 years and initially had a good response to l-DOPA therapy, however the disease course progressed rapidly with postural hypotension, personality changes and dementia by the age of 46 years. The two sporadic patients presented with typical PD with ages at onset of 50 and 65 years. These alternate clinical presentations demonstrate the phenotypic range of SNCA multiplication symptoms.
Table 1.
Presents a brief summary of the major clinical and neuropathological characteristics of SNCA multiplication families. MMSE: Mini Mental State Examination; SN: Substantia nigra, CTX: Cortex, HC: Hippocampus, NBM: Nucleus basalis of Meynert; LC: Locus coeruleus, LBs: Lewy bodies, GCIs: glial cytoplasmic inclusions; na - no data available.
| Kindred | Iowan | Lister-US | Lister-Swedish | French | Japanese–A | Japanese–B |
|---|---|---|---|---|---|---|
| SNCA multiplication | Triplication | Triplication | Duplication | Duplication | Duplication | Duplication |
| Number of patients with clinical data | 12 | 3 | 5 | 5 | 3 | 1 |
| Average age at onset (years) | 34 (20–48) | (31– early 40s) | 59 (40–71) | 48 (35–65) | 43 (38–77) | 47 |
| Rigidity | Yes | Severe, generalized | Yes | Yes | Yes | Yes |
| Bradykinesia | Yes | Yes | Yes | Mild to severe | Yes | Yes |
| Rest tremor | Some subjects: none to pronounced | Yes | Some subjects: mild to intermittent | Some subjects: none to pronounced | No | No |
| Postural tremor | Yes, not segregating with triplication | Yes | No | 3 of 5 | No | No |
| Postural instability | Yes, with falls | Mild to moderate | Pronounced, with falls | 3 of 5 | 2 of 3 | Yes |
| Myoclonus | Late | na | Late, in distal upper extremities | na | na | na |
| Response to 1-DOPA | Yes | Dramatic effect initially | Slight | Yes | None to good | Slight |
| Orthostatic hypotension | Yes, some subjects, sometimes requiring drug treatment | Moderate to severe, early in illness, partially requiring drug treatment | Early, symptomatic, required drug treatment | No | No | No |
| Other dysautonomia | Erectile, cardiac, and gastrointestinal dysfunctions | Urinary incontinence late in illness | Moderate urinary incontinence, dysphagia | No | No | No |
| Dementia / cognitive dysfunction | Memory loss, visuospatial dysfunction, decline of executive functions: may present with these features (LBD phenotype, or may be late in course (PD phenotype) | Early, severe | Not prominent (late) | Not prominent | No | Yes |
| Paranoia, anxiety | Not prominent | Early, pronounced | Yes | na | na | na |
| Depression | Yes, may precede parkinsonism by a decade or more. | History of depression between age 13 and 19, suicidal later in illness | Yes | 2 of 5 | Yes | na |
| Hallucinations | Partial: some have prominent visual when phenotype is LBD | Pronounced, visual, auditory and olfactory | Visual, olfactory and auditory | No | No | Yes |
| Other remarks | Weight loss may be seen. Rapidly-progressive | na | Rapidly-progressive disease | na | Psychosis in 1 of 2 | na |
| Neuropathology | Neuronal loss in SN and LC, extensive, pleomorphic and atypical LBs, GCIs, neuritic dystrophy, neuronal loss in HC (CA 2/3) | Neuronal loss in SN and LC (few LBs), NBM, CTX (widespread LBs), and HC (CA2/3) | na | na | na | Neuronal loss in SN, LC & HC (CA 2/3), Lewy neurites in the CA2, only a few LBs in SN and LC. |
The clinical phenotype in the SNCA triplication families is rapid, progressive parkinsonism with onset in the third and fourth decades. Movement disorder (resting tremor, bradykinesia and rigidity) occurs early in the course with autonomic dysfunction (including hyposmia and orthostatic hypotension) and neuropsychological impairments (hallucinations, anxiety, paranoia and depression), with subsequent cognitive decline and dementia. The neuropathology of SNCA triplication patients is reminiscent of diffuse Lewy body disease with numerous α-synuclein-positive Lewy bodies, Lewy neurites and glial cyoplasmic inclusions, with neuronal cell loss in the substantia nigra and locus ceruleus. Extensive neuronal loss is also observed in the hippocampus CA2/3 region and is a feature of both missense and multiplication SNCA mutations 6, 17.
