Abstract
Nephronophthisis (NPHP) comprises a group of autosomal recessive cystic kidney diseases, which constitute the most frequent genetic cause for end-stage renal failure in children and young adults. The most prominent histologic feature of NPHP consists of development of renal fibrosis, which, in chronic renal failure of any origin, represents the pathogenic event correlated most strongly to loss of renal function. Four gene loci for NPHP have been mapped to chromosomes 2q13 (NPHP1), 9q22 (NPHP2), 3q22 (NPHP3), and 1p36 (NPHP4). At all four loci, linkage has also been demonstrated in families with the association of NPHP and retinitis pigmentosa, known as “Senior-Løken syndrome” (SLS). Identification of the gene for NPHP type 1 had revealed nephrocystin as a novel docking protein, providing new insights into mechanisms of cell-cell and cell-matrix signaling. We here report identification of the gene (NPHP4) causing NPHP type 4, by use of high-resolution haplotype analysis and by demonstration of nine likely loss-of-function mutations in six affected families. NPHP4 encodes a novel protein, nephroretinin, that is conserved in evolution—for example, in the nematode Caenorhabditis elegans. In addition, we demonstrate two loss-of-function mutations of NPHP4 in patients from two families with SLS. Thus, we have identified a novel gene with critical roles in renal tissue architecture and ophthalmic function.
Introduction
Nephronophthisis (NPHP), an autosomal recessive cystic kidney disease, constitutes the most frequent genetic cause of end-stage renal disease (ESRD) in children and young adults, necessitating renal replacement therapy for survival (Smith and Graham 1945; Fanconi et al. 1951; Hildebrandt 1999). Three distinct gene loci for NPHP—NPHP1 (MIM 256100), NPHP2 (MIM 602088), and NPHP3 (MIM 604387)—have been mapped to chromosomes 2q13 (Antignac et al. 1993; Hildebrandt et al. 1993), 9q22 (Haider et al. 1998), and 3q22 (Omran et al. 2000), respectively. These disease variants share renal histology of interstitial infiltrations, renal tubular cell atrophy with cyst development, and renal interstitial fibrosis (Waldherr et al. 1982). The variants can be distinguished clinically by age at onset of ESRD. The most prominent histologic feature of NPHP is renal fibrosis, which, in chronic renal failure, regardless of origin, represents the pathogenic event that is correlated most strongly to loss of renal function (Zeisberg et al. 2001). Therefore, NPHP has been considered a model disease for the development of renal fibrosis.
We have previously identified, by positional cloning, the gene (NPHP1) for NPHP type 1 (Hildebrandt et al. 1997). Its gene product, nephrocystin, represents a novel docking protein that interacts with the signaling proteins p130Cas, tensin, focal adhesion kinase 2, and filamins A and B, which are involved in cell-cell and cell-matrix signaling of renal epithelial cells (Donaldson et al. 2000, 2002; Hildebrandt and Otto 2000; Benzing et al. 2001). The association between NPHP and autosomal recessive retinitis pigmentosa (RP) has been described as the so-called “Senior-Løken syndrome” (SLS [MIM 266900]) (Løken et al. 1961; Senior et al. 1961). In families with SLS, linkage has been demonstrated to the loci for NPHP1 and NPHP3 (Caridi et al. 1998; Omran et al. 2002). Very recently, we have localized a new gene locus (NPHP4 [MIM 606966]) for NPHP type 4 (Schuermann et al. 2002), and we have demonstrated linkage to this locus in a large kindred with SLS.
We here identified, by positional cloning, the gene (NPHP4) that causes NPHP type 4, through demonstration of nine likely loss-of-function mutations in six affected families. In addition, we detected two loss-of-function mutations in patients from two families with SLS. NPHP4 is a novel gene that is unrelated to any known gene families. It encodes a novel protein, nephroretinin. NPHP4, like NPHP1, is unique to the human genome, is conserved in Caenorhabditis elegans, and exhibits a broad expression pattern. It is therefore likely that both gene products, nephroretinin and nephrocystin, interact within a novel shared pathogenic pathway. Thus, we have identified a novel gene with critical roles in renal tissue architecture and ophthalmic function.
Subjects and Methods
Pedigree and Diagnosis
We obtained blood samples and pedigrees after receiving informed consent from patients with NPHP and their parents. Diagnostic criteria were (1) development of ESRD in addition to a history of polyuria, polydipsia, and anemia; and (2) renal ultrasound compatible with NPHP. In all families except F461, the diagnosis of NPHP was confirmed by renal biopsy. ESRD developed within a range of 6–35 years of age with a median age of 22 years (table 1). In SLS, the renal symptoms are associated with RP. Clinical data for family F3 (with SLS) have been published elsewhere (Polak et al. 1983; Schuermann et al. 2002). All three affected siblings had RP suggestive of Leber amaurosis congenita. Ophthalmologic data for family F60 have been published elsewhere (Fillastre et al. 1976) and comprise the following observations: In one individual (Fillastre et al. 1976), there was amblyopia and rotary nystagmus with grossly impaired vision starting at age 8 mo, and, on fundoscopy, there was retinochoroidal atrophy surrounded by pigment. In two individuals, there were abnormal ERG findings with diminished amplitude (Fillastre et al. 1976).
Table 1.
