Abstract
We have identified a missense mutation in the motor domain of the neuronal kinesin heavy chain gene KIF5A, in a family with hereditary spastic paraplegia. The mutation occurs in the family in which the SPG10 locus was originally identified, at an invariant asparagine residue that, when mutated in orthologous kinesin heavy chain motor proteins, prevents stimulation of the motor ATPase by microtubule-binding. Mutation of kinesin orthologues in various species leads to phenotypes resembling hereditary spastic paraplegia. The conventional kinesin motor powers intracellular movement of membranous organelles and other macromolecular cargo from the neuronal cell body to the distal tip of the axon. This finding suggests that the underlying pathology of SPG10 and possibly of other forms of hereditary spastic paraplegia may involve perturbation of neuronal anterograde (or retrograde) axoplasmic flow, leading to axonal degeneration, especially in the longest axons of the central nervous system.
The hereditary spastic paraplegias (HSPs) are a genetically heterogeneous group of neurodegenerative disorders characterized by progressive lower-limb spasticity and weakness, occurring in the absence (pure HSP) or presence (complicated HSP) of additional neurological abnormalities (Reid 1999). To date, 18 loci for HSP have been identified, including loci for autosomal dominant, autosomal recessive, and X-linked forms (Fink 2002). The pathology of HSP is characterized by axonal degeneration of motor and sensory neurons that is maximal at the distal ends of the longest axons of the CNS (McDermott et al. 2000).
Neurons are highly polarized cells that present an extreme example of the difficulties of proper intracellular trafficking of membranous organelles and protein complexes. The longest adult human neurons have axons of up to 1 m in length and axonal volumes that are as much as 1,000× the volume of the neuronal cell body (Goldstein 2001). The microtubule cytoskeleton provides structural support for the cell and serves as the molecular rail system for intracellular cargo trafficking. Recent data suggest that the most common form of HSP (SPG4 [MIM 182601]), caused by mutations in the gene encoding spastin (Hazan et al. 1999), is caused by impaired regulation of the microtubule cytoskeleton in long axons (Errico et al. 2002). Missense mutations in spastin were found to cause constitutive binding of spastin to microtubules and to lead to redistribution of the microtubule cytoskeleton, suggesting a pathogenic mechanism of disruption of microtubule dynamics (Errico et al. 2002).
The microtubule-associated protein families of kinesins and dyneins serve as the molecular motors that distribute intracellular cargo along microtubules, with kinesins working in the anterograde direction and dyneins working in the retrograde direction (Hirokawa 1998; Goldstein and Yang 2000). Along with genes encoding microtubule-associated proteins, those of the kinesin and dynein motor complexes make compelling candidate genes for the SPG loci in the context of the neuropathology associated with the distal ends of the longest axons of the CNS.
The kinesin heavy chain (KHC) proteins are part of a multisubunit complex (kinesin-I) that acts as a plus-end–directed microtubule motor involved in anterograde transport of membranous organelles in nerve axons (Xia et al. 1998; Goldstein and Yang 2000; Goldstein 2001). This family includes conventional kinesins KIF5A (MIM 602821), KIF5B (MIM 602809), and KIF5C (MIM 604593). Whereas KIF5B exhibits ubiquitous expression with prominent glial cell expression, KIF5A and KIF5C are expressed exclusively in neurons. Expression of KIF5C is restricted to a subset of neurons and includes developmentally regulated expression in the lower motor neurons. In contrast, KIF5A is expressed in all neurons and is found throughout the neuronal cytoplasm, including in the cell body, dendrites, and axon (Kanai et al. 2000).
An autosomal dominant locus for pure HSP (SPG10 [MIM 604187]) was originally established in a large family and was mapped to chromosome 12 (Reid et al. 1999). The 7-cM candidate interval, bordered by D12S270 and D12S355, is defined by crossovers in this single family (Reid et al. 2001). The gene for the neuronal KHC protein KIF5A maps within this interval. By virtue of its neuronal expression pattern and chromosomal map location, KIF5A makes a compelling candidate gene for SPG10.