In contrast, most patients in SNCA duplication families present with signs and symptoms that closely resemble idiopathic PD. Onset of motor symptoms is in the fifth to sixth decades, neither cognitive decline nor dementia are prominent and generally disease progression is slow with a sustained response to l-DOPA therapy 5, 7, 9. However, with each report, clinical variability within and among SNCA duplication families becomes more extensive. For example, Japanese A and B families are noted for reduced penetrance among carriers; patients may have a long duration of disease, may exhibit signs of cognitive decline and dementia, and have either a mild or excellent response to l-DOPA therapy 9. In contrast, affected carriers within Branch J of the Swedish Lister family initially present with dysautonomia (orthostatic hypotension and syncope) rather than motor problems but quickly develop rapidly progressive parkinsonism that is only poorly responsive to l-DOPA 11. Neuropathological examination of SNCA duplication highlights α-synuclein pathology reminiscent of diffuse Lewy body disease comparable to that observed in SNCA triplication patients 18. It is evident that disease onset in SNCA duplication carriers is several decades later than in SNCA triplication families 11. While SNCA dosage appears responsible and sufficient for disease, clinical variability may reflect the size of the duplicated segment and the aberrant expression of the additional genes 9, 11.
Genetic analysis: FISH and CHIPS
Fluorescent in situ hybridization (FISH) was performed on Epstein-Barr virus (EBV)-immortalized lymphocytes from one affected member of each family as previously described, with SNCA PAC 27M07 (146 kb; AF163864) labeled using FITC and SNCA promoter and intron 4 fragments (13 and 21 kb) labeled with rhodamine 4. Samples were considered duplicated/triplicated if they had 3/4 FISH probe signals in greater than 20% of interphase cells scored, from 100 interphase nuclei examined. To exclude the possibility of an artifact of EBV-immortalization, semi-quantitative PCR was performed on genomic DNA extracted from blood and confirmed a multiplication of the region of chromosome 4 containing SNCA in all families. Affymetrix 250k SNP microarrays (CHIPS) genotyping and SNP dosage analysis was then performed on 250ng of total genomic DNA samples for the probands of each family as previously described11. Copy number was estimated using dChipSNP software with GTYPE exported genotype calls and signal intensities ( .cel files) 19–21. This algorithm uses a rigorous ‘within and between array’ normalization method to compute estimates of the normal signal values for genotype calls observed with a set of arrays. Deviations from the normal signal values seen for any particular genotype in the set of abnormal DNA samples were compared with values observed for a set of 10 samples with normal 2N copy numbers throughout chromosome 4. Copy number changes in the probands and families were then inferred by median smoothing with a Hidden Markov Model applied. In our study, we present results based on a sliding window approach to average the inferred copy numbers across a continuous 250 kb stretch centered on each SNP, and for simplicity, only a single proband is shown for each nuclear family.
Using polymorphic microsatellite markers the centromeric and telomeric ends of the breakpoints were confirmed and further refined. Internal control peak height of heterozygous individuals were calculated and compared between patient samples, diploid, SNCA duplication and triplication samples to give copy numbers. These analyses confirmed the Affymetrix 250K SNP microarray results showing a different length of the multiplicated region in each family (Suppl Table 1).
