Abstract
1,2,3,4-diepoxybutane (DEB) is reported to be the most potent mutagenic metabolite of 1,3-butadiene, an important industrial chemical and environmental pollutant. DEB is capable of inducing the formation of monoalkylated DNA adducts, and DNA-DNA and DNA-protein crosslinks. We previously reported that DEB forms a conjugate with glutathione (GSH) and that the conjugate is considerably more mutagenic than several other butadiene-derived epoxides, including DEB, in the base pair tester strain Salmonella typhimurium TA1535 (Cho et. al., Chem. Res. Toxicol. 23, 1544–1546 (2010)). In the present study, we determined steady-state kinetic parameters of the conjugation of the three DEB stereoisomers— R,R-, S,S-, and meso- — (all formed by butadiene oxidation) with GSH by six GSH transferases. Only small differences (< 3-fold) were found in the catalytic efficiency of conjugate formation (kcat/Km) with all three DEB stereoisomers and the six GSH transferases. The three stereochemical DEB-GSH conjugates had similar mutagenicity. Six DNA adducts (N3-adenyl, N6-adenyl, N7-guanyl, N1-guanyl, N4-cytidyl, and N3-thymidyl) were identified in the reactions of DEB-GSH conjugate with nucleosides and calf thymus DNA using LC-MS and UV and NMR spectroscopy. N6-Adenyl and N7-guanyl GSH adducts were identified and quantitated in vivo in the livers of mice and rats treated with DEB i.p.. These results indicate that such DNA adducts are formed from the DEB-GSH conjugate, are mutagenic irregardless of sterochemistry, and therefore expected to contribute to the carcinogenicity of DEB.
INTRODUCTION
1,3-Butadiene is an important industrial chemical used in the synthesis of plastics and rubber and is also an environmental pollutant found in cigarette smoke and automobile exhaust.1,2 Butadiene was carcinogenic in inhalation experiments with B6C3F1 mice and Sprague-Dawley rats, particularly the former species apparently because of the increased formation of 1,2,3,4-diepoxybutane (DEB) and other DNA-reactive metabolites in the mouse.3–5 These findings, along with epidemiology studies, suggest that butadiene is a human carcinogen.6,7 Although butadiene is relatively unreactive by itself, it is converted by cytochrome P450 enzymes to three reactive epoxide metabolites that are capable of alkylating DNA: the mono-epoxide, the diol epoxide, and DEB.8,9 The modification of DNA bases caused by butadiene-derived epoxide metabolites has been reported.10–17 Among the three epoxide metabolites of butadiene, DEB is the most mutagenic because of its bis electrophilic properties and enhanced reactivity with DNA.18–20 DEB can induce DNA-protein cross-links as well as DNA-DNA cross-links, due to its bifunctional alkylating nature,21,22 and DNA-protein cross-links are considered to distort the helical structure of DNA and inhibit normal DNA metabolism.23,24
Three stereoisomers of DEB— R,R-, S,S-, and meso- —are generated by enzymatic oxidation of butadiene.25S,S-DEB has been regarded to be more genotoxic and cytotoxic, followed by R,R-DEB and then meso-DEB, although the differences are not strong and discrepancies have been reported in studies of rates of formation and reactivity.25–28
GSH conjugation is generally considered to be a detoxication process in the metabolism of xenobiotic chemicals.29 However, we previously reported that the mutagenicity of DEB is considerably increased in Salmonella typhimurium TA1535 by the cellular expression of rat GSH S-transferase (GST) 5-530 or human GST T1-1,31 and the enzymatically synthesized S-(2-hydroxy-3,4-epoxybutyl)GSH (DEB-GSH) conjugate is significantly more mutagenic than several other butadiene-derived epoxides, including DEB, in S. typhimurium TA1535.32 These results are attributed to the formation of cross-links between GSH and DNA induced by DEB.
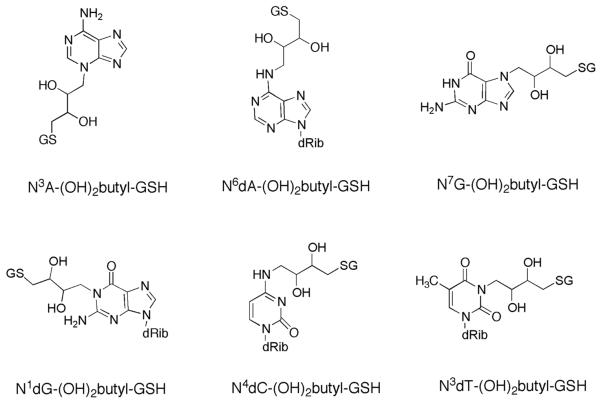
In the present work, we determined steady-state kinetic parameters of the conjugation of the three DEB stereoisomers with GSH by GSTs and investigated stereochemical aspects of DEB and GST selectivity. Six DNA adducts were identified in the reactions of DEB-GSH conjugate with nucleoside, using LC-MS and UV and NMR spectroscopy (Scheme 1), and calf thymus DNA. Two of the adducts were identified and quantitated in vivo in livers of mice and rats treated with DEB.
Scheme 1.
Structures of DNA Adducts Formed with DEB-GSH Conjugate
EXPERIMENTAL PROCEDURES
Materials
[glycine-13C2, 15N]-GSH was purchased from Cambridge Isotope Laboratories (Andover, MA). Racemic DEB, GSH, deoxyguanosine (dG), deoxyadenosine (dA), deoxycytidine (dC), deoxythymidine (dT), calf thymus DNA, and enzymes for digestion were purchased from Sigma Chemical Co. (St. Louis, MO). Rat GST 5-5 and human GSTs T1-1, A1-1, A3-3, M1-1, and P1-1 were purchased from Oxford Biomedical Res. (Oxford, MI). DEB steroisomers (S,S-, R,R-, and meso-) were synthesized as described in published procedures (Supporting Information Table S1).33–35 DEB-GSH conjugate was enzymatically synthesized and purified as described previously.32
Kinetic Analysis
A reaction mixture containing GSH (5 mM) and GST (10 μg/mL), with varying concentrations of DEB (0.05–3 mM), in 0.10 M Tris-HCl buffer (pH 7.7) was incubated for 1 h at 37 °C. After the reaction, CH3CN was added to 50% (v/v) to stop the reaction.
