Abstract
Background
WRKY genes encode one of the most abundant groups of transcription factors in higher plants, and its members regulate important biological process such as growth, development, and responses to biotic and abiotic stresses. Although the soybean genome sequence has been published, functional studies on soybean genes still lag behind those of other species.
Results
We identified a total of 133 WRKY members in the soybean genome. According to structural features of their encoded proteins and to the phylogenetic tree, the soybean WRKY family could be classified into three groups (groups I, II, and III). A majority of WRKY genes (76.7%; 102 of 133) were segmentally duplicated and 13.5% (18 of 133) of the genes were tandemly duplicated. This pattern was not apparent in Arabidopsis or rice. The transcriptome atlas revealed notable differential expression in either transcript abundance or in expression patterns under normal growth conditions, which indicated wide functional divergence in this family. Furthermore, some critical amino acids were detected using DIVERGE v2.0 in specific comparisons, suggesting that these sites have contributed to functional divergence among groups or subgroups. In addition, site model and branch-site model analyses of positive Darwinian selection (PDS) showed that different selection regimes could have affected the evolution of these groups. Sites with high probabilities of having been under PDS were found in groups I, II c, II e, and III. Together, these results contribute to a detailed understanding of the molecular evolution of the WRKY gene family in soybean.
Conclusions
In this work, all the WRKY genes, which were generated mainly through segmental duplication, were identified in the soybean genome. Moreover, differential expression and functional divergence of the duplicated WRKY genes were two major features of this family throughout their evolutionary history. Positive selection analysis revealed that the different groups have different evolutionary rates. Together, these results contribute to a detailed understanding of the molecular evolution of the WRKY gene family in soybean.
Background
The WRKY family is one of the largest transcription factor families in higher plants, but is absent in animals, extending throughout the entire green lineage. Recently, several WRKY genes were identified from non-plant eukaryotes, including Dictyostelium discoideum, a slime mold closely related to the animal and fungi lineages, and the green alga Chlamydomonas reinhardtii, an early branch of plants. This suggests that WRKY genes may have had an early origin in lower eukaryotes, which have since greatly expanded in plant species [1]. Since the first WRKY protein, SPF1, was cloned from sweet potato [2], more and more WRKY genes have been experimentally identified in various plant species [3-19]. Each WRKY protein in this family contains at least one WRKY domain of approximately 60 amino acids with the conserved amino acid sequence WRKYGQK at its N-terminus and a novel zinc finger motif, C2H2 (C–X4–5–C–X22–23–H–X–H) or C2HC (C–X7–C–X23–H–X–C), at the C-terminal region [20]. The WRKYGQK amino acid sequence forms a β-strand that facilitates binding to the promoters of target genes. Usually, the binding site is the W box, which is an element commonly found in the promoters of many stress-related plant genes [21]. WRKY proteins can be categorized into three groups based on their number of WRKY domains and the pattern of their zinc finger motif [22]. The first group contains two WRKY domains (N-terminal and C-terminal), including a C2H2 motif, whereas the other two groups have only one domain. Group III has a distinct zinc finger motif, C2HC rather than the C2H2 found in other groups. Group II proteins can be further subdivided into groups II a, II b, II c, II d, and II e based on the amino acid motifs contained outside the WRKY domain.
As transcription factors, plant WRKY proteins have been shown to be involved in responses to biotic and abiotic stresses, and in developmental processes [23]. It has been well documented that WRKY proteins play an important role in plant defense against biotic stresses, such as bacterial, fungal, and viral pathogens [24-27]. They are also key components in developmental processes, including embryogenesis [28], senescence [29], dormancy [30], trichome development [31], seed development [32], and some signal transduction processes mediated by plant hormones such as gibberellic acid [33], abscisic acid (ABA) [34], or salicylic acid [35]. Meanwhile, increasing evidence has revealed that WRKY proteins are involved in responses to various abiotic stresses [36]. In Arabidopsis, results of a microarray study demonstrated that the expressions of some WRKY transcripts are regulated in response to abiotic stresses, including salinity, drought, and cold [37-39]. In rice, under various abiotic and phytohormone treatments, the expression of WRKY genes showed significant differences [40]. In Poncirus trifoliate, a WRKY gene, PtrWRKY2, showed differential responses to cold and drought stresses [41], while in soybean, at least nine WRKY genes were found to be differentially expressed under abiotic stress [42]. Collectively, this evidence indicates that WRKY genes play important roles in various physiological processes in plants.
Soybean is one of the most important economic crops in the world. Genome and transcriptome sequencing of the palaeopolyploid soybean have been completed [43,44]. In the present study, we searched this genome sequence to identify WRKY proteins, and compared the structure of the encoded proteins with those of their putative homologous WRKY genes in Arabidopsis. In order to investigate tandem duplication events, soybean chromosome sequence information was applied to map WRKY transcripts to their corresponding genetic loci on chromosomes. A phylogenetic tree was constructed to evaluate evolutionary relationships among WRKY genes in the two plant species. In addition, we analyzed the transcriptome atlas of WRKY genes in different tissues under normal conditions, and found notable differential expression between groups, which indicated broad functional divergence in this family. Positive selection analysis revealed that evolutionary rates differed among the different groups. Moreover, evolutionary patterns of the WRKY gene family were examined in Arabidopsis, rice, and soybean. The results indicated that WRKY genes in soybean were duplicated mainly through segmental duplication, which differed from homologous genes in Arabidopsis and rice. These results provide useful information for future studies of molecular evolution of the WRKY gene family in soybean.
Results
Identification and distribution of the WRKY gene family in soybean
In plants, the dicot model organism Arabidopsis is commonly used to predict the function of a gene in a newly or partially sequenced organism that has a close phylogenetic relationship to Arabidopsis, such as soybean. Moreover, there are at least 72 WRKY family members in Arabidopsis, and most of these genes have been extensively studied and reported to be involved in many physiological and biochemical processes [20,22]. With the aim of defining the soybean protein-containing WRKY domains, we downloaded the 72 known Arabidopsis WRKY protein sequences from the Arabidopsis transcription factor database (AtTFDB; http://arabidopsis.med.ohio-state.edu/). In order to examine the structural features of each AtWRKY, we performed a multiple sequence alignment (data not shown). Two members, At4g12020 and At4g30930, were excluded from the analysis due to incomplete zinc fingers and the lack of WRKY domains. Therefore, 70 Arabidopsis WRKY protein sequences were used to BLAST the completed soybean genome sequences for genes that encode proteins containing the WRKY domain. The WRKY domain of each predicted protein was identified by searching against the SMART database. After manually removing overlapping genes, a total of 133 non-redundant genes in the soybean genome were identified as members of the WRKY family (Additional file 1). Among these members, annotation (predicted) of 23 proteins revealed that they have two complete WRKY domains each, which all belong to group I. In addition, physical positions of WRKY genes were obtained from the Phytozome database, and were used to map these genes onto their corresponding soybean chromosomes (Figure 1). Results showed that WRKY genes in soybean could be mapped on all chromosomes, from chromosome 1 to 20. Chromosome 8 had the highest density of WRKY genes with 11 members, while in chromosomes 10, 11, 12, and 20, no more than three WRKY genes could be found, respectively. Examination of the location of each WRKY gene revealed that all GmWRKY genes, except for Glyma02g39870, Glyma03g25770, Glyma04g05700, Glyma04g12830, Glyma05g20710, Glyma06g06530, Glyma06g13090, Glyma06g27440, Glyma06g47880, Glyma08g01430, Glyma09g09400, Glyma09g24080, Glyma09g37930, Glyma11g29720, Glyma12g23950, Glyma13g34280, Glyma14g17330, Glyma14g36430, Glyma14g37960, Glyma15g20990, Glyma16g05880, Glyma17g29190, Glyma18g39970, Glyma18g49140, Glyma19g02440, Glyma19g26400, and Glyma20g03410, originated from segmental duplications (102 of 133) or tandem duplications (18 of 133) (Figure 1). The 27 genes mentioned above might have been produced by retrotransposition instead.
Figure 1.

Chromosome distribution of soybean (Glycine max) WRKY genes. The size of a chromosome is indicated by its relative length. Red outlined boxes represent segmentally duplicated genes. Tandem duplicated genes are indicated with vertical green lines. The location information and chromosome information were obtained from Phytozome. The figure was produced using the MapInspector program.
Multiple sequence alignment and structure analysis
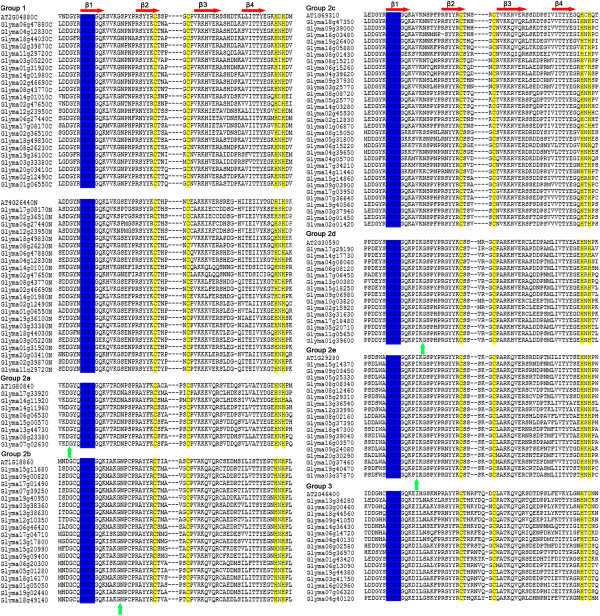
The phylogenetic relationship of GmWRKY proteins was examined by multiple sequence alignment of their WRKY domains, which span across approximately 60 amino acids (Figure 2). A comparison with soybean WRKY domains and several homologous Arabidopsis proteins resulted in a separation of the different groups and subgroups. For each group or subgroup, one Arabidopsis protein was selected randomly, which included At2g04880C, with only one C-terminal WRKY domain, At4g26440N, with only one N-terminal WRKY domain, At1g80840, At1g18860, At1g69310, At2g30590, At1g29280, and At2g46400. As shown in Figure 2, the sequences of soybean WRKY were found to be highly conserved.