Results
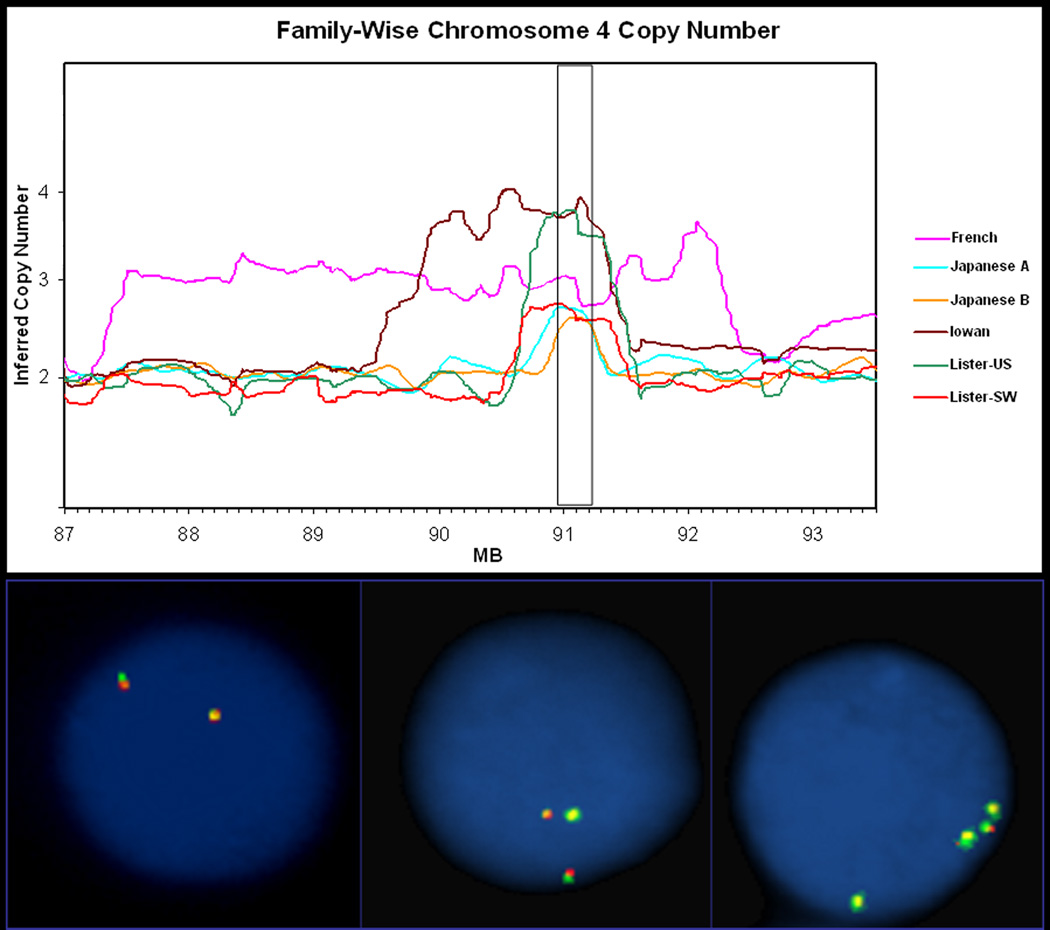
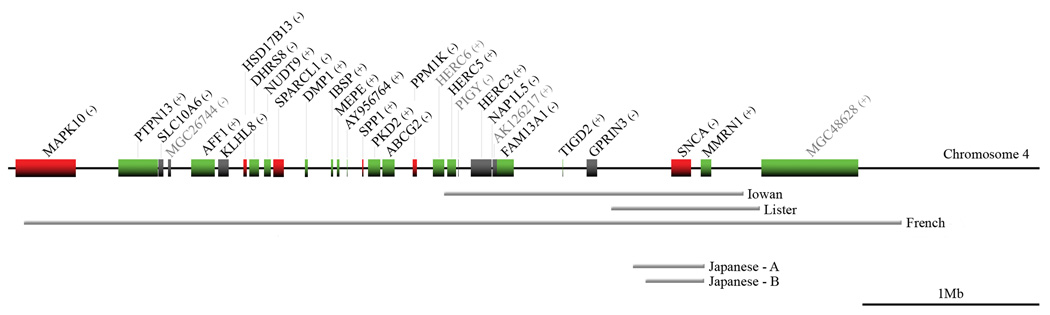
In each proband, all three SNCA FISH probes gave results that were consistent with Affymetrix dosage and microsatellite genotype analyses. Affymetrix CHIP dosage data was obtained from between 62 – 363 SNPs, within the chromosome 4q21 region of multiplication in each family. Illustrative results are shown for both SNCA duplication and triplication cell lines (FISH) and the proband of each family (CHIPS) (Figure 1). The longest region (4.93–4.97 Mb) is present in the French duplication family (also reported as FPD-131) and encompasses 31 transcripts, including genes associated with epileptic encephalopathy (MAPK10), type II dentinogensis imperfecta (DMP1 & SPP1), and polycystic kidney disease II (PKD2) (http://www.ncbi.nlm.nih.gov)(Figure 2). Five transcripts including SNCA are expressed at high levels in the brain (microarray expression data retrieved from UCSC website; http://genome.ucsc.edu/). In contrast, the shortest region (0.4 Mb) was observed in the Japanese B family with duplication of only SNCA and the 5’ region of the MMRN1 gene.
Figure 1. FISH and CHIPs.
A representation of fluorescent in-situ hybridization (FISH) and Affymetrix 250k SNP microarrays (CHIPS) that were used to examine the region of multiplication in the proband of each family. (a) Relative copy number estimates were plotted against physical genomic position on chromosome 4. Raw data is shown that has not been normalized with respect to integers. (b) FISH was performed on interphase cells with three labeled SNCA probes directed at the entire locus (PAC 27M07 in GREEN), with promoter and intron 4 fragments (visualized in RED). SNCA multiplication was confirmed in all samples using both methodologies.
Figure 2. A representation of the genes in the multiplication region in each family.