A Waters Acquity UPLC system (Waters, Milford, MA) interfaced to a Thermo-Finnigan LTQ mass spectrometer (ThermoElectron, Sunnyvale, CA), equipped with an ESI source, was used for analysis of DEB-GSH conjugates. Chromatographic separation was achieved with a Waters Acquity UPLC BEH C18 octadecylsilane column (2.1 mm × 100 mm, 1.7 μm). LC conditions were as follows: Mobile phase A was 0.1% CH3CO2H in H2O (v/v) and mobile phase B was 0.1% CH3CO2H in CH3CN (v/v). The following gradient program (v/v) was used with a flow rate of 300 μL/min: the gradient started with 2% B (v/v), increased to 10% B (v/v) at 2 min, to 30% B (v/v) at 6 min, and held at 30% B (v/v) for 1 min. The column was re-equilibrated for 3 min with 2% B (v/v). The temperature of the column was maintained at 40 °C. The MS conditions were as follows: ion spray voltage, 5 kV; capillary voltage, 20 V; capillary temperature, 350 °C; tube lens voltage, 50 V; and collision energy, 26%. A calibration curve was prepared to quantify the DEB-GSH conjugate in the concentration range 0.5–500 μM using DEB-[glycine-13C2, 15N]-GSH conjugate as an internal standard (ISTD) (Supporting Information Figure S1).
Kinetic analysis was done using nonlinear regression analysis (Michaelis-Menten fits) (Prism, GraphPad Software, La Jolla, CA).
Mutation Assays
Ames reversion assays were performed as previously described.32,36S. typhimurium TA1537 cells were purchased from Molecular Toxicology (Boone, NC). NaN3 was used a positive control for S. typhimurium TA1535 and 9-aminoacridine for S. typhimurium TA1537. Toxicity assays were done in the presence of histidine. The three DEB stereoisomer-GSH conjugates were dissolved in 200 mM sodium phosphate buffer (pH 7.4). A 100 μL aliquot of S. typhimurium 1535 was mixed with 50 μL of each conjugate for 5 min at room temperature. The solutions were then mixed with 2 mL of top agar and plated on minimal glucose plates. The plates were incubated for 48 h at 37 °C and revertants were counted.
Reaction of DEB-GSH Conjugate with Nucleosides
DEB-GSH conjugate (5 mM) was incubated with each of the four deoxyribonucleosides (2 mM) in 0.10 M Tris-HCl buffer (pH 7.7) for 12 h at 37 °C. Reactions were subsequently heated for 30 min at 90 °C under neutral conditions, to release some of the modified bases, and the products were separated by HPLC as described below.
Purification of DNA Adducts by HPLC
Separations of DNA adducts (formed from reaction of DEB-GSH conjugate with nucleosides) were performed by HPLC with a Hitachi L-7100 pumping system and LDC Analytical SpectroMonitor 3200 variable wavelength detector using a Phenomenex Prodigy octadecylsilane column (250 mm × 10 mm, 5 μm, ODS(3), 100Å). Mobile phase A was 20 mM NH4CH3CO2 in H2O (pH 6.5) and mobile phase B was 20 mM NH4CH3CO2 in CH3CN/H2O (30:70, v/v). The following gradient program (v/v) was used with a flow rate of 2 mL/min: the gradient started with 5% B (v/v), increased to 10% B (v/v) at 5 min, to 30% B (v/v) at 10 min, and held at 30% B (v/v) for 15 min. The column was re-equilibrated for 10 min with 2% B (v/v). The UV wavelength detector was set at 265 nm.
Characterization of DNA Adducts
DNA adducts formed from the reaction of DEB-GSH conjugate with nucleosides were characterized by LC-MS/MS and UV and NMR spectroscopy. LC-MS/MS analysis was performed by a Waters Acquity UPLC system (Waters) interfaced to a Thermo-Finnigan LTQ FT-Orbitrap mass spectrometer (ThermoElectron) equipped with an ESI source. Chromatographic separation was achieved with a Waters Acquity UPLC BEH C18 octadecylsilane column (2.1 mm × 100 mm, 1.7 μm). LC conditions were as follows: Mobile phase A was 0.1% CH3CO2H in H2O (v/v) and mobile phase B was 0.1% CH3CO2H in CH3CN (v/v). The following gradient program (v/v) was used with a flow rate of 300 μL/min: the gradient started with 5% B (v/v), increased to 15% B (v/v) at 2 min, to 30% B (v/v) at 6 min, and held at 30% B (v/v) for 1 min. The column was re-equilibrated for 3 min with 5% B (v/v). The temperature of the column was maintained at 40 °C. The MS conditions were as follows: ion spray voltage, 4.5 kV; capillary voltage, 20 V; capillary temperature, 350 °C; and tube lens voltage, 40 V.
UV spectra were obtained using a modified Cary14-OLIS spectrophotometer (On-Line Instrument Systems, Bogart, GA) after Raney nickel reaction for desulfurization of DNA adducts.37 Desulfurized DNA adducts were scanned under acidic (0.1 M HCl), neutral (H2O), and alkaline (0.1 M NaOH) conditions.
NMR experiments were acquired in the Vanderbilt facility using a 14.0 T Bruker magnet equipped with a Bruker AV-III console operating at 600.13 MHz. Samples were dissolved in D2O, and all spectra were acquired in 3 mm NMR tubes using a Bruker 5 mm TCI cryogenically-cooled NMR probe. For one-dimensional 1H NMR, typical experimental conditions included 32K data points, 13 ppm sweep width, a recycle delay of 1.5 s, and 645 receiver gain. For two-dimensional 1H-1H COSY, experimental conditions included a 2048 × 512 data matrix, 13 ppm sweep width, recycle delay of 1.5 s, and 912 receiver gain. The data were processed using squared sinebell window function, symmetrized, and displayed in magnitude mode.
Reaction of GSH-DEB Conjugate with Calf Thymus DNA
Calf thymus DNA (1.0 mg) was reacted with DEB-GSH conjugate (0.01–0.50 mM) in 1.0 mL of 50 mM Tris-HCl buffer (pH 7.0) for 12 h at 37 °C. The reactions were heated for 30 min at 90 °C under neutral conditions or digested by enzymes. The enzymatic digestion of calf thymus DNA was as follows: The reactions (0.5 mL) containing calf thymus DNA and DEB-GSH conjugate were incubated with DNase I (20 units), alkaline phosphatase (3 units), phosphodiesterase I type II (0.2 units), and nuclease P1 (4.25 units) for 48 h at 37 °C.
