Abstract
In insects, the steroid hormone 20-hydroxyecdysone (20E) coordinates major developmental transitions. While the first and the final steps of 20E biosynthesis are characterized, the pathway from 7-dehydrocholesterol to 5β-ketodiol, commonly referred as the “black box”, remains hypothetical and whether there are still unidentified enzymes is unknown. The black box would include some oxidative steps, which are believed to be mediated by P450 enzymes. To identify new enzyme(s) involved in steroid synthesis, we analyzed by small-scale microarray the expression of all the genes encoding P450 enzymes of the malaria mosquito Anopheles gambiae in active steroidogenic organs of adults, ovaries from blood-fed females and male reproductive tracts, compared to inactive steroidogenic organs, ovaries from non-blood-fed females. Some genes encoding P450 enzymes were specifically overexpressed in female ovaries after a blood-meal or in male reproductive tracts but only three genes were found to be overexpressed in active steroidogenic organs of both females and males: cyp307a1, cyp4g16 and cyp6n1. Among these genes, only cyp307a1 has an expression pattern similar to other mosquito steroidogenic genes. Moreover, loss-of-function by transient RNAi targeting cyp307a1 disrupted ecdysteroid production demonstrating that this gene is required for ecdysteroid biosynthesis in Anopheles gambiae.
Introduction
In insects and other arthropods, specific steroid hormones, called ecdysteroids, play a major role during growth, development and reproduction [1]–[5]. The prohormone ecdysone (E) is synthesized from dietary cholesterol (C) via a series of hydroxylation and oxidation steps in steroidogenic tissues, the prothoracic glands (PG) during post-embryonic development and the ovary of adults [4], [6]. E is further converted into the active hormone 20-hydroxyecdysone (20E) in target tissues. During the last decade, molecular genetic studies in Drosophila melanogaster have led to the identification and characterization of several genes involved in 20E biosynthesis (Figure 1). The first enzymatic step, i.e. the conversion of C into 7-dehydrocholesterol (7dC), is catalyzed by the Rieske-domain oxygenase Neverland (Nvd) [7]–[10]. The last four hydroxylation steps, from 5β-ketodiol to 20E, are catalyzed by four P450 enzymes (CYPs): CYP306A1 (Phantom; Phm) [11]–[12], CYP302A1 (Disembodied; Dib) [13]–[14], CYP315A1 (Shadow; Sad) [14] and CYP314A1 (Shade; Shd) [15]. The genes encoding these four P450 enzymes were identified from study of Drosophila embryonic lethal mutants, the Halloween mutants, which exhibit ecdysteroid deficiency [13], [16].
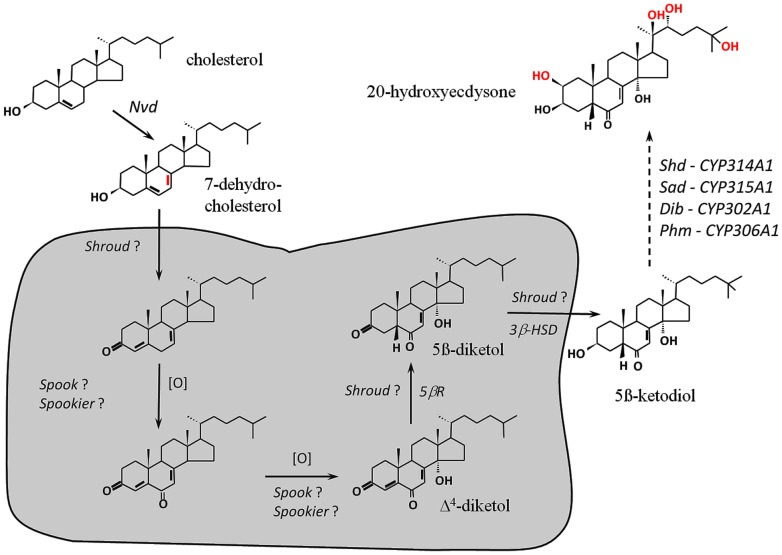
Figure 1. Biosynthetic pathway of ecdysteroids.