Displays the genes that are present in each of the multiplied regions of the families. The figure is drawn approximately to scale. The coding genomic DNA strand of each gene is indicated by (+) or (−). Genes are colored to represent their relative expression in brain according to the GNF Expression Atlas 2 (http://genome.ucsc.edu/), with red, black and green representing high, medium and low expression respectively. Gene symbols in gray text indicate hypothetical genes. The gray bars below the chromosome diagram show the regions of multiplication in each family.
Microsatellite genotype analysis revealed SNCA genomic duplication results from intra-allelic (segmental duplication) or inter-allelic recombination with unequal crossing-over, whereas both mechanisms are required for genomic SNCA triplication (Figure 3). The reason for genomic instability and chromosome 4q21 rearrangement remains unclear. Thus VISTA software 22 was employed for comparison of the DNA sequences, short and long interspersed repeats (SINE/LINE) at the centromeric and telomeric ends of the multiplicated region in each family. It is reported that Alu repeats constitute ~10% of the human genome 23, 24, and mobile elements make up over 45% of the human genome 23. Although our analysis identified a number of transposable repeat elements with over 70% conservation, the proportion of sequence occupied by SINE or LINE repeats in the breakpoint regions was not greater than observed within flanking sequence. Rather than one specific repeat there were a variety of Alu sub-types at the 5’ and 3’-ends of the SNCA multiplication regions (Suppl Table 2). Nevertheless, the presence of these genetically mobile elements can lead to genomic instability, unequal recombination and rearrangements that result in CNVs including multiplication or deletion 23, 25.
