Summary
The transforming growth factor beta (TGFβ) pathway is involved in embryonic development and several inherited and acquired human diseases. The gene for TGFβ3 (Tgfb3) encodes one of the three ligands for TGF b receptors. It is widely expressed in the embryo and its mutation or misexpression is found in human diseases. Tgfb3−/− mice die at birth from cleft palate, precluding functional studies in adults. Here, we generated mice in which exon 6 of Tgfb3 was flanked with LoxP sites (Tgfb3flox/flox). The adult mice were normal and fertile. EIIa-Cre-mediated deletion of exon 6 in Tgfb3flox/flox mice efficiently generated Tgfb3 conditional knockout (Tgfb3cko/cko) mice which died at birth from the same cleft palate defect as Tgfb3−/− mice, indicating that the conditional and knockout alleles are functionally equivalent. This Tgfb3cko allele will now enable studies of TGFβ3 function in different cell or tissue types in embryonic development and during adulthood. genesis 50:59-66, 2012.
Keywords: transforming growth factor beta, cardiovascular, cancer, craniofacial, wound healing, autoimmunity, neuromuscular
TGFb ligands are multifunctional proteins involved in tissue development and homeostasis and in tissue remodeling during disease pathogenesis and cancer (Azhar et al., 2003; Laverty et al., 2009; ten Dijke and Arthur, 2007). In mammals, there are three TGFb ligands (TGFβ1, TGFβ2 and TGFβ3). They are produced in latent forms, and upon activation they usually interact with TGFβR1 and TGFβR2 receptors in conjunction with TGFβR3 or Endoglin and induce phosphorylation of SMAD2 or SMAD3. These SMADs associate with SMAD4 and translocate to the nucleus, where in association with various co-repressors and activators they regulate the transcription of target genes. SMAD7 acts as an inhibitor of this canonical TGFb signaling. TGFβ signaling also occurs through SMAD-independent pathways.
Of the three TGFβ ligands, the role of TGFβ3 at the organismal level is somewhat less understood (Azhar et al., 2009a,b; Kaartinen et al., 1995; Kulkarni et al., 1993; Proetzel et al., 1995; Sanford et al., 1997; Shull et al., 1992). The importance of a better understanding of TGFβ3 function is underscored by several inherited and noninherited diseases in which it has been implicated. For example, mutations in the human TGFB3 gene causes familial arrhythmogenic right ventricular dysplasia type 1 (ARVD1) [MIM:107970], also known as arrhythmogenic right ventricular cardiomyopathy 1 (ARVC1) (Beffagna et al., 2005). ARVD is an autosomaldominant disease characterized by partial degeneration of the myocardium of the right ventricle, electrical instability, and sudden death. Moreover, gene association studies show an association of cleft lip and palate (CL/P) with TGFB3 (Lidral et al., 1998). Genetic polymorphisms are found in TGFB3 in patients with hypertension (Hu et al., 2010) and ossification of the posterior longitudinal ligament of the spine (OPLL) (Horikoshi et al., 2006). TGFB3 expression is increased in HELLP syndrome (hemolysis-elevated liver enzymes-low platelet count) (Emanuelli et al., 2008), and it is elevated in diseased canine mitral valves (Aupperle et al., 2008). TGFβ3 is critically involved in wound healing and is currently being used in a clinical trial for treatment of wounds (Laverty et al., 2009). It is involved in mammary gland development and post-lactational involution (Flanders and Wakefield, 2009), and it has been strongly implicated in the progression of various forms of cancer, including breast cancer (Laverty et al., 2009). In vitro, mouse keratinocytes are not protected against 12-O-tetradecanoylphorbol-13-acetate-induced cell death in its absence (Li et al., 1999). Finally, a recent study has identified an important role of TGFβ3 in immune tolerance and autoimmunity (Shah and Qiao, 2008). Thus, it is clear that TGFβ3 has important roles at the organismal level that need to be better understood.
Tgfb3 is expressed in several tissues during mouse embryonic development, including heart, lung, skin, and craniofacial structures. Tgfb3 is expressed in a partially overlapping fashion to Tgfb2 in the developing cardiovascular system (Azhar et al., 2003; Pelton et al., 1991). However, it is not clear if these ligands have any overlapping functions in cardiovascular development or function. Tgfb3 is expressed during palatogenesis and wound healing (Laverty et al., 2009). Consistent with its expression in the developing palate, Tgfb3 knockout mice (Tgfb3−/−) develop cleft palate and die soon after birth (Kaartinen et al., 1995; Proetzel et al., 1995), precluding any further determination of Tgfb3 function in adult mice. Thus, Tgfb3 conditional knockout mice are necessary to advance the understanding of Tgfb3 function not only during embryonic development, but also during tissue remodeling at postnatal and adult stages, and in the adult in cancer and in diseases of the cardiovascular and immune-related systems.