From cholesterol to 20-hydroxyecdysone, the active steroid hormone. Characterized steps: Nvd, neverland, Rieske-domain oxygenase; Phm, Phantom, CYP306A1, 25-hydroxylase; Dib, Disembodied, CYP302A1, 22-hydroxylase; Sad, Shadow, CYP315A1, 2-hydroxylase; Shd, Shade, CYP314A1, 20-hydroxylase. Chemical modifications are shown in red on molecules. Putative steps of the “black box”, from 7-dehydro-cholesterol to 5ß-ketodiol, are represented within the dark grey box. [O] indicates oxidative step that might be catalyzed by a CYP enzyme. Some steps could be catalyzed by shroud, spook (CYP307A1) or spookier (CYP307A2). 5ßR: 5ß-reductase. 3ßHSD: 3ß-hydroxysteroid-dehydrogenase. Modified from Lafont et al. [4].
While the above-mentioned steps of biosynthesis are well characterized, little is known about the conversion from 7dC to 5β-ketodiol, commonly referred as the “black box”, for which no stable intermediate has been identified. The hypothetic metabolic steps occurring in the black box imply modifications at multiple carbon positions (Figure 1, shaded part). This includes the oxidation of 3beta-alcohol to ketone, the oxidation of carbon 6 with concomitant loss of the 4beta- and 6-hydrogens to form the 6-keto group, and 14alpha-hydroxylation. Δ4-diketol would then be converted by a 5ß-reductase to 5ß-diketol further transformed in 5ß-ketodiol by a 3ß-reductase [4], [17]. The black box, and more particularly the oxidative steps, is thought to involve one or more P450 enzymes that still remain uncharacterized [6], [18]. Consistent with this hypothesis, CYP307A1 (Spook, Spo) and CYP307A2 (Spookier, Spok) have been proposed to catalyze one of the ecdysteroid biosynthesis oxidative steps [18]–[21]. The gene encoding CYP307A1 has been first described in the Drosophila Halloween mutants [13] and further identified in a differential display PCR screen in the PG of the Lepidoptera Bombyx mori [18]. In D. melanogaster, unlike other Halloween genes, cyp307a1 is expressed only in embryos and in the follicle cells of ovary but not during the larval stages. However, its paralog cyp307a2 is expressed within the PG cells during larval stages only and RNAi mediated reduction of its expression leads to developmental arrest at the first larval stage [19]. Ketotriol and ketodiol can rescue cyp307a1 mutant embryos and cyp307a2 knockdown larvae respectively, while C or 7dC do not, suggesting that cyp307a1 and cyp307a2 are likely to be components of the black box [19]. Recently, Niwa et al. [22] identified the non-molting glossy (nm-g)/shroud (sro) gene in B. mori and D. melanogaster, respectively. This gene encodes a short-chain dehydrogenase/reductase that seems to be also involved in the black box, as the application of ketodiol, but not C or 7dC, overcomes the larval arrest observed in nm-g/sro mutant animals. Similarly, cyp6t3 constitutes another candidate gene in the black box as its knockdown in Drosophila PG leads to E deficiency phenotypes that can be rescued by feeding larvae with E or one of several E biosynthetic precursors [23]. However, cyp6t3 has no clearly identifiable ortholog in other insect species, which is unusual compared to the characterized Halloween genes [23]–[24]. Even if these experiments tentatively place cyp307a1 and cyp307a2, along with cyp6t3 and Sro, inside the black box pathway, no specific enzymatic activity has been assigned yet to the corresponding proteins. Therefore, whether it remains unidentified enzymes responsible for ecdysteroid biosynthesis is still unknown.
In female mosquitoes, a blood meal triggers the ovaries to secrete high amounts of E, subsequently hydroxylated to 20E, which in turn activates the transcription of the vitellogenin (Vg) gene in the female fat body. This leads to the production and secretion of Vg proteins into the hemolymph, that are later incorporated into the growing oocytes [2], [25]. Among mosquitoes and more generally among insects, the malaria vector Anopheles gambiae appears so far unique because not only blood-fed (BF) females, but also males produce high amounts of 20E. In males, the steroid hormone is produced by and stored in the accessory glands (MAGs) to be further transferred to females during mating [26]. In both females and males, expression of the genes involved in the last steps of steroidogenesis is tightly correlated with ecdysteroid production [26] as described in several insect species [11]–[12], [27]–[31], as well as in crustaceans [32]. Taken together, all these results suggest that the timing of hormone production highly depends on transcriptional regulation of the enzymes involved in its biosynthesis. Due to the high steroidogenic capacities of A. gambiae females and males, this mosquito species then constitutes a good model to identify new genes involved in ecdysteroid biosynthesis. To uncover unidentified CYP(s) gene(s) involved in 20E biosynthesis in A. gambiae, we took advantage of a small-scale microarray, which was initially developed to study metabolic-based insecticide resistance in this malaria vector [33]. The microarray covers 230 genes of A. gambiae, including all members of the three main enzyme families involved in insecticide metabolism: the cytochrome P450 monooxygenases (CYPs), the gluthatione-S-transferases (GSTs) and the carboxylesterases (COEs). By comparing expression of A. gambiae CYP genes between a non active steroidogenic tissue, the ovaries from non blood-fed (NBF) females, and active steroidogenic tissues, ovaries from BF females or male reproductive tracts (MRTs), we identified 3 CYP genes significantly over-transcribed in both female and male steroidogenic tissues: cyp4g16, cyp6n1 and cyp307a1. We demonstrate that only cyp307a1 has the same expression pattern as other genes involved in steroid synthesis in A. gambiae [26]. Moreover, transient RNAi targeting cyp307a1 significantly decreases E production in A. gambiae females. Overall, our results demonstrate that cyp307a1 is required for ecdysteroid biosynthesis in A. gambiae.
