Abstract
Extensive molecular modeling with molecular dynamics simulations and van der Waals energy analyses were used to elucidate the striking finding that a mutagenic benzo[a]pyrene-derived DNA lesion, the base-displaced intercalated 10R-(+)-cis-anti-B[a]P-N2-dG (G*), manifests large differences in nucleotide excision repair (NER) efficiencies in DNA duplexes; that depend on the identities of the partner base opposite G*. The nature of the partner base causes marked differences in the extent of its major groove extrusion and dynamics, as well as energetic stability of the intercalation pocket that parallel the relative NER efficiencies.
Nucleotide excision repair (NER) (1) is an important cellular mechanism protecting the integrity of our genomic DNA from the deleterious effects of bulky DNA lesions derived from environmental tumorigens such as the tobacco carcinogen benzo[a]pyrene (B[a]P) (2). The NER machinery can recognize a broad range of DNA lesions (1). However, the repair efficiencies of chemically different DNA adducts can vary by several orders of magnitude, with some even being resistant to NER (3–5).The reasons for these large differences have been the subject of considerable interest. The impact of local sequence context surrounding a specific lesion on relative NER susceptibility in fully complementary duplex DNA has also been investigated (6, 7). However, the impact of mismatched bases in the complementary strand opposite the lesion that can occur in vivo because of error-prone translesion bypass (8–13) has not been adequately explored.
After metabolic activation of B[a]P to highly reactive (+)-7R,8S,9S,10R diol epoxide intermediates (14), a variety of stereoisomeric B[a]P diol epoxide-derived guanine adducts can form in DNA, that include the 10R-(+)-cis-anti-B[a]P-N2-dG (cis-B[a]P-dG) adduct (15) (Figure 1A). The replication of this and other stereoisomeric B[a]P-dG lesions is error-prone and all three of the non-canonical nucleotides are known to be inserted opposite the B[a]P-dG lesions with varying efficiencies (8–11, 13, 16). These mutational events produce modified base - non-canonical partner base mismatches that may have different structural features, and thus may be processed differently by the cellular NER apparatus. Although the cis-B[a]P-dG is efficiently excised in normal DNAduplexes with dC opposite this lesion, Hess et al. have shown that the same lesion paired with an adenine residue exhibits a ~ 10 times lower NER dual incision efficiency than the otherwise identical duplex with cytosine opposite the lesion (4). At the present time, there is no understanding of why replacing a single nucleotide opposite the same DNA lesion in otherwise completely identical duplexes should have such a dramatic impact on NER efficiencies. More recently, it has been reported (17) that the cis-B[a]P-dG damaged duplex with either T or G exhibits very different NER efficiencies as well. In these sets of experiments, the relative excision efficiencies with the different bases dC:dT:dA:dG opposite the cis-B[a]P-dG lesion are ~100:44:7:2 (K. Kropachev, M. Kolbanovskiy, and N.E. Geacintov, manuscript in preparation). These intriguing findings present a new opportunity for providing insights into the role of the partner strand in NER and more broadly for further elucidating the mechanisms of recognition of the lesion-induced helix distortions/destabilizations by the human XPC/RAD23B heterodimer that first recognizes the DNA damage (18). Our objective in the present study was to gain understanding of the remarkable observation that the same cis-B[a]P-dG lesion manifests different NER susceptibilities depending on the nature of the partner base, while all other base pairs in the 135-mer double-stranded DNA substrates remain unchanged (4, 17).
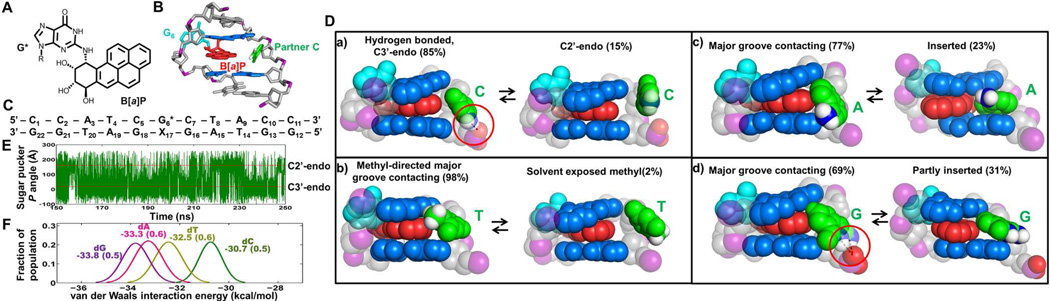
FIGURE 1.