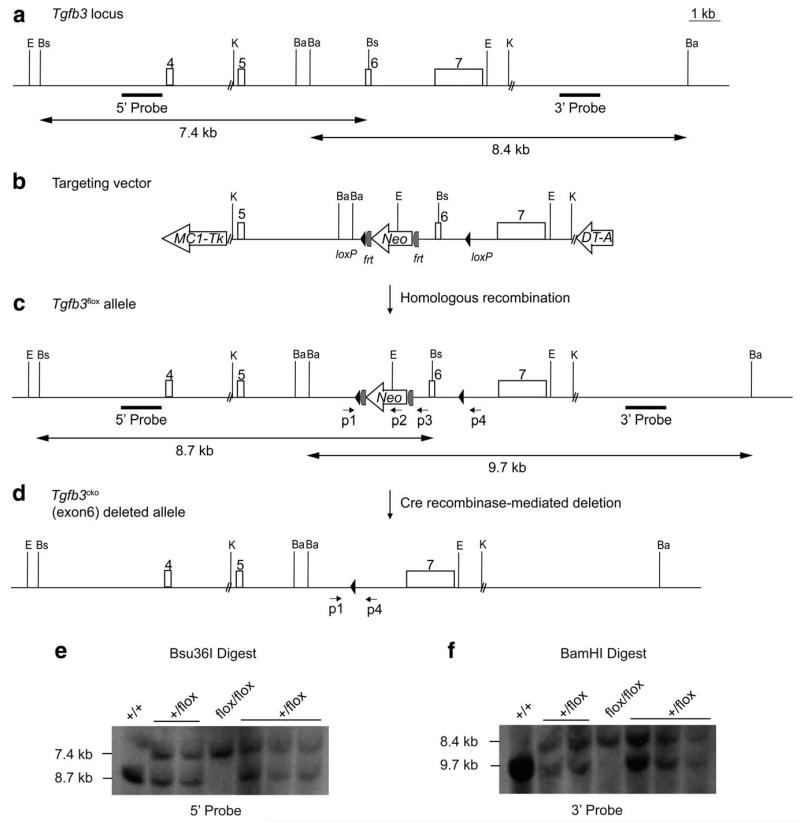
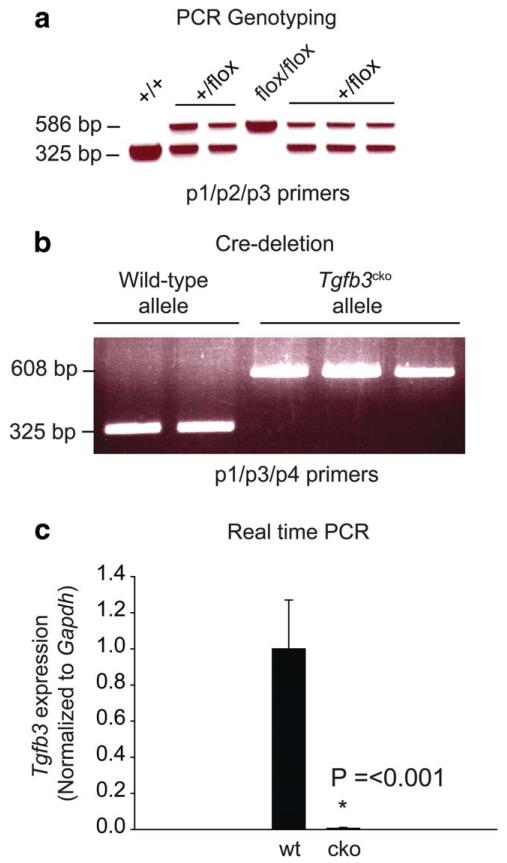
To circumvent the embryonic lethality exhibited by Tgfb3−/− mice, we generated Tgfb3cko/cko mice harboring conditional null alleles of Tgfb3. The Tgfb3 gene consists of seven exons spanning 21.7 kb on mouse Chromosome 12. A Cre-LoxP strategy was used to flank exon 6 for Cre-mediated deletion (Fig. 1a-d). This method has been successfully used by us (Azhar et al., 2009a) and numerous other researchers (Nagy et al., 2009). Exon 6 in Tgfb3 encodes the mature peptide of TGFb3. Total deletion of exon 6 in Tgfb3−/− embryos results in cleft palate (Kaartinen et al., 1995; Proetzel et al., 1995). Our conditional gene targeting of the Tgfb3 locus resulted in four out of 143 G418-resistant ES cell colonies which had undergone correct homologous recombination. Several mouse chimeras were produced which transmitted the Tgfb3flox allele to the germline as demonstrated by Southern hybridization (Fig. 1e-f). PCR genotyping was used to distinguish Tgfb3+/+, Tgfb3+/flox and Tgfb3flox/flox mice before weaning age (Fig. 2a). Tgfb3flox/flox mice that were born, were viable, fertile, and indistinguishable from wild-type littermates. Tgfb3flox/flox animals were born with near expected Mendelian ratio. Real time PCR analysis showed no difference in “wild-type” Tgfb3 expression between wild-type and Tgfb3flox/flox mice (not shown). The design of the Tgfb3 targeted allele permits removal of the MC1Neo cassette by mating to a Flp deleter mouse line (generating the Tgfb3flox allele) or removal of LoxP-flanked Tgfb3 genomic sequence (including exon 6 and the Frt-MC1NeopA-Frt cassette) by mating to a Cre deleter line (generating the Tgfb3cko allele). There are numerous publications detailing the unintended effects of the vigorously and ubiquitously expressed PGK promoter but not the MC1 promoter on flanking gene expression (Pham et al., 1996; Ren et al., 2002; Scacheri et al., 2001). However, we are currently analyzing these mice for possible hypomorphic effects. Collectively, these data are consistent with the observation that the presence of the LoxP or Frt-MC1NeopA-Frt cassette does not alter TGFb3 function in Tgfb3flox/flox mice. Consequently, we decided to delete exon 6 and the Frt-MC1NeopA-Frt cassette in order to produce Tgfb3cko/cko mice by breeding Tgfb3flox/flox mice with EIIa-Cre transgenic mice. Cre recombinase is expressed in oocytes and preimplantation embryos when under the control of the adenoviral (EIIa) promoter (Holzenberger et al., 2000). Hence, this Cre driver line eliminates genes in all cells and creates a complete gene knockout. The deletion of exon 6 in Tgfb3cko/cko mice was confirmed by PCR (Fig. 2b). Real-time PCR analysis of the palatal tissues showed that there is no detectable expression of “wild-type” Tgfb3 expression in Tgfb3cko/cko embryos when compared to control wild-type embryos (Fig. 2c).
FIG. 1.
