Abstract
Duane syndrome is a congenital eye movement disorder characterized most typically by absence of abduction, restricted adduction, and retraction of the globe on attempted adduction. Duane syndrome can be coinherited with radial ray anomalies as an autosomal dominant trait, referred to as “Okihiro syndrome” or “Duane radial ray syndrome” (DRRS). We ascertained three pedigrees with DRRS and mapped their disease gene to a 21.6-cM region of chromosome 20 flanked by markers D20S888 and D20S102. A new member of the SAL family of proposed C2H2 zinc finger transcription factors, SALL4, falls within the region. Mutation analysis of SALL4 in the three pedigrees revealed one nonsense and two frameshift heterozygous mutations. SALL4 represents the first identified Duane syndrome gene and the second malformation syndrome resulting from mutations in SAL genes and likely plays a critical role in abducens motoneuron development.
Several forms of congenital ophthalmoplegia are hypothesized to result from errors in the development of the ocular cranial nuclei (Nakano et al. 2001). The most common of these, Duane syndrome (MIM 126800 and MIM 604356), is characterized by restricted or absent abduction and variably restricted adduction with retraction of the globe on attempted adduction, and results from absence of the abducens (nVI) motoneurons and nerve, with sparing of the internuclear neurons (Hotchkiss et al. 1980; Miller et al. 1982). Two Duane syndrome loci (DURS1 and DURS2) have been mapped, but neither gene has been identified (Vincent et al. 1994; Appukuttan et al. 1999). Duane syndrome can be coinherited with radial anomalies in an autosomal dominant fashion (fig. 1A). In affected individuals, radial dysplasia ranges from hypoplasia of the thenar eminence to absence of the radial bone or forearm. Variable expression of sensorineural hearing loss and cardiac, renal, vertebral, and lower extremity malformations can occur. Described in the 1970s and referred to as “Okihiro syndrome” or “Duane radial ray syndrome” (DRRS) (included in MIM 126800) (Okihiro et al. 1977; Temtamy and McKusick 1978; Appukuttan et al. 1999), this distinct disorder partially overlaps clinically with Townes-Brocks and Holt-Oram syndromes, as well as acroreno-ocular, oculo-otoradial, cervico-oculoacostic, and oculoauriculovertebral syndromes.
Figure 1.
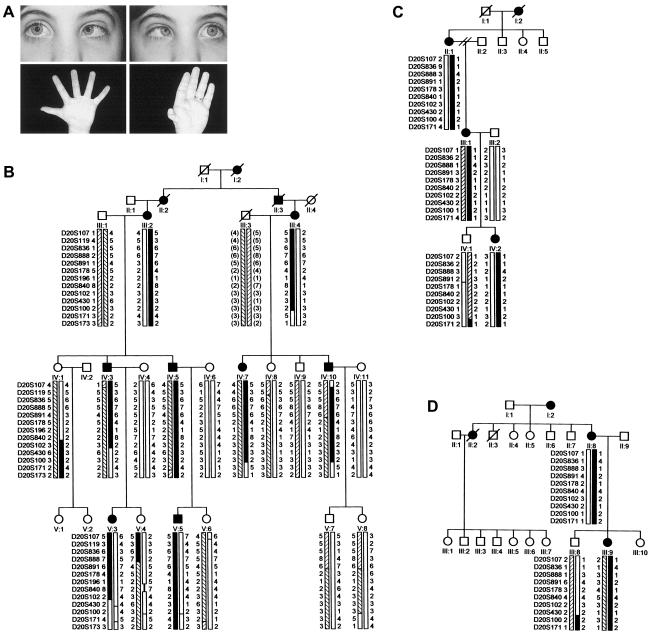
DRRS pedigrees. A, Top, attempted right and left gaze in an affected member of pedigree V (V:3) with Duane syndrome; bottom, thenar hypoplasia and thumb anomaly in affected members of pedigree V (V:3, left; IV:3, right). B, pedigree V. C, pedigree FN. D, pedigree DA. Affected individuals are indicated by filled symbols. Below the symbol for each participant are genotypes for chromosome 20q13.12-q13.31 markers. The disease-associated haplotype in each family is in black.