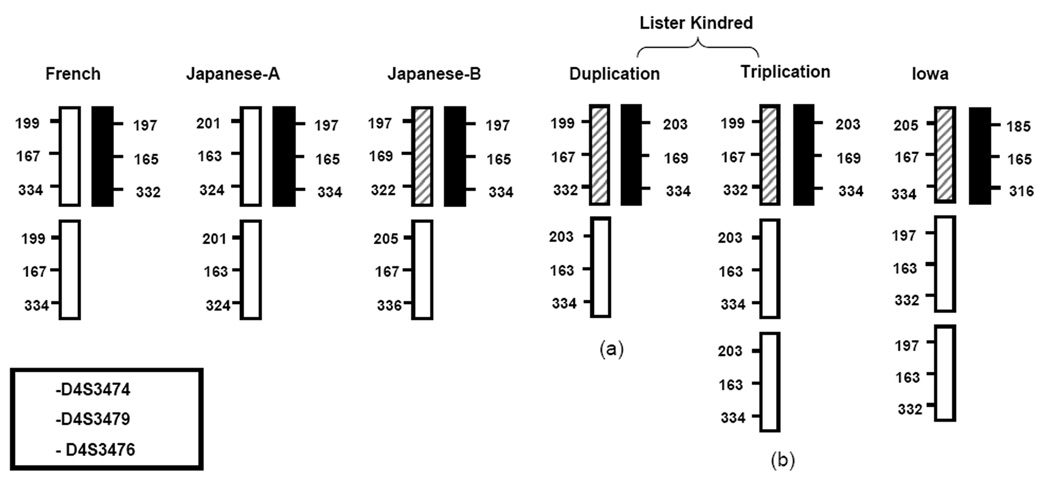
Figure 3. Legend.
The allele sizes and dosage for the chromosome 4 markers D4S3476, D4S3479 and D4S3474 are shown for each family. These data demonstrate an intra-allelic, segmental duplication in the French and Japanese-A families. However inter-allelic recombination occurred initially in Japanese-B and the in Lister kindred duplication, indicated by the presence of three different alleles at marker D4S3476 (163, 167 & 169)(a). A further segmental duplication is apparent in the Lister kindred branch with SNCA triplication (b). It is not possible to ascertain the sequence of events for the SNCA triplication in the Iowa kindred but the presence of three alleles sizes at all markers demonstrates a recombination event must have occurred.
Discussion
Multiplication of the SNCA locus is now reported to account for a greater number of families with autosomal dominant parkinsonism than the known pathogenic α-synuclein missense substitutions (A30P, E46K and A53T) 26. However this is still a small number of familial patients, given that ~10–15% of patients with PD report a family history of disease.
Only multiplication of α-synuclein (SNCA) appears necessary for parkinsonism as Japanese kindred B has only full-length SNCA and the 5’ end of multimerin1 (MMRN1). Deficiency of MMRN1, a specific platelet Factor V/Va binding protein is associated with an inherited bleeding disorder, Factor V Quebec, although haploinsufficiency does not appear to be associated with any phenotype 27. MMRN1 is increased in copy number in all other SNCA multiplication kindreds. It may be noteworthy that γ-synuclein (SNCG) and multimerin 2 (MMRN2) lie in the same orientation to each other on human chromosome 10 (murine chromosome 14) suggesting that these paralogs arose due to an evolutionary duplication event. Gamma-synuclein, also known as breast cancer-specific protein 1, is elevated in cancer and may play a role in disease 28, 29.
Given the instability of the SNCA-MMRN1 region, SNCG-MMRN2 multiplications/deletions may yet be identified. Limited expression and functional data is available on other genes within regions of chromosome 4q21 multiplication. Nevertheless, only SNCA dosage appears to specifically contribute to the variability in clinical observations among families. Genetic and genealogic studies recently identified a Swedish family with SNCA duplication and a US family of Swedish descent with a SNCA triplication as branches of the “Lister kindred” 11 30, 31. Within the families examined, this was the only example of copy number changes from one generation to another. Earlier onset, faster progression and more fulminant disease are associated with increasing SNCA copy number, suggestive of ‘genetic anticipation’, a clinical phenomenon usually confined to small simple repeats such as in spinocerebellar ataxias 32.