Clinical Details and Mutations Detected in Families with NPHP4
| Family | Numberof AffectedIndividuals | ESRDat Agea(years) | RP | Origin | ParentalConsanguinity | Exon | NucleotideChangeb | Effecton CodingSequence | Segregationc |
| F3d | 3 | 28, 30, 35 | Yes | Turkey | Yes | 18 | C2335T | Q779X | Hom |
| F24 | 2 | ND | No | Germany | No | 17 | G2260A | G754R | P |
| 17 | IVS16-1 G→C | Splice site | M | ||||||
| F30d | 3 | 18, 22, 22 | No | Germany | Yes | 23 | 3272delT | Stop at codon L1121 | Hom |
| F32 | 2 | 19, 20 | No | India | Yes | 11 | TC1334-1335AA | F445X | Hom |
| F60 | 4 | 6, 10, 17, 22 | Yes | France | Yes | 16 | C1972T | R658X | Hom |
| F444d | 2 | 23, 33 | No | Finland | No | 15 | IVS15+1 G→A | Splice site | M |
| 24 | IVS24+1 G→A | Splice site | P | ||||||
| F461d | 3 | ND | No | France | No | 16 | C2044T | R682X | P |
| 19 | C2542T | R848W | M | ||||||
| F622 | 2 | 8, 9 | No | Afghanistan | Yes | 18 | G2368T | E790X | Hom |
ND = no data available.
All mutations were absent from 92–96 unaffected control individuals.
M = maternal; P = paternal; Hom = homozygous mutation inherited from both parents.
In these four families, linkage to NPHP4 has been published elsewhere (Schuermann et al. 2002).
Haplotype and Mutational Analysis
The “screening markers” used for haplotype analysis consisted of microsatellites markers D1S2845, D1S2660, D1S2795, D1S2870, D1S2642, D1S214, D1S2663, and D1S1612 (in pter→cen orientation) (Dib et al. 1996). Novel microsatellite markers were generated by searching for di-, tri-, and tetranucleotide repeats, using the BLAST program on human genomic sequence in the interval between flanking markers D1S2660 and D1S2642. Preparation of genomic DNA and haplotype analysis were performed as described elsewhere (Schuermann et al. 2002). Mutational analysis was performed using exon-flanking primers, as described elsewhere (Hildebrandt et al. 1997). Primer sequences can be obtained from the authors.
Northern Blot Analysis
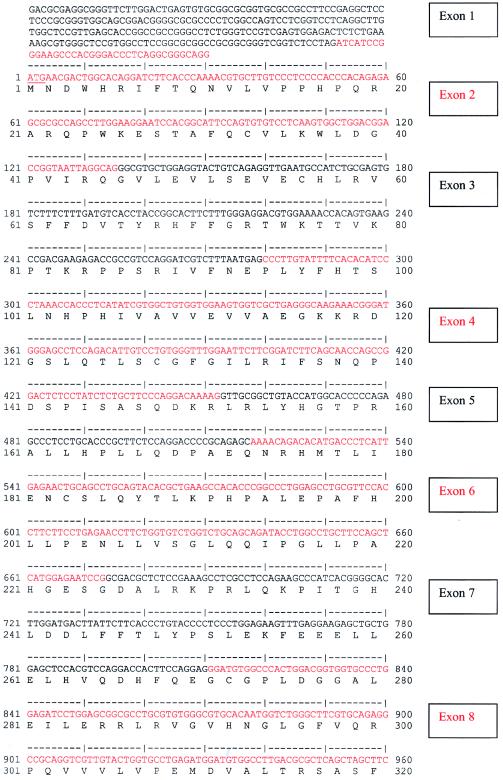
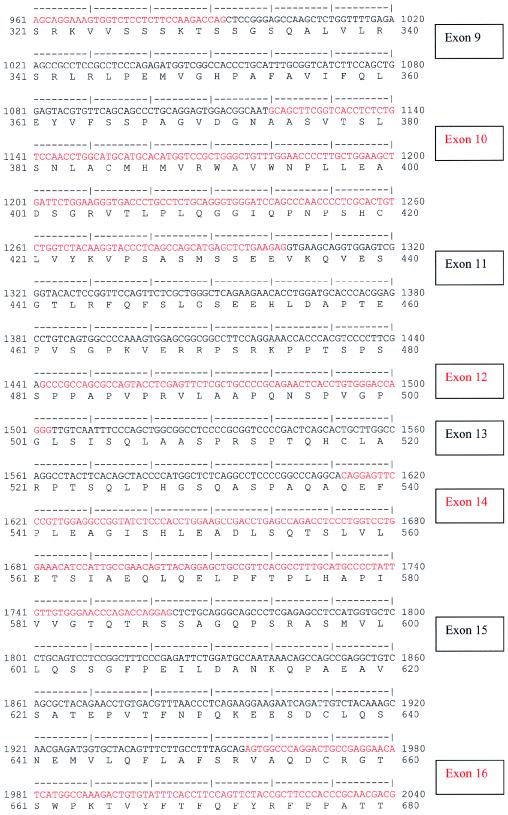
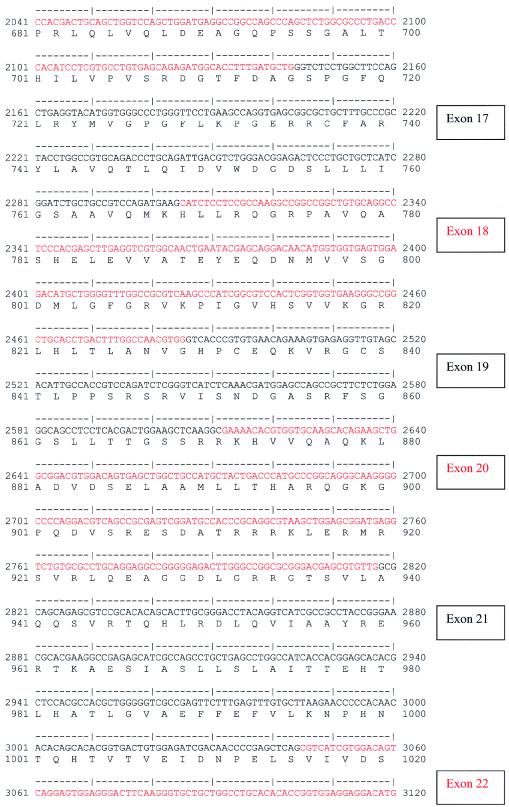
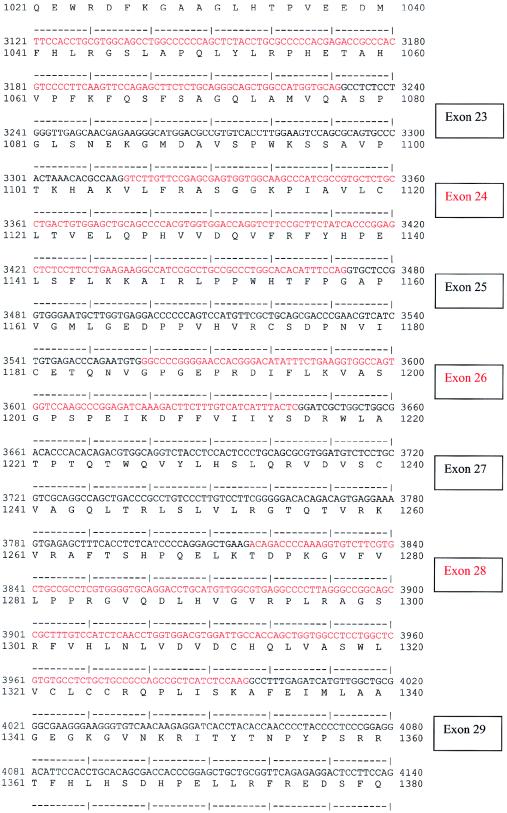
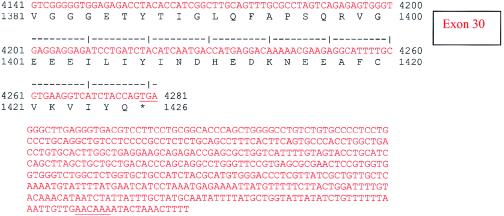
A multiple-tissue northern blot with human adult poly(A)+ RNA (MTN7760-1; Clontech) was hybridized with an NPHP4 DNA probe of 584 bp, derived from exon 30 (nucleotides 4141–4724; see fig. Afig. A [online only]) and generated by PCR amplification of human genomic DNA. The probe was labeled with [32P]dCTP by using Random Primers DNA Labeling System (Invitrogen). Hybridization was performed at 68°C by using ExpressHyb solution (Clontech). The final washing condition was 0.1 × saline sodium citrate, 0.1% SDS, at 55°C for 40 min.
Figure A.