The reactions followed by thermal hydrolysis or enzymatic digestion were filtered by 3K MWCO Centricon filters (Millipore Corp., Billercia, MA) and spiked with synthesized N6dA-(OH)2butyl-[glycine-13C2, 15N]-GSH and N7G-(OH)2butyl-[glycine-13C2, 15N]-GSH (prepared by the reaction of [glycine-13C2, 15N]-GSH-DEB conjugate with dA or dG). The resulting reactions were analyzed by LC-MS/MS described (vide supra).
Animals and Treatment
Sparague-Dawley rats and B6C3F1 mice were housed in plastic cages, with bedding, according to NIH guidelines. All procedures involving the use of animals were approved by the LRRI Institutional Animal Care and Use Committee. Eight rats and seven mice were treated with DEB (25 mg/kg, i.p., in corn oil) and two rats and two mice were administrated corn oil (i.p.) as controls. Animals were treated and killed 6 h later (treated rats: n = 4, treated mice: n = 3, and control per group: n = 1) and 48 h (treated rats: n = 4, treated mice: n = 4, and control per group: n = 1). Liver, kidney, and lung tissues were collected and processed immediately.
Quantitation of DNA Adducts in Animal Tissues
DNA from animal tissues was isolated as previously described,38 followed by thermal or acid-catalyzed hydrolysis or enzymatic digestion. The reactions were filtered and spiked with synthesized N6dA-(OH)2butyl-[glycine-13C2, 15N]-GSH, N7G-(OH)2butyl-[glycine-13C2, 15N]-GSH, and [18O]-N7G-(OH)3butane. The resulting reactions were analyzed by LC-MS/MS.
RESULTS
Kinetic Analysis
The time-dependence of the conjugation of racemic DEB with GSH by various GSTs was established (Supporting Information Figure S2). The concentration of the DEB-GSH conjugate (product) formed by six individual GSTs was increased up to an incubation time of 1 h, and this time was used for single time-point kinetic analysis. All GST-catalyzed reactions of DEB with GSH yielded DEB-GSH conjugates, analyzed by LC-MS/MS. LCMS/MS responses were linear over the range of 0.5–500 mM of DEB-GSH conjugate, with correlation coefficients (r2) > 0.99 (Supporting Information Figure S1). The limit of detection (LOD) of DEB-GSH conjugate was 0.2 μM.
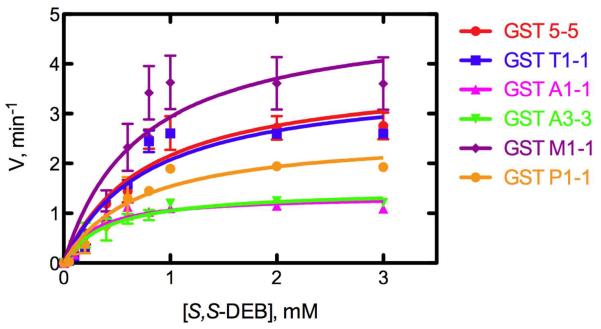
The catalytic efficiency of conjugate formation (kcat/Km) differed depending on the DEB stereoisomers and the GST, although all GSTs were active (Figure 1, Table 1). The kcat/Km values for substractes S,S-DEB and R,R-DEB revealed similar patterns with various GSTs and were the highest with GST M1-1, while the kcat/Km values for meso-DEB were the highest with GST T1-1. Among the three possible DEB stereoisomers, the kcat/Km value with S,S-DEB (3.9–7.0 mM−1 min−1) was the highest, followed by R,R-DEB (2.9–5.5 mM−1 min−1) and then meso-DEB (2.3–6.5 mM−1 min−1). Overall, the kcat/Km values only varied 3-fold.
Figure 1.
Production of GSH conjugate of S,S-DEB with GSH by GSTs (rat GST 5-5, and human GST T1-1, A1-1, A3-3, M1-1, and P1-1). Reactions were carried out at 37 °C for 1 h. Each point represents the mean ± SD (range) of duplicate experiments. Lines were fit to hyperbolic plots using non-linear regression (GraphPad Prism 5.0c).
Table 1.
Steady-state Kinetic Parameters for Conjugation of Three DEB Stereoisomers with GSH by GSTs
| S,S-DEB | R,R-DEB | meso-DEB | |||||||
|---|---|---|---|---|---|---|---|---|---|
| GST | kcat (min−1) | Km (mM) | kcat/Km (mM−1 min−1) | kcat (min−1) | Km (mM) | kcat/Km (mM−1 min−1) | kcat (min−1) | Km (mM) | kcat/Km (mM−1min−1) |
| 5-5 | 3.83 ± 0.50 | 0.77 ± 0.02 | 5.0 ± 0.8 | 3.44 ± 0.34 | 0.80 ± 0.06 | 4.3 ± 0.1 | 1.85 ± 0.03 | 0.83 ± 0.15 | 2.3 ± 0.4 |
| T1-1 | 3.71 ± 0.13 | 0.79 ± 0.03 | 4.7 ± 0.4 | 3.70 ± 0.17 | 0.93 ± 0.01 | 4.0 ± 0.2 | 2.42 ± 0.12 | 0.37 ± 0.01 | 6.5 ± 0.3 |
| A1-1 | 1.37 ± 0.02 | 0.30 ± 0.05 | 4.7 ± 0.7 | 1.31 ± 0.22 | 0.32 ± 0.01 | 4.1 ± 0.1 | 2.05 ± 0.04 | 0.54 ± 0.03 | 3.8 ± 0.1 |
| A3-3 | 1.52 ± 0.16 | 0.48 ± 0.33 | 4.0 ± 2.4 | 1.68 ± 0.05 | 0.51 ± 0.08 | 3.3 ± 0.4 | 1.81 ± 0.19 | 0.75 ± 0.12 | 2.4 ± 0.1 |
| M1-1 | 50.4 ± 0.96 | 0.73 ± 0.06 | 7.0 ± 1.9 | 4.44 ± 0.39 | 0.81 ± 0.05 | 5.5 ± 0.1 | 2.98 ± 0.35 | 0.92 ± 0.33 | 3.4 ± 0.8 |
| P1-1 | 2.64 ± 0.18 | 0.74 ± 0.33 | 3.9 ± 1.5 | 2.65 ± 0.01 | 0.91 ± 0.01 | 2.9 ± 0.1 | 1.59 ± 0.03 | 0.59 ± 0.15 | 2.8 ± 0.7 |
Mutagenicity of GSH Conjugates of S,S-, R,R-, and meso-DEB
The three possible DEB stereoisomer-GSH conjugates were synthesized enzymatically by the reaction of S,S-, R,R-, and meso-DEB with GSH and purified by HPLC, and mutagenicity assays of GSH conjugates of S,S-, R,R-, and meso-DEB were performed in S. typhimurium TA 1535 (Figure 2). The bacteria showed the greatest revertant response with S,S-DEB-GSH conjugate (7–802 revertants per plate), followed by R,R-DEB-GSH conjugate (7–534 revertants per plate) and then meso-DEB-GSH conjugate (8–530 revertants per plate), although the difference in the number of revertants per plate was less than 2-fold. The toxicity of these epoxides was not appreciable at these concentrations (Supporting Information Figure S3). None of the epoxides was mutagenic in the frameshift tester strain S. typhimurium TA1537 (Supporting Information Table S2), which is consistent with the lack of S. typhimurium TA1537 frameshift mutations reported previously for enzymatically-activated butadiene.39,40
Figure 2.