It is remarkable that both segmental intra-allelic duplication and inter-allelic recombination with unequal crossing-over appear to be responsible for SNCA multiplications. Microsatellelite genotyping clearly demonstrates both mechanisms operate; duplication does not necessarily precede unequal crossing-over and the opposite may occur. While our study identified a number of large repeat elements at either ends of the multiplicated regions, no single repeat was consistently identified at the breakpoints of all multiplications (Suppl Table 2). Thus a variety of transposable repeat elements including Alu and LINE repeats may promote instability causing irregular gene duplication and recombination events. Cloning the exact multiplication breakpoints across repeat elements may yet be insightful. Rovelet-Lecrux et al. have reported similar multiplication events on chromosome 21, that involve the amyloid precursor protein gene and result in Alzheimer disease 33. The regions duplicated in five of these families ranged from 0.58 to 6.37Mb in size and differed in their haplotypic structure suggesting these multiplication events are also independent.
Ahn and colleagues recently reported two sporadic patients with SNCA duplication suggestive of age-related penetrance, as observed for other mutations causing parkinsonism16. The frequency and the direct relevance of SNCA multiplication to the vast majority of PD patients remains to be determined. The hypothesis that α-synuclein overexpression contributes to disease susceptibility predates the discovery of SNCA multiplications. A number of classical association studies have examined the Rep1 microsatellite (D4S3481) in the SNCA promoter, a region implicated in transcript expression by in vitro luciferase assays 34–37. Combined, pooled analysis by the Genetic Epidemiology of Parkinson's Disease (GEO-PD) Consortium observed a significant association with increasing Rep1 allele size, 259<261<263 base-pairs 38. On-line meta-analysis of all published studies also highlight a SNP (rs356165) in the 3’ untranslated mRNA (www.pdgene.org). In contrast, genome-wide SNP association studies of PD have not highlighted common variants in SNCA suggesting their power to identify susceptibility genes is limited 39, 40. A reanalysis of genome-wide data highlighted copy number variants in PARKIN 41. The identification of heterozygous carriers and one homozygous early-onset patient demonstrates the method can detect both multiplications and deletions.
In vivo findings with respect to SNCA mRNA expression are inconsistent 42. We find a decrease in SNCA transcript levels in specific brain regions such as the surviving neurons of the substantia nigra, as well as the putamen and frontal cortex in subjects with PD (unpublished data) 43. Changes in mRNA expression in end-stage disease may compensate for the accumulation of α-synuclein protein but mRNA and protein expression levels have yet to be correlated within the same samples. Whether alternately spliced SNCA mRNA, predicted to lead to smaller isoforms (α-synuclein 98, 112 and 126) may also contribute to disease has yet to be determined.
The discovery of SNCA multiplication demonstrates aberrant α-synuclein expression is sufficient for parkinsonism and highlights a direct, dose response with age of onset, progression and symptom severity. Whether SNCA multiplication is a distinct entity, or a more aggressive form of typical PD, both are part of a spectrum of Lewy body disorders. The challenge is now to functionally translate genetic insights focused on SNCA into patient therapy.
Supplementary Material
Supplemental Table 1. The chromosome 4 breakpoints for each family harboring an SNCA multiplication event were characterized using quantitative PCR with SNPs and polymorphic microsatellite markers. Both the 5’ and 3’-end breakpoints were discriminated.
Supplemental Table 2. VISTA software was employed to characterize the conserved repeat DNA sequences located in the 5’ and 3’ breakpoint regions of the SNCA multiplication kindreds. At least one Alu repeat element was identified in each breakpoint of the five families.
