Abstract
Seventy-seven patients with aniridia, referred for cytogenetic analysis predominantly to assess Wilms tumor risk, were studied by fluorescence in situ hybridization (FISH), through use of a panel of cosmids encompassing the aniridia-associated PAX6 gene, the Wilms tumor predisposition gene WT1, and flanking markers, in distal chromosome 11p13. Thirty patients were found to be chromosomally abnormal. Cytogenetically visible interstitial deletions involving 11p13 were found in 13 patients, 11 of which included WT1. A further 13 patients had cryptic deletions detectable only by FISH, 3 of which included WT1. Six of these, with deletions <500 kb, share a similar proximal breakpoint within a cosmid containing the last 10 exons of PAX6 and part of the neighboring gene, ELP4. Two of these six patients were mosaic for the deletion. The remaining four had chromosomal rearrangements: an unbalanced translocation, t(11;13), with a deletion including the WAGR (Wilms' tumor, aniridia, genitourinary abnormalities, and mental retardation) region, and three balanced rearrangements with what appear to be position effect breakpoints 3′ of PAX6: (a) a t(7;11) with the 11p13 breakpoint ∼30 kb downstream of PAX6, (b) a dir ins(12;11) with a breakpoint >50 kb from PAX6, and (c) an inv(11)(p13q13) with a breakpoint >75 kb downstream of PAX6. The proportion and spectrum of chromosome anomalies in familial (4/14, or 28.5%) and sporadic (26/63, or 41%) cases are not significantly different. An unexpectedly high frequency of chromosomal rearrangements is associated with both sporadic and familial aniridia in this cohort.
Introduction
Aniridia (MIM 106200) is a rare developmental defect affecting the anterior segment of the eye, observed in ∼1 of 50,000–100,000 newborns (Nelson et al. 1984). Sporadic cases arise de novo but are subsequently inherited as an autosomal dominant phenotype, with virtually complete penetrance but variable expressivity. Aniridia generally occurs in isolation, but it also occurs, more rarely, as part of the WAGR (Wilms' tumor, aniridia, genitourinary abnormalities, and mental retardation [MIM 194072]) deletion syndrome. WAGR deletions are of variable size but always include part or all of chromosome band 11p13. WAGR-associated phenotypes were also found to be variable, even in patients with deletions that are apparently identical cytogenetically (Franke et al. 1979; Turleau et al. 1984). Subsequently, molecular studies were used to define the proximal and distal breakpoints of both visible and submicroscopic deletions in patients with isolated aniridia and WAGR syndrome–associated aniridia (van Heyningen et al. 1985; Davis et al. 1988; Mannens et al. 1991; Fantes et al. 1992, 1995b; Dreschler et al. 1994; Crolla et al. 1997; Kent et al. 1997).
Early analysis of such deletions led to the positional identification of PAX6 and WT1 as candidate genes for aniridia and Wilms tumor predisposition, respectively (Call et al. 1990; Gessler et al. 1990; Ton et al. 1991). WT1 and PAX6 map to a 700-kb interval in distal 11p13, which is within a fully sequenced region (GenBank accession number NT_009237). Mutations in PAX6, including deletions, have been identified in a high proportion of tested patients with either sporadic or familial aniridia (Fantes et al. 1992, 1995b; Glaser et al. 1992; Jordan et al. 1992; Hanson et al. 1993; Dreschler et al. 1994; Axton et al. 1997; Grønskov et al. 2001). Early studies showed that deletions that include the WT1 gene are correlated with a significant risk of Wilms tumor, perhaps as high as 60%. More-recent studies have attempted to quantify this empirical risk more accurately, on the basis of larger samples (Grønskov et al. 2001; Muto et al. 2002; authors' unpublished data).
A number of studies have confirmed that human aniridia is a haploinsufficiency disorder resulting from inactivation of one copy of the PAX6 gene, predominantly through intragenic mutations leading to premature termination of the protein (reviewed by Prosser and van Heyningen 1998). However, chromosomal abnormalities including deletions, both visible and cryptic (Mannens et al. 1991; Fantes et al. 1992, 1995a; Dreschler et al. 1994; Kent et al. 1997; Crolla et al. 1997; Grønskov et al. 2001; Muto et al. 2002), and inversions, translocations, and insertions (Simola et al. 1982; Moore et al. 1986; Mannens et al. 1991; Fantes et al. 1995b; Crolla et al. 1996, 1997; Llerena et al. 2000), have also been reported in association with both isolated sporadic and familial aniridia, as well as in patients with WAGR syndrome. More recently, cell hybrid studies demonstrated inactivation of two independent, aniridia-associated PAX6 alleles, each carrying an ∼1,000-kb submicroscopic deletion distal to PAX6, with the proximal breakpoints at ∼11 kb and ∼22 kb 3′ of the PAX6 polyA addition site (Lauderdale et al. 2000). Analysis of a region ∼150 kb downstream of the PAX6 transcriptional termination site revealed PAX6-specific DNAse hypersensitive sites and demonstrated a number of tissue-specific regulatory elements for PAX6 (Kleinjan et al. 2001; Griffin et al. 2002).
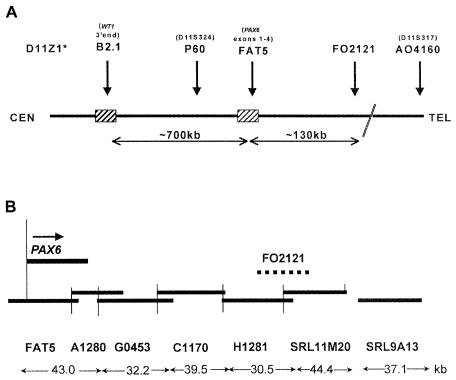
Following the early reports of cryptic 11p13 deletions, a diagnostic FISH service was set up in Salisbury in 1993, to screen patients presenting with aniridia (particularly sporadic cases) and identify those with interstitial deletions, so that the extent of the deletion and the associated empirical Wilms tumor risk could be assessed and reported clinically. Initially, the FISH analysis used only four cosmids (B2.1, P60, FAT5, and FO2121) (fig. 1A) (Crolla et al. 1997); however, after the identification of a patient with a deletion of FO2121 but not FAT5, an additional six cosmid probes contiguous with FAT5 were added to the panel (fig. 1B). A further seven flanking cosmids were used to give estimates of the overall sizes of the deletions (Fantes et al. 1995a). Patients 1–19 were initially reported by Crolla et al. (1997), and, for these patients, only the additional FISH investigations are presented here.