Conditional gene targeting scheme for generating Tgfb3flox mice. a: Schematic diagram of the Tgfb3 wild-type genomic locus depicting the genomic region from intron 4 to the 3′ untranslated region. Boxes with numbers on top represent exons. Both 5′ outside and 3′ outside probes and the expected band sizes used for Southern blot screening of targeted Tgfb3floxES cell colonies are indicated. Restriction enzymes that are used in Southern hybridization are also highlighted in abbreviated form: E, EcoRI; Bs, Bsu36I; K, KpnI; Ba, BamHI. b: Conditional gene targeting vector. The targeting vector consists of a KpnI genomic DNA fragment which contains Tk-MC1 negative selection gene, 5′ or left homology arm, a LoxP site followed by a Frt-MC1NeopA-Frt cassette in intron 6, another LoxP site in intron 7, 3′ or right homology arm, and a PGK-DTA expression cassette. c: Schematic representation of targeted Tgfb3flox allele. Southern hybridization probes and PCR screening and genotyping primers are indicated. d: Schematic diagram depicting Cre-mediated recombination of the Tgfb3flox allele to yield the Tgfb3cko allele. Primers that identify the Cre-deletion events are indicated. e: Southern hybridization screening showing successful generation of wild-type, Tgfb3+/flox and Tgfb3flox/flox animals. Genomic DNA from tail clips is digested with Bsu36I and probed with the external probe (5′ Probe). Bsu36I cuts in the 5′ outside region of Tgfb3 and inside the targeting vector. Wild-type mice show an expected 8.7 kb band. Both Tgfb3+/flox and Tgfb3flox/flox animals show expected and correctly targeted bands of 8.7 kb and 7.4 kb, and 7.4 kb, respectively. f: BamHI digest probed with an external probe (3′ Probe) confirms the presence of Tgfb3+/flox (9.7 kb and 8.4 kb) and Tgfb3flox/flox (8.4 kb) mice. Wild-type mice show the expected 9.7 kb band.
FIG. 2.
Generation of mice harboring Tgfb3cko allele. a: PCR genotyping of mice with Tgfb3flox allele. The primers used are: p1 (intron 6 forward primer), p2 (Neo reverse primer), and p3 (intron 6 reverse primer). The p1 and p3 primers produce a PCR product of 325 bp from the wild-type allele, whereas p1 and p2 primers give rise to a PCR product of 586 bp from the Tgfb3flox allele. Band size as measured by DNA size markers is indicated. +/+, wild-type; +/ flox, Tgfb3+/flox; flox/flox, Tgfb3flox/flox. b: Cre-mediated deletion in Tgfb3flox/flox EIIa-Cre embryos (i.e., embryos with Tgfb3cko alleles) at E18.5. Tgfb3+/+ EIIa-Cre embryos were used as wild-type controls. The primers used are p1 (intron 6 forward primer), p3 (intron 6 reverse primer), and p4 (intron 7 reverse primer). Genomic DNA extracted from the palate tissue is used for PCR analysis. Briefly, the p1 and p3 primers produce a PCR product of 325 bp from the wild-type allele, whereas p1 and p3 primers amplify a PCR product of 1246 bp from the undeleted Tgfb3flox allele. The p1 and p4 primers results in a PCR product of 608 bp from the Tgfb3cko allele. Notably, Tgfb3+/+ EIIa-Cre embryos show a PCR product of 325 bp (i.e., expected band size of a wild-type allele) and that Tgfb3flox/flox EIIa-Cre embryos give rise to a 608 bp PCR product (i.e., expected band size of a Tgfb3cko allele). There was no detectable 1246 bp band (i.e., expected band size of an undeleted Tgfb3flox allele) in the Tgfb3flox/flox EIIa-Cre embryos, indicating a complete deletion of both Tgfb3flox alleles (i.e., presence of Tgfb3cko alleles) in Tgfb3flox/flox EIIa-Cre embryos. c: Real time PCR analysis of “wild-type” Tgfb3 expression in wild-type control and Tgfb3cko embryos at E18.5. Total RNA from palate tissue is used to prepare the cDNA. Tgfb3 exon 4 forward and Tgfb3 exon 6 reverse primers are used for real time PCR analysis. Note that there is no detectable “wild-type” Tgfb3 expression in palate tissue of Tgfb3cko embryos (*P = <0.001, n = 5 for wild-type, n = 3 for Tgfb3cko). Expression levels are normalized to Gapdh and to the wild-type value.