Acknowledgements
We would like to thank all those who have contributed to our research, including Drs. Alexis Brice and Suzanne Lesage for helpful discussion. OAR is funded, in part, by a research grant for the American Parkinson Disease Association. This work is supported by a Morris K. Udall Parkinson's Disease Research Center of Excellence (NINDS P50 #NS40256), the National Institute of Aging (P01 #AG17216), German National Genome Network (NGFN, grant 01GS0116), PHRC 2005/1914 and the Draper Family Foundation. We would like to acknowledge The Swedish Parkinson Foundation. DNA and cell line samples and phenotypic data on the triplication subjects is available via the NINDS Repository, (http://ccr.coriell.org/ninds), (catalogue number ND00139, as are other affected and unaffected individual subject samples. The repository is supported by NINDS Contract # N01-NS-2-2349).
Footnotes
The authors declare no financial or other conflicts of interest.
References
- 1.Simon-Sanchez J, Scholz S, Fung HC, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 2.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong KK, Deleeuw RJ, Dosanjh NS, et al. A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.Chartier-Harlin MC, Kachergus J, Roumier C, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 6.Farrer M, Kachergus J, Forno L, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 7.Ibanez P, Bonnet AM, Debarges B, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 8.Lockhart PJ, Kachergus J, Lincoln S, et al. Multiplication of the alpha-synuclein gene is not a common disease mechanism in Lewy body disease. J Mol Neurosci. 2004;24:337–342. doi: 10.1385/JMN:24:3:337. [DOI] [PubMed] [Google Scholar]
- 9.Nishioka K, Hayashi S, Farrer MJ, et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson's disease. Ann Neurol. 2006;59:298–309. doi: 10.1002/ana.20753. [DOI] [PubMed] [Google Scholar]
- 10.Johnson J, Hague SM, Hanson M, et al. SNCA multiplication is not a common cause of Parkinson disease or dementia with Lewy bodies. Neurology. 2004;63:554–556. doi: 10.1212/01.wnl.0000133401.09043.44. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs J, Nilsson C, Kachergus J, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 12.Muenter MD, Forno LS, Hornykiewicz O, et al. Hereditary form of parkinsonism--dementia. Ann Neurol. 1998;43:768–781. doi: 10.1002/ana.410430612. [DOI] [PubMed] [Google Scholar]
- 13.Gajovic S, Mitrecic D, Augustincic L, et al. Unexpected rescue of alpha-synuclein and multimerin1 deletion in C57BL/6JOlaHsd mice by beta-adducin knockout. Transgenic Res. 2006;15:255–259. doi: 10.1007/s11248-006-0003-6. [DOI] [PubMed] [Google Scholar]
- 14.Specht CG, Schoepfer R. Deletion of multimerin-1 in alpha-synuclein-deficient mice. Genomics. 2004;83:1176–1178. doi: 10.1016/j.ygeno.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Hope AD, Myhre R, Kachergus J, et al. Alpha-synuclein missense and multiplication mutations in autosomal dominant Parkinson's disease. Neurosci Lett. 2004;367:97–100. doi: 10.1016/j.neulet.2004.05.100. [DOI] [PubMed] [Google Scholar]
- 16.Ahn TB, Kim SY, Kim JY, et al. {alpha}-Synuclein gene duplication is present in sporadic Parkinson disease. Neurology. 2007 doi: 10.1212/01.wnl.0000271080.53272.c7. [DOI] [PubMed] [Google Scholar]
- 17.Spira PJ, Sharpe DM, Halliday G, et al. Clinical and pathological features of a Parkinsonian syndrome in a family with an Ala53Thr alpha-synuclein mutation. Ann Neurol. 2001;49:313–319. [PubMed] [Google Scholar]
- 18.Obi T, Nishioka K, Ross OA, et al. Clinicopathologic study of a SNCA gene duplication patient with Parkinson disease and dementia. Neurology. 2008;70:238–241. doi: 10.1212/01.wnl.0000299387.59159.db. [DOI] [PubMed] [Google Scholar]
- 19.Janne PA, Li C, Zhao X, et al. High-resolution single-nucleotide polymorphism array and clustering analysis of loss of heterozygosity in human lung cancer cell lines. Oncogene. 2004;23:2716–2726. doi: 10.1038/sj.onc.1207329. [DOI] [PubMed] [Google Scholar]