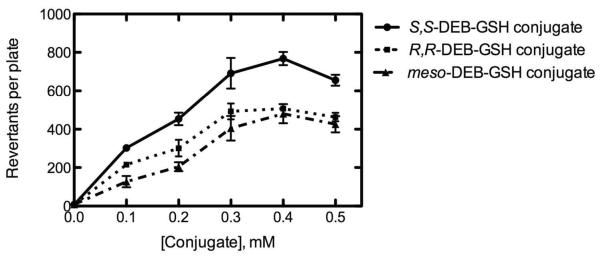
Mutagenicity of GSH conjugates of S,S-, R,R-, and meso-DEB in S. typhimurium TA1535. Each point represents the mean ± SD of duplicate experiments. Lines were fit to hyperbolic plots using non-linear regression (GraphPad Prism 5.0c).
Quantitation of DNA Adducts in Calf Thymus DNA
Six major DNA adducts were identified in the reaction of the DEB-GSH conjugate with nucleosides followed by neutral thermal hydrolysis (Scheme 1) using LC-MS (Supporting Information Figure S4, S5) and UV (Supporting Information Figure S6) and NMR spectroscopy (Supporting Information Table S3) (after purification by HPLC) (Supporting Information Figure S7). LC-MS/MS methods were developed to quantify DNA adducts formed from the DEB-GSH conjugate. In this approach, DNA is spiked with N6dA-(OH)2butyl-[glycine-13C2 15N]-GSH and N7G-(OH)2butyl-[glycine-13C2, 15N]-GSH as ISTD. The calibration curves showed good linearity in the range of 0.5–200 μM of DNA adducts, with r2 > 0.99 for all of the DNA adducts (Supporting Information Figure S8). The LOD and the limit of quantitation (LOQ) of DNA adducts were 0.1–0.3 and 0.3–0.9 mM, respectively.
All six characterized DNA adducts were identified in the reaction of DEB-GSH conjugate (0.01–0.50 mM) with calf thymus DNA (Figure 3). Among the DNA adducts, N6dA-(OH)2butyl-GSH (2.3–46.2 adducts per 106 bases) and N7G-(OH)2butyl-GSH adducts (2.1–43 adducts per 106 bases) were highest, followed by N3A-(OH)2butyl-GSH (0.8–21 adducts per 106 bases), and N3dT-(OH)2butyl-GSH (0.1–8.3 adducts per 106 bases), N4dC-(OH)2butyl-GSH (0.1–4.2 adducts per 106 bases), and N1dG-(OH)2butyl-GSH (0.1–3.0 adducts per 106 bases).
Figure 3.
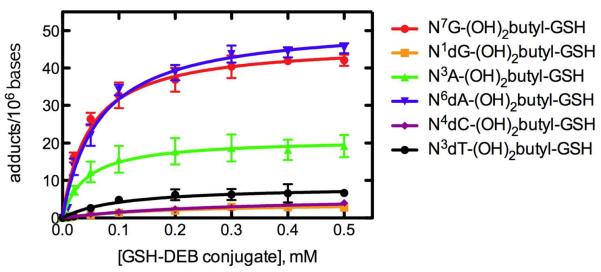
Quantitative analysis of DNA adducts in calf thymus DNA treated with varying concentrations of DEB-GSH conjugate. Each point represents the mean ± SD (range = 0.10–46.21 adducts/106 bases) of duplicate experiments. Lines were fit to hyperbolic plots using non-linear regression (GraphPad Prism 5.0c).
Quantitation of DNA Adducts in Vivo
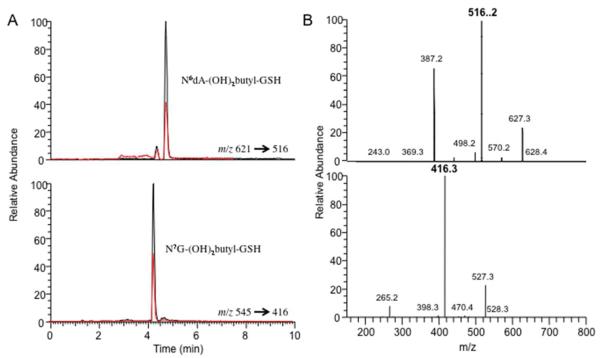
Rats and mice were treated with DEB and the six known GSH-DEB DNA adducts were determined in tissues (liver, kidney, and lung) by LC-MS/MS (Figure 4, Table 2). 18O-N7G-(OH)3butane was used as ISTD for quantitation of the “direct” DEB DNA adducts (non-GSH) N7G-DEB, N3A-DEB, and N6A-DEB, which were synthesized as described previously (Supporting Information Figure S9).11
Figure 4.
HPLC-ESI+-MS/MS chromatograms (A) and MS/MS spectra (B) for N6dA-(OH)2butyl-GSH and N7G-(OH)2butyl-GSH adducts in mouse liver (red line) and standards (black line).
Table 2.