Figure 1.
A, Map showing the position of the four cosmids used in the original FISH deletion analysis. D11Z1 is the centromeric alphoid repeat probe used to identify the 11 homologues, and D11S317 is a cosmid A4160, which lies ∼400 kb telomeric of FO2121 (see fig. 2). B, Higher-resolution map showing the relative positions of the cosmid contig covering PAX6 and the ∼180-kb region telomeric of the gene.
Subjects and Methods
The Study Population
Seventy-seven patients, falling into four principal categories, were referred for aniridia/WAGR FISH testing (table 1):
-
1.
Sporadic isolated aniridia: 43 patients, of whom 38 (88%) were ⩽5 years old at referral (mean ± SD 13.2±13.4 mo). The mean age at referral for all 43 cases was 43.0±117 mo. Patient 94 is an MZ twin referred with bilateral isolated aniridia and optic atrophy. His twin brother is phenotypically normal.
-
2.
WAGR syndrome: 12 patients had aniridia and one or more of the other features associated with WAGR syndrome (table 2). Eight were ⩽5 years old at referral (mean 29.9±26.7 mo). The mean age at referral for all 12 patients was 96±151 mo.
-
3.
Aniridia and additional anomalies (not WAGR): Six of these eight patients had eye and/or other abnormalities in addition to aniridia (table 2). All patients were ⩽2 years old at referral (mean 10.4±9.1 mo).
-
4.
Familial aniridia, all independent pedigrees: the age at referral for the probands in the 14 patients with familial aniridia ranged from a 38-wk-gestation stillborn infant, who had inherited aniridia from the mother, to a 57-year-old man. All patients with familial aniridia were from pedigrees showing aniridia in two or more generations.
Table 1.
Patients by Referral Category and Rates of Abnormality
| Referral Category | Total No. (%) | No. (%) Abnormal |
| Isolated sporadic aniridia | 43 (56) | 16 (37) |
| WAGR | 12 (15) | 8 (66.6) |
| Aniridia plus other abnormalities (not WAGR) | 8 (10.5) | 2 (25) |
| Familial aniridia | 14 (18.5) |
4 (28.6) |
| Total | 77 | 30 (39) |
Table 2.
Clinical Features of Patients with Aniridia and Associated Anomalies
|
Clinical Features |
|||||||
| Disorder andPatient Number | Age at Referral(XX/XY Status) | W | A | G | R | Other Abnormalities | Deletion(V/C)a |
| WAGR syndrome: | |||||||
| 11b | 4 years (XY) | − | + | − | + | Nystagmus, glaucoma | − |
| 12b | 1 mo (XY) | − | + | + | − | Fallot's tetralogy, 1st and 2nd ribs absent | − |
| 14b | 2 mo (XY) | − | + | + | − | Single kidney | V |
| 16b | 5 years (XY) | + | + | + | − | C | |
| 32 | 2 mo (XY) | − | + | − | + | − | |
| 33 | 10 mo (XY) | − | + | − | + | V | |
| 34 | 7 years (XY) | − | + | + | + | Bilateral cataracts | V |
| 36 | 32 years (XY) | + | + | + | + | Nystagmus | V |
| 41 | 4 years (XX) | − | + | − | + | Immunodeficiency | V |
| 44 | 5 years (XY) | − | + | − | + | Bilateral colobomata | − |
| 47 | 37 years (XY) | − | + | + | + | V | |
| 96 | 18 mo (XX) | + | + | − | − | C | |
| Aniridia and other anomalies: | |||||||
| 13b | 4 mo (XY) | − | + | − | − | Optic atrophy | V |
| 15b | 4 mo (XX) | − | + | − | − | Facial dysmorphism, glaucoma | V |
| 29 | 2 years (XX) | − | + | − | − | Left microphthalmia, anterior segment dysgenesis | − |
| 45 | 2 years (XY) | − | + | − | − | Glaucoma, bicuspid valve, mild stenosis | − |
| 50 | 1 year (XX) | − | + | − | − | Facial dysmorphism | − |
| 68 | 10 mo (XY) | − | + | − | − | Glaucoma | − |
| 69 | 4 mo (XY) | − | + | − | − | Coloboma | − |
| 77 | 1 mo (XY) | − | + | − | − | Exomphalos, glaucoma | − |
V = visible deletion; C = cryptic deletion; − = no deletion.
Previously reported by Crolla et al. (1997).
Clinical outcomes have been collated to determine the empirical risk of developing Wilms tumor in patients with isolated aniridia who are shown by FISH to have cryptic or visible constitutional deletions that include PAX6 and WT1. The results of these studies will be presented as part of a larger study (authors' unpublished data).
Chromosome Preparation, Analysis, and Probe Labeling
Chromosome preparations were made using conventional techniques. All cases referred for FISH were examined by the referring laboratories with G banding at a minimum of 550-band resolution and were reported using the system recommended by the International System for Human Cytogenetic Nomenclature (table 3). Cytogenetic breakpoints for visible deletions and rearrangements are as reported by the referring laboratories.
Table 3.