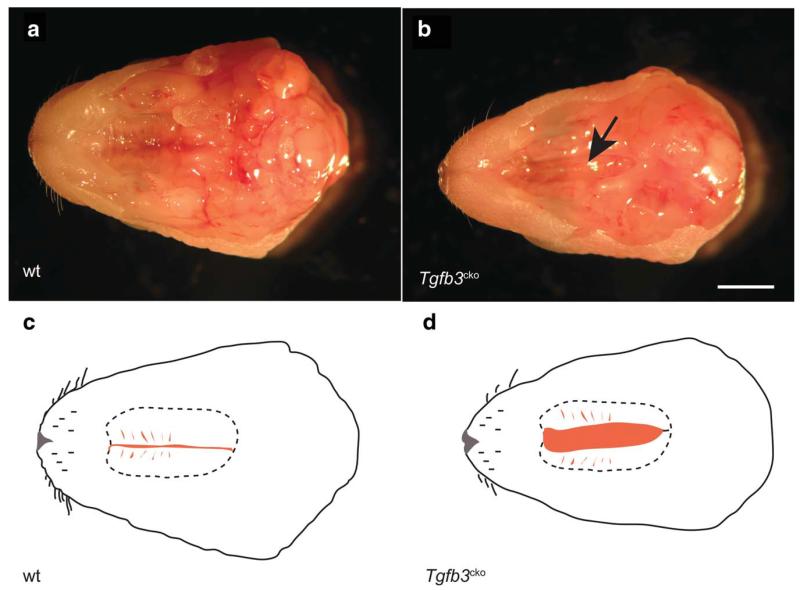
Intercrossing of Tgfb3+/flox EIIa-Cre mice did not produce viable Tgfb3cko mice. Closer examination of five different litters showed that Tgfb3cko/cko mice were born in the expected Mendelian ratio but they gasped to death. PCR genotyping confirmed these findings. Tgfb3cko/cko pups could not suckle well and their stomachs had little milk. Consequently, Tgfb3cko/cko and wild-type littermate embryos were collected at E18.5 of gestation and carefully examined for cleft palate (n = 10 for Tgfb3cko/cko, n = 30 for wild-type control). Interestingly, gross morphological examination of E18.5 embryos under a dissecting microscope revealed complete cleft palate or severe partial cleft palate in all ofthe Tgfb3cko/cko embryos (Fig. 3). Similar cleft palate defects were seen in Tgfb3cko/cko new born pups (not shown). There was no cleft palate in wild-type control embryos (Fig. 3a,c). Interestingly, Tgfb3cko/cko embryos exhibit the same cleft palate defects which were seen in Tgfb3−/− mice when on a C57BL/6 genetic background (Fig. 3b-d), (Kaartinen et al., 1995; Proetzel et al., 1995). This observation is consistent with the fact that we backcrossed Tgfb3flox/flox mice on to the C57BL/6 background for two generations before crossing with EIIa-Cre mice which were also on a C57BL/6 background. Thus, our data indicate that the Tgfb3cko allele is functionally equivalent to the Tgfb3 null allele.
FIG. 3.
Conditional deletion of Tgfb3 causes cleft palate in embryos. a: Gross morphological examination of a wild-type embryo (E18.5) showing normally fused palatal shelves. b: A Tgfb3flox/flox EIIa-Cre (i.e., Tgfb3cko) embryo at E18.5 exhibits a cleft palate (arrow). c,d: Diagrammatic representation of normal palatal shelves of a wild-type (C) and the cleft palate of a Tgfb3cko (D) embryo.