Quantitative Analysis of DNA Adducts in Liver DNA of Sprague-Dawley Rats and B6C3F1 Mice at 6 and 48 h after DEB Treatment (25 mg/kg, i.p.)
| adducts per 107 bases | Rats | Mice | ||||
|---|---|---|---|---|---|---|
| controla (n = 2) | after 6 h (n = 4) | after 48 h (n = 4) | controla (n = 2) | after 6 h (n = 3) | after 48 h (n = 4) | |
| N3A-(OH)2butyl-GSH | ND | ND | ND | ND | ND | ND |
| N6dA-(OH)2butyl-GSH | ND | 0.19 ± 0.04 | 0.15 ± 0.05 | ND | 0.21 ± 0.03 | 0.17 ± 0.05 |
| N7G-(OH)2butyl-GSH | ND | 0.24 ± 0.05 | 0.16 ± 0.05 | ND | 0.27 ± 0.06 | 0.16 ± 0.04 |
| N1dG-(OH)2butyl-GSH | ND | ND | ND | ND | ND | ND |
| N4dC-(OH)2butyl-GSH | ND | ND | ND | ND | ND | ND |
| N3dT-(OH)2butyl-GSH | ND | ND | ND | ND | ND | ND |
| N7G-DEB | NDb | ND | ND | ND | ND | ND |
| N3A-DEB | ND | ND | ND | ND | ND | ND |
| N6A-DEB | ND | ND | ND | ND | ND | ND |
Controls: not treatment with DEB.
ND, not detected (limit of detection 0.03 adducts/107 bases).
DNA adducts were not detected in kidney or lung tissues of the rodents (< LOD, 0.03 adducts/107 bases). The two previously identified adducts N6dA-(OH)2butyl-GSH (0.10–0.24 adducts per 107 bases) and N7G-(OH)2butyl-GSH (0.10–0.33 adducts per 107 bases) were identified in liver tissues of both rats and mice treated with DEB (Table 2). The amounts of N6dA-(OH)2butyl-GSH and N7G-(OH)2butyl-GSH adducts found in rats and mice were similar and were slightly decreased after 48 h.
DISCUSSION
Six DNA adducts were formed from the reaction of the GSH conjugate with DEB with calf thymus DNA and rigorously characterized. Among these adducts, N6dA-(OH)2butyl-GSH and N7G-(OH)2butyl-GSH adducts were identified and quantitated in vivo in livers of mice and rats treated with DEB (Figure 4, Table 2). Because the mutagenicity of the GSH-DEB conjugate is considerably greater than that of DEB, the results must be considered in the context of the role of these conjugates in butadiene carcinogenicity in animals and humans.
The high mutagenic activity of DEB is likely a result of its bifunctional nature, based on comparison with the mono-epoxide,20 DEB forms DNA-DNA and DNA-protein cross-links.21,22 DNA-protein cross-links can be deleterious to cells because they are bulky, helix-distorting lesions that block the binding and progression of protein complexes and interfere with normal DNA metabolism.23,24 DNA-protein cross-links induced by DEB were first reported in liver tissue of B6C3F1 mice exposure to butadiene.41 Recently, cross-linking studies induced by DEB with purified recombinant proteins have been reported in vitro including O6-alkylguanine DNA alkyltransferase (AGT),42 glyceradehyde 3-phosphate dehydrogenase (GAPDH),43 and histone H3.44 Although GAPDH and histone H3 with DEB form DNA-protein cross-links, the adducts are not mutagenic.43,44 We previously reported a dramatic enhancement of base pair mutagenicity of DEB following GST expression in Salmonella typhimurium TA1535,30,31 and subsequently a synthetic DEB-GSH conjugate was formed to be considerably more mutagenic than DEB or several other butadiene-derived epoxides in S. typhimurium TA1535.32
In order to understand the stereochemical aspects of GSH conjugate of DEB by GSTs and GST selectivity, steady-state kinetic parameters were measured for the conjugation of the three DEB stereoisomers with GSH by GSTs (Figure 1, Table 1) and the mutagenicity of the three DEB stereoisomer-GSH conjugates was determined (Figure 2). Relatively small differences in mutagenicity (and catalytic efficiency of formation) of the three DEB stereoisomer-GSH conjugates were found, in agreement with the mutagenesis and reactivity of DEB stereoisomers that have been reported.27,45 These results indicate that stereochemistry is a minor issue in GSH-mediated DEB mutagenicity. Also, kcat/Km values differed depending on the GST although the differences were not major (S,S- and R,R-DEB, < 2-fold; meso-DEB, < 3-fold) (Table 1). These results may be relevant to the mutagenicity of DEB in different organs, although tissue-selective expression of specific GSTs is not the considered to be a major issue.
DEB preferentially reacts at the N7 atom of guanine to yield 2-hydroxyl-3,4-epoxybutane adduct, which can either be hydrolyzed to a 2,3,4-trihydroxybutane adduct or, less frequently, form cross-links with other nucleophiles.11 Identification and quantitation of DEB adducts have been reported for reactions with calf thymus DNA:11,12,45,46 N7G-DEB (5.9 adducts per 102 bases),11 N3A-DEB (1.3 adducts per 102 bases),12 and N6A-DEB (5.0 adducts per 103 bases)12 (as mono-adducts were determined in calf thymus DNA treated with DEB, 0.3 M for guanine adduct and 0.23 M for adenine adducts) and bis-(guan-7-yl)-2,3-butanediol as the most abundant DNA-DNA cross link adduct by DEB.45 Four adenine-guanine cross-linked adducts—(1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol, 1-(guan-7-yl)-4-(aden-3-yl)-2,3-butanediol, 1-(guan-7-yl)-4-(aden-7-yl)-2,3-butanediol, and 1-(guan-7-yl)-4-(aden-6-yl)-2,3-butanediol)46—were also quantified in calf thymus DNA treated with DEB (0.01–0.50 mM for guanine-guanine cross-linked adducts and 0–1.0 mM for the adenine-guanine cross-linked adducts). The bis-(guan-7-yl)-2,3-butanediol adduct was found in the range of 0.001 to 3.0 adducts per 104 bases under the specified conditions, and four adenine-guanine cross-links adducts were ~10 times less abundant than bis-(guan-7-yl)-2,3-butanediol. In this study, we identified six DNA adducts in calf thymus DNA