Conventional Karyotypes of Patients
| PatientNumber | Age at Referral | Cytogenetic Result (ISCN) |
| 1 | 2 years | 46,XX,t(7;11)(q31.2;p13) |
| 2 | 1 year | 46,XY |
| 6 | 6 mo | 46,XX |
| 13 | 4 mo | 46,XY,del(11)(p13;p15.1) |
| 14 | 2 mo | 46,XY,del(11)(p13;p14.2) |
| 15 | 4 mo | 46,XX,del(11)(p11.2;p14.1) |
| 16 | 5 years | 46,XY |
| 19a | 29 years | 46,XX |
| 21 | 2 years | 46,XX,t(11;13)(p?13;q?) de novo |
| 25 | 4 mo | 46,XY |
| 26 | 3 years | 46,XY |
| 33 | 10 mo | 46,XY,del(11)(p13p14.2) |
| 34 | 7 years | 46,XY,del(11)(p13p14.3) |
| 36 | 32 years | 46,XY,del(11)(p11.2p14) |
| 41 | 4 years | 46,XX,del(11)(p12p15.1) |
| 42 | 1 years | 46,XX,del(11)(p11.2p13) |
| 47 | 37 years | 46,XY,del(11)(p13p14.3) |
| 51 | 2 years | 46,XX,del(11)(p12p14) |
| 61a | 2 years | 46,XX,inv(11)(p13q13)mat |
| 64 | 6 mo | 46,XY |
| 70a | 38 wk gestation | 46,XX |
| 73 | 4 mo | 46,XX,del(11)(p11.2p13) |
| 74 | 8 mo | 46,XX,del(11)(p13p15.1) |
| 75a | 1 years | 46,XX |
| 81 | 23 years | 46,XX |
| 83 | 5 mo | 46,XY |
| 85 | 1 mo | 46,XY,dir ins(12;11)(q24.11;p13p15.1) de novo |
| 94 | 2 mo | 46,XY |
| 95 | 3 mo | 46,XX,t(2;11)(q33;q25)del(11)(p?12p?13) de novo |
| 96 | 18 mo | 46,XX |
Patient with familial aniridia.
Whenever possible, spare material from the cytogenetic and/or FISH analyses was stored either at −20°C (cell suspensions in 3:1 methanol:acetic acid fixative) or as lymphoblastoid cell lines. The FISH technique and analyses were performed as described elsewhere (Crolla et al. 1997). In brief, cosmids were labeled by nick translation with digoxigenin and the chromosome 11 centromere probe with biotin. The probe and chromosomal target DNAs were codenatured at 75°C for 5 min and were incubated overnight at 37°C, and, after two 5-min stringent washes at 42°C in 1×SSC/50% formamide, the sites of hybridization were visualized using antibodies: one layer each of anti-digoxigenin-TRITC (red fluorescence) and avidin-FITC (green fluorescence). Images were captured using a cooled CCD camera and were digitized for analysis through use of PSI Powergene software. A minimum of 10 metaphases were examined after hybridization with each probe; for patients 2 and 94, 30 metaphases were scored.
All cases were initially studied using four cosmids (from centromere to telomere: B2.1, P60, FAT5, and FO2121; see fig 1A) (Fantes et al. 1995a). Additional studies were prompted by the results obtained for patient 64, who was not deleted for FAT5 but was hemizygous for the more telomeric probe, FO2121 (∼100 kb 3′ of PAX6). All cases previously reported as normal were subsequently reexamined using the contig of fully sequenced cosmids covering the ∼180-kb region 3′ of PAX6 (fig. 1B). The FISH reexamination with this cosmid contig was performed mostly with stored fixed cells. Three cases were examined using fresh chromosome preparations derived from stored lymphoblastoid cell lines. For patient 2, the result was confirmed using chromosome preparations derived from a fresh peripheral blood sample. In four cases, FISH reexamination was not possible because no stored material was available, and these have been excluded from the results.
Anchoring of Cosmids onto the Chromosome 11 Map
Public STSs, gene segments, or sequence data associated with each cosmid used (fig. 2) are as follows: DO8153 (rapsyn, RAPSN), c1-11-474 (D11S1103), c1-11-458 (D11S1776), B2.1 (WT1), P60 (D11S324), FAT5 (PAX6, European Molecular Biology Laboratory [EMBL] accession number Z95332), A1280 (EMBL accession number Z83307), G0453 (EMBL accession number Z83301), C1170 (EMBL accession number Z83306), H1281 (EMBL accession number Z83309), FO2121 (overlaps H1281 and SRL11M20), SRL11M20 (EMBL accession number Z83308), SRL9A13 (EMBL accession number Z86001), AO4160 (D11S317), CO8160 (D11S151, KCNA4), F1238 (D11S1446), and EO6182 (D11S1062).
Figure 2.
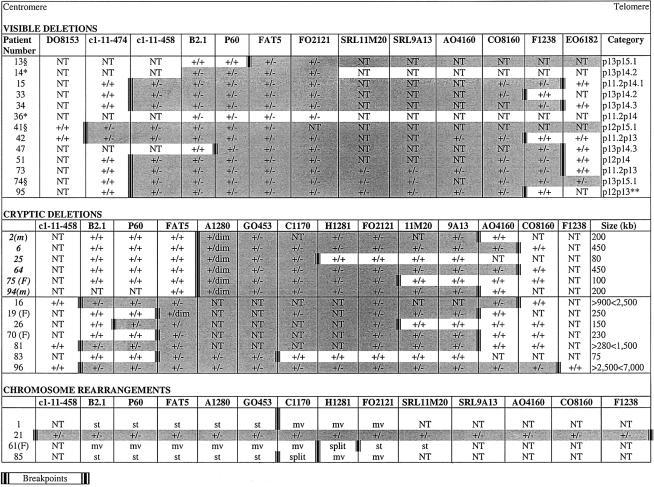
Details of the FISH results in the chromosomally abnormal patients. Patient numbers in boldface italic type are those with a proximal breakpoint in A1280. Shaded areas define the approximate size of the deletion. Size (kb) is the deletion size estimated using known mapping and sequence data. m = mosaic for deletion; F = familial; +/+ = signal on both homologues; +/− = one homologue deleted; +/dim = one homologue with diminished signal; mv = cosmid signal moved; st = cosmid signal stays on der(11); split = cosmid signal split; NT = not tested for this probe; § = cytogenetic breakpoint 11p15 (FISH on distal breakpoint not performed); * = no stored material for additional FISH; ** = also had t(2;11)(q33;q25) de novo.
The cosmids B2.1 and P60 and the PAX6 contig covering the ∼180-kb downstream region (fig. 1B) are available on request from J.A.C.
Results
The overall frequency of chromosomal abnormalities found in this cohort of patients with aniridia is summarized in tables 1 and 3 and in figure 2.