Tgfb3cko/cko mice offer several opportunities. They will be useful in directly testing the role of TGFβ3 in susceptibility and pathogenesis of ARVD1, CL/P, hypertension, OPLL, valve disease, craniofacial diseases, and HELLP syndrome. These mice will facilitate TGFβ3 research in wound healing at postnatal stages. Tgfb3cko/cko mice will open up avenues to investigate the tissue-specific role of TGFb3 in mammary gland development and in post-lactational involution and breast cancer progression in postnatal mice. In addition, these mice will provide new opportunities to evaluate TGFβ3 function in various other forms of cancer. Finally, Tgfb3cko/cko mice can be used to formally test the potential involvement of TGFβ3 in the pathogenesis of numerous inherited and noninherited diseases in which mutations or dysregulation of downstream TGFβ signaling components has been implicated. For instance, genetic mutations and/or dysregulation of TGFβ pathway genes are involved in cardiovascular and muscular (valve disease, cardiac hypertrophy, cardiac fibrosis, aneurysm, hypertension, atherosclerosis) (Dietz, 2010; Shimizu et al., 2011; Teekakirikul et al., 2010; van I et al., 2011), skeletal and craniofacial (Lidral et al., 1998), ocular (Saika et al., 2009), neurodegenerative, and neuromuscular (Katsuno et al., 2011), and in fibrotic and immunological disorders and diseases (Pohlers et al., 2009; Shah and Qiao, 2008). In most cases it remains unclear as to which TGFβ ligand(s) are responsible for these diseases, so these mice will be useful for determining their ligand/signaling specificity.
In conclusion, we have produced a Tgfb3cko allele and showed that it causes perinatal lethality due to cleft palate defects that phenocopy those of Tgfb3β/β mice. Thus, Tgfb3cko/cko mice provide a novel mouse strain to determine the tissue or cell-type specific roles of TGFβ3 during embryogenesis and in adult life and to investigate the mechanisms underlying TGFβ3 dysregulation in human disease.
METHODS
Generation and Phenotypic Analysis of Mice Carrying Tgfb3cko Allele
All procedures are approved by the Institutional Animal Care and Use the Committee at University of Arizona. Conditional gene targeting vector for the Tgfb3flox/flox mice was produced from a mouse genomic library clone (129/J). The targeting strategy was to conditionally delete exon 6 of Tgfb3 by using a Cre-LoxP strategy. A similar approach targeting exon 6 had been used previously to successfully produce Tgfb3β/β mice (Proetzel et al., 1995). A 6.5 kb KpnI genomic DNA fragment containing exon 5-7 of Tgfb3 was subcloned in a pAC7-P7 plasmid and used for building the targeting vector (Fig. 1a-d). The left homology arm of the targeting vector was 3.1 kb. The right homology arm was 2.4 kb. Two LoxP sites were placed in similar orientation. The first LoxP site is located in intron 6. A FrtMC1NeopA-Frt selection cassette with two flanking Frt sites was inserted after this LoxP site in intron 6. The MC1NeopA cassette was flanked by Frt sites for removal by Flp recombinase (Farley et al., 2000). The second LoxP site was inserted 344 bp into intron 7. A diphtheria toxin cassette (PGK-DTA) at the 3′ terminus and an MC1 promoter-herpes simplex virus-thymidine kinase cassette (MC1TK) at the 5′ terminus of the gene targeting vector was introduced for negative selection against random integration of the targeting vector into the embryonic stem (ES) cell genome. The accuracy of the entire targeting vector was confirmed by DNA sequenc-ing. The PI PspI-linearized targeting vector was electroporated into KG-1 (129/SvEv) ES cells. ES cells were treated with G418 (225 lg/ml) and remained in this selection for the entire time they were in culture. Counter selection with Ganciclovir (2 mM) was applied for 3 days. A total of 143 ES cell colonies were picked and screened by PCR and southern blotting. Two targeted ES clones were injected into C57BL/6 x DBA2 blastocysts for generating chimeric mice. Germline transmission of the Tgfb3flox allele was established by breeding the male ES cell chimeric mice to Black-Swiss females according to previously described methods (Azhar et al., 2009a). Germline transmission of the Tgfb3flox allele was confirmed by both genomic PCR and southern blotting. The germline chimeric mice were backcrossed to C57BL/6J mice for two generations. Tgfb31/flox mice were intercrossed to yield Tgfb3flox/flox mice. Tgfb3+/floxEIIa-Cre mice (129/J/Black-Swiss/C57BL/6) were generated by crossing Tgfb3flox/flox (129/Black-Swiss/ C57BL/6) to EIIa-Cre mice (C57BL/6) (Jax Lab, Bar harbor, ME). Timed-pregnant Tgfb31/flox EIIa-Cre intercrossed mice were used to collect E18.5 wild-type and Tgfb3flox/flox EIIa-Cre embryos in which the Tgfb3flox allele should have been converted to the Tgfb3cko allele. All embryos were examined for cleft palate under a stereozoom dissecting microscope (Zeiss Inc.) and digitally photographed. Tail clips were used to extract genomic DNA (Invitrogen Inc.) for PCR genotyping of embryos. Non-transgenic EIIa-Cre littermates which were Tgfb3flox/flox or Tgfb3+/+ EIIa-Cre were used as control animals. PCR analysis on tail clip DNA was used for genotyping Tgfb3cko/cko mice. The following PCR primers were used for genotyping: AGATAAACAATGGAGTCT GTCATGG (Tgfb3 forward primer, p1), TTCTGGATTC ATCGACTGTGG (Neomycin, reverse primer, p2), GTC TCATATGTGTCTTCCTGTCTCC (Tgfb3 reverse primer, p3); PCR conditions (Denaturation, 95°C/90 sec; Amplification, 57°C/50 sec, 72°C/60 sec, 95°C/30 sec for 34 cycles, 57°C/50 sec, 72°C/5 min, 28°C/10 min).