treated with the DEB-GSH conjugate (0.01–0.50 mM): N3A-(OH)2butyl-GSH, N6dA-(OH)2butyl-GSH, N7A-(OH)2butyl-GSH, N1dG-(OH)2butyl-GSH, N4 dC-(OH)2butyl-GSH, and N3dT-(OH)2butyl-GSH adducts (Figure 3). N6dA-(OH)2butyl-GSH (0.23–4.6 adducts per 105 bases) and N7G-(OH)2butyl-GSH adducts (0.21–4.3 adducts per 105 bases) were found at higher levels than other GSH-DNA cross-linked adducts. Although N6dA-(OH)2butyl-GSH and N7G-(OH)2butyl-GSH adducts were less abundant than the bis-(guan-7-yl)-2,3-butanediol adduct following reaction with high concentrations (> 0.1 mM) of DEB or DEB-GSH conjugate, they were similar to the level of the bis-(guan-7-yl)-2,3-butanediol adduct following reaction with a low concentration (< 0.1 mM) of DEB or DEB-GSH conjugate (Figure 3). Formation of GSH-DNA cross-linked adducts was rapidly saturated, with regard to the concentration of GSH-DEB conjugate (Figure 3), while DNA-DNA cross-links adducts were reported to increase linearly depending on increase of concentration of DEB.45,46
In in vivo experiments, N6dA-(OH)2butyl-GSH (0.10–0.24 adducts per 107 bases) and N7G-(OH)2butyl-GSH adducts (0.10–0.33 adducts per 107 bases) were identified and quantitated in livers of Sparague-Dawley rats and B6C3F1 mice (Figure 4, Table 2). Several DNA-DNA cross-links adducts by DEB—meso bis-(guan-7-yl)-2,3-butanediol (0.62 ± 0.06 adducts per 107 bases), racemic bis-(guan-7-yl)-2,3-butanediol (3.95 ± 0.89 adducts per 107 bases), 1-(guan-7-yl)-4-(aden-1-yl)-2,3-butanediol (0.27 ± 0.01 adducts per 107 bases), and N6-(hydroxymethylhydroxypropan-1,3-diyl)-deoxyadenosine (0.04 ± 0.01 adducts per 107 bases)—were previously quantified in liver tissues of mice exposed by inhalation to 625 ppm butadiene for 2 weeks.47,48 The levels of these adducts in livers of rats exposed to butadiene were significantly less than that in mice because of differences in butadiene metabolism in mice and rats.22,46 However, we measured similar amounts of N6dA-(OH)2butyl-GSH (mice, 0.17–0.24 adducts per 107 bases; rats, 0.10–0.22 adducts per 107 bases) and N7G-(OH)2butyl-GSH (mice, 0.11–0.33 adducts per 107 bases; rats, 0.10–0.28 adducts per 107 bases) adducts in mice and rats, presumably due to direct administration of DEB and the similarity of GSH conjugation rates of DEB in livers of mice and rats.49
In the present study, we characterized the chemical cross-linking between GSH and DNA induced by DEB. In vitro studies demonstrate a lack of major effects on GSH conjugation and mutagenicity due to DEB stereochemistry or GST selectivity. Six DNA adducts derived from the DEB-GSH conjugate were characterized, and two of these were detected at levels of ~1/107 bases in livers DNA in mice and rats treated (i.p.) with DEB. We conclude that i) the formation of the GSH-conjugates (by GSTs) and their mutagenesis are not influenced considerably by stereochemistry, ii) at least six DNA are formed (in vitro), and iii) two of these DNA adducts can be found in livers of mice and rats administered DEB. This level of biology goes beyond that in the literature for most other DNA adducts reported in the literature. Further studies on the mechanism of base pair mutagenicity and the in vivo relevance of GSH conjugation of DEB are under investigation.
Supplementary Material
Acknowledgments
Funding Support. This work was supported in part by United States Public Health Service Grants R01 ES010546 and P30 ES00267.
Abbreviations
- AGT
O6-alkylguanine DNA alkyltransferase
- COSY
correlated (NMR) spectroscopy
- DEB
1,2,3,4-diepoxybutane
- DEB-GSH conjugate
S-(2-hydroxy-3,4-epoxybutyl)GSH
- ESI
electrospray ionization
- GAPDH
glyceradehyde 3-phosphate dehydrogenase
- GST
GSH S-transferase
- N3A-(OH)2butyl-GSH
S-[4-(N3-adenyl)2,3-dihydroxybutyl)GSH
- N6dA-(OH)2butyl-GSH
S-[4-(N6-deoxyadenosinyl)2,3-dihydroxybutyl)GSH
- N7G-(OH)2butyl-GSH
S-[4-(N7-guanyl)2,3-dihydroxybutyl)GSH
- N1dG-(OH)2butyl-GSH
S-[4-(N1-deoxyguanosinyl)2,3-dihydroxybutyl)GSH
- N4dC-(OH)2butyl-GSH
S-[4-(N4-deoxycytidinyl)2,3-dihydroxybutyl)GSH
- N3dT-(OH)2butyl-GSH
S-[4-(N3-tymidinyl)2,3-dihydroxybutyl)GSH
- N7G-DEB
N7-(2,3,4-trihydroxybutyl)guanine; N3A-DEB, N3-(2,3,4-trihydroxybutyl)adenine
- N6A-DEB
N6-(2,3,4-trihydroxybutyl)adenine
Footnotes
Supporting Information Available: NMR characterization of S,S-, R,R- and meso-DEB, calibration curve for DEB-GSH-conjugate, time dependence of conjugation of DEB with GSH by various GSTs, lack of toxicity of butadiene-derived epoxides in S. typhimurium TA1535 and lack of mutagenicity in S. typhimurium TA1537, LC-MS/MS characterization of six DNA adducts by DEB-GSH conjugate, UV absorption spectra of six DNA adducts by DEB-GSH conjugate, NMR characterization of six DNA adducts by DEB-GSH conjugate, HPLC separation of DNA adducts by DEB-GSH conjugate, calibration curves of DNA adducts by DEB-GSH conjugate, and synthesis of [18O]-N7G-(OH)3butane and LC-MS/MS characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Pelz N, Dempster NM, Shore PR. Analysis of low molecular weight hydrocarbons including 1,3-butadiene in engine exhaust gases using an aluminum oxide porous-layer open-tubular fused-silica column. J. Chromatogr. Sci. 1990;28:230–235. doi: 10.1093/chromsci/28.5.230. [DOI] [PubMed] [Google Scholar]
- (2).Morrow NL. The industrial production and use of 1,3-butadiene. Environ. Health Perspect. 1990;86:7–8. doi: 10.1289/ehp.90867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Owen PE, Glaister JR, Gaunt IF, Pullinger DH. Inhalation toxicity studies with 1,3-butadiene. 3. Two year toxicity/carcinogenicity study in rats. Am. Ind. Hyg. Assoc. J. 1987;48:407–413. doi: 10.1080/15298668791384959. [DOI] [PubMed] [Google Scholar]