Abnormalities by Referral Category
Sporadic isolated aniridia
Of the 43 patients in this group, 16 (37%) had chromosome abnormalities that accounted for their aniridia. Thirteen of the abnormalities were interstitial deletions—eight cryptic and five cytogenetically visible. Two of the three remaining patients (patients 1 and 85) had apparently balanced chromosome rearrangements; the third was patient 21, the child with a t(11;13) with a deletion at the 11p breakpoint. All five visible deletions involved WT1, compared with only one (patient 81) of the eight cryptic deletions. Five of the six cryptic deletions involving a proximal breakpoint in cosmid A1280 were ascertained in patients with sporadic isolated aniridia.
WAGR
Eight (66.6%) of the 12 patients with WAGR (tables 2 and 3) had deletions—six visible and two cryptic. Both cryptic deletions included WT1 (in patients 16 and 96), as did five of the six visible deletions. The one visible deletion not involving WT1 (in patient 47) had a proximal breakpoint between WT1 and P60 (D11S324).
Aniridia and other abnormalities
Two (25%) of the eight patients (tables 2 and 3) were chromosomally abnormal, both with visible deletions; the deletion in one patient included WT1 (patient 15), whereas the deletion in the other patient did not include WT1, with a proximal breakpoint between P60 (D11S324) and PAX6.
Familial aniridia
Four (28.6%) of the 14 patients with familial aniridia were chromosomally abnormal; three cryptic deletions were observed (patients 19, 70, and 75), the last being one of the patients with a proximal breakpoint in cosmid A1280, and a fourth patient had a pericentric inversion of chromosome 11.
Visible Deletions
Thirteen (17%) of the patients, all with sporadic aniridia, had visible deletions, only two of which (in patients 13 and 47) did not include WT1. The size of the deletions varied (fig. 2), but the two patients with WT1 not deleted had proximal breakpoints between P60 and FAT5 (patient 13), and WT1 and P60 (patient 47). Seven of the nine deletions tested with the centromeric FISH markers had a breakpoint between c1-11-474 and c1-11-458. Four of these seven shared a similar distal breakpoint between F1238 and EO6182 (patients 15, 34, 51, and 73); two had breakpoints between CO8160 and F1238 (patients 33 and 95), and patient 74 had a more distal breakpoint beyond EO6182.
Cryptic Deletions
Thirteen (17%) of the 70 patients had cryptic 11p deletions that were visible only when FISH was used (fig. 2). Five of these were not detected in the original analysis, because the deletion did not include cosmid FAT5 (PAX6 exons 1–4). These deletions were subsequently identified using the ∼180-kb cosmid contig starting with A1280. Three of the 13 cryptic deletions included WT1 (patients 16, 81, and 96). Patient 16 presented with Wilms tumor at age 17 mo, and patient 96 received a diagnosis of Wilms tumor at age 18 mo. Patient 81 was referred at age 23 years and, apart from visual problems associated with her aniridia, she was reported to be well and intellectually normal.
The extent of cryptic deletions was estimated using information from chromosome 11 mapping (Fantes et al. 1995a) and sequencing around the PAX6 region (Ensembl Genome Browser). Eleven of the 13 cryptic deletions ranged in size from ∼75 to <1,500 kb (fig. 2). In patients 16 and 96, the deletion is deduced to cover 900–2,500 kb and 2,500–7,000 kb, respectively. Subsequent conventional reanalysis of these two cases showed that the deletions were not visible through use of light microscopy.
The cryptic deletions can be further subdivided into two groups:
-
1.
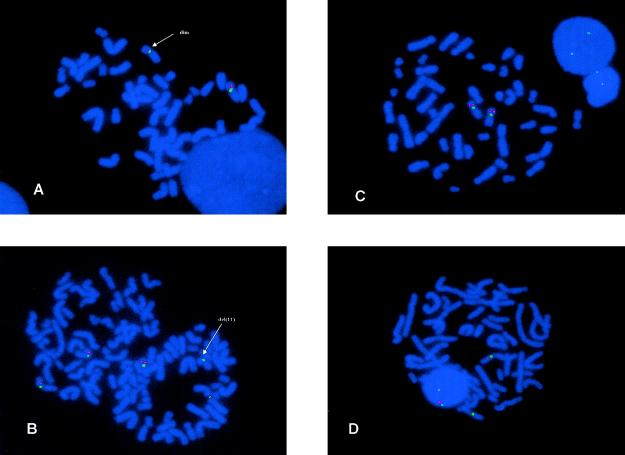
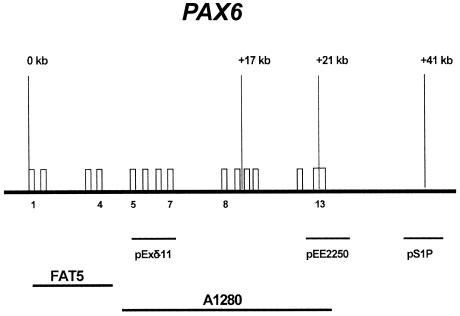
Six patients had an apparently recurrent common proximal breakpoint in cosmid A1280, resulting in a diminished signal on one chromosome 11 homologue (fig. 3A). These deletions ranged in size from ∼80 kb (patient 25) to ∼450 kb (patients 6 and 64). In addition, two unrelated patients, 2 and 94, were mosaics for the deletion, both of which were ∼200 kb in size and present in ∼50% (patient 2) and 60% (patient 94) of peripheral blood metaphases (fig. 3B). The diminished A1280 signal suggested that the proximal breakpoint had occurred either within exons 5–13 of PAX6, including the ∼1 kb 3′ UTR (PAX6 3′ UTR), or immediately distal to this region. In an attempt to resolve this, FISH with three plasmid probes from the 3′ end of the gene and the 3′ UTR (fig. 4), was performed on five of the six patients. Use of the plasmids individually did not yield FISH signals, but use of all three as pooled FISH probes gave signal on both 11p13 regions in approximately half of the metaphases examined (fig. 3C and 3D). Binding of the plasmid pool suggests that the telomeric region of PAX6 is still present, but higher-resolution techniques are required to define these breakpoints with more precision.
Cosmid A1280 is 34,614 bp long, with its centromeric end at nucleotide 1 in the sequence given for EMBL Z83307 and the telomeric end >12 kb into the sequence for G0453 (EMBL Z83001). Exons 5–13 of PAX6 cover a little more than 15 kb at the centromeric end of A1280. The final exon (exon 10) of the ELP4 neighboring gene, transcribed on the opposite strand to PAX6, stretches over ∼400 bp at 20 kb from the centromeric end of A1280.