Figure 2.
Alignment of multiple soybean WRKY genes and selectedAtWRKY domain amino acid sequences. Alignment was performed using Clustal W. The suffixes ‘N’ or ‘C’ denote the N-terminal and C-terminal WRKY domains from Group I WRKY proteins, respectively. The amino acids forming the zinc-finger motif are highlighted in yellow. The conserved WRKY amino acid signature is highlighted in blue. The four β-strands are shown in red. The position of a conserved intron is indicated by an arrowhead.
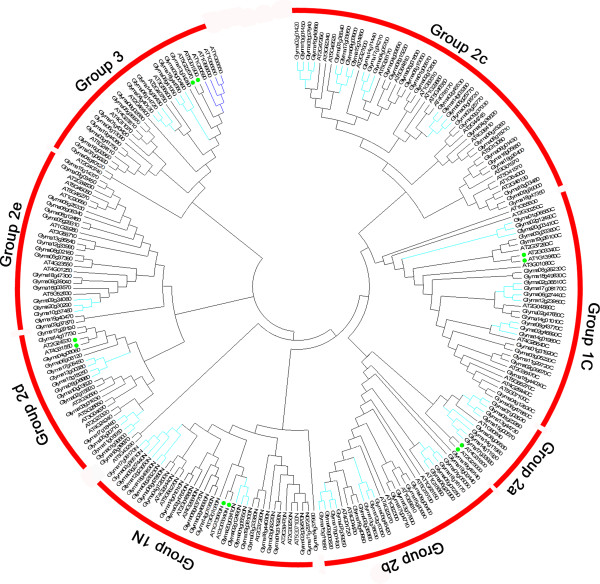
In order to better separate the groups and examine the evolutionary relationships of the WRKY family in soybean and Arabidopsis, an unrooted phylogenetic tree was constructed from alignments of their domain protein sequences, which resulted in the formation of three distinct clusters: group I, group II, and group III (Figure 3). WRKY proteins of Arabidopsis and soybean were present in all clusters. This classification was consistent with results of Rushton et al. [20], who suggested that WRKY domains could be classified into three large groups corresponding to groups I, II, and III of Arabidopsis. Notably, AtWRKY members were more similar to those in the same class in divergent species than they were to other WRKY proteins in the same species. In order to examine the syntenic relationship of the WRKY gene family between the genomes of soybean and Arabidopsis, each WRKY gene within the family in Arabidopsis was searched in the PGDD (http://chibba.agtec.uga.edu/duplication/) (data no shown). During the course of this analysis we found that synteny was relatively well conserved between soybean and Arabidopsis proteins. For example, in subgroup II a (Figure 3), several GmWRKYs (Glyma06g06530, Glyma07g02630, Glyma08g23380 Glyma13g44730, and Glyma15g00570) located on different chromosomes are orthologs of a same AtWRKY gene (At1g80840). Additionally, it is worth noting that the structure and phylogenetic tree of the GmWRKY domain clearly indicated that group II proteins could be divided into five distinct subgroups (II a-e).
Figure 3.
Phylogenetic tree of WRKY domains among soybean andArabidopsis. The amino acid sequences of the WRKY domain of soybean and Arabidopsis were aligned with Clustal W and the phylogenetic tree was constructed using the maximum likelihood method in MEGA 5.0. Group 1 proteins with the suffix ‘N’ or ‘C’ indicate the N-terminal WRKY domains or the C-terminal WRKY domains, respectively. Genes with similar functions clustered together are indicated by filled green circles. Gene expansion in soybean and Arabidopsis are indicated by coloring the subclade with the same color as the leaf label. The red arcs indicate different groups (or subgroups) of WRKY domains.
The phylogenetic classification was found to be consistent with the motif composition among group or subgroup. Differences between groups or subgroups were observed in not only the type of motifs in one WRKY protein, but also in the motif number in one WRKY protein. As displayed schematically in Additional file 2 and Additional file 3, nine types of motifs were detected, including three types of WRKY motifs. The majority of the proteins of subgroups I (73.9%; 17 of 23), II c (93.8%; 30 of 32) and II d (93.3%; 14 of 15), together with those of group III (87.5%; 14 of 16), share a unique WRKY motif, which is shown in red color in Additional file 3. Subgroups II a and II b have the same motif components, suggesting a close phylogenetic relationship. The motif number in each WRKY protein ranged from two to six, and this difference is apparent in groups or subgroups of the WRKY family. For example, all members of group III and the majority of subgroups II e and II d members have two motifs, including a WRKY motif. Interestingly, the relative motif positions in different groups or subgroups also vary significantly. Therefore, motif composition can shed light on phylogenetic relationships of the WRKY family.
Comparison of full-length cDNA sequences with corresponding genomic DNA sequences suggested that the exon number of soybean WRKY genes ranged from two to eight. The results of intron/exon structure identification (Additional file 4) showed that most of the WRKY domain-containing protein genes in different groups or subgroups have very conservative exon numbers. Members of group II d have three exons, and 14 of the 16 genes in group III have three exons. Notably, except for their differences in exon numbers, the relative exon positions in different groups or subgroups also vary significantly. The exon/intron analysis showed clear differences in both exon positions and exon numbers across the different groups or subgroups.
Together, these results indicated that soybean WRKY domains could be classified into three large groups: group I, group II, including group II a-e, and group III. Basic information of all soybean WRKY family members, including conserved heptapeptide, zinc-finger type, domain number, group, coding sequence (CDS) length, and gene length, is provided in Additional file 1.
Transcriptome atlas and promoter analysis
Since the transcriptome sequencing of soybean was completed, the availability of the soybean gene expression atlas facilitates additional studies on the basic biology of soybean [44]. The recently developed RNA-Seq web-based tools, which include gene expression data across multiple tissues and organs, allow for characterization and comparisons of the gene transcriptome atlas in soybean. Consequently, distinct transcript abundance patterns are readily identifiable in the RNA-Seq atlas data set for all 133 GmWRKYs. Similar to other genes that encode transcription factors, many of the GmWRKYs exhibited low transcript abundance levels, as determined by the RNA-Seq atlas analysis. Furthermore, we observed that most of the genes had very broad expression spectra. However, six GmWRKYs, including Glyma04g05700, Glyma04g39650, Glyma04g40120, Glyma08g01430, Glyma01g05050, and Glyma14g11440, lacked expression information, which possibly indicated that these were pseudogenes or were expressed only at specific developmental stages or under special conditions. We observed that accumulation of WRKY gene transcripts was associated with different tissues, and expression patterns differed among each WRKY gene member (Figure 4). In soybean, 33.1% (44 of 133) of the analyzed WRKYs were constitutively expressed in all of the seven tissues tested, which suggested that GmWRKYs play regulatory roles at multiple developmental stages. By contrast, most GmWRKYs showed preferential expression. RNA-Seq atlas data revealed that the majority (92 of 133; 69.2%) of GmWRKYs exhibited transcript abundance profiles with marked peaks in only a single tissue. This result suggests that these WRKY proteins function as tissue-specific regulators and are limited to discrete cells or organs. Approximately 45 of these 133 (33.8%) GmWRKYs showed the highest transcript accumulation level in root tissue, 20 (15.0%) showed the highest transcript accumulation in flower tissue, 13 (9.8%) showed the highest transcript accumulation level in nodule tissue, and surprisingly, only one showed the highest transcript accumulation level in seed tissue. The wide expression of these genes suggests that WRKY genes from soybean are involved in the development of all organs or tissues under normal conditions. In addition to groups of genes that exhibited similar transcript abundance profiles but were relatively phylogenetically distinct, several phylogenetic clades shared the same transcript abundance profile to a large extent. For example, in subgroup II b, most of the GmWRKYs were preferentially expressed in root tissue. Interestingly, in Arabidopsis, according to the experimental results of Dong et al. [45], more than half of the members in subgroup II b showed similar preferential expression in the root tissue under normal conditions, which indicated the conserved functional role of subgroup II b in root development between the two species. The expression of members of group I in soybean was detectable in flower tissue, which suggested their conserved roles in flower formation. Members of group I also showed similar expression patterns in nodule and root development. Furthermore, GmWRKYs with high sequence similarity and shared expression profiles represent good candidates for evaluation of gene functions in soybean. The transcriptome atlas indicted that differential expression was extended to all groups or subgroups of the soybean WRKY gene family, which was further verified by the promoter analysis. Because transcription factors bind to corresponding transcription factor binding sites (TFBSs) upstream of genes of interest, profiles of cis-acting elements may thus provide useful information related to the regulatory mechanism of gene expression. A computational tool, PlantCARE [46], was adopted to identify putative TFBSs in the 1500-bp DNA sequence upstream of the translation initiation codon of WRKY genes in soybean. Four types of cis-acting elements were found to be significantly abundant in the promoter region of GmWRKY genes (Additional file 5). The first type of cis-acting element enriched in the promoter region is the light responsive elements, such as G-box [47,48], GAG-motif [49], Box I [50], and Box 4 [51], etc. The G-box element appears to be more abundant in subgroup II a, in which each member contains at least two copies. In addition, the mean number of G-box copies was 3.625 in subgroup II a, which is higher than that in other subgroups or in the whole WRKY gene family. This result indicates that the G-box element seems to be enriched in subgroup II a. All but seven (94.7%; 126 of 133, Group I; 2, Group II b; 2, Group II c; 1, Group II d; 1, and Glyma14g37960; 1) have at least one Box 4 element copy. Its mean number of copies (3.744) in the whole WRKY gene family was apparently higher than that of other types of cis-acting elements except for TATA-box, CAAT-box, and unnamed-4. This result suggests that the Box 4 element tends to be enriched in the soybean WRKY gene family. As one part of a conserved DNA module involved in light responsiveness, previous studies showed that the Box 4 element is frequently found in promoter regions of different genes from various species [51,52]. However, it is noteworthy that the Box 4 element was found in high frequency in the soybean WRKY gene family, which suggests that the Box 4 element may be important for light-controlled transcriptional activity [53]. Plant hormone responsive elements, such as ABRE [54], P-box [55], as well as the TCA-element [56], constitute the second class. ABRE (51.9%; 69 of 133) appears to be one of the most abundant hormone-related cis-acting element in soybean, suggesting that abscisic acid (ABA) regulates the expression of some GmWRKYs, whereas such elements were rarely detected in Group II d (20.0%; 3 of 15). By contrast, the salicylic acid responsive TCA-element was frequently found in groups or subgroups. These observations suggest that GmWRKYs in different groups are likely to be significantly regulated by different types of hormones. The third most abundant cis-acting element class consisted of elements that respond to external environmental stresses. We observed that most of the GmWRKYs examined appeared to contain MBS (72.2%; 96 of 133) [57], heat shock element (HSE) (77.4%; 103 of 133) [58], and TC-rich repeat elements (71.4%; 95 of 133) [59]. MBS is an element involved in drought induction, and HSE is also enriched in the promoter. With a few exceptions, GmWRKYs contain more than two copies of this element. Circadian elements, which are involved in circadian control [60], is the fourth type of cis-acting element that was abundantly found in the promoter regions of soybean WRKY genes. PlantCARE identified 98 (73.7%; 98 of 133) GmWRKY genes containing circadian elements, which may be responsible for its distinct diurnal expression pattern. The presence of a diversity of cis-acting elements in the upstream regions of GmWRKYs indicates that GmWRKYs may function in a relatively wide range of activities.