To investigate the role that the DRRS gene plays in abducens development, we analyzed two white pedigrees and one Japanese pedigree (V, FN, and DA, respectively) segregating DRRS as an autosomal dominant trait with apparent full penetrance (fig. 1B–1D). The study was approved by Children's Hospital Boston Review Board, and appropriate informed consent was obtained from all participants. The clinical features of pedigrees DA and FN have been described elsewhere (Okihiro et al. 1977; Chun et al. 2001). Of the 13 affected participants in the three families, 12 have Duane syndrome. Among them, the Duane syndrome phenotype can be unilateral or bilateral and includes individuals with marked or complete limitation of abduction with minimal or no limitation of adduction (Duane syndrome type 1) and individuals with marked or complete limitation of both abduction and adduction (Duane syndrome type 3). None of the participants had marked or complete limitation of adduction with minimal or no limitation of abduction (Duane syndrome type 2). The presence of more than one Duane syndrome type within a single pedigree was also described in a family whose disease gene maps to the DURS2 locus (Chung et al. 2000) and may result from variable expression of the primary gene mutations or from different patterns of secondary aberrant innervation. Twelve of the affected participants have radial dysplasia. The radial dysplasia can also be unilateral or bilateral and ranges from hypoplasia of the thenar eminence to foreshortening of the forearm with absent radial bone, radial pulse, and thumb. Only one participant has unilateral Duane syndrome and unilateral radial anomalies on opposite sides. Two affected participants have sensorineural hearing loss. Facial weakness, abnormal pinna, fused cervical vertebrae, pectoralis muscle aplasia, and Hirschsprung disease were each detected once. No participants have known renal or cardiac anomalies. The clinical findings are summarized in table 1.
Table 1.
Clinical Synopsis of DRRS in Participating Family Members[Note]
|
Finding for Characteristic |
||||||||||
| PedigreeandIndividual | DuaneType | ThenarHypoplasia | ThumbDefect | AbsentRadialPulse | ClubWrist | ShortForearm | ShoulderDislocation | ShortNeck | FusedCervicalVertebrae | SensorineuralHearingLoss |
| Pedigree V: | ||||||||||
| III:2 | L (1) | L | L | + | B | L | + | + | ||
| IV:3 | B (3) | B | B | + | B | + | ||||
| IV:5 | R (1) | L | L | L | L | L | L | |||
| V:3 | B (1) | B | B | + | + | + | ||||
| V:5 | B (1) | B | B | B | ||||||
| III:4 | R (1) | R | ||||||||
| IV:7 | B (3) | B | B | B | B | L | L | |||
| IV:10 | L (1) | − | − | |||||||
| Pedigree FN: | ||||||||||
| II:1 | − | B | ||||||||
| III:1 | R (1) | B | R | |||||||
| IV:2 | R (1) | B | ||||||||
| Pedigree DA: | ||||||||||
| II:8 | B | R | ||||||||
| III:9 | B (3) | B | B | |||||||
Note.— L = left, R = right, B = bilateral, + = present, − = absent, (1) = Duane type 1, (3) = Duane type 3.
We conducted an ∼10-cM genomewide linkage screen of pedigree V's phenotype, using the ABI Prism Linkage Mapping Set Panels (Version 2; PE Applied Biosystems) and calculated two-point LOD scores as reported elsewhere (Engle et al. 1994). Adjacent screening markers D20S178, D20S196, and D20S100 revealed LOD scores of 2.6, 2.9, and 3.0 at θ=0. No other screening marker yielded a score >2. Using radioactive methods, we analyzed 18 additional markers between D20S119 and D20S171 and refined the locus to an ∼22-cM region of chromosome 20q13.12–q13.31 flanked by D20S888 and D20S102 (fig. 1B). We then analyzed FN and DA and found that their phenotypes were consistent with linkage (fig. 1C and 1D). Maximum LOD scores were 3.57, 0.6, and 0.3 at θ=0 in V, FN, and DA, respectively, and a maximum combined LOD score of 4.47 was obtained at the fully informative marker, D20S891 (table 2).
Table 2.