In addition to KIF5A (table 1), we sequenced 29 other genes (table 2) that mapped within the SPG10 candidate interval. Although a number of polymorphic variants (i.e., those also observed in normal population control individuals) were identified in these 30 genes, only the KIF5A gene exhibited a potential mutation. We identified an N256S missense mutation in the KIF5A gene in all affected members of the family in which the SPG10 locus was originally identified (Reid et al. 1999, 2001) but not in 220 normal control individuals (figs. 1 and 2). DNA was also available from five family members who were not classified as definitely affected after clinical examination. Two of these subjects were clinically normal at ages 54 and 31 years, and one had bilateral lower-limb hyperreflexia only, at age 17 years. None of these three subjects had the N256S mutation. The two remaining subjects both had the N256S mutation. One of these had bilateral lower-limb hyperreflexia, a unilateral extensor plantar reflex, no clonus, and no gait abnormality at age 22 years, whereas the other had bilateral lower-limb hyperreflexia and nonsustained clonus at age 54 years. This latter patient was likely to be an obligate carrier, since one of her offspring (not available for study) was reported to have bilateral hyperreflexia, bilateral clonus, and bilateral extensor plantar reflexes.
Table 1.
Primers Used for KIF5A Amplification and DNA Sequencing
|
Primer |
Annealing Temperaturea(°C) |
|||
| Exon(s) | Forward | Reverse | Buffer 1 | Buffer 2 |
| 1 | tctgtccccagagactgagcacctg | gaagaggatgaaggatgagcgg | 60 | |
| 2–6 | ggccattctcccacatttc | ccctaggcacatctcattcc | 55 | |
| 4b | aagcaatggaagtccaaagg | |||
| 5b | cttgactccctttccggtta | |||
| 7 | gctgagattcttgacactgcac | ccacagacaaggggtcctc | 55 | |
| 8 | gaggaggaccccttgtctgt | atcgtgccactgcaaacc | 55 | |
| 9 | ccagccaagcatctctgtta | tcaacagggaatcagaaaggat | 55 | |
| 10 | ccatatgatcatgccccaat | ctgggtgaggagaagaggaa | 55 | |
| 11 | aagtccaccctgatgtttgg | cagaaacacagcccctcttc | 55 | |
| 12 | cctcccatactcccaaaagg | tccacatttcaagcataagca | 55 | |
| 13 | gcttcaaccgatcttcctgt | ggagtgattaatccccaagc | 55 | |
| 14 | aggcaatgtgcccaccta | tcacaatgcactgaacagca | 55 | |
| 15 | ctgcactttcccctcacagt | gcgcccaacctagaatgtat | 55 | |
| 16 | ggcggaggaatttaatgtga | cccagcttttctgtccttga | 55 | |
| 17 | aaaggaagagccccatagga | gagctacatgcaaaaccttgg | 55 | |
| 18 and 19 | cccttgttctctgttcctgct | gcattgttgagggaatgaca | 55 | |
| 20 | aatgccacaacttcgtagcc | cgtccctaccccatcatattt | 55 | |
| 21 | gtctgtggtcccagatgctt | gccagataccaggtctgtcc | 55 | |
| 22 and 23 | ttttgcagccattctgtagc | agggaaagtaaaagcgaacag | 55 | |
| 24 and 25 | ggccttttgaggaaatgaca | gcatgaagttgagaaagggttt | 55 | |
| 26 | gtggccatgtttgttttcct | ggctgagtgcagtggctatt | 55 | |
| 27 | taagatgggagagggtttgc | gggcacaagggaaatttaatc | 55 | |
| 28 | aatagtccctgccgtttgtg | ggaagcagcctgacactacc | 55 | |
Buffer 1: 10× buffer (100 mM Tris, 500 mM KCl, 10 mM MgCl2, 1% Triton X-100, and 0.1% gelatin). Buffer 2: 10× buffer (100 mM Tris, 15 mM MgCl2, and 500 mM KCl).
Nested sequencing primers for exons 4 and 5.
Table 2.