Figure 4.
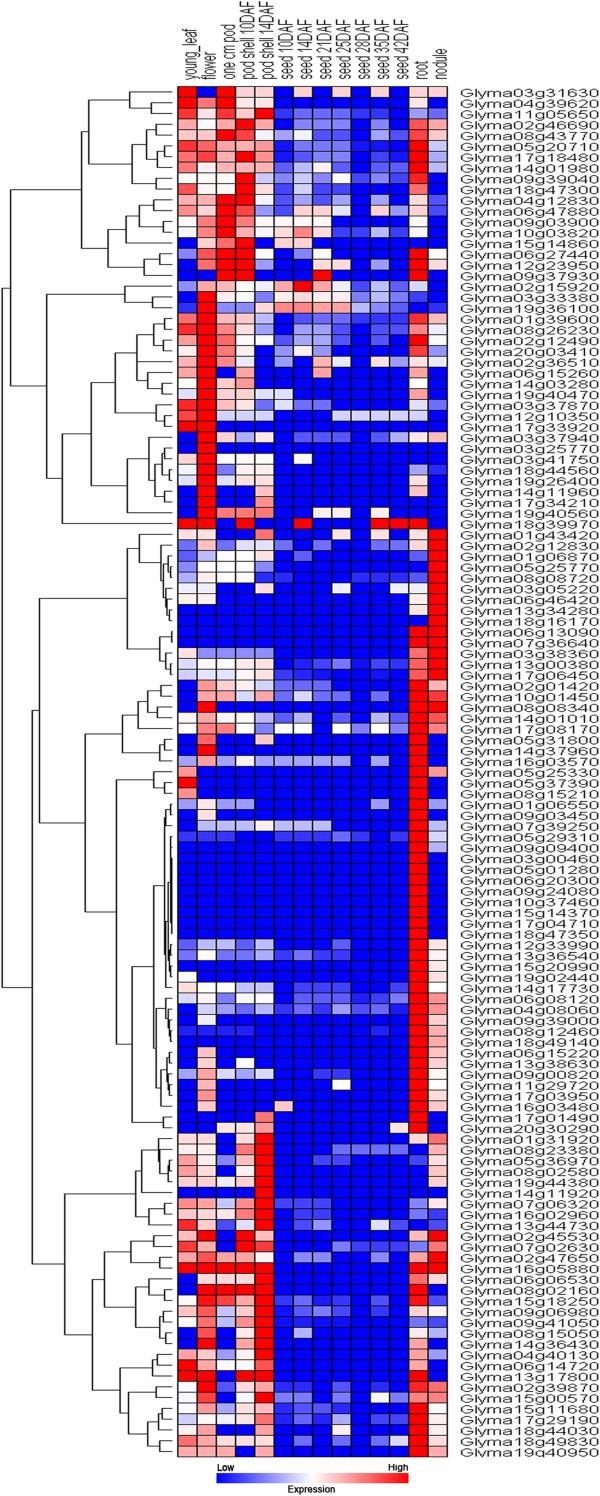
Expression profiles of 127 soybean WRKY genes. The hierarchical cluster color code: the largest values are displayed as the reddest (hot), the smallest values are displayed as the bluest (cool), and the intermediate values are a lighter color of either blue or red. Pearson correlation clustering was used to group the developmentally regulated genes. Six genes were excluded from the analysis due to no expression in an organ or a period.
The above results indicated that the 133 WRKY genes in soybean display differential expression, either in their transcript abundance or in their expression patterns under normal growth conditions in different groups or subgroups.
Detection of positive selection and functional divergence analysis (FDA)
Site models and branch-site models in PAML [61] were used to detect positive selection in the WRKY gene family of soybean. Substitution rate ratios of non-synonymous (dN or Ka) versus synonymous (dS or Ks) mutations (dN/dS or ω) were calculated. A Ka/Ks ratio of 1 indicates genes that are subject to neutral selection, <1 indicates genes subject to negative selection, and >1 indicates genes subject to positive selection [62]. Additional file 6 lists parameter estimates and log-likelihood values for each site model. Two pairs of models (M0/M3 and M7/M8) were selected and compared. The site-homogeneous model, M0 (one-ratio), assumes one ω for all sites, whereas M3 (discrete) assumes a general discrete distribution. Two other models used were M7 (beta), which assumes a beta distribution of ω that is limited to the range (0, 1), and M8 (beta & ω), which adds an extra site class with ω estimated [63]. In addition, to test for variable omega ratios among lineages, we conducted the likelihood ratio test (LRT) to compare the two extreme models. The log likelihood values under the one-ratio model and the discrete model were determined to be −8608.409 and −8272.457, respectively. Twice the log likelihood difference value, 2ΔlnL = 335.95, was found to be strongly statistically significant (p < 0.01), thus revealing a heterogeneous selective pressure among lineages. Moreover, the log likelihood value under the beta model and the beta & ω were −8253.022 and −10197.202, respectively. Twice the log likelihood difference, 2ΔlnL = 1944.18, was also strongly statistically significant (p < 0.01). The comparison of M3 versus M0 revealed that none of the codon sites appeared to be under the influence of positive selection (ω > 1). By contrast, comparing the M7 model to the M8 model indicated that ~0.001% of codons fell within an estimated ω value of 2.638, suggesting positive selection. We also used Bayes empirical Bayes (BEB) estimation methods in model M8 [64] to identify sites under positive selection. We found only one positive selection site at the 0.05 significance level, and three sites at the 0.01 significance level. Together, these results indicate that no strong positive selection sites could be detected under the site model in the soybean WRKY gene family.
Branch-site models allow ω to vary both among sites in the protein and across branches on the tree, and aim to detect positive selection affecting a few sites along particular lineages [64]. The branches being tested for positive selection are referred to as the foreground branches, and all other branches on the tree are referred to as background branches. The BEB method was implemented to calculate posterior probabilities (Qks) for site classes if the LRT suggested the presence of codons under positive selection on the foreground branch [65]. In our study, group I, group II a-e, and group III were selected as foreground branches, respectively, while the other groups were selected as the background branches. It is notable that no positive selection sites were observed in groups II a, II b, or II d. In contrast, positive selection sites detected by the branch-site model (Table 1) were distributed in groups II e and III at the 0.01 significance level. This result demonstrated that the groups have different evolutionary rates. Group II e and group III appeared to be confronted with strong positive Darwinian selection, as many highly significant positive sites were present, whereas evolution in the other groups appeared to be more conservative.
Table 1.