-
2.
Seven patients had a proximal breakpoint centromeric of PAX6. Three of these shared a proximal breakpoint between c1-11-458 and B2.1 (patients 16, 81, and 96). The proximal and distal breakpoints of the remaining four were heterogeneous (fig. 2).
Figure 3.
A, Result from use of cosmid A1280 on patient 2. The normal and abnormal homologue (indicated by the arrow) show a different size of hybridization signal, showing that only part of the genomic sequence contained in this cosmid binds to the deleted 11. The green signal represents the D11Z1 centromere probe. B, Result from use of cosmid FO2121 on metaphases from the mosaic patient 2. The metaphase on the left shows FO2121 signal on both 11 homologues, whereas the metaphase on the right has cosmid signal on only one of the 11 homologues. The green signal represents the D11Z1 centromere probe. C, FISH results in patient 64 with the contig of plasmid probes pExδ-11, pEE2250, and pS1P, showing signals on both homologues. D, FISH results in patient 64. In contrast to C, signal is only seen on the normal homologue.
Figure 4.
Map showing the position of the plasmid and cosmid probes used for FISH
Chromosomal Rearrangements
Four patients were identified with chromosomal rearrangements involving one breakpoint in distal 11p13 (fig. 2). Patient 1 has an apparently balanced t(7;11) (Crolla et al. 1996), where the 11p breakpoint apparently lies between adjacent cosmids, with the G0453 signal present on the der(11) and C1170 on the der(7), placing the breakpoint ∼30 kb 3′ of PAX6. Patient 61, with familial aniridia, has a pericentric inversion of chromosome 11 (p13q13) with the 11p breakpoint identified by a split in cosmid H1281, placing the breakpoint >75 kb downstream of PAX6. Patient 85 has an apparently balanced rearrangement defined as a dir ins(12;11) where the 11p breakpoint splits cosmid C1170 ∼50 kb 3′ of PAX6. These three chromosomal rearrangements almost certainly represent position effects leading to heterozygous loss of PAX6 function. The overall frequency of position effects in this series of patients with aniridia is 3/77 (3.8%).
The fourth chromosomal rearrangement, an apparently balanced t(11;13), was found by FISH to have an ∼7.5-Mb deletion at the breakpoint on the der(11) (Llerena et al. 2000).
Discussion
The Need for Deletion Analysis and Choice of Probes
Since the original identification of PAX6 as the aniridia candidate gene, intragenic mutations and chromosomal deletions, translocations, and inversions have been associated with a high proportion (∼80%) of aniridia cases studied in detail (Axton et al. 1997; Crolla et al. 1997; Grønskov et al. 2001). Most of the intragenic mutations lead to premature protein truncation (Hanson et al. 1993; Axton et al. 1997; Prosser and van Heyningen, 1998; Chao et al. 2000) and haploinsufficiency. FISH analysis is the most efficient way of identifying the extent of aniridia-associated deletions, both visible and cryptic. The aim was to ascertain whether patients with aniridia, particularly children younger than ∼7 years of age with de novo phenotypes, are deleted for WT1 and therefore require monitoring for increased risk of Wilms tumor (Fantes et al. 1992; Crolla 1997; Kent et al. 1997). FISH analysis, using cosmids B2.1, P60, FAT5, and FO2121, was also ideal for defining the position of chromosomal breakpoints associated with translocations and inversions leading to aniridia (Fukushima et al. 1993; Fantes et al. 1995b; Crolla et al. 1996). Once it became clear that chromosomal rearrangements with breakpoints outside the PAX6 transcription unit could also lead to classical aniridia phenotypes, the cosmid contig used for FISH studies was extended ∼180 kb downstream of the PAX6 polyA addition site (fig. 1B).
Cosmid contigs are ideal probes for use in fine deletion analysis, since the signal is strong enough to be present reliably, but the genomic insert size (∼30 kb) is small enough to detect small deletions by loss or diminution of signal. Some smaller structural changes may be detectable only through pulsed field gel or Southern blot analysis.
After the setting up of the molecular cytogenetic service in Salisbury, through use of the four-cosmid set for the identification of aniridia-associated deletions, an affected individual was identified who had an intact PAX6 cosmid (FAT5) but was deleted for the more distant FO2121 (fig. 1A). Subsequently, the extended cosmid contig (fig. 1B) was used for all new FISH studies and in a retrospective analysis of earlier samples. Subtle rearrangements were defined, with breakpoint positions identified more accurately.
Unexpectedly High Frequency of Rearrangements Seen
The spectrum of patients with aniridia sent by clinicians for FISH studies was broad: the largest category comprised patients with isolated sporadic aniridia (43); patients with syndromic aniridia, including 12 with the WAGR phenotype, accounted for 20 patients in total, and analysis was also requested for 14 patients with familial isolated aniridia (table 1). That 8/12 WAGR phenotypes were found to be associated with deletions is not surprising. In contrast, the identification of cryptic deletions in 8/44 sporadic isolated aniridia cases, (86% of patients were ⩽5 years old at referral), with no evidence of associated anomalies, was unexpected. A further eight of the submitted patients with sporadic isolated aniridia carried previously identified structural rearrangements or visible deletions. However, one of the structural rearrangements and all eight cryptic deletions were only identified in the course of this analysis. Overall, 9/44 (20%) of patients with sporadic isolated aniridia were found to have unsuspected underlying chromosomal rearrangements.
Perhaps most surprising was the finding of chromosomal rearrangements in four independent pedigrees among the 14 patients with familial aniridia studied, with three cryptic deletions and a previously identified cosegregating pericentric inversion. The frequency of unsuspected aberrations (3/14 [21%]) among patients with familial aniridia is similar to the frequency in the sporadic category. In contrast, Grønskov et al. (2001), using only FAT5 and B2.1 as FISH probes, found no chromosomal anomalies among 19 patients with familial aniridia, but they identified intragenic point mutations in 17 patients. However, some small deletions may not be identifiable when only these two cosmids are used.