Results
Expression of steroidogenic genes is increased in steroidogenic active versus steroidogenic inactive tissues
To identify new CYP(s) involved in ecdysteroid synthesis, changes in A. gambiae CYP transcription levels in gonads associated with steroidogenesis were assessed using the “Anopheles detox chip microarray” which contains probes for the major A. gambiae detoxification genes [33]. Because steroidogenic CYPs genes are usually up-regulated in active steroidogenic tissues [11]–[12], [26]–[32], we compared gene expression between ovaries of NBF females, which do not produce ecdysteroids, and ovaries at different times after the blood meal (5, 16, 22 h PBM) or MRTs, tissues which actively produce ecdysteroids [26]. Gene expression results obtained for the four steroidogenic cyps previously identified, i.e. cyp306a1, cyp302a1, cyp315a1 and cyp314a1, are given in Table 1. In ovaries, the transcription of cyp306a1 and cyp302a1 is significantly increased at 16 h and 22 h PBM. By contrast, cyp315a1 and cyp314a1, involved in the last two steps of ecdysteroid biosynthesis, are downregulated at 16 h and 22 h PBM when ecdysteroid production peaks. In MRTs, cyp306a1, cyp302a1 and cyp314a1 are strongly overexpressed compared to ovaries of NBF females while cyp315a1 is not significantly differently transcribed between MRTs and NBF ovaries. Overall, the earlier the steroidogenic genes are in the 20E biosynthetic pathway, the more they are up-regulated in steroidogenic active tissues. These results are in agreement with previous RT-PCR results [26] and validate the use of this microarray to identify genes encoding the early steps of steroidogenesis from the so-called “black box”.
Table 1. Expression of genes encoding steroidogenic CYP in steroidogenic ovaries and MRTs compared to non steroidogenic ovaries.
| Steroidogenic | Ovaries | Ovaries | Ovaries | MRTs |
| gene | 5 h PBM | 16 h PBM | 22 h PBM | |
| CYP306A1 | 0.98 - 9.45E-01 | 1.55 - 1.24E-02 | 2.05 - 1.29E-02 | 18.77 - 3.82E-08 |
| CYP302A1 | 1.01 - 9.04E-01 | 1.11 - 2.04E-01 | 1.29 - 3.54E-02 | 5.44 - 1.27E-02 |
| CYP315A1 | 0.93 - 2.57E-01 | 0.79 - 4.88E-02 | 0.67 - 2.40E-02 | 0.86 - 0.65E-01 |
| CYP314A1 | 0.98 - 9.46E-01 | 0.97 - 9.88E-01 | 0.86 - 7.96E-01 | 2.86 - 4.21E-05 |
Expression ratios and p values (italic) for the genes encoding CYPs previously characterized as steroidogenic CYPs, in ovaries of blood-fed females at 5 h, 16 h, 22 h post blood-meal (PBM) and in male reproductive tracts (MRTs) compared to ovaries from non blood-fed females. CYP genes are listed according to their position in the 20E biosynthesis pathway. Genes showing a significant over transcription (ratio >1.5 and P<0.05) are shown in bold.