Human NPHP4 cDNA sequence and deduced nephroretinin amino acid sequence. Exons are numbered at right. Exon sequence alternates in color. Start codon, stop codon, and the variant polyadenylation signal AACAAA are underlined.
Results
We have previously mapped, by total-genome search for linkage, a gene locus (NPHP4) for NPHP type 4 within a 2.1-Mb interval delimited by flanking markers D1S2660 and D1S2642 (Schuermann et al. 2002). To establish compatibility with linkage to NPHP4 in further kindreds, we selected 20 families with multiple affected children or parental consanguinity, in whom no mutation was present in the NPHP1 gene. In 12 families, there was NPHP only, and, in 8 families, there was an association between NPHP and RP. Haplotype analysis using eight microsatellite markers covering the critical NPHP4 region (Schuermann et al. 2002) was compatible with linkage to NPHP4 in nine families. To further refine the critical genetic interval of 2.1 Mb, we performed high-resolution haplotype analysis in these nine families and in the seven families with linkage to NPHP4 published elsewhere (Schuermann et al. 2002). In a total of three families, NPHP was associated with RP. These were two families (F18 and F60) from the new set of nine linked families and one family (F3) that has been described elsewhere (Schuermann et al. 2002). Eight published (Dib et al. 1996) and 38 newly generated microsatellite markers (table Atable A [online only]) were used at an average marker density of one marker per 45 kb within the interval of flanking markers D1S2660 and D1S2642 (fig. 1). Haplotype analysis, by the criterion of minimization of recombinants, clearly revealed erroneous inversion of sequence between markers D1S2795 and D1S244 in human genomic sequence databases (see the Ensembl Genome Browser Web site) (data available from the authors); this inversion was not described in the recent high-resolution genetic map of the human genome (Kong et al. 2002). Using high-resolution haplotype data, we established the correct marker order at the NPHP4 locus as pter-D1S2660-D1S2795-D1S2633-D1S2870-D1S253-D1S2642-D1S214-D1S1612-D1S2663-D1S244-cen (flanking markers to NPHP4 are underlined) (Schuermann et al. 2002). A 22-kb sequence gap remaining in the interval D1S2660-D1S2795 was filled by use of Celera human genomic sequence. In haplotype analysis, three consanguineous kindreds yielded new key recombinants by the criterion of homozygosity by descent (Lander and Botstein 1987) (fig. 1). We thus refined the NPHP4 critical genetic interval to <1.2 Mb within secure borders that were based on a large kindred, and, in addition, to <700 kb within suggestive borders that were based on two small families (figs. 1, 2A, and 2B).