Pairwise LOD Scores of Chromosome 20 Markers with DRRS
|
Recombination Fraction (θ) |
||||||
| LocusandPedigree | .00 | .05 | .10 | .20 | .30 | .40 |
| D20S119: | ||||||
| V | −∞ | 2.36 | 2.31 | 1.86 | 1.24 | .55 |
| D20S836: | ||||||
| V | −∞ | 1.83 | 1.84 | 1.54 | 1.09 | .56 |
| FN | .60 | .56 | .51 | .41 | .29 | .16 |
| DA | .26 |
.22 |
.19 |
.11 |
.05 |
.01 |
| Total | −∞ | 2.61 | 2.54 | 2.06 | 1.43 | .73 |
| D20S888: | ||||||
| V | −∞ | 2.36 | 2.31 | 1.86 | 1.24 | .55 |
| FN | .60 | .56 | .51 | .41 | .29 | .16 |
| DA | −.78 |
−.49 |
−.33 |
−.15 |
−.06 |
−.01 |
| Total | −∞ | 2.43 | 2.49 | 2.12 | 1.47 | .7 |
| D20S891: | ||||||
| V | 3.57 | 3.25 | 2.91 | 2.21 | 1.47 | .71 |
| FN | .60 | .56 | .51 | .41 | .29 | .16 |
| DA | .30 |
.26 |
.21 |
.13 |
.06 |
.02 |
| Total | 4.47 | 4.07 | 3.63 | 2.75 | 1.82 | .89 |
| D20S178: | ||||||
| V | 2.63 | 2.55 | 2.36 | 1.83 | 1.20 | .55 |
| FN | .30 | .26 | .21 | .13 | .06 | .02 |
| DA | .30 |
.26 |
.21 |
.13 |
.06 |
.02 |
| Total | 3.23 | 3.07 | 2.78 | 2.09 | 1.32 | .59 |
| D20S196: | ||||||
| V | 2.90 | 2.58 | 2.25 | 1.57 | .90 | .33 |
| D20S840: | ||||||
| V | 3.57 | 3.20 | 2.82 | 2.03 | 1.23 | .51 |
| FN | .60 | .56 | .51 | .41 | .29 | .16 |
| DA | .00 |
.00 |
.00 |
.00 |
.00 |
.00 |
| Total | 4.17 | 3.76 | 3.33 | 2.44 | 1.52 | .67 |
| D20S102: | ||||||
| V | −∞ | 2.30 | 2.24 | 1.80 | 1.21 | .57 |
| FN | .00 | .00 | .00 | .00 | .00 | .00 |
| DA | −.78 |
−.49 |
−.33 |
−.15 |
−.06 |
−.01 |
| Total | −∞ | 1.81 | 1.91 | 1.65 | 1.15 | .56 |
| D20S430: | ||||||
| V | −∞ | .56 | .90 | .96 | .74 | .40 |
| FN | .30 | .26 | .21 | .13 | .06 | .02 |
| DA | .30 |
.26 |
.21 |
.13 |
.06 |
.02 |
| Total | −∞ | 1.08 | 1.32 | 1.22 | .86 | .44 |
| D20S100: | ||||||
| V | 3.04 | 2.76 | 2.47 | 1.88 | 1.26 | .62 |
| FN | .60 | .56 | .51 | .41 | .29 | .16 |
| DA | −∞ |
−.72 |
−.44 |
−.19 |
−.08 |
−.02 |
| Total | −∞ | 2.60 | 2.54 | 2.10 | 1.47 | .76 |
| D20S171: | ||||||
| V | −∞ | .52 | .88 | .95 | .72 | .37 |
| FN | −∞ | −.72 | −.44 | −.19 | −.08 | −.02 |
| DA | −.78 |
−.49 |
−.33 |
−.15 |
−.06 |
−.01 |
| Total | −∞ | −.69 | .11 | .61 | .5 | .34 |
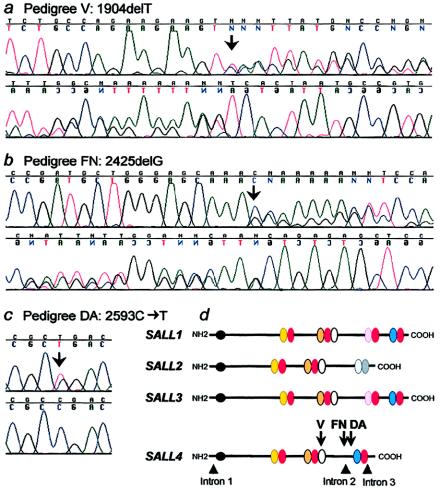
We reviewed positional candidate genes on the basis of the UCSC Human Genome Project Working Draft and chose to analyze a previously unreported gene, LOC57167, that has been annotated by GenBank as a new member of the SAL family (SALL4) and maps between D20S196 and D20S840. This gene was particularly interesting because heterozygous truncating mutations in SALL1 cause Townes-Brocks (Kohlhase et al. 1998), a syndrome characterized by the distinct but overlapping complex of supernumerary/triphalangeal thumbs, hearing loss, malformed ears, and imperforate anus. We aligned the SALL4 genomic and 3,162-bp cDNA sequences, identified four exons, amplified these exons and their flanking introns with 16 sets of primers (table A [online only]), and directly sequenced the PCR products. In the affected members of pedigrees V and FN, we detected heterozygous exon 2 single–base-pair deletions 1904delT and 2425delG, respectively (fig. 2a and 2b). Both these mutations are predicted to result in frameshifts leading to a stop codon after 4 (pedigree V) and 46 (pedigree FN) altered amino acids. In pedigree DA, we detected a heterozygous 2593C→T change in exon 3, resulting in the transition of an arginine to a stop codon (R865X; fig. 2c). Five SALL4 polymorphisms were also detected (table B [online only]). All three mutations cosegregate with the DRRS phenotype in each family, are not present on at least 174 chromosomes of mixed ethnicity (including 70 of Japanese ethnicity for the DA mutation), and, like SALL1 mutations, are predicted to result in premature termination of the SALL4 protein (fig. 2d).