Parameters estimation and likelihood ratio tests for the branch-site models
|
Branch-site model | |||||
|---|---|---|---|---|---|
|
Foreground branches |
Estimates of parameter |
Positive selection sites (BEB) 4 |
|||
| Site class10 | Site class 1 | Site class 2a | Site class 2b | ||
| Group 1 |
P0 = 0.75959 |
P1 = 0.05958 |
P2a = 0.16768 |
P2b = 0.01315 |
261G*,262S*,282H*, |
| ω0(b)2 = 0.05978 |
ω1(b) = 1.00000 |
ω2a(b) = 0.05978 |
ω2b(b) = 1.00000 |
288D*, 292 M* |
|
| ω0(f)3 = 0.05978 |
ω1(f) = 1.00000 |
ω2a(f) = 232.95169 |
ω2b(f) = 232.95169 |
|
|
| Group 2a |
P0 = 0.00000 |
P1 = 0.00000 |
P2a = 0.92727 |
P2b = 0.07273 |
None |
| ω0(b) = 0.06077 |
ω1(b) = 1.00000 |
ω2a(b) = 0.06077 |
ω2b(b) = 1.00000 |
|
|
| ω0(f) = 0.06077 |
ω1(f) = 1.00000 |
ω2a(f) = 999.00000 |
ω2b(f) = 999.00000 |
|
|
| Group 2b |
P0 = 0.00001 |
P1 = 0.00000 |
P2a = 0.92726 |
P2b = 0.07273 |
None |
| ω0(b) = 0.06039 |
ω1(b) = 1.00000 |
ω2a(b) = 0.06039 |
ω2b(b) = 1.00000 |
|
|
| ω0(f) = 0.06039 |
ω1(f) = 1.00000 |
ω2a(f) = 1.00000 |
ω2b(f) = 1.00000 |
|
|
| Group 2c |
P0 = 0.80507 |
P1 = 0.06312 |
P2a = 0.12223 |
P2b = 0.00958 |
261G**,275R** |
| ω0(b) = 0.06042 |
ω1(b) = 1.00000 |
ω2a(b) = 0.06042 |
ω2b(b) = 1.00000 |
|
|
| ω0(f) = 0.06042 |
ω1(f) = 1.00000 |
ω2a(f) = 167.23585 |
ω2b(f) = 167.23585 |
|
|
| Group 2d |
P0 = 0.00000 |
P1 = 0.00000 |
P2a = 0.92727 |
P2b = 0.07273 |
None |
| ω0(b) = 0.06118 |
ω1(b) = 1.00000 |
ω2a(b) = 0.06118 |
ω2b(b) = 1.00000 |
|
|
| ω0(f) = 0.06118 |
ω1(f) = 1.00000 |
ω2a(f) = 981.94932 |
ω2b(f) = 981.94932 |
|
|
| Group 2e |
P0 = 0.76730 |
P1 = 0.06008 |
P2a = 0.16008 |
P2b = 0.01253 |
248E**, 249Y**, 286A*, |
| ω0(b) = 0.06061 |
ω1(b) = 1.00000 |
ω2a(b) = 0.06061 |
ω2b(b) = 1.00000 |
288D**, 298E**, 299G** |
|
| ω0(f) = 0.06061 |
ω1(f) = 1.00000 |
ω2a(f) = 999.00000 |
ω2b(f) = 999.00000 |
|
|
| Group 3 |
P0 = 0.63465 |
P1 = 0.04978 |
P2a = 0.29262 |
P2b = 0.02295 |
258P*, 260 K**, 263P**, 275R*, |
| ω0(b) = 0.06176 |
ω1(b) = 1.00000 |
ω2a(b) = 0.06176 |
ω2b(b) = 1.00000 |
293 L*, 294I**, 298E**, 303H** |
|
| ω0(f) = 0.06176 | ω1(f) = 1.00000 | ω2a(f) = 999.00000 | ω2b(f) = 999.00000 | ||
Note: * p < 0.05 and ** p < 0.01 (x2 test).
1 The sites in the sequence evolve according to the same process, the transition probability matrix is calculated only once for all sites for each branch.
2 Background ω.
3 Foreground ω.
4 The number of amino acid sites estimated to have undergone positive selection; BEB: Bayes Empirical Bayes.
Type I functional divergence (shifted evolutionary rate) and Type II functional divergence (altered amino acid physicochemical property) between gene clusters of the WRKY gene family were estimated by posterior analysis using the program DIVERGE v2.0 [66,67]. Because these methods are not sensitive to saturation of synonymous sites, they have been extensively applied in research of various gene families [68-70]. The estimation was based on the WRKY protein neighbor-joining tree, in which seven major clades were clearly present. Pairwise comparisons of paralogous WRKY genes from group I, group II a-e, and group III were carried out, and the rate of amino acid evolution at each sequence position was estimated. Our results (Additional file 7) indicated that with nine exceptions (group pairs II d/II e, II d/III, II d/II a, II d/I, II e/III, III/II a, and II b/I), the coefficients of Type-I functional divergence (θML) between WRKY groups were moderately statistically significant (p < 0.05), with θML values ranging from 0.201 to 0.395, or strongly statistically significant (p < 0.01) with θ ML values ranging from 0.311 to 0.618. These results indicated significant site-specific altered selective constraints on some members of the WRKY family, leading to subgroup-specific functional evolution after diversification. Additionally, Type-I functional divergence was not evident in the comparison of group II d with the other four groups, which suggests that group II d may be the most conservative clade. Type-II functional divergence was evident in all groups or subgroups (Additional file 8), with θ-II values ranging from 0.033 to 0.288 (p < 0.05), indicating a radical shift of amino acid properties. These results suggest that the relative importance of Type-I and Type-II functional divergence might be associated with specific functional classes of the WRKY gene family in soybean.
Furthermore, some critical amino acid residues responsible for functional divergence were predicted with suitable cut-off values derived from the Qk of each comparison. In order to reduce false positives, Qk > 0.8 was used as the cutoff to identify Type-I and Type-II functional divergence-related residues in all comparisons between the seven WRKY groups or subgroups. Results showed distinct differences in the number and distribution of predicted sites for functional divergence within each pairwise comparison. However, some critical amino acid sites still showed evidence of both Type-I and Type-II functional divergence in corresponding pairs. For example, five critical residues were predicted for the subgroup II e/II c (248E, 258P, 264Y, 275R, 295 V) and II e/II b (248E, 258P, 275R, 276G, 298E) pairwise comparisons, whereas three critical amino acids sites were predicted for the subgroup II e/I (248E, 264Y, 275R) and III/II b (264Y, 295 V, 298E) pairwise comparisons, respectively. Similar cases were found in other subgroup pairwise comparisons. Shifted evolutionary rates and altered amino acid physicochemical properties co-occurred at the same amino acid sites, revealing that these sites have played important roles in functional divergence during the process of evolution.
Expansion pattern of the WRKY family in soybean
Gene duplication events are important for gene family evolution, because duplicated genes provide the raw materials for the generation of new genes, which in turn facilitate the generation of new functions. Three principal evolutionary patterns were attributed to gene duplications: segmental duplication, tandem duplication, and transposition events such as retroposition and replicative transposition [71]. Among these, segmental duplication occurs most frequently in plants because most plants are diploidized polyploids and retain numerous duplicated chromosomal blocks within their genomes [72]. Previous studies reported several rounds of whole-genome duplication (WGD) in both the Arabidopsis and rice genomes [73,74]. The occurrence of large-scale gene duplication events was also demonstrated in soybean [43]. For this analysis, we focused on the tandem and segmental duplication modes. Tandem duplications were characterized as multiple members of one family occurring within the same intergenic region or in neighboring intergenic regions, where genes were clustered together with a maximum of 10 extra genes between them [40]. We searched for contiguous WRKY genes in both the sharing region and neighboring regions. Eighteen of the 133 genes (13.5%) in this family were found to be located as tandem repeats in soybean (Figure 1), indicating that tandem duplications contributed to the expansion of this family. We also tested the hypothesis that segmental duplication events played a large role in the evolution of the WRKY gene family in soybean. For each WRKY gene, we tallied the number of flanking protein-coding genes with a best non-self match to a protein-coding gene neighboring its paralog. According to our results, 76.7% (102 of 133) of genes showed high conservation, indicating that these WRKY genes were formed through segmental duplication in soybean (Table 2). Intriguingly, comparison of the 102 segmental duplicated genes in our study to the results of Du et al. [75] suggested that 91 (89.2%; 91 of 102) WRKY genes originated from WGDs, and the duplication status of the remaining 11 (10.7%; 11 of 102) WRKY genes, including Glyma01g05050, Glyma01g43420, Glyma02g15920, Glyma02g45530, Glyma03g00460, Glyma03g31630, Glyma08g15210, Glyma10g03820, Glyma13g38630, Glyma18g16170, and Glyma18g44030, was singleton, which indicated that these segmental duplication genes may be derived from independent duplication events. These results indicated that most of the WRKY genes in soybean were retained after WGDs. Edger et al. [76] stated that dosage-sensitive genes, including transcription factors, were preferentially retained following WGDs, which is compatible with the present study. We did not find evidence that other pairs of paralogous genes in soybean originated from segmental duplication. These results indicate that the soybean WRKY family arose mainly through segmental duplications.
Table 2.