Table A.
Primer Sequences (5′→3′) Used in Exon Amplification for Mutational Analysis of NPHP4
| Exon | Forward Primer | Reverse Primer | Product Size(bp) |
| 1 | gtcggacatgcaaatcagg | aggctctggccaacactg | 439 |
| 2 | aagccttcaggattgctgtg | catccatctgttaactggaagc | 319 |
| 3 | acatggcctgccagtgac | cctggacccacaagtctgag | 346 |
| 4 | acgtgtaggaaggcggtctc | gacgagcagttaaaccaccatag | 649 |
| 5 | gaggcctccatgtgctttc | gctaaaggtggggaacactc | 209 |
| 6 | tgaccctcattgagaactgc | gtgccttcaaggtttcactg | 217 |
| 7 | ttgtgctctgtctgggagtc | catcagatgcggggtctc | 439 |
| 8 | ctcccccagggacttctg | cctgacatgcacaaatgacc | 335 |
| 9 | ttctgacagtggtcgacgtg | tgcccactacatttatcctcac | 279 |
| 10 | cactgttgatttcccctctcc | gcaaacatatttgtgaacttttgc | 343 |
| 11 | ttcctggttggatcgttctg | cgacgattatcttacaaatgtgg | 329 |
| 12 | aggcctgtggagacctgac | ggggacagagggttttcttg | 232 |
| 13 | catgttgggagctttgtgg | gacaggcacagtgcaaaaac | 262 |
| 14 | atctgagcaccgttggttg | gggttcacaaggtccaacag | 295 |
| 15 | ggtttccacagggaggtg | aggtcagaacctcagcgaag | 345 |
| 16 | accatcccctatgcaaacac | gcactggtcaccgtatgattc | 409 |
| 17 | gaccagagctgaaatctcttcc | acgctggaagcgtgactc | 315 |
| 18 | cacagtggctttcctgctg | cgagggagcccacactctac | 358 |
| 19 | tgtggtgggttgatctgtttc | cactgacagcaccacgaatg | 332 |
| 20 | ccctggtgtctgctcctg | gaggcagggaaaggatgtg | 351 |
| 21 | agcaatagccccttgtggag | tctcgggcagaattcgag | 386 |
| 22 | tctctcccactcctctgagc | agggacactggtggagactg | 377 |
| 23 | tggcagtggtgtctctaagc | aggaggggagagaaggacac | 251 |
| 24 | ttggcaacagtggagatacg | catgaggccatctgtcacc | 342 |
| 25 | tcttgctgagcacctgtgac | aggatacccgtggggaag | 282 |
| 26 | cactcgctgcgtgtattagtc | caagcccactttcaatccac | 268 |
| 27 | ccttgttggcctctcgtg | ccagctgaatgcccactg | 318 |
| 28 | ggaaccacccatgaccttg | cagtggtccgagtcacagg | 388 |
| 29 | cagggaatacttggaggaagg | gaggaactcgctcctaaatgc | 310 |
| 30 | gcagagaggttgctggtgag | accgggcttgtgctgtag | 738 |
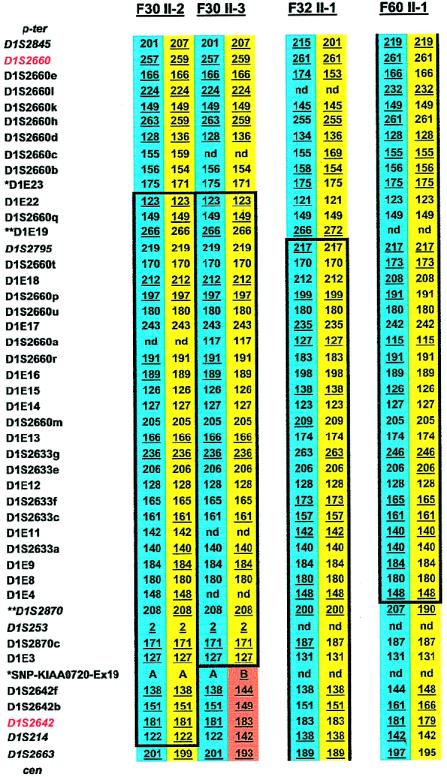
Figure 1.