Genes Sequenced in the SPG10 Candidate Interval
| Gene | Name/Description | Reference Sequencea |
| KIF5A | Kinesin heavy chain 5A, neuronal | NM_004984 |
| INHBC | Inhibin β C chain preproprotein | NM_005538 |
| GALGT | β-1,4-N-acetylgalactosaminyltransferase | NM_001478 |
| OS-9 | Similar to mouse neurofilament protein | NM_006812 |
| AVIL | Advillin | NM_006576 |
| MYO1A | Myosin IA | NM_005379 |
| TAC3 | Tachykinin 3 | NM_013251 |
| GDF11 | Growth differentiation factor 11 | NM_005811 |
| RAB5B | Member RAS oncogene family | NM_002868 |
| ERBB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 | NM_001982 |
| NPFF | FMFRamide-related protein precursor | NM_003717 |
| LOC51644 | Copz1 mouse ortholog | NM_016057 |
| ATP5G2 | ATP synthase, H+ transporting, mitochondrial F0 | NM_005176 |
| PPP1R1A | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | NM_006741 |
| PFDN5 | Prefoldin 5 | NM_002624 |
| AAAS | Aladin | NM_015665 |
| PDE1B | Phosphodiesterase IB, calmodulin-dependent | NM_000924 |
| FLJ22055 | Similar to chain B, phosphatidylinositol phosphate kinase type II β | NM_024779 + BG720492 |
| KIAA0747 | Mbc2 mouse ortholog | NM_015292 |
| PA2G4 | Proliferation-associated 2G4, 38kD | NM_006191 |
| DKFZp564J157 | Similar to mouse formin 4 | NM_018457 |
| KIAA1536 | Similar to rat myosin heavy chain, neuronal | NM_020898 |
| CCT2 | Chaperonin containing TCP1, subunit 2 (β) | NM_006431 |
| LOC115557 | Similar to triple functional domain (PTPRF interacting) | NM_133483 |
| NEUROD4 | Neurogenic differentiation 4 | NM_021191 |
| NAB2 | NGFI-A binding protein 2 | NM_005967 |
| MY050 | Hypothetical brain protein | NM_032624 |
| GLI | Glioma-associated oncogene homolog (zinc finger protein) | NM_005269 |
| DCTN2 | Dynactin 2 (p50) | NM_006400 |
| MAP3K12 | Mitogen-activated protein kinase kinase kinase 12 | NM_006301 |
Reference sequences are available from the working draft human genome sequence database on the UCSC Genome Bioinformatics Web site.
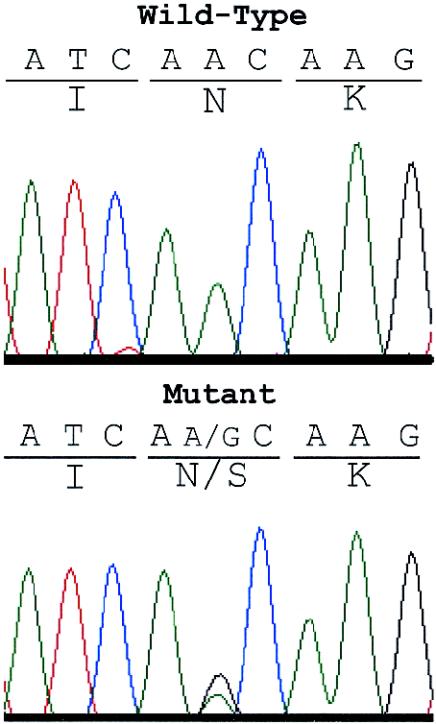
Figure 1.
Electropherograms of sequences in the motor domain of the KIF5A gene. The affected individual from the family in which the SPG10 locus was originally identified (Reid et al. 1999, 2001) has both A (wild-type) and G (mutant) at nucleotide 767. The mutation changes the invariant asparagine at position 256 to a serine in the switch II loop/helix region (see fig. 3).
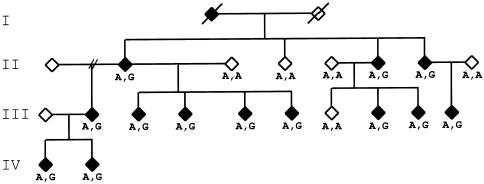
Figure 2.
Cosegregation of the A767G mutation with the spastic paraplegia phenotype in the family in which the SPG10 locus was originally identified (Reid et al. 1999, 2001). Standard pedigree symbols are used. Genotypes at the mutation are listed as either wild-type A/A or heterozygous mutant A/G.