- (4).Melnick RL, Huff JE. 1,3-Butadiene induces cancer in experimental animals at all concentrations from 6.25 to 8000 parts per million. IARC Sci. Publ. 1993:309–322. [PubMed] [Google Scholar]
- (5).Henderson RF, Thornton-Manning JR, Bechtold WE, Dahl AR. Metabolism of 1,3-butadiene: species differences. Toxicology. 1996;113:17–22. doi: 10.1016/0300-483x(96)03422-1. [DOI] [PubMed] [Google Scholar]
- (6).Rice JM, Boffetta P. 1,3-Butadiene, isoprene and chloroprene: reviews by the IARC monographs programme, outstanding issues, and research priorities in epidemiology. Chem.-Biol. Interact. 2001;135–136:11–26. doi: 10.1016/s0009-2797(01)00175-2. [DOI] [PubMed] [Google Scholar]
- (7).Himmelstein MW, Acquavella JF, Recio L, Medinsky MA, Bond JA. Toxicology and epidemiology of 1,3-butadiene. Crit. Rev. Toxicol. 1997;27:1–108. doi: 10.3109/10408449709037482. [DOI] [PubMed] [Google Scholar]
- (8).Malvoisin E, Roberfroid M. Hepatic microsomal metabolism of 1,3-butadiene. Xenobiotica. 1982;12:137–144. doi: 10.3109/00498258209046787. [DOI] [PubMed] [Google Scholar]
- (9).Himmelstein MW, Turner MJ, Asgharian B, Bond JA. Metabolism of 1,3-butadiene: inhalation pharmacokinetics and tissue dosimetry of butadiene epoxides in rats and mice. Toxicology. 1996;113:306–309. doi: 10.1016/0300-483x(96)03462-2. [DOI] [PubMed] [Google Scholar]
- (10).Zhang XY, Elfarra AA. Characterization of 1,2,3,4-diepoxybutane-2'-deoxyguanosine cross-linking products formed at physiological and nonphysiological conditions. Chem. Res. Toxicol. 2006;19:547–555. doi: 10.1021/tx0503395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tretyakova N, Sangaiah R, Yen TY, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N-7-guanine adducts of diepoxybutane. Chem. Res. Toxicol. 1997;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- (12).Tretyakova N, Sangaiah R, Yen TY, Gold A, Swenberg JA. Adenine adducts with diepoxybutane: isolation and analysis in exposed calf thymus DNA. Chem. Res. Toxicol. 1997;10:1171–1179. doi: 10.1021/tx9700681. [DOI] [PubMed] [Google Scholar]
- (13).Seneviratne U, Antsypovich S, Goggin M, Dorr DQ, Guza R, Moser A, Thompson C, York DM, Tretyakova N. Exocyclic deoxyadenosine adducts of 1,2,3,4-diepoxybutane: synthesis, structural elucidation, and mechanistic studies. Chem. Res. Toxicol. 2010;23:118–133. doi: 10.1021/tx900312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Selzer RR, Elfarra AA. Characterization of four N-3-thymidine adducts formed in vitro by the reaction of thymidine and butadiene monoxide. Carcinogenesis. 1997;18:1993–1998. doi: 10.1093/carcin/18.10.1993. [DOI] [PubMed] [Google Scholar]
- (15).Selzer RR, Elfarra AA. Characterization of N1- and N6-adenosine adducts and N1-inosine adducts formed by the reaction of butadiene monoxide with adenosine: evidence for the N1-adenosine adducts as major initial products. Chem. Res. Toxicol. 1996;9:875–881. doi: 10.1021/tx960039a. [DOI] [PubMed] [Google Scholar]
- (16).Fernandes PH, Hackfeld LC, Kozekov ID, Hodge RP, Lloyd RS. Synthesis and mutagenesis of the butadiene-derived N3 2'-deoxyuridine adducts. Chem. Res. Toxicol. 2006;19:968–976. doi: 10.1021/tx060016o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Citti L, Gervasi PG, Turchi G, Bellucci G, Bianchini R. The reaction of 3,4-epoxy-1-butene with deoxyguanosine and DNA in vitro: synthesis and characterization of the main adducts. Carcinogenesis. 1984;5:47–52. doi: 10.1093/carcin/5.1.47. [DOI] [PubMed] [Google Scholar]
- (18).Tice RR, Boucher R, Luke CA, Shelby MD. Comparative cytogenetic analysis of bone marrow damage induced in male B6C3F1 mice by multiple exposures to gaseous 1,3-butadiene. Environ. Mutagen. 1987;9:235–250. doi: 10.1002/em.2860090303. [DOI] [PubMed] [Google Scholar]
- (19).Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: II. Mutational spectra of butadiene, 1,2-epoxybutene and diepoxybutane at the hprt locus in splenic T cells from exposed B6C3F1 mice. Carcinogenesis. 1994;15:719–723. doi: 10.1093/carcin/15.4.719. [DOI] [PubMed] [Google Scholar]
- (20).Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2-butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- (21).Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J. Proteome Res. 2010;9:4356–4367. doi: 10.1021/pr1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Goggin M, Swenberg JA, Walker VE, Tretyakova N. Molecular dosimetry of 1,2,3,4-diepoxybutane-induced DNA-DNA cross-links in B6C3F1 mice and F344 rats exposed to 1,3-butadiene by inhalation. Cancer Res. 2009;69:2479–2486. doi: 10.1158/0008-5472.CAN-08-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- (24).Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- (25).Krause RJ, Elfarra AA. Oxidation of butadiene monoxide to meso- and (+/−)-diepoxybutane by cDNA-expressed human cytochrome P450s and by mouse, rat, and human liver microsomes: evidence for preferential hydration of meso-diepoxybutane in rat and human liver microsomes. Arch. Biochem. Biophys. 1997;337:176–184. doi: 10.1006/abbi.1996.9781. [DOI] [PubMed] [Google Scholar]
- (26).Nieusma JL, Claffey DJ, Ruth JA, Ross D. Stereochemical aspects of the conjugation of epoxide metabolites of butadiene with glutathione in rat liver cytosol and freshly isolated rat hepatocytes. Toxicol. Sci. 1998;43:102–109. doi: 10.1006/toxs.1998.2461. [DOI] [PubMed] [Google Scholar]