Haplotype results on chromosome 1p36 performed for refinement of the NPHP4 locus in affected offspring from three consanguineous families with NPHP. Family, generation, and individual numbers are indicated above haplotypes. Paternal haplotypes are shown on blue background, and maternal haplotypes are shown on yellow background. A recombination in the maternal haplotype, which was directly observed in parent-to-child transmission in this pedigree, is shown on orange background. At left, 8 published microsatellites are given in italic, and 38 newly generated microsatellites are given in roman. Flanking markers to the published 2.1-Mb critical NPHP4 interval are depicted in red. Informative alleles are underlined. Haplotypes homozygous in continuity are encased in boxes. Homozygosity mapping revealed a p-terminal recombinant for marker D1E23 (asterisks) on the basis of heterozygosity in individuals F30 II-2 and F30 II-3 and an observed recombinant for marker SNP-KIAA0720-Ex19 (asterisks) in individual F30 II-3, thus refining the critical genetic region to a secure interval <1.2 Mb. On the basis of a significant LOD score yielded for F30 alone (Schuermann et al. 2002), this refinement yields secure borders. Further refinement was achieved through heterozygosity for markers D1E19 and D1S2870 (double asterisks) in individuals F32 II-1 and F60 II-1, respectively. This refines the critical genetic region to a suggestive interval <700 kb. Because of the presence of only two affected individuals in each family, this refinement is only suggestive. p-ter = Telomeric; cen = centromeric; nd = not done.
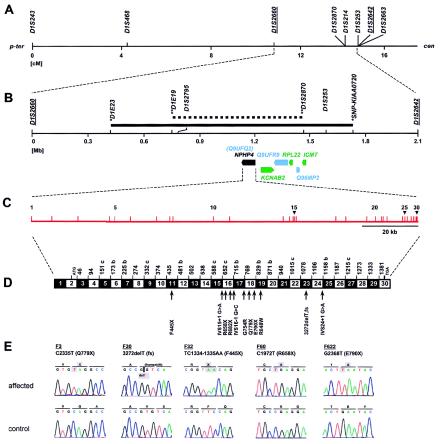
Figure 2.
Positional cloning strategy for the NPHP4 gene, on human chromosome 1p36. A, Genetic map position for microsatellites used in linkage mapping of NPHP4 (see fig. 1). Sex-averaged genetic distance (in cM) from the Marshfield map was used. Published flanking markers are underlined (Schuermann et al. 2002). p-ter = Telomeric; cen = centromeric. B, Physical map distances of critical microsatellites relative to D1S2660. The secure 1.2-Mb critical interval (solid bar) and the 700-kb suggestive critical interval (stippled bar) are delimited by the newly identified secure flanking markers (asterisks) and suggestive flanking markers (double asterisks) defined by haplotype analysis (see fig. 1). Below the axis, known genes (green), predicted unknown genes (blue), and the NPHP4 gene (also known as “Q9UFQ2”) are represented as arrows in the direction of transcription. C, Genomic organization of NPHP4, with exons indicated by vertical hatches and numbered. D, Exon structure of NPHP4 cDNA. Blackened and unblackened boxes represent the 30 exons encoding nephroretinin. The number of the first codon of each exon is indicated; exons beginning with the second or third base of a codon are indicated by “b” or “c,” respectively. At bottom, locations of the 11 different mutations identified in eight kindreds with NPHP4 mutations are shown. fs = Frameshift. E, NPHP4 mutations occurring homozygously in affected individuals from five consanguineous families (underlined). Compound heterozygous mutations are not shown. Mutated nucleotides and altered amino acids are depicted on gray background.
Within the 700-kb critical interval for NPHP4, there mapped three known genes (KCNAB2, RPL22, and ICMT) and three unknown genes (Q9UFQ2, Q9UFR9, and Q96MP2) (fig. 2B). In addition, in the interval between Q9UFQ2 and flanking marker D1E19 (fig. 2B), the program GENESCAN predicted ∼40 nonannotated exons (see the Ensembl Genome Browser Web site) (data not shown). We performed mutational analysis in affected individuals from the 16 families compatible with linkage to NPHP4, examining all 79 exons of the three known and three unknown genes by direct sequencing of the forward strands of exon-PCR products. Although no mutations were detected in five of these genes, in Q9UFQ2, we detected 11 distinct mutations in 8 of the 16 families with NPHP (table 1). In families F3 and F60, NPHP is associated with RP. In the affected individuals from all eight families, mutations were shown to segregate from both parents (table 1). All of these mutations were absent from 92–96 unaffected control individuals from a similar ethnic background. Of the 11 mutations detected, 9 represent very likely loss-of-function mutations: 5 were stop-codon mutations, 1 was a frameshift mutation, and 3 were obligatory splice-consensus mutations (table 1 and fig. 2D). We thus identified Q9UFQ2 as the gene that causes NPHP type 4. The gene was termed “NPHP4,” and the respective gene product was called “nephroretinin” for its role in NPHP and RP. In the five consanguineous families—F3, F30, F32, F60, and F622—all mutations occurred in the homozygous state and represented stop-codon mutations and one frameshift mutation, truncating the protein in exons 18, 23, 11, 16, and 18, respectively (table 1 and figs. 2D and 2E). In the three nonconsanguineous families, we found six distinct compound heterozygous mutations. Four represented stop-codon or obligatory splice-consensus mutations, truncating the gene product in exons 15, 16, 17, and 24. The missense mutations R848W and G754R affect amino acid residues conserved in mouse and cow. No mutations were detected in eight families.