The N256S mutation occurs at an invariant asparagine residue in the switch II loop/helix motif within the motor domain of the protein (fig. 3). Intriguingly, a Saccharomyces cerevisiae mutant isolated as a suppressor of mutations in plus-end–directed mitotic kinesins has a mutation in the homologous amino acid position (N650K) in Kar3, the minus-end–directed motor of the mitotic spindle (Hoyt et al. 1993). The mutant allele exhibits dominance over the wild-type KAR3 allele, suggesting a possible dominant-negative mechanism. This mutation was subsequently shown to decouple nucleotide and microtubule binding of the motor, resulting in a block in microtubule-dependent stimulation of motor ATPase activity (Song and Endow 1998). The Ncd kinesin motor of Drosophila, engineered with the homologous mutation (N600K), shows a similar inhibitory effect on microtubule-stimulated ATPase activity and, in addition, demonstrates a lack of microtubule-stimulated motility in a gliding assay (Song and Endow 1998). The recently determined crystal structures of the wild-type Kar3 protein and the N650K mutant illustrate the critical role of this amino acid residue in kinesin motor function (Yun et al. 2001). Upon binding to microtubules, the N650 residue participates in the activation of ATPase, via hydrogen bonding and side chain interactions with the switch I and switch II regions, which form a characteristic salt bridge in the nucleotide-binding cleft (Yun et al. 2001).
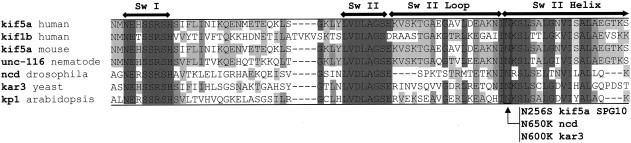
Figure 3.
Kinesin protein homologies in the motor region. Multiple alignment showing conserved residues (shaded), nucleotide-binding motifs Switch I (Sw I) and Switch II (Sw II), and the switch II loop/helix region (Sack et al. 1999; Song et al. 2001). Identical residues are shaded in dark gray, strongly similar residues in medium gray, and weakly similar residues in light gray. The KIF5A mutation in the family with SPG10, as well as the NCD and KAR3 mutations, are indicated by the arrow. Sequence alignments of the various kinesin proteins were performed using ClustalW (Thompson et al. 1994; EMBL-EBI European Bioinformatics Institute Web site).
To date, only one other known family with autosomal dominant HSP exhibits linkage to chromosome 12 (Ashley-Koch et al. 2001). In this family, we were unable to identify a mutation in the KIF5A gene, despite sequencing the entire coding region, ∼2 kb upstream of the transcriptional start site, and ⩾50 nts of intronic sequence flanking each exon. Southern blot analysis using full-length kinesin cDNA as a probe did not reveal evidence of gross rearrangement, and separation of the wild-type and mutant chromosomes (as determined by disease haplotype) in somatic cell hybrids did not reveal deletion of any of the KIF5A exons. In these hybrid lines, the KIF5A transcripts from both the wild-type and mutant chromosomes were expressed at equivalent levels. Finally, the transcript derived from the mutant chromosome did not show a novel splice variant that might be caused by an undiscovered mutation lying deep within an intron. Although a mutation affecting message stability or an unknown alternatively spliced exon cannot be excluded, this family may instead harbor a mutation in another gene mapping to the same chromosome. In support of this conjecture, the candidate interval for this second family is >40 cM, extending from D12S1052 (which maps to 12q) to beyond D12S1057 (which maps to 12p). This interval includes but greatly exceeds the 7-cM interval defined in the original family. In addition, the clinical presentations of the two families are dissimilar (full clinical descriptions for each family can be found in the articles by Reid et al. [1999] and Ashley-Koch et al. [2001]). Most notable is an apparent age-at-onset difference between the two families. Members of the family in which the SPG10 locus was originally identified were noted to have a juvenile-onset form of the disease, with a mean age at onset of 11 years (10.8±9.6 years; Reid et al. 1999, 2001). In contrast, members of the second family exhibit an adult onset of the phenotype, with a mean age at onset of 30 years (30.4±13.4 years; Ashley-Koch et al. 2001). Finally, only 2 of the 13 affected members of the original family developed even a mild amyotrophy (muscle wasting), whereas older affected members of the second family exhibited moderate-to-severe amyotrophy. This degree of amyotrophy is unusual for pure HSP. The phenotypic differences between the two families are consistent with the existence of a distinct locus that maps within the >40-cM candidate interval defined by the other family showing linkage to chromosome 12.