Recurrent Breakpoint Region in Cosmid A1280
Six patients were identified (four retrospectively and two prospectively) with a diminished A1280 signal (figs. 2 and 3A) and a deletion involving two or more of the telomeric cosmids in the contig. Cosmid A1280 carries 10 of the 14 PAX6 exons and >15 kb of downstream region beyond the predicted polyA addition site, which also contains the final exon of the gene neighboring PAX6, now known to be the human homologue of transcriptional elongation factor ELP4, first defined in yeast (Kleinjan et al. 2002). The latest gene prediction programs also define a predicted additional single-exon 138–amino acid ORF sequence only 2 kb beyond the PAX6 polyA addition site (like ELP4, transcribed on the opposite strand). In addition to the six deletion breakpoints within A1280 among the current cohort, the centromeric end of one of the deletions described by Lauderdale et al. (2000) is ∼8 kb from the telomeric end of A1280 (Lauderdale et al. 2000). Sequence comparison reveals no region-specific low-copy repeats (Stankiewicz and Lupski 2002) that might be associated with recurrent breakpoints, although the last 2 kb at the telomeric end of A1280 contain two different 50–60-bp segments that are also found, perfectly repeated, on several other chromosomes, though not on chromosome 11. The second deletion breakpoint described by Lauderdale et al. (2000) is within cosmid G0453. In view of the high frequency of breakpoints within this region, the use of this contig (fig. 1B) may identify aniridia-associated breakpoints in cases in which conventional sequence-based mutation analysis fails to find point mutations.
Mosaicism for Cryptic Deletion in Two Patients with the Aniridia Phenotype
Two of the patients who have a de novo cryptic deletion (patients 2 and 94) with a reduced A1280 signal show the deletion in ∼50% of the peripheral blood metaphases (fig. 3B), with the remaining metaphases showing normal signals on both chromosome 11 homologues. In both patients, the telomeric breakpoint of the mosaic deletion is between cosmids SRL9A13 and AO4160 (fig. 2). The results from the use of the pool of the three plasmid markers over PAX6 exons 10–13 (fig. 4) suggest reduced FISH signals in these patients, implying truncation of the PAX6 transcription unit by the mosaic deletion. In both patients, chromosomes have been analyzed only from peripheral blood lymphocytes, so that the degree of mosaicism in other tissues is not known. Both of the babies were referred (at ages 12 mo and 2 mo) with bilateral aniridia and optic atrophy, without any other clinical complications. Optic atrophy is a phenotype that has not previously been associated with PAX6 mutations. Interestingly, the second patient (patient 94) is an MZ twin with an unaffected brother, which suggests that somatic mutation in the affected twin occurred after the two-blastomere stage, which is the earliest point at which MZ twinning can occur. The presence of the normal cell lines in both patients would have made it very unlikely for the deletions to be detected using conventional DNA mutation-detection techniques, so that overall mosaicism involving cryptic deletions may be underrepresented among patients with aniridia. Furthermore, the fact that we observed mosaicism in 2 of the 26 deletions overall (2/13 of the microdeletions), suggests that mosaicism may be underrepresented as a mutational mechanism in microdeletion syndromes (Pearson 2002).
Somatic mosaicism for structural chromosome abnormalities is a well-recognized but rarely reported phenomenon and has been described for a number of syndromes, including 22q13 deletion (Riegel et al. 2000; Phelan et al. 2001), neurofibromatosis type 1 (Ainsworth et al. 1997; Tinschert et al. 2000), 22q11.3 deletion (Kasprzak et al. 1998), dystrophin (Bunyan et al. 1995), 1p36.33 deletion (Eugster et al. 1997), Angelman syndrome 15q11-13 deletion (Tekin et al. 2000), and 19q13.33 deletion (Mikelsaar et al. 2001). Molecular confirmations of germline mosaicisms for deletions of 22q11.2 (Hatchwell et al. 1998) and Williams-Beuren syndrome (Kara-Mostefa et al. 1999) have also been reported.
The observation of the aniridia phenotype in two mosaic cases is also surprising, in the light of previous findings that cells carrying heterozygous Pax6 mutations cannot contribute to lens tissue in artificial mouse chimeras (Collinson et al. 2001).
Position Effects as a Mutational Mechanism
Position effects, in which loss or alteration of gene function results from chromosomal rearrangements outside the transcribed region of a gene, have been described for a number of human disease loci (Kleinjan and van Heyningen 1998). On the basis of the cases reported here and those described in previous studies (Lauderdale et al. 2000; Kleinjan et al. 2001, 2002), it is clear that position-effect disruptions of PAX6 regulatory element(s) affecting the chromosomal domain as distant as 180 kb telomeric of the 3′ end of the gene are rare but important causes of human sporadic and familial aniridia. In addition to position-effect breakpoints, we have observed a wide variety of interstitial deletions varying in size from 75 kb to ⩾7 Mb, which suggests that this region, particularly the ∼180 kb immediately telomeric to PAX6, may be a region of genomic instability.
FISH Analysis Is the Ideal First Choice for Assessing the Molecular Cause of Sporadic Aniridia
The present study illustrates the importance of FISH for the characterization of mutations associated with both familial and sporadic aniridia. The high frequency of submicroscopic deletions detected suggests that FISH should be used as the primary screening procedure, particularly in the neonatal period, in patients ascertained with sporadic isolated aniridia. Our results, in combination with those of Lauderdale et al. (2000), also show that, in order to detect all deletions, it is necessary to use a contig of FISH probes covering the ∼180-kb region 3′ of PAX6. This is particularly important in young patients with sporadic isolated aniridia who are reported to have a normal karyotype by conventional analysis. In these cases, we recommend that cosmids (listed in order from centromere to telomere) B2.1, P60, FAT5, A1280, GO453, C1170, H1281, and FO2121 should be used as the initial FISH screen. Intragenic mutation-screening protocols can then be used on those patients reported to be normal when tested by conventional cytogenetics and FISH.
Additional Data Reported Since Submission of the Present Article
FISH analysis of the phenotypically normal MZ brother of patient 94 has shown that he is also a mosaic for the same sized (∼200 kb) cryptic deletion but that in the peripheral blood, the proportion of deleted cells is ∼30%, compared with ∼60% in the affected twin brother. This implies that mosaicism arose before the splitting of the embryo, or that the twins arose from a chimeric embryo.