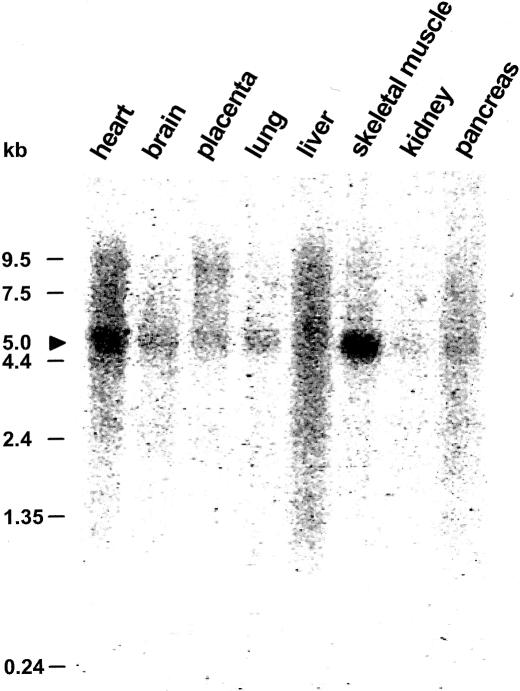
NPHP4 expression studies by northern blot analysis revealed a 5.0-kb transcript expressed strongly in human skeletal muscle, weakly in kidney, and in six additional tissues studied (fig. 3). Northern dot blot analysis (data not shown) confirmed a widespread expression pattern in human adult and fetal tissues, including testis. This broad expression pattern, with strong expression in skeletal muscle and testis, corresponds well with the expression pattern described for the NPHP1 gene (Otto et al. 2000).
Figure 3.
Northern blot analysis of the NPHP4 expression pattern. A multiple-tissue northern blot with human adult poly(A)+ RNA was hybridized with a 584-bp NPHP4 human DNA probe. Expression of a 5.0-kb transcript (arrowhead) is apparent in all tissues studied, with highest expression being in skeletal muscle.
Human genomic sequence of NPHP4 was assembled using the Homo sapiens chromosome 1 working draft sequence segment NT_028054, which predicted 25 exons. Five additional 5′ exons were identified using additional working draft sequence—in particular, the mRNA KIAA0673 and 57 human ESTs from UniGene cluster Hs.106487. The genomic structure shown in figures 2C, 2D, and AA was confirmed by human/mouse total-genomic-sequence comparison. The NPHP4 gene contains 30 exons encoding 1,426 amino acids and extends over 130 kb, with splice sites that confirm to the canonical consensus gt-ag. An exception was found in intron 24, with gc-ag splicing, which occurs in 0.5% of mammalian splice sites (Burset et al. 2001). A polymorphism is known to be present at the intron 20 splice acceptor (tg for ag). The presence of exon 20 is supported by three human EST clones. Ten different splice variants have been suggested for KIAA0673 (see the AceView Web site).
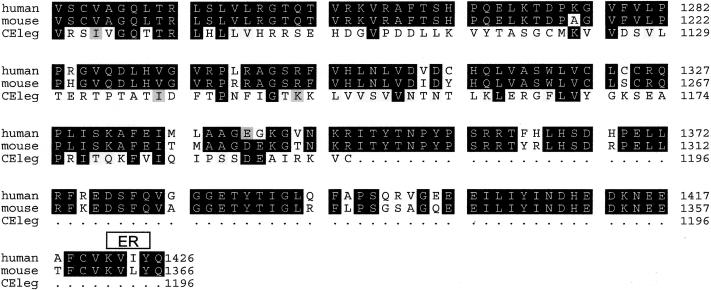
The NPHP4 cDNA (fig. Afig. A) and deduced nephroretinin protein sequences were found to be novel, without any sequence similarity to known human cDNA or protein sequences. Therefore, NPHP4 encodes a hitherto unknown protein. As shown for the NPHP1 gene product nephrocystin (Hildebrandt et al. 1997; Otto et al. 2000), there was, however, strong sequence conservation for nephroretinin in evolution, with 23% amino acid identity in a protein of C. elegans (fig. Bfig. B [online only]). Translated ESTs also demonstrated evolutionary conservation in mouse, cow, pig, zebrafish, Xenopus laevis, Ascaris suum, and Halocynthia roretzi. Sequence identity of the murine ortholog was 78% (fig. Bfig. B). Analysis of nephroretinin amino acid sequence (see the ExPASy Molecular Biology Server Web site) provided no signal sequence, conserved domains, or predicted transmembrane regions. Instead, in the N-terminal half, there was a putative nuclear localization signal, a glutamate-rich domain, and a proline-rich domain. The latter two domains have also been found in nephrocystin (Otto et al. 2000). No sequence similarity to nephrocystin was present. In addition, two serine-rich sequences and a C-terminal endoplasmic reticulum membrane domain, were found in human and murine nephroretinin sequences. Encoded by exons 15 and 16, there was, in nephroretinin, a domain of unknown function (DUF339) with evolutionary conservation dating back to prokaryotes and a 63-amino-acid stretch with 30% sequence identity to a gas-vesicle protein of Halobacterium salinarium (fig. Bfig. B).
Figure B.