In addition to the biochemical evidence that specific mutations in kinesin orthologues cause loss of kinesin motor function, loss-of-function mutations in related KHC genes of various species cause movement defects that are reminiscent of those observed in human spastic paraplegias. Drosophila larvae harboring either homozygous or hemizygous mutations in the KHC exhibit behavioral and movement defects, including a progressive gradient of dorsoventral and anteroposterior paralysis (Saxton et al. 1991; Hurd and Saxton 1996). Analysis of the neurons in the mutant larvae shows aberrant accumulation of membranous organelles in so-called “organelle jams.” Similarly, Caenorhabditis elegans mutants lacking either the unc-104 or the unc-116 kinesin orthologues exhibit uncoordinated and slow movement (Hall and Hedgecock 1991; Patel et al. 1993). Animals with severe unc-104 alleles assume a tightly coiled posture with periods of muscle contraction that persist for several minutes at a time (Hall and Hedgecock 1991). In this case, the neuron cell bodies of unc-104 mutants accumulate synaptic vesicles in the cytoplasm. In the mouse, disruption of the gene encoding the kinesin motor KIF1Bβ, implicated in mitochondrial transport, results in multiple neurological abnormalities in homozygous animals and a “staggering” gait defect in adult heterozygotes (Zhao et al. 2001). The human KIF1Bβ gene was subsequently shown to be mutated in a family with Charcot-Marie-Tooth disease type 2A (MIM 118210), an autosomal dominant peripheral neuropathy characterized by weakness and atrophy of distal muscles, depressed tendon reflexes, and mild sensory loss (Zhao et al. 2001). Finally, deletion of the related kif5B gene in the mouse results in embryonic lethality and abnormal perinuclear clustering of mitochondria in cells derived from the extraembryonic membranes (Tanaka et al. 1998).
On the basis of the genetic and biochemical studies of kinesin proteins in diverse species, we propose that the KIF5A mutation identified in SPG10 will result in a loss-of-function or dominant-negative version of the neuronal kinesin-I motor. It is perhaps surprising that the mutation we identified affects an amino acid residue previously shown to be crucial to motor function in both genetic and biochemical studies of kinesin proteins. Although we cannot exclude serendipity, we favor the hypothesis that this human mutation represents a naturally occurring dominant-negative allele, which would represent the type of mutation identified in genetic screens and employed in biochemical studies requiring a stable but nonfunctional protein. In this context, KIF5A is thought to participate in a heterotetrameric complex that constitutes the kinesin motor (Goldstein and Yang 2000). A dominant-negative allele is also consistent with the autosomal dominant inheritance of SPG10, although other mechanisms clearly are possible (Wilkie 1994). In any case, we postulate that this specific mutation results in aberrant neuronal intracellular anterograde trafficking of membranous organelles, possibly mitochondria, especially affecting the longest axons of the CNS, consistent with the phenotype and pathology of HSP.
Prior to the present study, the genes for 6 of the 18 SPG loci had been identified, but no unifying theme of disease pathogenesis had emerged from these discoveries. An emerging connection involving mitochondrial function has been provided by the identification of mutations in genes encoding two mitochondrial proteins: HSP60 (MIM 118190), in SPG13 (MIM 605280), and paraplegin, in SPG7 (MIM 602783). The discovery of a mutation in a kinesin motor protein in SPG10 suggests a potential unifying theme for the pathogenesis of HSP—namely, disruption of mitochondrial function in the distal ends of the longest axons of the CNS. In some cases, including SPG10, this might be due to impairment of neuronal axoplasmic transport of mitochondria. In support of this, the gene encoding spastin, mutated in the most common form of autosomal dominant HSP (SPG4), may also be involved in axonal transport, based on its role in microtubule dynamics (Errico et al. 2002). The underlying pathology for a number of forms of HSP and other long-axon neuropathies may involve a common physiological pathway of anterograde and/or retrograde axoplasmic flow that, when perturbed, leads to axonal degeneration, especially in the longest axons of the CNS.
Note added in proof.—A 19th locus for HSP has recently been identified, by Patel et al. (2002).