- 20.Lieberfarb ME, Lin M, Lechpammer M, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res. 2003;63:4781–4785. [PubMed] [Google Scholar]
- 21.Lin M, Wei LJ, Sellers WR, et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 22.Frazer KA, Pachter L, Poliakov A, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Eskin E, Pevzner PA. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 27.Hayward CP, Rivard GE, Kane WH, et al. An autosomal dominant, qualitative platelet disorder associated with multimerin deficiency, abnormalities in platelet factor V, thrombospondin, von Willebrand factor, and fibrinogen and an epinephrine aggregation defect. Blood. 1996;87:4967–4978. [PubMed] [Google Scholar]
- 28.Liu H, Liu W, Wu Y, et al. Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65:7635–7643. doi: 10.1158/0008-5472.CAN-05-1089. [DOI] [PubMed] [Google Scholar]
- 29.Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192–198. doi: 10.3748/wjg.v12.i2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gosal D, Ross OA, Toft M. Parkinson's disease: the genetics of a heterogeneous disorder. Eur J Neurol. 2006;13:616–627. doi: 10.1111/j.1468-1331.2006.01336.x. [DOI] [PubMed] [Google Scholar]
- 31.Mjönes H. Paralysis agitans: clinical and genetic study. Acta Psychiatr Neurol Scand Suppl. 1949;54:1–195. [Google Scholar]
- 32.Manto MU. The wide spectrum of spinocerebellar ataxias (SCAs) Cerebellum. 2005;4:2–6. doi: 10.1080/14734220510007914. [DOI] [PubMed] [Google Scholar]
- 33.Rovelet-Lecrux A, Hannequin D, Raux G, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 34.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 35.Kruger R, Vieira-Saecker AM, Kuhn W, et al. Increased susceptibility to sporadic Parkinson's disease by a certain combined alpha-synuclein/apolipoprotein E genotype. Ann Neurol. 1999;45:611–617. doi: 10.1002/1531-8249(199905)45:5<611::aid-ana9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 36.Tan EK, Tan C, Shen H, et al. Alpha synuclein promoter and risk of Parkinson's disease: microsatellite and allelic size variability. Neurosci Lett. 2003;336:70–72. doi: 10.1016/s0304-3940(02)01178-3. [DOI] [PubMed] [Google Scholar]
- 37.Farrer M, Maraganore DM, Lockhart P, et al. alpha-Synuclein gene haplotypes are associated with Parkinson's disease. Hum Mol Genet. 2001;10:1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- 38.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. Jama. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 39.Fung HC, Scholz S, Matarin M, et al. Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 40.Maraganore DM, de Andrade M, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon-Sanchez J, Scholz S, Del Mar Matarin M, et al. Genomewide SNP assay reveals mutations underlying Parkinson disease. Hum Mutat. 2007 doi: 10.1002/humu.20626. [DOI] [PubMed] [Google Scholar]
- 42.Dachsel JC, Lincoln SJ, Gonzalez J, et al. The ups and downs of alpha-synuclein mRNA expression. Mov Disord. 2007;22:293–295. doi: 10.1002/mds.21223. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, James M, Middleton FA, Davis RL. Transcriptional analysis of multiple brain regions in Parkinson's disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2005;137:5–16. doi: 10.1002/ajmg.b.30195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. The chromosome 4 breakpoints for each family harboring an SNCA multiplication event were characterized using quantitative PCR with SNPs and polymorphic microsatellite markers. Both the 5’ and 3’-end breakpoints were discriminated.
Supplemental Table 2. VISTA software was employed to characterize the conserved repeat DNA sequences located in the 5’ and 3’ breakpoint regions of the SNCA multiplication kindreds. At least one Alu repeat element was identified in each breakpoint of the five families.