Alignment of human, mouse, and C. elegans deduced nephroretinin sequences. The order from top to bottom is human, mouse, and C. elegans (GenBank accession numbers BankIt 486643, BankIt 486675, and Z81579, respectively). Identities are shown in reverse (white on black), and similarities between all three sequences are shown on gray background. There was 82% amino acid identity between human and mouse sequences and 23% amino acid identity between human and C. elegans sequences. Boxes above the sequence delimit motifs as predicted by the programs given in parenthesis: NLS = nuclear localization domain (ACEVIEW); ER = endoplasmic reticulum membrane domain (ACEVIEW); E-rich = glutamic acid–rich domain (SMART); S-rich = serine-rich domain (SMART); P-rich = proline-rich domain (MOTIF SCAN); and DUF339 = PFAM (protein families database of alignments) domain of unknown function (MOTIF SCAN). In H. salinarium, the program BLAST detected a 30% sequence identity with gas-vesicle protein gvpL of H. salinarium (GenBank accession number P33964).
Discussion
Our conclusion that we have identified the gene causing NPHP type 4 is based on the identification, in eight families with NPHP, of nine distinct truncating mutations and two missense mutations, none of which occurred in >92 unaffected control individuals. The absence of mutations in eight additional families with NPHP is not surprising, since linkage in kindreds with only two affected individuals can be fortuitous and since NPHP in these families may be caused by mutations in the NPHP3 gene, which has not yet been identified (Omran et al. 2000). This finding may also indicate further genetic locus heterogeneity for NPHP. Here we also demonstrate the presence of two homozygous truncating mutations in two families with SLS (F3 and F60). Interestingly, a small percentage of patients also exhibit SLS in families with NPHP1 mutations (Caridi et al. 1998) and in families with linkage to NPHP3 (Omran et al. 2002). For all three genes, no distinction can be made on the basis of allelic differences between the NPHP phenotypes with and without RP. Therefore, it seems likely that modifier genes may be responsible for the occurrence of RP in NPHP types 1, 3, and 4. NPHP4, like NPHP1, is unique to the human genome, is conserved in C. elegans, and exhibits a broad expression pattern. It is possible that their gene products, nephroretinin and nephrocystin, respectively, interact within a novel shared pathogenic pathway. Since identification of the NPHP1 gene (Hildebrandt et al. 1997) had revealed nephrocystin as a novel docking protein that interacts with p130Cas (Donaldson et al. 2000; Hildebrandt and Otto 2000), tensin, focal adhesion kinase 2 (Benzing et al. 2001), and filamins A and B (Donaldson et al. 2002), which are involved in cell-cell and cell-matrix signaling, further studies will have to determine the functional role that nephroretinin plays within those interactions. Studies into the function of nephroretinin will provide new insights into disease mechanisms of renal fibrosis, cyst development, and visual function.
Acknowledgments
We thank the patients and their families, for participation; S. Ala-Mello, C. Antignac, K. E. Bonzel, J. Brodehl, K. Devriendt, H. Felten, P. Gissen, M. J. Grosso, L. Guay-Woodford, E. Maher, H. P. H. Neumann, B. C. P. Polak, and U. Vossmerbaeumer, for contribution of patient material; Anita Imm, for outstanding technical assistance; and R. H. Lyons, for excellent large-scale sequencing. F.H. and R.B. are supported by Sonderforschungsbereich 592 from the German Research Foundation.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- AceView, http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ (for genomic structure)
- Ensembl Genome Browser, http://www.ensembl.org/ (for genomic structure of NPHP4)
- ExPASy Molecular Biology Server, http://www.expasy.ch/ (for Tmpred)
- Hits Home, http://hits.isb-sib.ch/ (for MOTIF SCAN program)
- Marshfield Medical Research Foundation Home Page, http://research.marshfieldclinic.org/ (for map of microsatellite markers)
- NCBI Blast Home Page, http://www.ncbi.nlm.nih.gov/BLAST/ (for sequence comparisons through BLASTP/N/X programs)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NPHP1 [MIM 256100], [NPHP2 MIM 602088], NPHP3 [MIM 604387], NPHP4 [MIM 606966], and SLS [MIM 266900])
- SMART, http://smart.embl-heidelberg.de/ (for secondary-structure prediction)
- Submit to GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for full-length NPHP4 cDNAs in human, mouse, and C. elegans [accession numbers BankIt 486643, BankIt 486675, and Z81579] and gas-vesicle protein gvpL of H. salinarium [accession number P33964])
References
- Antignac C, Arduy C, Beckmann JS, Benessy F, Gros F, Medhioub M, Hildebrandt F, Dufier JL, Kleinknecht C, Broyer M (1993) A gene for familial juvenile nephronophthisis (recessive medullary cystic disease) maps to chromosome 2p. Nat Genet 3:342–345 [DOI] [PubMed] [Google Scholar]