Table A.
SALL4 Sequencing Primers[Note]
| Exon | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | bp | Tm | Qa |
| 1 | ggggtaaatttcccaactcc | aatctcggctcctgaatttg | 329 | 54 | + |
| 2A | gattatagatgtgagcgacggtgc | GGACTGGTGGGCTGGTGG | 334 | 63 | + |
| 2B | GAGGATGAAGCCACAGTAAAGCG | GTGCCCCGTAGTGCCTGC | 395 | 63 | + |
| 2C | GTCACAGGGAGAATGGCGG | GTGGAGGGCGTGGGAGG | 350 | 63 | + |
| 2D | CGGTGAATCAGCGGAGCG | CAGAGTGAAGGGTGCCAGCC | 371 | 51 | + |
| 2E | The same as 2D forward | The same as 2F reverse | 870 | 58 | + |
| 2E-seq | CAACATCCCTTCTGCCACCAG | TCAGTCCCAAAAACCTTGCTACAG | 291 | – | – |
| 2F | CAAGAAAGGGAAGGGGAAGCC | AGGTGGTTACAAGGACAGGTTTGC | 377 | 58 | + |
| 2G | GTGGATGTCAAACCCAAAGACGA | ctctccgtttgtaaagttcaaccca | 1443 | 64 | – |
| 2G-nested | GCTGTTTGCCGAGTTCCAGG | GTCAATGTTCTCCACCAACTGCTG | 358 | 58 | – |
| 2H | GACCTCACGGGTGGCTCC | CGGCATTAGTGAACTTCTTCTGG | 427 | 58 | + |
| 2I | ATGTCTCATTTGCCACCGAGTC | GCTCAGAACCCGTAAAGTCACAG | 308 | 62 | – |
| 2J | TGCCAGAAGAAGTTCACTAATGCC | AGGATGAGTCGTTGGTCAAGCC | 403 | 62 | – |
| 2K | CACTCGGCATCACCCACG | ttgtaaagttcaacccaggctcc | 394 | 62 | – |
| 3A | tgaaggttggatgataaaggtctgg | GGCTGGGCTGCTAACAAAGG | 175 | 62 | – |
| 3B | GCCCCGCCGACCTATGTC | caaaggaggaatggaaggatgg | 351 | 62 | – |
| 4A | cataaccccgctgtaagtcaagg | TTTCCGTCCGTACCTAACAGAGC | 238 | 62 | – |
| 4B | ACACGGGGCGAACAATAACTC | gtgtctgcattgctccttccac | 440 | 58 | – |
Note.— Primers located in introns are lowercase and in exons are uppercase. Exon 2 was screened by sequencing of 11 overlapping amplicons. All primers are used for both PCR and direct sequencing with the exception of 2E and 2G. Amplicon 2E was amplified using 2D forward and 2F reverse primers and sequenced using 2E-seq primers. Amplifications of amplicon 2G were performed as nested PCR (initial PCR using 2G, subsequent PCR using 2G-nested primers). The products were then sequenced using 2G-nested primers.
PCR amplification was performed using HotStarTaq with (+) or without (−) 1x Q-solution (QIAGEN Inc.). Initial denaturation at 95°C for 15 min, followed by 35 cycles at 95°C for 30 sec, Tm°C for 60 sec, 72°C for 60 sec, and final extension at 72°C for 10 min. The exception to this was the extension time for amplicon 2G, which was 100 sec.