Estimates of the dates for the segmental duplication events of WRKY family in soybean
| Segment pairs | Number of anchors | KS (mean ± s.d.) | Estimated time (mya) |
|---|---|---|---|
| Glyma01g05050 & Glyma18g16170 |
4 |
0.60 ± 0.20 |
49 |
| Glyma01g06550 & Glyma02g12490 |
3 |
0.17 ± 0.06 |
14 |
| Glyma01g06870 & Glyma02g12830 |
4 |
0.16 ± 0.08 |
13 |
| Glyma01g31920 & Glyma03g05220 |
5 |
0.19 ± 0.06 |
16 |
| Glyma01g31920 & Glyma18g44030 |
3 |
0.71 ± 0.22 |
58 |
| Glyma01g39600 & Glyma11g05650 |
17 |
0.17 ± 0.07 |
14 |
| Glyma01g43420 & Glyma05g36970 |
5 |
0.70 ± 0.19 |
57 |
| Glyma01g43420 & Glyma08g02580 |
4 |
0.68 ± 0.17 |
56 |
| Glyma02g01420 & Glyma03g37940 |
8 |
0.67 ± 0.11 |
55 |
| Glyma02g01420 & Glyma10g01450 |
19 |
0.20 ± 0.11 |
16 |
| Glyma02g01420 & Glyma19g40560 |
7 |
0.72 ± 0.17 |
59 |
| Glyma02g15920 & Glyma03g31630 |
6 |
0.60 ± 0.16 |
49 |
| Glyma02g15920 & Glyma10g03820 |
11 |
0.15 ± 0.11 |
12 |
| Glyma02g36510 & Glyma17g08170 |
18 |
0.13 ± 0.05 |
11 |
| Glyma02g45530 & Glyma14g03280 |
6 |
0.12 ± 0.05 |
10 |
| Glyma02g46690 & Glyma08g43770 |
8 |
0.61 ± 0.19 |
50 |
| Glyma02g46690 & Glyma14g01980 |
16 |
0.13 ± 0.07 |
11 |
| Glyma02g47650 & Glyma14g01010 |
21 |
0.14 ± 0.08 |
11 |
| Glyma03g00460 & Glyma09g41050 |
5 |
0.51 ± 0.12 |
42 |
| Glyma03g31630 & Glyma10g03820 |
6 |
0.55 ± 0.10 |
45 |
| Glyma03g33380 & Glyma19g36100 |
21 |
0.19 ± 0.17 |
16 |
| Glyma03g37870 & Glyma19g40470 |
18 |
0.15 ± 0.07 |
12 |
| Glyma03g37940 & Glyma10g01450 |
9 |
0.73 ± 0.16 |
60 |
| Glyma03g37940 & Glyma19g40560 |
17 |
0.15 ± 0.07 |
12 |
| Glyma03g38360 & Glyma19g40950 |
11 |
0.16 ± 0.09 |
13 |
| Glyma03g41750 & Glyma07g06320 |
5 |
0.66 ± 0.23 |
54 |
| Glyma03g41750 & Glyma16g02960 |
3 |
0.62 ± 0.26 |
51 |
| Glyma03g41750 & Glyma19g44380 |
19 |
0.18 ± 0.13 |
15 |
| Glyma04g08060 & Glyma06g08120 |
14 |
0.18 ± 0.12 |
15 |
| Glyma04g39620 & Glyma06g15260 |
14 |
0.23 ± 0.19 |
19 |
| Glyma04g39650 & Glyma05g31800 |
4 |
0.72 ± 0.19 |
59 |
| Glyma04g39650 & Glyma06g15220 |
15 |
0.22 ± 0.19 |
18 |
| Glyma04g39650 & Glyma08g15050 |
4 |
0.66 ± 0.19 |
54 |
| Glyma04g40130 & Glyma06g14720 |
18 |
0.25 ± 0.25 |
20 |
| Glyma05g01280 & Glyma06g20300 |
6 |
0.57 ± 0.21 |
47 |
| Glyma05g25330 & Glyma08g08340 |
12 |
0.21 ± 0.19 |
17 |
| Glyma05g25770 & Glyma08g08720 |
14 |
0.20 ± 0.15 |
16 |
| Glyma05g29310 & Glyma08g12460 |
18 |
0.16 ± 0.07 |
13 |
| Glyma05g31800 & Glyma06g15220 |
4 |
0.63 ± 0.22 |
52 |
| Glyma05g31800 & Glyma08g15050 |
19 |
0.14 ± 0.08 |
11 |
| Glyma05g36970 & Glyma08g02580 |
17 |
0.22 ± 0.15 |
18 |
| Glyma05g37390 & Glyma08g02160 |
17 |
0.14 ± 0.08 |
11 |
| Glyma06g15220 & Glyma08g15050 |
5 |
0.68 ± 0.23 |
56 |
| Glyma06g15260 & Glyma08g15210 |
3 |
0.71 ± 0.21 |
58 |
| Glyma06g46420 & Glyma12g10350 |
9 |
0.23 ± 0.19 |
19 |
| Glyma06g46420 & Glyma13g38630 |
6 |
0.72 ± 0.11 |
59 |
| Glyma07g02630 & Glyma08g23380 |
15 |
0.22 ± 0.18 |
18 |
| Glyma07g02630 & Glyma13g44730 |
6 |
0.56 ± 0.18 |
46 |
| Glyma07g02630 & Glyma15g00570 |
6 |
0.54 ± 0.15 |
44 |
| Glyma07g06320 & Glyma16g02960 |
12 |
0.16 ± 0.07 |
13 |
| Glyma07g06320 & Glyma19g44380 |
5 |
0.61 ± 0.17 |
50 |
| Glyma07g36640 & Glyma15g14860 |
5 |
0.59 ± 0.21 |
48 |
| Glyma07g36640 & Glyma17g03950 |
13 |
0.23 ± 0.23 |
19 |
| Glyma07g36640 & Glyma09g03900 |
5 |
0.73 ± 0.18 |
60 |
| Glyma07g39250 & Glyma09g00820 |
5 |
0.63 ± 0.15 |
52 |
| Glyma07g39250 & Glyma15g11680 |
7 |
0.65 ± 0.18 |
53 |
| Glyma07g39250 & Glyma17g01490 |
23 |
0.17 ± 0.11 |
14 |
| Glyma08g23380 & Glyma13g44730 |
4 |
0.49 ± 0.18 |
40 |
| Glyma08g23380 & Glyma15g00570 |
4 |
0.52 ± 0.17 |
43 |
| Glyma08g26230 & Glyma18g49830 |
8 |
0.21 ± 0.08 |
17 |
| Glyma08g43770 & Glyma14g01980 |
7 |
0.56 ± 0.13 |
46 |
| Glyma09g00820 & Glyma15g11680 |
15 |
0.20 ± 0.14 |
16 |
| Glyma09g00820 & Glyma17g01490 |
5 |
0.67 ± 0.21 |
55 |
| Glyma09g03450 & Glyma15g14370 |
14 |
0.20 ± 0.15 |
16 |
| Glyma09g03900 & Glyma15g14860 |
12 |
0.25 ± 0.21 |
20 |
| Glyma09g03900 & Glyma17g03950 |
3 |
0.59 ± 0.17 |
48 |
| Glyma09g06980 & Glyma13g00380 |
5 |
0.69 ± 0.18 |
57 |
| Glyma09g06980 & Glyma15g18250 |
7 |
0.22 ± 0.21 |
18 |
| Glyma09g06980 & Glyma17g06450 |
5 |
0.56 ± 0.11 |
46 |
| Glyma09g39000 & Glyma18g47350 |
11 |
0.19 ± 0.10 |
16 |
| Glyma09g39040 & Glyma18g47300 |
13 |
0.18 ± 0.10 |
15 |
| Glyma09g41050 & Glyma18g44560 |
12 |
0.16 ± 0.06 |
13 |
| Glyma10g01450 & Glyma19g40560 |
6 |
0.70 ± 0.17 |
57 |
| Glyma10g37460 & Glyma20g30290 |
11 |
0.13 ± 0.05 |
11 |
| Glyma11g05650 & Glyma17g18480 |
3 |
0.71 ± 0.23 |
58 |
| Glyma12g10350 & Glyma13g38630 |
4 |
0.73 ± 0.10 |
60 |
| Glyma12g33990 & Glyma13g36540 |
12 |
0.23 ± 0.20 |
19 |
| Glyma13g00380 & Glyma17g06450 |
17 |
0.19 ± 0.13 |
16 |
| Glyma13g17800 & Glyma17g04710 |
17 |
0.20 ± 0.17 |
16 |
| Glyma13g44730 & Glyma15g00570 |
13 |
0.12 ± 0.06 |
10 |
| Glyma14g11440 & Glyma17g34210 |
6 |
0.32 ± 0.19 |
26 |
| Glyma14g11920 & Glyma17g33920 |
5 |
0.17 ± 0.05 |
14 |
| Glyma15g11680 & Glyma17g01490 |
7 |
0.67 ± 0.20 |
55 |
| Glyma15g14860 & Glyma17g03950 |
5 |
0.58 ± 0.24 |
48 |
| Glyma15g18250 & Glyma17g06450 |
4 |
0.68 ± 0.16 |
56 |
| Glyma16g02960 & Glyma19g44380 | 3 | 0.58 ± 0.17 | 48 |
Abbreviation: mya, million years ago.
We also used Ks, as the proxy for time, and the conserved flanking protein-coding genes to estimate the dates of the segmental duplication events. The mean Ks values and the estimated dates for all segmental duplication events corresponding to WRKY genes are listed in Table 2. The segmental duplicated events in soybean appear to have occurred recently, and focus on two periods, 10–20 mya and 40–60 mya, which is consistent with the ages of the genome duplication events [43]. Taken together, our analyses suggested that segmental duplication is the main mechanism for expansion of this WRKY gene family, accompanied by tandem duplications.
Discussion
Identification, classification, and phylogenetic analysis of the soybean WRKY gene family
The genome sequence and transcriptome profiles of soybean provide a large amount of useful data to explore functional diversity from multiple perspectives. In this study, we identified 133 WRKY members in the soybean genome. A phylogenetic tree including 133 distinct protein sequences clearly demonstrated that these genes could be divided into three groups. This classification was further supported by the results of motifs and exon/intron analyses. The topology of our phylogenetic tree constructed from WRKY genes of two species (soybean and Arabidopsis) is largely consistent with that derived from Arabidopsis alone. All of the evidence obtained suggested that the classification was reasonable and reliable.
WRKY transcription factors have their evolutionary origin in ancient eukaryotes, whose genomes contain a single WRKY gene with two WRKY domains. The presence of a group I WRKY protein in these ancient organisms suggests that group I WRKY genes represent the ancestral form, with other groups arising later through losses and gains of WRKY domains [22]. In the present study, phylogenetic analysis based on the relationship of different groups, indicated that domain gain and loss has indeed been a driving force in the expansion of the WRKY gene family. For example, subgroups II a, II b, and II c were phylogenetically closer to the C-terminal WRKY domain of group I.
Glyma14g37960 and Glyma18g39970 were not assigned to any groups or subgroups. Glyma14g37960 has one WRKY domain; however, it is phylogenetically closer to group I N. Thus, Glyma14g37960 may have arisen from a two-domain WRKY protein that lost one of its WRKY domains during evolution, whereas in Glyma18g39970, a mutation in the sequence outside of the WRKY domain may have occurred before or after the domain loss.