Acknowledgments
We thank the members of the families, for their participation, and J. Heitman and S. Endow (Duke University), for helpful comments. This work was supported by National Institute of Neurological Disorders and Stroke program project grant PO1 NS 26630 (to M.A.P.-V.) and by grants from the Wellcome Trust and Action Research (both to E.R.). D.C.R. is a Wellcome Trust Senior Fellow in Clinical Research.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- EMBL-EBI European Bioinformatics Institute, http://www.ebi.ac.uk/clustalw/ (for ClustalW)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SPG4 [MIM 182601], SPG7 [MIM 602783], SPG10 [MIM 604187], SPG13 [MIM 605280], Charcot-Marie-Tooth disease type 2A [MIM 118210], KIF5A [MIM 602821], KIF5B [MIM 602809], KIF5C [MIM 604593], and HSP60 [MIM 118190])
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/ (for working draft human genome sequence database)
References
- Ashley-Koch A, Bonner ER, Gaskell PC, West SG, Tim R, Wolpert CM, Jones R, Farrell CD, Nance M, Svenson IK, Marchuk DA, Boustany RM, Vance JM, Scott WK, Pericak-Vance MA (2001) Fine mapping and genetic heterogeneity in the pure form of autosomal dominant familial spastic paraplegia. Neurogenetics 3:91–97 [DOI] [PubMed] [Google Scholar]
- Errico A, Ballabio A, Rugarli, EI (2002) Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet 11:153–163 [DOI] [PubMed] [Google Scholar]
- Fink JK (2002) Hereditary spastic paraplegia: the pace quickens. Annals of Neurology 51:669–672 [DOI] [PubMed] [Google Scholar]
- Goldstein LSB (2001) Kinesin molecular motors: transport pathways, receptors, and human disease. Proc Natl Acad Sci USA 98:6999–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LSB, Yang Z (2000) Microtubule-based transport systems in neurons: the roles of kinesins and dynamins. Annu Rev Neurosci 23:39–71 [DOI] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM (1991) Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65:837–847 [DOI] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Durr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder JM, Prud'homme JF, Brice A, Fontaine B, Heilig B, Weissenbach J (1999) Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet 23:296–303 [DOI] [PubMed] [Google Scholar]
- Hirokawa N (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279:519–526 [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Totis L, Saunders WS (1993) Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 135:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM (1996) Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics 144:1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N (2000) KIF5C, a novel neuronal kinesin enriched in motor neurons. J Neurosci 20:6374–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott CJ, White K, Bushby K, Shaw PJ (2000) Hereditary spastic paraparesis: a review of new developments. J Neurol Neurosurg Psychiatry 69:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H, Cross H, Proukakis C, Hershberger R, Bork P, Ciccarelli FD, Patton MA, McKusick VA, Crosby AH (2002) SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet 31:347–348 [DOI] [PubMed] [Google Scholar]
- Patel N, Thierry-Mieg D, Mancillas JR (1993) Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proc Natl Acad Sci USA 90:9181–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E (1999) The hereditary spastic paraplegias. J Neurol 246:995–1003 [DOI] [PubMed] [Google Scholar]
- Reid E, Dearlove AM, Rhodes M, Rubinsztein DC (1999) A new locus for autosomal dominant “pure” hereditary spastic paraplegia mapping to chromosome 12q13, and evidence for further genetic heterogeneity. Am J Hum Genet 65:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Escayg A, Dearlove AM, Meisler MH, Rubinsztein DC (2001) The spastic paraplegia SPG10 locus: narrowing of critical region and exclusion of sodium channel gene SCN8A as a candidate. J Med Genet 38:65–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack S, Kull FJ, Mandelkow E (1999) Motor proteins of the kinesin family: structures, variations, and nucleotide binding sites. Eur J Biochem 262:1–11 [DOI] [PubMed] [Google Scholar]
- Saxton WM, Hicks J, Goldstein LSB, Raff EC (1991) Kinesin heavy chain is essential for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell 64:1093–1102 [DOI] [PubMed] [Google Scholar]
- Song H, Endow SA (1998) Decoupling of nucleotide- and microtubule-binding sites in a kinesin mutant. Nature 396:587–590 [DOI] [PubMed] [Google Scholar]
- Song YH, Marx A, Muller J, Woehlke G, Schliwa M, Krebs A, Hoenger A, Mandelkow E (2001) Structure of a fast kinesin: implications for ATPase mechanism and interactions with microtubules. EMBO J 20:6213–6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TanakaY, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N (1998) Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell 93:1147–1158 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO (1994) The molecular basis of genetic dominance. J Med Genet 31:89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Rahman A, Yang ZH, Goldstein LSB (1998) Chromosomal localization reveals three kinesin heavy chain genes in the mouse. Genomics 52:209–213 [DOI] [PubMed] [Google Scholar]
- Yun M, Zhang X, Park CG, Park HW, Endow SA (2001) A structural pathway for activation of the kinesin motor ATPase. EMBO J 20:2611–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hayashi Y, Hirokawa N (2001) Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell 105:587–597 [DOI] [PubMed] [Google Scholar]