- (27).Kim MY, Tretyakova N, Wogan GN. Mutagenesis of the supF gene by stereoisomers of 1,2,3,4-diepoxybutane. Chem. Res. Toxicol. 2007;20:790–797. doi: 10.1021/tx700003b. [DOI] [PubMed] [Google Scholar]
- (28).Bianchi A, Contin M. Mutagenic activity of isomeric forms of diepoxybutane in maize. J. Hered. 1962;53:277–281. [Google Scholar]
- (29).Meister A, Anderson ME. Glutathione. Annu. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- (30).Thier R, Muller M, Taylor JB, Pemble SE, Ketterer B, Guengerich FP. Enhancement of bacterial mutagenicity of bifunctional alkylating agents by expression of mammalian glutathione S-transferase. Chem. Res. Toxicol. 1995;8:465–472. doi: 10.1021/tx00045a019. [DOI] [PubMed] [Google Scholar]
- (31).Thier R, Pemble SE, Kramer H, Taylor JB, Guengerich FP, Ketterer B. Human glutathione S-transferase T1-1 enhances mutagenicity of 1,2-dibromoethane, dibromomethane and 1,2,3,4-diepoxybutane in Salmonella typhimurium. Carcinogenesis. 1996;17:163–166. doi: 10.1093/carcin/17.1.163. [DOI] [PubMed] [Google Scholar]
- (32).Cho SH, Loecken EM, Guengerich FP. Mutagenicity of a glutathione conjugate of butadiene diepoxide. Chem. Res. Toxicol. 2010;23:1544–1546. doi: 10.1021/tx100304f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Robbins MA, Devine PN, Oh T. Synthesis of chiral non-racemic diols from (S,S)-1,2,3,4-diepoxybutane: (2S,3S)-dihydroxy-1,4-diphenylbutane (2,2'-bioxirane, [S-(R,R)]- and 2,3-butanediol, 1,4-diphenyl-, [S-(R,R)]-) Org. Synth. 1999;76:101–109. [Google Scholar]
- (34).Mash EA, Nelson KA, Van Deusen S, Hemperly SB. 1,4-Di-O-alkyl threitols from tartaric acid: 1,4-di-O-benzyl-L-threitol. Org. Synth. 1990;68:92–103. [Google Scholar]
- (35).Claffey DJ. Multigram-scale stereoselective synthesis of meso-1,3-butadiene bisepoxide. Synth. Commun. 2002;32:3041–3045. [Google Scholar]
- (36).Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat. Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- (37).Hecht SM, Kirkegaard LH, Bock RM. Chemical modifications of transfer RNA species. Desulfurization with Raney nickel. Proc. Natl. Acad. Sci. U S A. 1971;68:48–51. doi: 10.1073/pnas.68.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kim DH, Guengerich FP. Formation of the DNA adduct S-[2-(N7-guanyl)ethyl]glutathione from ethylene dibromide: effects of modulation of glutathione and glutathione S-transferase levels and lack of a role for sulfation. Carcinogenesis. 1990;11:419–424. doi: 10.1093/carcin/11.3.419. [DOI] [PubMed] [Google Scholar]
- (39).Arce GT, Vincent DR, Cunningham MJ, Choy WN, Sarrif AM. In vitro and in vivo genotoxicity of 1,3-butadiene and metabolites. Environ. Health Perspect. 1990;86:75–78. doi: 10.1289/ehp.908675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Araki A, Noguchi T, Kato F, Matsushima T. Improved method for mutagenicity testing of gaseous compounds by using a gas sampling bag. Mut. Res. 1994;307:335–344. doi: 10.1016/0027-5107(94)90307-7. [DOI] [PubMed] [Google Scholar]
- (41).Jelitto B, Vangala RR, Laib RJ. Species differences in DNA damage by butadiene: role of diepoxybutane. Arch. Toxicol. Suppl. 1989;13:246–249. doi: 10.1007/978-3-642-74117-3_42. [DOI] [PubMed] [Google Scholar]
- (42).Loeber R, Rajesh M, Fang Q, Pegg AE, Tretyakova N. Cross-linking of the human DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of 1,2,3,4-diepoxybutane. Chem. Res. Toxicol. 2006;19:645–654. doi: 10.1021/tx0600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Loecken EM, Guengerich FP. Reactions of glyceraldehyde 3-phosphate dehydrogenase sulfhydryl groups with bis-electrophiles produce DNA-protein cross-links but not mutations. Chem. Res. Toxicol. 2008;21:453–458. doi: 10.1021/tx7003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Loecken EM, Dasari S, Hill S, Tabb DL, Guengerich FP. The bis-electrophile diepoxybutane cross-links DNA to human histones but does not result in enhanced mutagenesis in recombinant systems. Chem. Res. Toxicol. 2009;22:1069–1076. doi: 10.1021/tx900037u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Park S, Anderson C, Loeber R, Seetharaman M, Jones R, Tretyakova N. Interstrand and intrastrand DNA-DNA cross-linking by 1,2,3,4-diepoxybutane: role of stereochemistry. J. Am. Chem. Soc. 2005;127:14355–14365. doi: 10.1021/ja051979x. [DOI] [PubMed] [Google Scholar]
- (46).Park S, Hodge J, Anderson C, Tretyakova N. Guanine-adenine DNA cross-linking by 1,2,3,4-diepoxybutane: potential basis for biological activity. Chem. Res. Toxicol. 2004;17:1638–1651. doi: 10.1021/tx0498206. [DOI] [PubMed] [Google Scholar]
- (47).Goggin M, Seneviratne U, Swenberg JA, Walker VE, Tretyakova N. Column switching HPLC-ESI(+)-MS/MS methods for quantitative analysis of exocyclic dA adducts in the DNA of laboratory animals exposed to 1,3-butadiene. Chem. Res. Toxicol. 2010;23:808–812. doi: 10.1021/tx900439w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Goggin M, Sangaraju D, Walker VE, Wickliffe J, Swenberg JA, Tretyakova N. Persistence and repair of bifunctional DNA adducts in tissues of laboratory animals exposed to 1,3-butadiene by inhalation. Chem. Res. Toxicol. 2011;24:809–817. doi: 10.1021/tx200009b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Boogaard PJ, Sumner SC, Bond JA. Glutathione conjugation of 1,2:3,4-diepoxybutane in human liver and rat and mouse liver and lung in vitro. Toxicol. Appl. Pharmacol. 1996;136:307–316. doi: 10.1006/taap.1996.0037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.