Different chemical structures of partner nucleotides in cis-B[a]P-dG-modified duplexes produce different extents of interaction with the major groove. (A) cis-B[a]P-dG adduct chemical structure. (B) NMR solution structure of the cis-B[a]P-dG:dC duplex viewed from the major groove (24). (C) Base sequence of simulated 11-mers. G6* is the lesion-containing base, and X17 is the partner base C, T, A, or G. (D) Central trimers of the most representative structures in CPK. a) Fully extruded partner dC. b) The hydrophobic methyl group of partner dT avoids solvent and promotes close contact of the base with the groove surface. c) Partner dA and d) partner dG are in contact with the groove surface in its dominant conformation and inserted or partly-inserted into the helix in a minor population. Hydrogen bonds (dashed lines) are circled in red. (E) The dynamics of the sugar pucker pseudo-rotation phase angle (P) (32) of partner dC. (F) Population distribution of van der Waals interaction energies at the intercalation sites of cis-B[a]P-dG-modified duplexes. Mean values with standard deviations are given.
The human XPC/RAD23B complex, a key element in lesion recognition, is required for the recruitment of the factors involved in the subsequent steps of NER (1, 19). A crystal structure of Saccharomyces cerevisiae Rad4/Rad23, a yeast ortholog of the mammalian XPC/RAD23B, in complex with a DNA duplex containing a cyclobutane pyrimidine dimer (CPD) lesion, revealed that a β-hairpin, BHD3, is inserted into the DNA helix, separating the damaged and partner strands, while the two bases opposite the CPD are flipped out of the helix and interact with residues in the protein (20). This structure supports the hypothesis that local thermodynamic stability plays an important role in determining lesion recognition through DNA XPC/RAD23B binding and successful β-hairpin insertion with partner base flipping (3, 20–23).
According to previous NMR studies (24), the cis-B[a]P-dG-modified DNA duplex with normal partner dC (cis-B[a]P-dG:dC) adopts a base-displaced intercalated conformation with the lesion-containing guanine (G*) displaced into the minor groove, and the B[a]P aromatic ring system is intercalated into the duplex; the partner base dC is displaced into the major groove (Figure 1B). Although no NMR solution structures are available for the cis-B[a]P-dG-modified DNA duplexes with mismatched partner bases (cis-B[a]P-dG:dT, cis-B[a]P-dG:dA and cis-B[a]P-dG:dG), UV spectroscopic (17) studies indicate that the aromatic B[a]P ring system is intercalated as shown by red-shifted UV absorption maxima; an example for the dT case is shown in Figure S1 of the Supporting Information. However, nothing is known about the positions and orientations of the partner bases in mismatched duplexes. Our hypothesis is that the physical size and nature of the partner base opposite the lesion in the complementary strand has a strong impact on its orientation in the duplex that affects both its interactions with neighboring residues and its dynamic properties. We propose that these differences give rise to the striking variations in NER efficiencies observed when different partner bases are positioned opposite the same cis-B[a]P-dG adduct in duplex DNA. To identify the preferred conformations, we extensively surveyed the sterically and energetically accessible conformations using cis-B[a]P-dG:dC, cis-B[a]P-dG:dT, cis-B[a]PdG: dA and the cis-B[a]P-dG:dG pairs (Figure 1C) with molecular modeling approaches. We modeled an intercalated aromatic B[a]P ring system as revealed in the characteristic red-shifted UV absorption spectra of the bulky B[a]P residues (25) in the different mismatched duplexes (Figure S1 of the Supporting Information and unpublished observations). We created four different initial positions for the partner nucleotide that were evenly rotated around the DNA backbone at ~90° intervals (Figure S2 and Table S1 of the Supporting Information). We then performed long scale molecular dynamics (MD) simulations (at least 200 ns), and free energy calculations for energy ranking of different conformers (Table S2 of the Supporting Information), utilizing the AMBER9 program suite (26). Full details of the modeling, MD simulations and energetic analyses are provided in the Supporting Information. Our analyses of the dynamic ensembles of these duplexes with four different complementary bases opposite the lesion revealed a set of preferred conformations that differed with the identity of the partner base. In all cases, the B[a]P rings remain intercalated in the DNA helix, with the damaged guanine (G*) displaced into the minor groove as in the NMR solution structure with dC as the partner base. These results are provided in full details in Figures S3–S11 of the Supporting Information.