Southern Hybridization, DNA Sequencing, and PCR Analyses
For southern hybridization, mouse tail genomic DNA samples (30 μg) were digested with restriction endonucleases and electrophoresed in 0.7% agarose gels. DNA was transferred to positively charged nylon membrane (Roche Applied Science Inc.) by capillary blotting and crosslinked by UV irradiation. For probe labeling, 5′-end and 3′-end digoxygenin (DIG)-labeled southern blot probes were prepared by PCR procedure using Taq DNA polymerase and incorporating DIG-11-dUTP according to the procedures described by the manufacturer (Roche Applied Science Inc.). The following primers were used to amplify the 5′-end (1.0 kb) probe: AATTGAACTCTGCTCTATTGCTTGC (forward primer) and GGAAGTGAGTTATATTCAGAGTCATGG (reverse primer). For the 3′-end (0.9 kb probe), the following primers were used: AGCTTAGATGTGCTTCTCAATG ACC (forward primer) and CTTCAGCAGACCTAGTCAT TGTAGTCC (reverse primer). Genomic DNA (50 ng) was used as template in the reaction. The synthesis of the labeled probes was examined on a gel. The labeled probe was clearly distinguished from the unlabeled probe since it migrated slower than the unlabeled probe. The manufacturer’s recommended probe concentration of 10-20 ng/ml was used in the blot hybridizations. Following prehybridization in 10 ml of DIG EasyHyb solution at 42°C for 1 h, hybridization was carried out at 42°C overnight in a hybridization oven. The membranes were then washed 2-3 times in 2x standard sodium citrate (SSC), 0.1% sodium dodecyl sulfate (SDS) at room temperature for 5 min each and twice in 0.1x SSC, 0.1% SDS at 68°C for 15-20 min each. Detection of the hybridized probe DNA was carried out as described in the Manufacturer’s User Guide. CSPD chemiluminescent substrate was used and signals were visualized on X-ray film after 5-30 min.
Automated DNA sequencing of PCR-amplified products was used to confirm the sequence accuracy and correct orientation of the Frt-MC1NeopA-Frt cassette and LoxP and Frt sequences in all targeted ES cell clones and Tgfb3cko/cko mice. The following combination of PCR and DNA sequencing primers were used to confirm the sequence orientation of LoxP and FrtMC1NeopA-Frt cassette located in intron 6: For PCR amplification, GTCACACCTTTCAGCCCAAT (forward primer); CGTGCTATGGGTTGTGTCTG (reverse primer); sequencing primers, TCGCCTTCTTGACGAGTTCT; AAA ACCACACTGCTCGACAT; AGGATCTCCTGTCATCTCAC CTTGCTCCTG. The following primers for amplification and sequencing of the second LoxP located downstream to exon 6 were used: For PCR amplification, AAATGGGT CCACGAACCTAA (forward primer); CGTGCTATGGGTT GTGTCTG (reverse primer); sequencing primer, ATGCT TAGTGTGTGCCATGC.
Genomic PCR was used to detect the Tgfb3cko allele after Cre-mediated excision of exon 6 in the Tgfb3flox allele. Specific primers that were used for the PCR amplification included: AGATAAACAATGGAGTCTGTCATGG (forward primer, p1), GTCTCATATGTGTCTTCCTGTCT CC (Tgfb3 reverse primer, p3), AAGGTCTTCCTGTCTG TAAGTTTCC (Tgfb3 reverse primer, p4); PCR conditions (denaturation: 95°C/3 min, annealing and amplification 95°C/30 sec, 64°C/1 min, 72°C/2 min for 35 cycles, 72°C/10 min).