Cyp4g16, cyp6n1 and cyp307a1 are significantly up-regulated in steroidogenic tissues
To identify candidate genes that could be involved in steroidogenesis, any CYP satisfying all of the following criteria was selected: (i) the gene is up-regulated with both a transcription ratio >1.5-fold and P value<0.05, (ii) the gene is up-regulated both in ovaries after a blood meal at any time point and in MRTs compared to ovaries of NBF females. Selecting only CYPs up-regulated in both steroidogenic ovaries and MRTs removed genes that may be involved in sex-specific gonad functions. Moreover, as steroidogenic CYPs are well conserved in insects while detoxification CYPs are not, using results obtained with two different strains (Yaoundé and Kisumu) of A. gambiae appears to be a good criteria for identifying steroidogenic CYPs. Such candidates are expected to be regulated in the same way in two different strains contrary to detoxification enzyme which might be differently regulated between two strains [34]–[35]. Of 103 P450 genes represented on the microarray, only 3 candidate CYPs met our screening criteria: cyp4g16, cyp6n1 and cyp307a1 (Table 2).
Table 2. CYP genes over transcribed in steroidogenic ovaries and/or in steroidogenic MRTs compared to non steroidogenic ovaries.
| Up-regulated genes | Ovaries | Ovaries | Ovaries | MRTs |
| 5 h PBM | 16 h PBM | 22 h PBM | ||
| CYP12F1 | 1.52 - 1.74E-01 | 1.15 - 6.30E-01 | 0.85 - 7.61E-01 | 1.92 - 2.09E-02 |
| CYP12F2 | 0.76 - 1.25E-02 | 0.73 - 7.25E-02 | 0.61 - 2.78E-02 | 2.85 - 6.41E-07 |
| CYP12F3 | ND | ND | ND | 14.96 - 1.87E-05 |
| CYP12F4 | 0.83 - 1.90E-01 | 0.91 - 7.36E-01 | 0.76 - 2.00E-01 | 16.67 - 6.20E-04 |
| CYP302A1 | 1.01 - 9.04E-01 | 1.11 - 2.04E-01 | 1.29 - 3.54E-02 | 5.44 - 1.27E-02 |
| CYP305A2 | 1.50 - 6.20E-03 | 1.15 - 2.98E-01 | 1.00 - 9.99E-01 | ND |
| CYP306A1 | 0.98 - 9.45E-01 | 1.55 - 1.24E-02 | 2.05 - 1.29E-02 | 18.77 - 3.82E-08 |
| CYP307A1 | 1.32 - 7.77E-02 | 1.28 - 1.01E-01 | 1.51 - 4.41E-02 | 11.61 - 9.30E-10 |
| CYP314A1 | 0.98 - 9.46E-01 | 0.97 - 9.88E-01 | 0.86 - 7.96E-01 | 2.86 - 4.21E-05 |
| CYP4AR1 | 0.96 - 9.46E-01 | 1.07 - 7.90E-01 | 1.11 - 8.76E-01 | 1.95 - 1.83E-02 |
| CYP4D15 | ND | 1.31 - 6.19E-02 | 1.27 - 5.58E-01 | 2.14 - 4.05E-03 |
| CYP4D22 | 0.89 - 8.690E-01 | 0.99 - 9.99E-01 | ND | 8.46 - 3.89E-08 |
| CYP4G16 | 2.00 - <2.00E-16 | 2.32 - 6.30E-03 | 2.22 - 2.00E-04 | 3.31 - 2.16E-02 |
| CYP4J5 | 0.93 - 7.50E-01 | 0.90 - 8.76E-01 | 0.84 - 7.61E-01 | 3.38 - 5.97E-03 |
| CYP4K2 | 0.82 - 1.95E-01 | 0.87 - 4.97E-01 | 0.86 - 7.61E-01 | 2.28 - 2.00E-04 |
| CYP6AF1/2 | 1.70 - 7.70E-03 | 1.02 - 9.93E-01 | 0.97 - 9.83E-01 | ND |
| CYP6AG1 | 2.33 - 4.10E-03 | 1.82 - 6.30E-03 | 0.62 - 7.96E-01 | ND |