- Benzing T, Gerke P, Hildebrandt F, Kim E, Walz G (2001) Nephrocystin forms a multimeric protein complex with Pyk2, p130cas and tensin, and triggers phosphorylation and activation of Pyk2. Proc Natl Acad Sci USA 98:9784–9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Seledtsov IA, Solovyev VV (2001) SpliceDB database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res 29:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi G, Murer L, Bellantuono R, Sorino P, Caringella DA, Gusmano R, Ghiggeri GM (1998) Renal-retinal syndromes: association of retinal anomalies and recessive nephronophthisis in patients with homozygous deletion of the NPH1 locus. Am J Kidney Dis 32:1059–1062 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach, J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Donaldson JC, Dempsey PJ, Reddy S, Bouton AH, Coffey RJ, Hanks SK (2000) Crk-associated substrate p130(Cas) interacts with nephrocystin and both proteins localize to cell-cell contacts of polarized epithelial cells. Exp Cell Res 256:168–178 [DOI] [PubMed] [Google Scholar]
- Donaldson JC, Dise RS, Ritchie MD, Hanks SK (2002) Nephrocystin-conserved domains involved in targeting to epithelial cell-cell junctions, interaction with filamins, and establishing cell polarity. J Biol Chem 277:29028–29035 [DOI] [PubMed] [Google Scholar]
- Fanconi G, Hanhart E, Albertini A, Uhlinger E, Dolivo G, Prader A (1951) Die familiäre juvenile Nephronophthise. Helv Paediatr Acta 6:1–49 [PubMed] [Google Scholar]
- Fillastre JP, Guenel J, Riberi P, Marx P, Whitworth JA, Kunh JM (1976) Senior-Løken syndrome (nephronophthisis and tapetoretinal degeneration): a study of 8 cases from 5 families. Clin Nephrol 5:14–19 [PubMed] [Google Scholar]
- Haider NB, Carmi R, Shalev H, Sheffield VC, Landau D (1998) A Bedouin kindred with infantile nephronophthisis demonstrates linkage to chromosome 9 by homozygosity mapping. Am J Hum Genet 63:1404–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F (1999) Juvenile nephronophthisis. In: Avner E, Holliday M, Barrat T (eds) Pediatric nephrology. Williams & Wilkins, Baltimore [Google Scholar]
- Hildebrandt F, Otto E (2000) Molecular genetics of the nephronophthisis-medullary cystic disease complex. J Am Soc Nephrol 11:1753–1761 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Otto E, Rensing C, Nothwang HG, Vollmer M, Adolphs J, Hanusch H, Brandis M (1997) A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet 17:149–153 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Singh-Sawhney I, Schnieders B, Centofante L, Omran H, Pohlmann A, Schmaltz C, Wedekind H, Schubotz, C, Antignac C, Weber JL, Brandis M (1993) Mapping of a gene for familial juvenile nephronophthisis: refining the map and defining flanking markers on chromosome 2. Am J Hum Genet 53:1256–1261 [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- Løken AC, Hanssen O, Halvorsen S, Jølster NJ (1961) Hereditary renal dysplasia and blindness. Acta Paediatr 50:177–184 [DOI] [PubMed] [Google Scholar]
- Omran H, Fernandez C, Jung M, Häffner K, Fargier B, Villaquiran A, Waldherr R, Gretz N, Brandis M, Rüschendorf F, Reis A, Hildebrandt F (2000) Identification of a new gene locus for adolescent nephronophthisis, on chromosome 3q22 in a large Venezuelan pedigree. Am J Hum Genet 66:118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H, Sasmaz G, Häffner K, Volz A, Olbrich H, Melkaoui R, Otto E, Wienker T, Korinthenberg R, Brandis M, Antignac C, Hildebrandt F (2002) Identification of a gene locus for Senior-Løken syndrome in the region of the nephronophthisis type 3 gene. J Am Soc Nephrol 13:75–79 [DOI] [PubMed] [Google Scholar]
- Otto E, Kispert A, Schaetzle S, Lescher B, Rensing C, Hildebrandt F (2000) Nephrocystin: gene expression and sequence conservation between human, mouse, and Caenorhabditis elegans. J Am Soc Nephrol 11:270–282 [DOI] [PubMed] [Google Scholar]
- Polak BCP, van Lith FHM, Delleman JW, van Balen ATM (1983) Carrier detection in tapetoretinal degeneration in association with medullary cystic disease. Am J Ophthalmol 95:487–494 [DOI] [PubMed] [Google Scholar]
- Schuermann M, Otto E, Becker A, Saar K, Rüschendorf F, Polak BC, Ala-Mello S, Hoefele J, Wiedensohler A, Haller M, Omran H, Nürnberg P, Hildebrandt F (2002) Mapping of gene loci for nephronophthisis type 4 and Senior-Løken syndrome, to chromosome 1p36. Am J Hum Genet 70:1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior B, Friedmann AI, Braudo JL (1961) Juvenile familial nephropathy with tapetoretinal degeneration: a new oculorenal dystrophy. Am J Ophthalmol 52:625–633 [DOI] [PubMed] [Google Scholar]
- Smith CH, Graham JB (1945) Congenital medullary cysts of the kidneys with severe refractory anemia. Am J Dis Child 69:369–377 [Google Scholar]
- Waldherr R, Lennert T, Weber HP, Fodisch HJ, Scharer K (1982) The nephronophthisis complex: a clinicopathologic study in children. Virchows Arch A Pathol Anat Histol 394:235–254 [DOI] [PubMed] [Google Scholar]
- Zeisberg M, Strutz F, Muller GA (2001) Renal fibrosis: an update. Curr Opin Nephrol Hypertens 10:315–320 [DOI] [PubMed] [Google Scholar]