Real-Time PCR Analysis
Various tissues (heart, lung, liver, intestine, kidney, lower and upper jaws, and skin) were collected in RNA. Later RNA stabilization solution (Qiagen Inc., Valencia, CA) from both control and Tgfb3flox/flox EIIa-Cre-generated embryos at E18.5. Total RNA was extracted by RNeasy Mini kit (Cat# 74104: Qiagen, Valencia, CA). RNA was DNase treated with TURBO-DNA free kit (Ambion). cDNA was transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, Richmond, CA). cDNA concentration was measured using fluorometry (Turner Biosystems) after staining with Quant-iT™ OliGreen® ssDNA Assay Kit (# O11492, Molecular Probes). Equal amounts of cDNA samples from more than three control or experimental embryos were assessed (each in triplicate) by both RT-PCR and real time PCR analyses, as described (Azhar et al., 2009a,b). FastStart SYBR Green Master (Roche) mix was used for real time PCR analysis. Real-time PCR analysis was carried out in the Rotor Gene 3000 System from Corbett Research. Analysis of the data was carried out using the Rotor Gene 6 software. All real-time PCR results were normalized to the housekeeping gene Gapdh. Primers that were used for real time PCR amplification of Tgfb3 included: Tgfb3 exon 4 forward, GTCACACCTTTCAGCCCAAT; Tgfb3 exon 6 reverse, CGTGCTATGGGTTGTGTCTG; Gapdh forward, TGACCACAGTCCATGCCATC; Gapdh reverse, GACGGACACATTGGGGGTAG. Microsoft Excel was used for managing the data. Findings were reported as means ± SD of the mean, and two-tailed Student’s t-test (SigmaPlot, Systat Software, Inc., CA) was used for comparing groups. P-values were calculated, and a P < 0.05 was considered significant.
ACKNOWLEDGMENTS
We thank The Gene Targeted Mouse Service at the University of Cincinnati College of Medicine and The Genetically Engineered Mouse Modeling Core at the BIO5 Institute of the University of Arizona for generating chimeric Tgfb3flox animals and PCR/Southern blot screening of Tgfb3flox mice, respectively. Supported by NIH grants AI067903, HL092508 and CA084291 to TD.
Contract grant sponsor: National Institutes of Health Grants; Contract grant numbers: HL070174 and HL92508; Contract grant sponsor: Arizona Biomedical Research Commission; Contract grant numbers: ABRC #0901; Contract grant sponsor: The Stephen Michael Schneider/The William J. “Billy” Gieszl Award
Footnotes
Current address for Hongqi Li: DesigneRX Pharmaceuticals Inc., Vacaville, CA 95688.
LITERATURE CITED
- Aupperle H, Marz I, Thielebein J, Schoon HA. Expression of transforming growth factor-beta1, - beta2 and -beta3 in normal and diseased canine mitral valves. J Comp Pathol. 2008;139:97–107. doi: 10.1016/j.jcpa.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Azhar M, Runyan RB, Gard C, Sanford LP, Miller ML, Andringa A, Pawlowski S, Rajan S, Doetschman T. Ligand-specific function of transforming growth factor beta in epithelial-mesenchymal transition in heart development. Dev Dyn. 2009b;238:431–442. doi: 10.1002/dvdy.21854. PMC2805850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar M, Schultz JE, Grupp I, Dorn GW, Meneton P, Molin DG, Gittenberger-de Groot AC, Doetschman T. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003;14:391–407. doi: 10.1016/s1359-6101(03)00044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar M, Yin M, Bommireddy R, Duffy JJ, Yang J, Pawlowski SA, Boivin GP, Engle SJ, Sanford LP, Grisham C, Singh RR, Babcock GF, Doetschman T. Generation of mice with a conditional allele for transforming growth factor beta 1 gene. Genesis. 2009a;47:423–431. doi: 10.1002/dvg.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, Danieli GA, Rampazzo A. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res. 2005;65:366–373. doi: 10.1016/j.cardiores.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dietz HC. TGF-beta in the pathogenesis and prevention of disease: A matter of aneurysmic proportions. J Clin Invest. 2010;120:403–407. doi: 10.1172/JCI42014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli M, Giannubilo SR, Landi B, Sartini D, Pierella F, Corradetti A, Tranquilli AL. Placental overexpression of transforming growth factor-beta3 in the HELLP syndrome. Gynecol Obstet Invest. 2008;65:1–5. doi: 10.1159/000106497. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Flanders KC, Wakefield LM. Transforming growth factor-(beta)s and mammary gland involution: Functional roles and implications for cancer progression. J Mammary Gland Biol Neoplasia. 2009;14:131–144. doi: 10.1007/s10911-009-9122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Lenzner C, Leneuve P, Zaoui R, Hamard G, Vaulont S, Bouc YL. Cre-mediated germline mosaicism: A method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res. 2000;28:E92. doi: 10.1093/nar/28.21.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi T, Maeda K, Kawaguchi Y, Chiba K, Mori K, Koshizuka Y, Hirabayashi S, Sugimori K, Matsumoto M, Kawaguchi H, Takahashi M, Inoue H, Kimura T, Matsusue Y, Inoue I, Baba H, Nakamura K, Ikegawa S. A large-scale genetic association study of ossification of the posterior longitudinal ligament of the spine. Hum Genet. 2006;119:611–616. doi: 10.1007/s00439-006-0170-9. [DOI] [PubMed] [Google Scholar]