Figure 2.
SALL4 mutations and predicted SAL protein schematics. Forward (top) and reverse (bottom) strand direct sequence of an affected member of (a) pedigree V, showing the heterozygous 1904delT and of (b) pedigree FN, showing the heterozygous 2425delG. c, Direct sequence of an affected member of pedigree DA (top) and control (bottom) showing the heterozygous 2593C→T mutation. d, Predicted SALL1–4 protein structures and location of SALL4 DRRS mutations. SALL4 contains one NH2-terminus C2HC-type zinc finger (ZF; black circle) and 7 C2H2-type ZF (oval) domains. The latter are arranged as double ZFs (DZF), with a third finger associated with the second doublet (white oval). The first finger of each pair contains a somewhat variable sequence (yellow, orange, and blue ovals), whereas the second finger of each pair contains the conserved SAL-box sequence, FT/STKGNLK (red ovals). The homologies to SALL1–3 are shown. SALL2 also contains three DZF motifs; the first two are highly similar to the other proteins, but the third is divergent (gray ovals). In contrast, SALL1 and SALL3 each contain three DZF motifs highly similar to SALL4 with an additional fourth DZF motif located between the second and third DZF in SALL4. The positions of the SALL4 splice junctions and the three DRRS mutations are superimposed on the schematic protein. The three mutations are predicted to result in unstable mRNAs or truncated proteins disrupting the fifth and/or lacking the sixth and seventh DZF motifs. Protein structures and similarities were predicted by alignment in MacVector 6.5.3 and by the prediction programs SWISS-PROT and TrEMBL on the ExPASy Molecular Biology Server.
Table B.
Single Nucleotide Polymorphisms Detected in SALL4
| Change | Position | Amino Acid | |
| 1a | 540T→C | Exon 2 | N180N |
| 2 | 1520T→G | Exon 2 | L507R |
| 3 | 1860A→G | Exon 2 | T620T |
| 4 | 2037C→T | Exon 2 | T679T |
| 5 | 2392A→C | Exon 2 | I798L |
| 6 | 2640G→C | Exon 3 | S880S |
Probable error in reference sequence
SALL4 is predicted to encode a 1,053 aa protein with eight zinc finger motifs, three with SAL boxes, arranged in a characteristic fashion similar to SALL1–3 (fig 2d). Thus, it is the fourth member of a group of human C2H2-type zinc finger proteins proposed to encode organ-specific transcription factors closely related to the Drosophila melanogaster spalt (sal) gene (Kohlhase et al. 1996, 1999). The SALL4 protein shares 49%, 39%, 36%, and 29% identity Xsal-3, SALL3, SALL1, and SALL2, respectively. SALL1 and mSall1 were recently demonstrated to be transcriptional repressors (Netzer et al. 2001; Kiefer et al. 2002) that localize to pericentromeric heterochromatin (Netzer et al. 2001). It is interesting that only the first 130 amino acids (including the amino-terminal H2CH-ZF) of mSall1 are required for repression (Kiefer et al. 2002). This region is upstream of the known SALL1 and SALL4 mutations, suggesting that the truncated proteins may have a dominant negative action as upregulated repressors (Kiefer et al. 2002). SALL1–3 are reportedly expressed in restricted patterns in the embryonic nervous system (Kohlhase et al. 1996, 1999), with SALL2 exclusively and SALL3 selectively expressed in the fetal pons (potentially in the developing abducens motoneurons). Thus, SALL2 and SALL3 are candidates for other forms of Duane syndrome. On the basis of our findings, SALL4 is likely to have an important role in abducens motoneuron development and may serve as a unique marker for these developing neurons. This complements our previous report of ARIX mutations in CFEOM2, a much rarer form of congenital strabismus that selectively targets the developing oculomotor and trochlear motoneurons (Nakano et al. 2001). We hope that continued studies of these genes will lead to a definition of the cascade of factors that determine ocular motoneuron identity.