The solution structure of the cis-B[a]P-dG:dC pair, the best NER substrate among the four considered here, has been well characterized by NMR methods. While the dC residue was found to be displaced into the major groove, its location could not be precisely ascertained (24). Consistently, our MD simulations suggest that the dC base, with only one aromatic ring, is highly dynamic and is extruded into the major groove while remaining in the anti glycosidic bond conformation (Figure 1D). This conformation is consistent with the observed NMR solution structure (Figure 1B) which supports the feasibility of using the same strategy for analyzing the structures and dynamics of the mismatched cis-B[a]P-dG: nucleotide pairs. The simulations indicate that a dynamic hydrogen bond between the cytosine amino group and a pendant phosphate oxygen atom 5’ to the dC (Figure 1D) forms and breaks continuously; overall it is present 85% of the time (Table S3 and Figure S5 of the Supporting Information). This dynamic hydrogen bond regulates the position of the partner dC: when it is present (85% of the time), the dC, usually in C3’-endo sugar conformation, is oriented closer to the backbone; when it is lost (15% of the time) the dC residue, usually in C2’-endo sugar conformation, is displaced further away from the backbone and is thus even more extruded (Figure 1D, E and Figures S5, S6 and Movie S1 of the Supporting Information).
The cis-B[a]P-dG:dT duplex is a weaker NER substrate than cis-B[a]P-dG:dC by a factor of ~ 2. Both these pyrimidines are similar in size with one aromatic ring, but thymine possesses its characteristic hydrophobic methyl group. Our structural analyses of the MD ensemble reveal that the methyl group avoids aqueous solvent by positioning itself in the major groove, forming contacts with its surface and with the edge of the intercalated B[a]P ring system, with rare dynamic excursions into an extruded position (2% of the time) (Figure 1D and Figure S7 of the Supporting Information); this movement, due to transient crowding between the methyl and a backbone oxygen atom, entails rotation about the glycosidic bond from the syn domain in the predominant conformation, to the anti domain in the minor extruded conformation, and is correlated with C2’-endo to C3’-endo sugar re-puckering (Figure S7 and Movie S2 of the Supporting Information).
The cis-B[a]P-dG:dA and cis-B[a]P-dG:dG duplexes are NER-resistant. The purine partners with two aromatic rings are more bulky than the pyrimidine partners, and provide more extensive opportunities for stabilizing van der Waals interactions. With dA as the partner, the predominant conformation has van der Waals contacts with the major groove surface. In the minor conformer (23%), the aromatic dA residue is inserted into the helix (Figures S8 of the Supporting Information), and there are no dynamic excursions into extruded orientations (Figure 1D and Movie S3 of the Supporting Information). The glycosidic bond conformation of the dA is in the syn domain in both cases (Figure 1D), and the sugar pucker remains in the C1’-exo region, without oscillating between domains (Figure S9 of the Supporting Information).
With dG as the partner base, the dominant population is also in van der Waals contact with the major groove surface (Figure 1D). In this orientation, there is a restraining bifurcated hydrogen bond involving the dG amino/imino protons and a pendant phosphate oxygen atom on its 5’ side (Table S3 of the Supporting Information) that adds stability. There is also a minor population (31%) that is partially inserted into the helix (Figure 1D, Figure S8 and Movie S4 of the Supporting Information). There are no dynamic excursions to extruded orientations. The glycosidic bond of the dG is syn in both cases. The sugar pucker of dG oscillates stably around C1’-exo (Figure S10 of the Supporting Information).
In order to gain further insights into the impact of the different partner bases opposite the cis-B[a]P-dG adduct on NER efficiencies, we computed van der Waals interaction energies in the intercalation pocket. All interactions involving the B[a]P aromatic ring system, the partner base, and the adjacent base pairs were included. Figure 1F shows the interaction energies for each duplex. The lowest van der Waals interaction energy is manifested in the case of the cytosine partner base. Among the four partner bases, cytosine is the most extruded and dynamic one, followed by thymine which is in better contact with the major groove surface. The adenine and guanine partner bases, with two aromatic rings, give rise to similar interaction energies. In the dominant populations in both cases the purine base contacts the major groove surface, and there are also minor inserted or partially inserted populations with similar van der Waals interaction energies for the groove-bound and inserted states (Figure S11 of the Supporting Information). The possibly slightly better interaction energy in the case of the partner dG base may stem from the episodic hydrogen bond formation involving its amino/imino group and the DNA backbone, which adds stability to the dG base in the major groove.
The van der Waals interaction energies (Figure 1F) and the relative NER efficiencies are well correlated with one another, supporting the hypothesis that local thermodynamic properties are strongly affected by the local base sequence context and that stabilizing van der Waals interactions between a given lesion and surrounding nucleotide residues can lead to NER resistance or even complete abrogation of NER (21, 27–30). In addition, the Rad4/Rad23 crystal structure revealed that the BHD3 β-hairpin intrusion is accompanied by flipping out of two bases opposite the lesion in the unmodified partner strand (20). This suggests that partner bases that are more extruded could more readily promote productive interactions with the NER recognition proteins Rad4/Rad23 or the human XPC/RAD23B. It is interesting that the extent of base extrusion and the dynamics of the partner base in our series also parallels the repair efficiencies, with dC being the most extruded partner base and the lesion in the cis-B[a]P-dG:dC duplex being the most susceptible to NER. The possibility that partner base flippability is a component of lesion recognition has recently been explored (31). In summary, changing only the partner base to the same cis-B[a]P-dG lesion, affects the thermodynamic stabilities of the lesion sites that may be sensed by the XPC/RAD23B lesion recognition factor and perhaps subsequent downstream events in NER that affect overall NER efficiencies (19).