Acknowledgments
The authors gratefully acknowledge the technical help of Sarah Beal (Wessex Regional Genetics Laboratory) for skillful preparation of the FISH probes used in the study. We thank Dr. Dirk-Jan Kleinjan for providing the plasmid probes used to characterize the 3′ PAX6 UTR. We are grateful to Professor Pat Jacobs for her reading of the manuscript and constructive criticisms of this work. We are indebted to the many referring clinicians and their patients.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Ensembl Genome Browser, http://www.ensembl.org/Homo_sapiens/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for aniridia [MIM 106200] and WAGR syndrome [MIM 194072])
References
- Ainsworth PJ, Chakraborty PK, Weksberg R (1997) Example of somatic mosaicism in a series of de novo neurofibromatosis type 1 cases due to a maternally derived deletion. Hum Mutat 9:452–457 [DOI] [PubMed] [Google Scholar]
- Axton R, Hanson I, Danes S, Sellar G, van Heyningen V, Prosser J (1997) The incidence of PAX6 mutation in patients with simple aniridia: an evaluation of mutation detection in 12 cases. J Med Genet 34:279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunyan DJ, Crolla JA, Collins AL, Robinson DO (1995) Fluorescence in situ hybridisation studies provide evidence for somatic mosaicism in de novo dystrophin gene deletions. Hum Genet 95:43–45 [DOI] [PubMed] [Google Scholar]
- Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH, Jones C, Houseman DE (1990) Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell 60:509–520 [DOI] [PubMed] [Google Scholar]
- Chao LY, Huff V, Strong LC, Saunders GF (2000) Mutation in the PAX6 gene in twenty patients with aniridia. Hum Mutat 15:332–339 [DOI] [PubMed] [Google Scholar]
- Collinson JM, Quinn JC, Buchanan MA, Kaufman MH, Wedden SE, West JD, Hill RE (2001) Primary defects in the lens underlie complex anterior segment abnormalities of the Pax6 heterozygous eye. Proc Natl Acad Sci USA 98:9688–9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crolla JA, Cawdery JE, Oley CA, Young ID, Gray J, Fantes J, van Heyningen V (1997) A FISH approach to defining the extent and possible clinical significance of deletions at the WAGR locus. J Med Genet 34:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crolla JA, Cross I, Atkey N, Wright M, Oley CA (1996) FISH studies in a patient with sporadic aniridia and t(7;11) (q31.2;p13). J Med Genet 33:66–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LM, Stallard R, Thomas GH, Couillin P, Junien C, Nowak NJ, Shows TB (1988) Two anonymous DNA segments distinguish the Wilms' tumor and aniridia loci. Science 241: 840–842 [DOI] [PubMed] [Google Scholar]
- Dreschler M, Meijers-Heijboer EJ, Schneider S, Schurich B, Grond-Ginsbach C, Tariverdian G, Kantner G, Blankenagel A, Kaps D, Schroeder-Kurth T, Royer-Pokora B (1994) Molecular analysis of aniridia patients for deletions involving the Wilms' tumor gene. Hum Genet 94:331–338 [DOI] [PubMed] [Google Scholar]
- Eugster EA, Berry SA, Hirsch B (1997) Mosaicism for deletion 1p36.33 in a patient with obesity and hyperphagia. Am J Med Genet 70:409–412 [DOI] [PubMed] [Google Scholar]
- Fantes JA, Bickmore WA, Fletcher JM, Ballesta F, Hanson IM, van Heyningen V (1992) Submicroscopic deletions at the WAGR locus, revealed by nonradioactive in situ hybridization. Am J Hum Genet 51:1286–1294 [PMC free article] [PubMed] [Google Scholar]
- Fantes JA, Oghene K, Boyle S, Danes S, Fletcher JM, Bruford EA, Williamson K, Seawright A, Scheld A, Hanson I, Zehetner G, Bhogal R, Lehrach H, Gregory S, Williams J, Little PFR, Sellar GC, Hoovers J, Mannens M, Weissenbach J, Junien C, van Heyningen V, Bickmore W (1995a) A high-resolution integrated physical, cytogenetic, and genetic map of human chromosome 11: distal p13 to proximal p15.1. Genomics 25:447–461 [DOI] [PubMed] [Google Scholar]
- Fantes J, Redeker B, Breen M, Boyle S, Brown J, Fletcher J, Jones S, Bickmore W, Fukushima Y, Mannens M, Danes S, van Heyningen V, Hanson I (1995b) Aniridia-associated cytogenetic rearrangements suggest that a position effect may cause the mutant phenotype. Hum Mol Genet 4:415–422 [DOI] [PubMed] [Google Scholar]
- Francke U, Holmes LB, Atkins L, Riccardi VM (1979) Aniridia-Wilms' tumor association: evidence for specific deletion of 11p13. Cytogenet Cell Genet 24:185–192 [DOI] [PubMed] [Google Scholar]
- Fukushima Y, Hoovers J, Mannens M, Wakui K, Ohashi H, Ohno T, Ueoka Y, Niikawa N (1993) Detection of a cryptic paracentric inversion within band 11p13 in familial aniridia by fluorescence in situ hybridization. Hum Genet 91:205–209 [DOI] [PubMed] [Google Scholar]
- Gessler M, Poustka A, Cavanee W, Neve RL, Orkin SH, Bruns GAP (1990) Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature 343:774–778 [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL (1992) Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet 2:232–239 [DOI] [PubMed] [Google Scholar]
- Griffin C, Kleinjan DA, Doe B, van Heyningen V (2002) New 3′ elements control Pax6 expression in the developing pretectum, neural retina, and olfactory region. MechDev 112: 89–100 [DOI] [PubMed] [Google Scholar]
- Grønskov K, Olsen JH, Sand A, Pedersen W, Carlsen N, Bak Jylling AM, Lyngbye T, Brondum-Nielsen K, Rosenberg T (2001) Population-based risk estimates of Wilms tumor in sporadic aniridia: a comprehensive mutation screening procedure of PAX6 identifies 80% of mutations in aniridia. Hum Genet 109:11–18 [DOI] [PubMed] [Google Scholar]