Acknowledgments
We thank the family members for their participation and Patrick J. Dillon for technical assistance. This work was supported by the Fight for Sight research division of Prevent Blindness America, National Institutes of Health grants R01 EY012498 and R01 EY013583, and by the Children's Hospital Mental Retardation Research Center (Boston) (P30 HD 18655).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- ExPASy Molecular Biology Server, http://us.expasy.org/ (for prediction programs SWISS-PROT and TrEMBL)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SALL4 genomic, NT_011362; SALL4 cDNA, NM_020436; SALL4, NP_065169; SALL1, NP_002959; SALL2, XP_033473; SALL3, AAK18311; Xsal-3, BAA85900)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DURS1, including DRRS, [MIM 126800] and DURS2 [MIM 604356])
- UCSC Human Genome Project Working Draft, http://genome.cse.ucsc.edu/ (for working drafts for the mouse genome and the human genome)
References
- Appukuttan B, Gillanders E, Juo SH, Freas-Lutz D, Ott S, Sood R, Van Auken A, Bailey-Wilson J, Wang X, Patel RJ, Robbins CM, Chung M, Annett G, Weinberg K, Borchert MS, Trent JM, Brownstein MJ, Stout JT (1999) Localization of a gene for Duane retraction syndrome to chromosome 2q31. Am J Hum Genet 65:1639–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun BB, Mazzoli RA, Raymond WR (2001) Characteristics of Okihiro syndrome. J Pediatr Ophthalmol Strabismus 38:235–239 [DOI] [PubMed] [Google Scholar]
- Chung M, Stout JT, Borchert MS (2000) Clinical diversity of hereditary Duane's retraction syndrome. Ophthalmology 107:500–503 [DOI] [PubMed] [Google Scholar]
- Engle EC, Kunkel LM, Specht LA, Beggs AH (1994) Mapping a gene for congenital fibrosis of the extraocular muscles to the centromeric region of chromosome 12. Nat Genet 7:69–73 [DOI] [PubMed] [Google Scholar]
- Hotchkiss MG, Miller NR, Clark AW, Green WG (1980) Bilateral Duane's retraction syndrome: a clinical-pathological case report. Arch Ophthalmol 98:870–874 [DOI] [PubMed] [Google Scholar]
- Kiefer SM, McDill BW, Yang J, Rauchman M (2002) Murine Sall1 represses transcription by recruiting a histone deacetylase complex. J Biol Chem 277:14869–14876 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Hausmann S, Stojmenovic G, Dixkens C, Bink K, Schulz-Schaeffer W, Altmann M, Engel W (1999) SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics 62:216–222 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Schuh R, Dowe G, Kuhnlein RP, Jackle H, Schroeder B, Schulz-Schaeffer W, Kretzschmar HA, Kohler A, Muller U, Raab-Vetter M, Burkhardt E, Engel W, Stick R (1996) Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics 38:291–298 [DOI] [PubMed] [Google Scholar]
- Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W (1998) Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet 18:81–83 [DOI] [PubMed] [Google Scholar]
- Miller NR, Kiel SM, Green WR, Clark AW (1982) Unilateral Duane's retraction syndrome (type 1). Arch Ophthalmol 100:1468–1472 [DOI] [PubMed] [Google Scholar]
- Nakano M, Yamada K, Fain J, Sener EC, Selleck CJ, Awad AH, Zwaan J, Mullaney PB, Bosley TM, Engle EC (2001) Homozygous mutations in ARIX(PHOX2A) result in congenital fibrosis of the extraocular muscles type 2. Nat Genet 29:315–320 [DOI] [PubMed] [Google Scholar]
- Netzer C, Rieger L, Brero A, Zhang CD, Hinzke M, Kohlhase J, Bohlander SK (2001) SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/PIN2 and localizes to pericentromeric heterochromatin. Hum Mol Genet 10:3017–3024 [DOI] [PubMed] [Google Scholar]
- Okihiro M, Tasaki T, Nakano K, Bennett B (1977) Duane syndrome and congenital upper-limb anomalies. Arch Neurol 34:174–179 [DOI] [PubMed] [Google Scholar]
- Temtamy SA, McKusick VA (1978) The genetics of hand malformations. Birth Defects Orig Art Ser 1–619 [PubMed] [Google Scholar]
- Vincent C, Kalatzis V, Compain S, Levillers J, Slim R, Graia F, de Lurdes Pereia M, Nivelon A, Croquette M-F, Lacombe D, Vigneron J, Helias J, Broyer M, Callen DF, Haan EA, Weissenbach J, Lacroix B, Bellane-Chantelot C, Le Paslier D, Cohen D, Petit C (1994) A proposed new contiguous gene syndrome on 8q consists of branchio-oto-renal (BOR) syndrome, Duane syndrome, a dominant form of hydrocephalus and trapeze aplasia: implications for the mapping of the BOR gene. Hum Mol Genet 3:1859–1866 [DOI] [PubMed] [Google Scholar]