Supplementary Material
ACKNOWLEDGMENT
This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (NSF) Grant MCB060037, and the high performance computing resources of New York University (NYU-ITS).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting Figures S1–S11, Tables S1–S3, material and methods, results, and Movies S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
FUNDING SOURCES
This research was supported by National Institutes of Health Grants R01-CA28038 (to S.B.) and 1R01-CA168469 (to N.E.G.). Computational infrastructure and systems management were partially supported by R01-CA75449 (S.B).
The authors declare no competing financial interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- 1.Schärer OD, Campbell AJ. Mechanisms of base excision repair and nucleotide excision repair. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Weinheim, Germany: Wiley-VCH; 2010. pp. 239–260. [Google Scholar]
- 2.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 3.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 4.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 6.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/174252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, Geacintov NE, Broyde S. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012;40:9675–9690. doi: 10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes A, Liu T, Amin S, Geacintov NE, Grollman AP, Moriya M. Mutagenic potential of stereoisomeric bay region (+)- and (−)-cis-anti-benzo[a]pyrene diol epoxide-N2-2'-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 10.Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Repair (Amst) 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO. J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 13.Shukla R, Jelinsky S, Liu T, Geacintov NE, Loechler EL. How stereochemistry affects mutagenesis by N2-deoxyguanosine adducts of 7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene: configuration of the adduct bond is more important than those of the hydroxyl groups. Biochemistry. 1997;36:13263–13269. doi: 10.1021/bi971195z. [DOI] [PubMed] [Google Scholar]
- 14.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 15.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem. Res. Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki N, Ohashi E, Kolbanovskiy A, Geacintov NE, Grollman AP, Ohmori H, Shibutani S. Translesion synthesis by human DNA polymerase kappa on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-N(2)-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Kropachev KY, Liu Z, Kolbanovsky M, Geacintov NE. Human NER recognition mechanisms involving the undamaged strand of the double helix. Presented at the 240th American Chemical Society National Meeting; August 24th; Boston, MA. 2010. Poster TOXI 89. http://abstracts.acs.org/chem/240nm/program/view.php?obj_id=38524&terms= [Google Scholar]
- 18.Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 19.Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Hofer T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schärer OD. Achieving broad substrate specificity in damage recognition by binding accessible nondamaged DNA. Mol. Cell. 2007;28:184–186. doi: 10.1016/j.molcel.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 24.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 25.Huang W, Amin S, Geacintov NE. Fluorescence characteristics of site-specific and stereochemically distinct benzo[a]pyrene diol epoxide-DNA adducts as probes of adduct conformation. Chem. Res. Toxicol. 2002;15:118–126. doi: 10.1021/tx010135y. [DOI] [PubMed] [Google Scholar]
- 26.Case DA, Darden TA, Cheatham TE, Simmerling CL, III, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 27.Kropachev K, Kolbanovskiy M, Liu Z, Cai Y, Zhang L, Schwaid AG, Kolbanovskiy A, Ding S, Amin S, Broyde S, Geacintov NE. Adenine-DNA adducts derived from the highly tumorigenic dibenzo[a,l]pyrene are resistant to nucleotide excision repair while guanine adducts are not. Chem. Res. Toxicol. 2013;26:783–793. doi: 10.1021/tx400080k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves DA, Mu H, Kropachev K, Cai Y, Ding S, Kolbanovskiy A, Kolbanovskiy M, Chen Y, Krzeminski J, Amin S, Patel DJ, Broyde S, Geacintov NE. Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions. Nucleic Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai Y, Geacintov NE, Broyde S. Nucleotide excision repair efficiencies of bulky carcinogen - DNA adducts are governed by a balance between stabilizing and destabilizing interactions. Biochemistry. 2012;51(7):1486–1499. doi: 10.1021/bi201794x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO. J. 2009;28:2387–2399. doi: 10.1038/emboj.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai Y, Zheng H, Ding S, Kropachev K, Schwaid AG, Tang Y, Mu H, Wang S, Geacintov NE, Zhang Y, Broyde S. Free energy profiles of base flipping in intercalative polycyclic aromatic hydrocarbon-damaged DNA duplexes: Energetic and structural relationships to nucleotide excision repair susceptibility. Chem. Res. Toxicol. 2013;26:1115–1125. doi: 10.1021/tx400156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.