Transcriptome atlas, positive selection, and FDA of soybean WRKY proteins
The transcriptome atlas revealed differential expression of the WRKY gene family under normal growth conditions. Furthermore, results of the promoter analysis were compatible with differential expression patterns. The elements were distributed across three main functional categories, including biotic and abiotic stresses and developmental processes. Surprisingly, Skn-1 motif elements, which are required for high levels of endosperm expression in cooperative interaction with other motifs (AACA, GCN4, ACGT) [77], were found to be abundant in all groups or subgroups. This result appears to contradict with the expression analysis, in which only one gene showed the highest transcript accumulation level in seed tissue. Since the function of cis-acting elements is to regulate gene expression, we speculated that the reason for this phenomenon might be due to the deficiency of Skn-1 motif element partners, AACA and ACGT elements, which were rarely detected in our study. On the other hand, according to the transcriptome atlas of the soybean WRKY gene family, the majority of GmWRKYs showed relatively reduced expression in seed development compared to other organs, which suggests that the expression of genes can be significantly affected when the Skn-1 motif lacks its partners. To further investigate the reason for this differential expression, we performed a positive selection analysis and a functional divergence analysis.
We used both site models and branch-site models to detect positive selection. The results of site models indicated that one category of ω did not fit the data well to describe the variability in selection pressures across amino acid sites. Therefore, the branch-site models, which allowed ω ratios to vary among sites and lineages simultaneously, appeared to be most suitable for describing evolutionary processes of the WRKY gene family. The branch-site analysis revealed that several sites were under positive selection. Along the group II e clade, the following sites were identified to be under positive selection: 248E, 249Y, 286A, 288D, 298E, and 299G. Similar results were found in the group I, III, and II c clades.
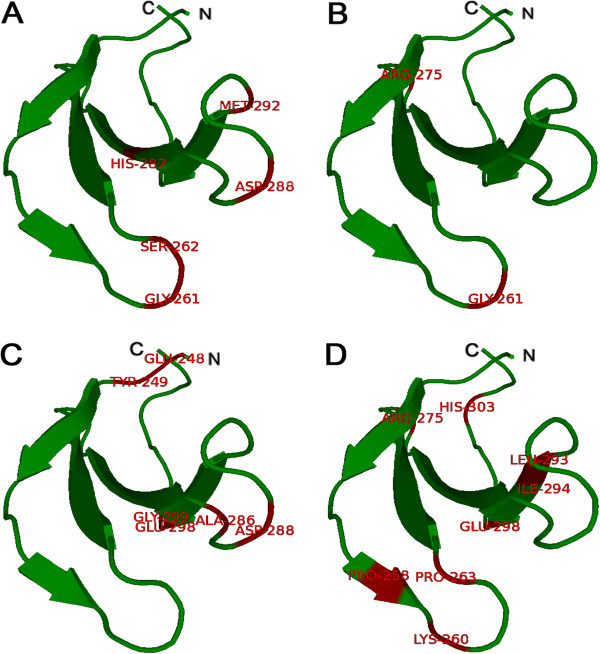
Figure 5 shows the locations of amino acid sites detected by PAML 4 in the 3D structure. Interestingly, with the exception of four amino acid sites (position 258, 282, 293, and 294), sites in different groups or subgroups were all located in the loop regions. Duan et al. [78] suggested that the DNA-binding ability of AtWRKY was mediated through the beta-hairpin regions between β2 and β3, and similar results were reported by Maeo et al. [79]. These results confirmed the theory proposed by Church et al. [80] for non-helical DNA binding. Furthermore, previous work on DNA binding of the WRKY family revealed that the conserved WRKYGQK region was important for DNA binding [79]. According to our results, the amino acid residues of bridging loops between β-strand regions may have been adapted for new functional roles during the process of evolution.
Figure 5.
Model building of the 3D structure of the soybean WRKY protein (Glyma13g00380) based on similarity to theAtWRKY4-C domain (Protein Data Bank (PDB) code: 2lexA). The ensemble of the selected structures in stereo view (A), (B), (C), and (D) positive selection sites detected by the branch-site model presented in group I, group II c, group II e, and group III, respectively. The sites with red color indicate amino acid residues under statistically significant (p < 0.05) positive selection, as calculated by Bayes Empirical Bayes estimation methods. The presented region is Asp247–Pro306, excluding the N-terminal region. The figure was produced using the Swiss-model and pyMOL programs.
Moreover, we further compared the number of WRKY genes in different groups or subgroups among Arabidopsis, rice, and soybean (Table 3). We observed that the number of members in different groups or subgroups was approximately doubled in soybean than in corresponding groups or subgroups in Arabidopsis and rice, which can be attributed to the more recent two genome duplication events in the soybean genome [43]. The key difference is that the number of group III members in soybean is roughly the same as that in Arabidopsis, but half of that in rice. This result may indirectly reflect the fact that group III in the dicotyledons may be confronted with strong positive Darwinian selection, whereas the evolution of this subgroup may be more conservative in the monocotyledons.
Table 3.
Number of WRKY genes in Arabidopsis, rice, and soybean
| Group1 | Group2a | Group2b | Group2c | Group2d | Group2e | Group3 | |
|---|---|---|---|---|---|---|---|
|
AtWRKY |
13 |
4 |
7 |
18 |
7 |
9 |
14 |
|
OsWRKY |
15 |
4 |
8 |
15 |
7 |
11 |
36 |
| GmWRKY※ | 23 | 8 | 19 | 32 | 15 | 18 | 16 |
Note: ※ the WRKY proteins of soybean (Glycine max).
Functional innovations include subfunctionalization [81], neofunctionalization [82], and subneofunctionalization [83]. Gene duplication may result in altered functional constraints between the gene clusters of a gene family. The results of the functional divergence analysis suggested that WRKY genes should be significantly functionally divergent from each other, especially with respect to the four amino acid residues (248E, 275R, 288D and 298E) identified by both PAML 4 and DIVERGE 2.0 analyses, which were inferred to have played important roles during evolution. On the other hand, functional divergence might reflect the existence of long-term selective pressures.
The soybean WRKY gene family arose mainly through segmental duplication
The dramatic variation we observe in gene family size and distribution may have resulted owing to many processes, including tandem duplication with high rates of birth and death and gene duplication resulting from larger scale genome events such as polyploidy or duplications of chromosomal regions (here referred to as “segmental duplications”).
The current investigation revealed the duplication pattern of the soybean WRKY gene family. One hundred two genes were found to evolve from segmental duplication, suggesting that segmental duplication likely played a pivotal role in WRKY gene expansion in the soybean genome. The genome sequencing results revealed two genome duplication events in soybean, aging ~13 mya and ~57 mya [43], which is consistent with results of the present study. We inferred that the expansion of the WRKY gene family occurred along with genome duplication events, and that these genes were retained during evolution. The structural similarity and variation between genes located on the same chromosome and the phylogenetic analysis might help to explain the order of duplication events of the soybean WRKY genes on the same chromosome. For example, Glyma02g01420, Glyma02g12830, and Glyma02g15920, which are located in different duplication blocks of the same chromosome, all have two introns flanked by three exons. However, phylogenetic analysis showed that Glyma02g01420 was more similar to Glyma10g01450, and Glyma02g15920 was more similar to Glyma10g03820, whereas Glyma02g12830, which is located relatively close to Glyma02g15920, had no duplicate genes on chromosome 10. It is possible that the duplication of the same ancestral gene on chromosome 2 resulted in Glyma02g12830 and the ancestor of Glyma10g01450 and Glyma10g03820, which then evolved independently. The intron and exon sequences of the ancestor gene might have elongated or shorten because of various reasons after it split into Glyma02g01420 and Glyma02g15920. Through segmental duplication, the two chromosome segments, one contained in Glyma02g12830 and the other contained in Glyma02g01420 and Glyma02g15920, were independently copied to different parts of chromosome 10. During subsequent evolution, the counterpart of Glyma02g12830 was lost and structures for the counterparts of Glyma02g01420 and Glyma02g15920 changed by deletion or insertion of other fragments or partial sequence repeat variations. Moreover, we noticed that both the Arabidopsis and rice genomes underwent recent duplication events, which also resulted in large-scale expansion of the WRKY gene family in their genomes. Therefore, we also examined the duplicated pattern of WRKY genes in these model species.
The complete sequencing of the Arabidopsis genome revealed numerous large-scale segmental duplications [84]. Previous studies concluded that at least two rounds of duplications probably occurred in the Arabidopsis genome, with many losses and rearrangements leaving a mosaic of “segmental duplications” or “duplication blocks” [74,84]. Most duplication blocks appear to have originated from one round of polyploidy, as estimated by using various methods, that occurred 20–40 mya, before the evolution of the genus Brassica but after the separation of Brassicaceae from other closely related eudicot families [74]. Results of the present study showed that no apparent tandem duplication events, and rare segmental duplication events (six pairs), exist in the Arabidopsis WRKY gene family. Furthermore, the estimated time of the six pairs of segmental duplicated genes focus on the period of 24–27 mya (Additional file 9). Cannon at el. [72] found nine distinct pairs of duplicated segments and no tandem duplication events in the Arabidopsis WRKY family, which is compatible with our study. Comparison of six pairs of segmental duplicated genes in our study with the results of Blanc et al. [85] suggested that only one pair of genes (At1g13960 and At2g033400) originated from polyploidy in Arabidopsis. Consequently, we speculated that the other five segmental duplicated genes might have derived from independent segmental duplication events. The long period of time over which genome evolution has occurred has provided many opportunities for functional divergence in the genes that arose from duplications. Our results did not reveal evidence that other pairs of WRKY genes in Arabidopsis originated from duplicated blocks. Therefore, most of the Arabidopsis WRKY genes may have lost their paralogous genes after genome duplication [74].