- Hanson IM, Seawright A, Hardman K, Hodgson S, Zaletayev D, Fekete G, van Heyningen V (1993) PAX6 mutations in aniridia. Hum Mol Genet 2:915–920 [DOI] [PubMed] [Google Scholar]
- Hatchwell E, Long F, Wilde J, Crolla J, Temple K (1998) Molecular confirmation of germ line mosaicism for a submicroscopic deletion of chromosome 22q11. Am J Med Genet 78:103–106 [DOI] [PubMed] [Google Scholar]
- Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V (1992) The human PAX6 gene is mutated in two patients with aniridia. Nat Genet 1:328–332 [DOI] [PubMed] [Google Scholar]
- Kara-Mostefa A, Raoul O, Lyonnet S, Amiel J, Munnich A, Vekemans M, Magnier S, Ossareh B, Bonnefont JP (1999) Recurrent Williams-Beuren syndrome in a sibship suggestive of maternal germ-line mosaicism. Am J Hum Genet 64:1475–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzak L, Der Kaloustian VM, Elliott AM, Shevell M, Lejtenyi C, Eydoux P (1998) Deletion of 22q11 in two brothers with different phenotype. Am J Med Genet 75:288–291 [PubMed] [Google Scholar]
- Kent J, Lee M, Schedl A, Boyle S, Fantes J, Powell, M, Rushmere N, Abbott C, van Heyningen V, Bickmore WA (1997) The reticulocalbin gene maps to the WAGR region in man and to Small eyeHarwell deletion in mouse. Genomics 42:260–267 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Elgar G and van Heyningen V (2002) Characterisation of a novel gene adjacent to PAX6, revealing synteny conservation with functional significance. Mamm Genome 13:102–107 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Schedl A, Quinlan RA, Danes S, van Heyningen V (2001) Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet 10:2049–2059 [DOI] [PubMed] [Google Scholar]
- Kleinjan DJ, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 [DOI] [PubMed] [Google Scholar]
- Lauderdale JD, Wilensky JS, Oliver ER, Walton DS, Glaser T (2000) 3′ deletions cause aniridia by preventing PAX6 gene expression. Proc Natl Acad Sci USA 97:13755–13759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llerena JC, Cabral de Almeida JC, Bastos E, Crolla JA (2000) FISH studies in a girl with sporadic aniridia and an apparently balanced de novo t(11;13)(p13;q33) translocation detect a microdeletion involving the WAGR region. Genet Mol Biol 23:535–539 [Google Scholar]
- Mannens M, Hoovers JM, Bleeker-Wagemakers EM, Redeker E, Bliek J, Overbeeke-Melkert M, Saunders G, Williams B, van Heyningen V, Junien C, Haber D, Speleman F, Heyting C, Slater RM, Leschot NJ, Westerveld A (1991) The distal region of 11p13 and associated genetic diseases. Genomics 11:284–293 [DOI] [PubMed] [Google Scholar]
- Mikelsaar RV, Varb K, Suvari A, Schinzel A (2001) Mosaic terminal del(19)(q13.33:) in a girl with seizures and mental retardation. J Med Genet 38:E2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JW, Hyman S, Antonarakis SE, Mules EH, Thomas GH (1986) Familial isolated aniridia associated with a translocation involving chromosomes 11 and 22 [t(11;22)(p13;q12.2)]. Hum Genet 72:297–302 [DOI] [PubMed] [Google Scholar]
- Muto R, Yamamori S, Ohashi H, Osawa M (2002) Prediction by FISH analysis of the occurrence of Wilms tumor in aniridia patients. Am J Med Genet 108:285–289 [DOI] [PubMed] [Google Scholar]
- Nelson LB, Spaeth GL, Nowinski TS, Margo CE, Jackson L (1984) Aniridia: a review. Surv Ophthalmol 28:621–642 [DOI] [PubMed] [Google Scholar]
- Pearson H (2002) Dual identities. Nature 417:10–11 [DOI] [PubMed] [Google Scholar]
- Phelan MC, Brown EF, Rogers RC (2001) Prenatal diagnosis of mosaicism for deletion 22q13.3. Prenat Diagn 21:1100 [DOI] [PubMed] [Google Scholar]
- Prosser J, van Heyningen V (1998) PAX6 mutations reviewed. Hum Mutat 11:93–108 [DOI] [PubMed] [Google Scholar]
- Riegel M, Baumer A, Wisser J, Acherman J, Schinzel A (2000) Prenatal diagnosis of mosaicism for a del(22)(q13). Prenat Diagn 20:76–79 [DOI] [PubMed] [Google Scholar]
- Simola KOJ, Knuutila S, Kaitila I, Pirkola A, Pohja P (1982) Familial aniridia and translocation t(4;11)(q22;p13) without Wilms' tumor. Hum Genet 63:158–161 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- Tekin M, Jackson-Cook C, Buller A, Ferreira-Gonzalez A, Pandya A, Garrett CT, Bodurtha J (2000) Fluorescence in situ hybridization detectable mosaicism for Angelman syndrome with biparental methylation. Am J Med Genet 95:145–149 [DOI] [PubMed] [Google Scholar]
- Tinschert S, Naumann I, Stegmann E, Buske A, Kaufmann D, Thiel G, Jenne DE (2000) Segmental neurofibromatosis is caused by somatic mutation of the neurofibromatosis type 1 (NF1) gene. Eur J Hum Genet 8:455–459 [DOI] [PubMed] [Google Scholar]
- Ton CCT, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie N, Meijers-Heijboer H, Dreschler M, Royer-Pokora B, Collins F, Swaroop A, Strong LC, Saunders GF (1991) Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell 67:1059–1074 [DOI] [PubMed] [Google Scholar]
- Turleau C, de Grouchy J, Nihoul-Fekete C, Dufier JL, Chavin-Colin F, Junien C (1984) Del 11p13/nephroblastoma without aniridia. Hum Genet 67:455–456 [DOI] [PubMed] [Google Scholar]
- van Heyningen V, Boyd PA, Seawright A, Fletcher JM, Fantes JA, Buckton KE, Spowart G, Porteous DJ, Hill RE, Newton MS, Hastie ND (1985) Molecular analysis of chromosome 11 deletions in aniridia-Wilms tumor syndrome. Proc Natl Acad Sci USA 82:8592–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]