- Hu BC, Li L, Sun RH, Gao PJ, Zhu DL, Wang JG, Chu SL. The association between transforming growth factor beta3 polymorphisms and left ventricular structure in hypertensive subjects. Clin Chim Acta. 2010;411:558–562. doi: 10.1016/j.cca.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Banno H, Suzuki K, Tanaka F, Sobue G. Transforming growth factor-beta signaling in motor neuron diseases. Curr Mol Med. 2011;11:48–56. doi: 10.2174/156652411794474356. [DOI] [PubMed] [Google Scholar]
- Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor beta 1 nullmutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty HG, Wakefield LM, Occleston NL, O’Kane S, Ferguson MW. TGF-beta3 and cancer: A review. Cytokine Growth Factor Rev. 2009;20:305–317. doi: 10.1016/j.cytogfr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Foitzik K, Calautti E, Baden H, Doetschman T, Dotto GP. TGF-beta3, but not TGF-beta1, protects keratinocytes against 12-O-tetradecanoylphorbol-13-acetate-induced cell death in vitro and in vivo. J Biol Chem. 1999;274:4213–4219. doi: 10.1074/jbc.274.7.4213. [DOI] [PubMed] [Google Scholar]
- Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, Semina EV, Johnson LR, Machida J, Burds A, Parnell TJ, Rubenstein JL, Murray JC. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol Biol. 2009;530:365–378. doi: 10.1007/978-1-59745-471-1_19. [DOI] [PubMed] [Google Scholar]
- Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. Immunohistochemical localization of TGF beta 1, TGF beta 2, and TGF beta 3 in the mouse embryo: Expression patterns suggest multiple roles during embryonic development. J Cell Biol. 1991;115:1091–1105. doi: 10.1083/jcb.115.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci USA. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlers D, Brenmoehl J, Loffler I, Muller CK, Leipner C, Schultze-Mosgau S, Stallmach A, Kinne RW, Wolf G. TGF-beta and fibrosis in different organs - Molecular pathway imprints. Biochim Biophys Acta. 2009;1792:746–756. doi: 10.1016/j.bbadis.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren SY, Angrand PO, Rijli FM. Targeted insertion results in a rhombomere 2-specific Hoxa2 knock-down and ectopic activation of Hoxa1 expression. Dev Dyn. 2002;225:305–315. doi: 10.1002/dvdy.10171. [DOI] [PubMed] [Google Scholar]
- Saika S, Yamanaka O, Okada Y, Tanaka S, Miyamoto T, Sumioka T, Kitano A, Shirai K, Ikeda K. TGF beta in fibroproliferative diseases in the eye. Front Biosci (Schol Ed) 2009;1:376–390. doi: 10.2741/S32. [DOI] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de GA, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non- overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri PC, Crabtree JS, Novotny EA, Garrett-Beal L, Chen A, Edgemon KA, Marx SJ, Spiegel AM, Chandrasekharappa SC, Collins FS. Bidirectional transcriptional activity of PGK-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis. 2001;30:259–263. doi: 10.1002/gene.1072. [DOI] [PubMed] [Google Scholar]
- Shah S, Qiao L. Resting B cells expand a CD41CD251Foxp31 Treg population via TGF-beta3. Eur J Immunol. 2008;38:2488–2498. doi: 10.1002/eji.200838201. [DOI] [PubMed] [Google Scholar]
- Shimizu C, Jain S, Davila S, Hibberd ML, Lin KO, Molkara D, Frazer JR, Sun S, Baker AL, Newburger JW, Rowley AH, Shulman ST, Davila S, Burgner D, Breunis WB, Kuijpers TW, Wright VJ, Levin M, Eleftherohorinou H, Coin L, Popper SJ, Relman DA, Fury W, Lin C, Mellis S, Tremoulet AH, Burns JC. Transforming growth factor-{beta} signaling pathway in patients with Kawasaki disease. Circ Cardiovasc Genet. 2011;4:16–25. doi: 10.1161/CIRCGENETICS.110.940858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, Doetschman T. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teekakirikul P, Eminaga S, Toka O, Alcalai R, Wang L, Wakimoto H, Nayor M, Konno T, Gorham JM, Wolf CM, Kim JB, Schmitt JP, Molkentin JD, Norris RA, Tager AM, Hoffman SR, Markwald RR, Seidman CE, Seidman JG. Cardiac fibrosis in mice with hypertrophic cardiomyopathy is mediated by non-myocyte proliferation and requires Tgf-beta. J Clin Invest. 2010;120:3520–3529. doi: 10.1172/JCI42028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- van dL I, Oldenburg RA, Pals G, Roos-Hesselink JW, de Graaf BM, Verhagen JM, Hoedemaekers YM, Willemsen R, Severijnen LA, Venselaar H, Vriend G, Pattynama PM, Collee M, Majoor-Krakauer D, Poldermans D, Frohn-Mulder IM, Micha D, Timmermans J, Hil-horst-Hofstee Y, Bierma-Zeinstra SM, Willems PJ, Kros JM, Oei EH, Oostra BA, Wessels MW, Bertoli-Avella AM. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]