With respect to rice, the expansion patterns of WRKY gene family have been clearly demonstrated. Ramamoorthy et al. [40] predicted 103 genes encoding WRKY transcription factors in rice, and the majority of rice WRKY genes (77.7%; 80 of 103) were detected on duplicated blocks. Of the WRKY genes, 45.6% (47 of 103) of WRKY genes were found to have corresponding coordinates generated by segmental duplications. Furthermore, 35.0% (36 of 103) of the WRKY genes were clustered together with a maximum of 10 extra genes between them, and were regarded as tandemly duplicated genes. The results above were confirmed by Jiang et al. [86]. That is, that both tandem and segmental duplication significantly contributed to the expansion of the WRKY gene family in rice.
All of the evidence suggests that the evolutionary patterns of the WRKY gene family differ between soybean, rice, and Arabidopsis. Species-specific expansion played an important role in the evolution of this family in plants. Segmental duplication appears to be the dominant mechanism for the generation of duplicated genes in soybean, whereas segmental duplication and tandem duplication may play similar roles in the expansion of the rice WRKY gene family. Moreover, although Arabidopsis may have a tetraploid ancestor, the majority of its duplicated genes appear to have been lost throughout evolutionary processes.
Conclusion
Previous studies have demonstrated that members of the WRKY gene family play important roles in the regulation of several plant developmental processes and in responses to various biotic and abiotic stresses. Results of the present study indicate that segmental duplication has likely been the dominant mechanism of gene amplification during the expansion of the WRKY family in soybean. Furthermore, positive selection could be the main driving forces for the functional divergence of duplicated genes, which may have played a critical role in the responses of plants to various stresses throughout their evolutionary history. The results of this study will not only further our understanding of the evolutionary processes of soybean WRKY genes, but will also help to enhance functional genomics studies of WRKY transcription factors in an important model system.
Methods
Sequence collection
Seventy WRKY protein sequences downloaded from AtTFDB (http://arabidopsis.med.ohio-state.edu/AtTFDB/) were used to BLAST against the soybean genome database, Phytozome v8.0 (http://www.phytozome.net/soybean), using the BLASTP program. Sequences were selected as candidate proteins if their E value was ≤ 1e-10. For each query sequence, information of the location on chromosomes, genomic sequences, full coding sequences (CDS), and protein sequences were collected from Phytozome, and redundant genes were removed manually. The Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) was used to confirm each predicted WRKY member.
Phylogenetic tree construction and sequence analysis
The SMART program was used to extract the protein sequences of the WRKY domain for each protein. Multiple sequence alignment of domain sequences of 133 WRKY family proteins from soybean and 70 protein sequences from Arabidopsis was performed using the Clustal X 1.83 program with default parameters, and a phylogenetic tree was generated and viewed using MEGA Version 5.0. Exon and intron organizations of soybean WRKY genes were determined by comparing predicted CDS with their corresponding genomic sequences using GSDS (http://gsds.cbi.pku.edu.cn/) software. Motifs of paralogous WRKY proteins were identified statistically using MEME with default settings, except that the maximum number of motifs to find was set at 10.
RNA-Seq atlas and promoter analysis
RNA-Seq data were introduced to further analyze the expression of GmWRKY genes. Data was normalized using a variation of the reads/Kb/Million method, and Z-score analysis was obtained from SoyBase (http://soybase.org/soyseq/) [44,87]. A heat map was generated using the GenePattern program (http://www.broadinstitute.org/cancer/software/genepattern/index.html). The cis-acting elements that regulate gene expression are distributed in 300–3000 bp upstream of the coding region, also take into consideration of sequence restriction in PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and the methods described by Liu et al. [68], therefore, 1500 bp upstream of the coding region were selected as promoter sequence and were downloaded from Phytozome (http://www.phytozome.net) and Soybean Functional Genomics Database (bioinformatics.cau.edu.cn). Then these sequences were submitted to PlantCARE for in silico analysis.
Positive selection and functional divergence
A maximum likelihood method in PAML was applied to test the hypothesis of positive selection in the WRKY gene family [63] under the site model and branch-site model. In the site model, two pairs of models were contrasted to test the selective pressures at codon sites. First, models M0 (one ratio) and M3 (discrete) were compared, using a test for heterogeneity between codon sites in the dN/dS ratio value, ω. The second comparison was M7 (beta) versus M8 (beta + ω > 1). Meanwhile, we introduced the likelihood ratio test (LRT) to compare the two extreme models. When the LRT suggested positive selection, the Bayes empirical Bayes (BEB) method was used to calculate the posterior probabilities that each codon was from the site class of positive selection under models M3 and M8.
The branch-site model assumes that the ω ratio varies between codon sites and that there are four site classes in the sequence. The first class of sites is highly conserved in all lineages with a small ω ratio, ω0. The second class includes neutral or weakly constrained sites for which ω = ω1, where ω1 is near or smaller than 1. In the third and fourth classes, the background lineages show ω0 or ω1, but foreground branches have ω2, which may be greater than 1. When constructing the LRTs, the null hypothesis fixes ω2 = 1, allowing sites evolving under negative selection in the background lineages to be released from constraint and to evolve neutrally on the foreground lineage; the alternative hypothesis constrains ω2 ≥ 1 [64]. Posterior probabilities (Qks) were calculated using the BEB method [65].
The software DIVERGE was used to reveal the functional divergence between members of the WRKY protein family. The coefficients of Type-I and Type-II functional divergence (θ-I and θ-II, respectively) between any two clusters of interest were calculated. A θ-I or θ-II significantly > 0 indicates site-specific altered selective constraints or a radical shift of amino acid physicochemical properties after gene duplication and/or speciation [66]. Moreover, Qk was used to predict critical amino acid residues that were responsible for functional divergence. In this study, we screened the codons (Qk > 0.8) as potential sites that were crucial for functional divergence.
Analysis of WRKY gene expansion patterns and dating the duplication events
In this study, we focused on two patterns of gene expansion: tandem duplication and segmental duplication. Tandem duplications were characterized as multiple members of this family occurring within the same or neighboring intergenic regions, where the WRKY genes were clustered together with a maximum of 10 extra genes between them [40]. Segmental duplications of each WRKY gene within the family in soybean and Arabidopsis genomes were searched in the PGDD (http://chibba.agtec.uga.edu/duplication/). Within the range of 100 kb, the anchors with synonymous substitution rates (Ks) values greater than 1.0 were discarded because of the risk of saturation. Assuming a molecular clock, the Ks values of duplicated genes are expected to be similar over time [88]. Therefore, we used Ks values to estimate the dates of the segmental duplication events. The mean Ks value was calculated for each pair of genes within a duplicated block and was then used to date the duplication events. The approximate date of the duplication event was then calculated using the mean Ks values (T = Ks/2λ), assuming clock-like rates (λ) of synonymous substitution of 1.5 × 10−8 substitutions/synonymous site/year for Arabidopsis[89] and 6.1 × 10−9 for soybean [90].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GY and HX for bioinformatics analysis and writing of the manuscript. SX and YQ for the discussion of the evolutionary pattern of WRKY genes. YL, YY, GY and YH for discussion and comments on an earlier version of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
WRKY gene family in soybean.
Schematic diagram of amino acid motifs of soybean WRKY proteins from different groups (or subgroups). Motif analysis was performed using MEME, as described in the Methods. The grey solid line represents the corresponding WRKY protein and its length. The different-colored boxes represent different motifs and their position in each WRKY sequence. A detailed motif introduction is shown in Additional file 3.
Schematic diagram of WRKY protein motifs. The schematic diagram was derived from MEME. The order of motifs of WRKY proteins in the diagram was automatically generated by MEME according to scores.
Exon/intron structures of soybean WRKY genes. The boxes and lines represent exons and introns, respectively. The bold, dark blue lines indicate the 3’ downstream region. The WRKY genes were separated according to group or subgroup.
Promoter analysis of the soybean WRKY gene family. The locus names and cis-acting element names are listed.
Tests for positive selection among codons of WRKY genes using site models.
Maximum likelihood estimates of the coefficient of Type-I functional divergence (θ) from pairwise comparisons between WRKY groups. Posterior probability (PP) of the site-specific Type-I functional divergence is provided.
Estimates of the coefficient of Type-II functional divergence (θ).
Estimates of the dates for the segmental duplication events of WRKY family in Arabidopsis.
Contributor Information
Guangjun Yin, Email: yinguangjundejia@gmail.com.
Hongliang Xu, Email: xuxxx812@umn.edu.
Shuyang Xiao, Email: susanshaw@Yeah.net.
Yajuan Qin, Email: qyj718897021@126.com.
Yaxuan Li, Email: lyx1006@sohu.com.
Yueming Yan, Email: yanym2004@163.com.
Yingkao Hu, Email: yingkaohu@yahoo.com.
Acknowledgements
Authors would like to thank the National Natural Science Foundation of China (30971783) and the Natural Science Foundation of Beijing, China (5132005) for financial support. GY would like to thank Wan Ping, associate professor from school of life sciences, Capital Normal University, for all the help with bioinformatics.
References
- Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro N. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. Members of a new family of DNA binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol. 1995;29:691–702. doi: 10.1007/BF00041160. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Cormack RS, Eulgem T, Rushton PJ, Köchner P, Hahlbrock K, Somssich IE. Leucine zipper containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim Biophys Acta. 2002;1576:92–100. doi: 10.1016/S0167-4781(02)00298-1. [DOI] [PubMed] [Google Scholar]
- De Pater S, Greco V, Pham K, Memelink J, Kijne J. Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 1996;24:4624–4631. doi: 10.1093/nar/24.23.4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova KS, Conger BV. Isolation of two somatic embryogenesis-related genes from orchardgrass (Dactylis glomerata) Plant Sci. 2002;162:301–307. doi: 10.1016/S0168-9452(01)00571-4. [DOI] [Google Scholar]
- Chen C, Chen Z. Isolation and characterization of two pathogen-and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol Biol. 2000;42:387–396. doi: 10.1023/A:1006399311615. [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet. 2000;263:30–37. doi: 10.1007/PL00008673. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yang P, Fan B, Chen Z. An oligo selection procedure for identification of sequence-specific DNA-binding activities associated with the plant defence response. Plant J. 1998;16:515–522. doi: 10.1046/j.1365-313x.1998.00311.x. [DOI] [PubMed] [Google Scholar]
- Yoda H, Ogawa M, Yamaguchi Y, Koizumi N, Kusano T, Sano H. Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol Genet Genomics. 2002;267:154–161. doi: 10.1007/s00438-002-0651-z. [DOI] [PubMed] [Google Scholar]
- Ashida Y, Nishimoto M, Matsushima A, Watanabe J, Hirata T. Molecular cloning and mRNA expression of geraniol-inducible genes in cultured shoot primordia of Matricaria chamomilla. Biosci Biotechnol Biochem. 2002;66:2511–2514. doi: 10.1271/bbb.66.2511. [DOI] [PubMed] [Google Scholar]
- Kim CY, Lee SH, Park HC, Bae CG, Cheong YH, Choi YJ, Han C, Lee SY, Lim CO, Cho MJ. Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol Plant Microbe Interact. 2000;13:470–504. doi: 10.1094/MPMI.2000.13.4.470. [DOI] [PubMed] [Google Scholar]
- Lambais MR. In silico differential display of defenserelated expressed sequence tags from sugarcane tissues infected with diazotrophic endophytes. Genet Mol Biol. 2001;24:103–111. doi: 10.1590/S1415-47572001000100015. [DOI] [Google Scholar]
- Huang T, Duman JG. Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bitter sweet night shade Solanum dulcamara. Plant Mol Biol. 2002;48:339–350. doi: 10.1023/A:1014062714786. [DOI] [PubMed] [Google Scholar]
- Beyer K, Binder A, Boller T, Collinge M. Identification of potato genes induced during colonization by Phytophthora infestans. Mol Plant Pathol. 2001;2:125–134. doi: 10.1046/j.1364-3703.2001.00059.x. [DOI] [PubMed] [Google Scholar]
- Dellagi A, Helibronn J, Avrova AO, Montesano M, Palva ET, Stewart HE, Toth IK, Cooke DE, Lyon GD, Birch PR. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol Plant Microbe Interact. 2000;13:1092–1101. doi: 10.1094/MPMI.2000.13.10.1092. [DOI] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. A Novel WRKY Transcription Factor, SUSIBA2, Participates in Sugar Signaling in Barley by Binding to the Sugar-Responsive Elements of the iso1 Promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Mzid R, Deluc L, Barrieu F, Pirrello J, Gauthier A, Corio-Costet MF, Regad F, Cailleteau B, Hamdi S, Lauvergeat V. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. J Exp Bot. 2007;58:1999–2010. doi: 10.1093/jxb/erm062. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 2013;18:267–276. doi: 10.1016/j.tplants.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol. 2012;15:431–437. doi: 10.1016/j.pbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009;4:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagacé M, Matton DP. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta. 2004;219:185–189. doi: 10.1007/s00425-004-1253-2. [DOI] [PubMed] [Google Scholar]
- Hinderhofer K, Zentgraf U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta. 2001;213:469–473. doi: 10.1007/s004250000512. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002;31:319–330. doi: 10.1046/j.1365-313X.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci U S A. 2005;48:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004;134:1500–1513. doi: 10.1104/pp.103.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, Chen X, Finer JJ, Shen QJ, Rushton PJ. WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnology J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- Du L, Chen Z. Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J. 2000;24:837–847. doi: 10.1046/j.1365-313x.2000.00923.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta. 1819;2012:120–128. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M. et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008;49:865–879. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- Şahin-Çevik M. A WRKY transcription factor gene isolated from Poncirus trifoliata shows differential responses to cold and drought stresses. Plant Omics. 2012;5:438–445. [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stress in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- Schmutz J. et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;14:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Saedler H. Structure of the chalcone synthase gene of Antirrhinum majus. Mol Gen Genet. 1986;202:429–434. doi: 10.1007/BF00333273. [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/S0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Werneke JM, Chatfield JM, Ogren WL. Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell. 1989;1:815–825. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello-Astorga GR, Herrera-Estrella LR. Ancestral Multipartite Units in Light-Responsive Plant Promoters Have Structural Features Correlating with Specific Phototransduction Pathways. Plant Physiol. 1996;3:1151–1166. doi: 10.1104/pp.112.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois R, Dietrich A, Hahlbrock K, Schulz W. A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheem O, Botha CE, Bradley G. In silico analysis of cis-acting regulatory elements in 5’ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput Biol Chem. 2010;34:268–283. doi: 10.1016/j.compbiolchem.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003;33:259–270. doi: 10.1046/j.1365-313X.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- Kim JK, Cao J, Wu R. Regulation and interaction of multiple protein factors with the proximal promoter regions of a rice high pI α-amylase gene. Mol Gen Genet. 1992;232:383–393. doi: 10.1007/BF00266241. [DOI] [PubMed] [Google Scholar]
- Pastuglia M, Roby D, Dumas C, Cock JM. Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase in Brassica oleracea. Plant Cell. 1997;9:49–60. doi: 10.1105/tpc.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Arabidopsis DNA Encoding Two Desiccation-Responsive rd29 Genes. Plant Physiol. 1993;101:1119–1120. doi: 10.1104/pp.101.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas FZ, Bertolini MC. Genomic organization of the Neurospora crassa gsn gene: possible involvement of the STRE and HSE elements in the modulation of transcription during heat shock. Mol Genet Genomics. 2004;272:550–561. doi: 10.1007/s00438-004-1086-5. [DOI] [PubMed] [Google Scholar]
- Klotz KL, Lagrimini LM. Phytohormone control of the tobacco anionic peroxidase promoter. Plant Mol Biol. 1996;31:565–573. doi: 10.1007/BF00042229. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Bernatzky R, Tanksley SD, Breidenbach RB, Kausch AP, Cashmore AR. Molecular characterization and genetic mapping of two clusters of genes encoding chlorophyll a/b-binding proteins in Lycopersicon esculentum (tomato) Gene. 1985;40:247–258. doi: 10.1016/0378-1119(85)90047-2. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML4: A program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum Likelihood Estimation on Large Phylogenies and Analysis of Adaptive Evolution in Human Influenza Virus A. J Mol Evol. 2000;51:423–432. doi: 10.1007/s002390010105. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang H, Zhang Z, Wu J, Feng Y, Zhu Z. Divergence in function and expression of the NOD26-like intrinsic. BMC Genomics. 2009;10:313. doi: 10.1186/1471-2164-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhu H. Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene. 2008;409:1–10. doi: 10.1016/j.gene.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Wang M, Wang Q, Zhao H, Zhang X, Pan Y. Evolutionary selection pressure of forkhead domain and functional divergence. Gene. 2009;432:19–25. doi: 10.1016/j.gene.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, dePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–885. doi: 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. et al. The genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tian Z, Sui Y, Zhao M, Song Q, Cannon SB, Cregan P, Ma J. Pericentromeric Effects Shape the Patterns of Divergence, Retention, and Expression of Duplicated Genes in the Paleopolyploid Soybean. Plant Cell. 2012;24:21–32. doi: 10.1105/tpc.111.092759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Wu C, Washida H, Onodera Y, Harada K, Takaiwa F. Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. Plant J. 2000;23:415–421. doi: 10.1046/j.1365-313x.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- Duan MR, Nan J, Liang YH, Mao P, Lu L, Li L, Wei C, Lai L, Li Y, Su XD. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 2007;35:1145–1154. doi: 10.1093/nar/gkm001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotechnol Biochem. 2001;65:2428–2436. doi: 10.1271/bbb.65.2428. [DOI] [PubMed] [Google Scholar]
- Church GM, Sussman JL, Kim SH. Secondary structural complementarity between DNA and proteins. Proc Natl Acad Sci U S A. 1977;74:1458–1462. doi: 10.1073/pnas.74.4.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The Probability of Duplicate Gene Preservation by Subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization ccompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Iniative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;14:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional Divergence of Duplicated Genes Formed by Polyploidy during Arabidopsis Evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, González JM, Ramachandran S. Comparative genomic and transcriptomic analysis of tandemly and segmentally duplicated genes in rice. PLoS One. 2013;8:e63551. doi: 10.1371/journal.pone.0063551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ. et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010;10:160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
WRKY gene family in soybean.
Schematic diagram of amino acid motifs of soybean WRKY proteins from different groups (or subgroups). Motif analysis was performed using MEME, as described in the Methods. The grey solid line represents the corresponding WRKY protein and its length. The different-colored boxes represent different motifs and their position in each WRKY sequence. A detailed motif introduction is shown in Additional file 3.
Schematic diagram of WRKY protein motifs. The schematic diagram was derived from MEME. The order of motifs of WRKY proteins in the diagram was automatically generated by MEME according to scores.
Exon/intron structures of soybean WRKY genes. The boxes and lines represent exons and introns, respectively. The bold, dark blue lines indicate the 3’ downstream region. The WRKY genes were separated according to group or subgroup.
Promoter analysis of the soybean WRKY gene family. The locus names and cis-acting element names are listed.
Tests for positive selection among codons of WRKY genes using site models.
Maximum likelihood estimates of the coefficient of Type-I functional divergence (θ) from pairwise comparisons between WRKY groups. Posterior probability (PP) of the site-specific Type-I functional divergence is provided.
Estimates of the coefficient of Type-II functional divergence (θ).
Estimates of the dates for the segmental duplication events of WRKY family in Arabidopsis.