| CYP6M2 | 1.09 - 2.93E-01 | 0.87 - 2.22E-01 | 0.83 - 2.57E-01 | 6.95 - 2.14E-05 |
| CYP6M3 | 1.22 - 1.44E-02 | 0.82 - 1.19E-01 | 0.67 - 1.29E-02 | 1.61 - 1.36E-02 |
| CYP6M4 | 1.25 - 1.63E-01 | 1.43 - 1.79E-01 | 1.17 - 7.77E-01 | 6.61 - 1.60E-04 |
| CYP6N1 | 0.91 - 4.15E-01 | 1.71 - <2.00E-16 | 0.52 - 1.29E-02 | 5.99 - 2.70E-04 |
| CYP6P3 | 0.92 - 6.19E-01 | 1.23 - 8.28E-01 | 0.89 - 8.10E-01 | 2.03 - 7.58E-03 |
| CYP6S1 | 0.76 - 6.40E-03 | 0.78 - 4.88E-02 | 0.91 - 5.76E-01 | 7.43 - 2.91E-07 |
| CYP6S2 | 0.91 - 2.930E-01 | 1.04 - 8.93E-01 | 1.19 - 1.72E-01 | 8.66 - 1.54E-09 |
| CYP6Z1 | ND | ND | ND | 3.04 - 6.20E-04 |
| CYP6Z2 | 1.30 - 9.08E-02 | 1.08 - 8.72E01 | 0.85 - 7.96E-01 | 12.67 - 2.25E-05 |
| CYP9J5 | 0.87 - 2.01E-01 | 0.75 - 1.79E-01 | 0.65 - 1.57E-01 | 2.52 - 1.88E-03 |
| CYP9K1 | 0.92 - 2.95E-01 | 0.90 - 4.81E-01 | 0.92 - 7.61E-01 | 2.25 - 6.85E-03 |
Only CYP genes overexpressed in ovaries of blood-fed females at least in one time point and/or in MRTs compared to ovaries from NBF females are listed. Values in bold indicate a ratio >1.5 and a p value<0.05. ND: Not detected or detected in less than 2 arrays. CYP genes previously characterized as involved in steroidogenesis are underlined. Candidate genes, overexpressed in ovaries of BF females, at least in one time point, and in MRTs compared to ovaries from NBF females, are in bold. PBM: post-blood meal. MRTs: Male reproductive tracts.
Only cyp307a1 shows a typical steroidogenic enzyme expression pattern
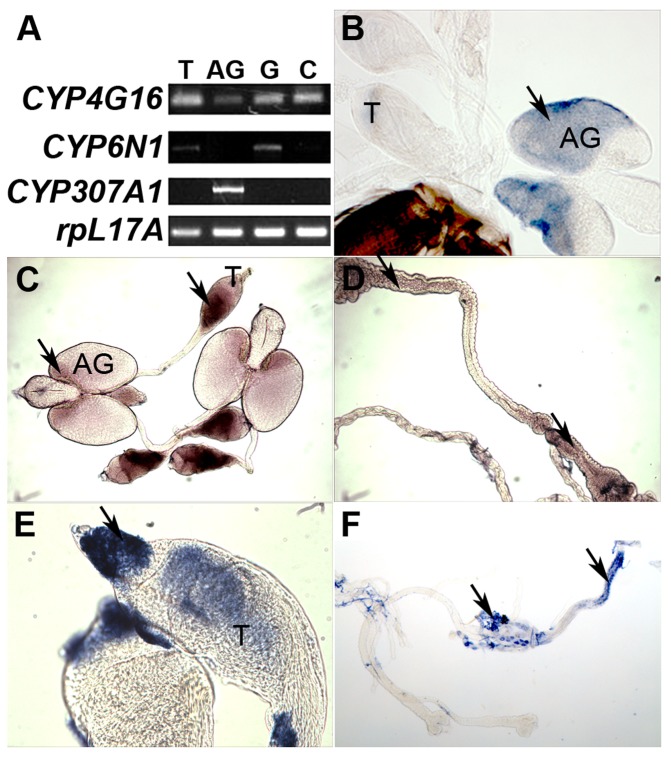
Expression of the three candidate genes was further analyzed by RT-PCR and in situ hybridization in different tissues of adult males (Figure 2) as steroidogenic enzyme gene expression is strictly restricted to the anterior part of MAGs in Anopheles males contrary to a broader expression in female tissues [26]. As shown in Figure 2A, cyp4g16 is expressed in testes, MAGs, gut and carcass. In situ hybridization revealed that cyp4g16 is expressed in the posterior part of the testicular follicular sheath and in the posterior part of the MAGs (Figure 2C), as well as in the anterior and posterior midgut (Figure 2D). Cyp6n1 is mainly expressed in the testes and in the gut (Figure 2A, 2E, 2F). In contrast with cyp4g16, cyp6n1 is expressed in the spermatogonies during early stages of spermatogenesis (Figure 2E). Unlike the two other candidate genes, expression of cyp307a1 was restricted to the MAGs and more precisely to the anterior part of the glands (Figure 2B), as observed for other steroidogenic genes [26]. In conclusion, only cyp307a1 shows a typical steroidogenic CYP expression pattern in Anopheles adult male and therefore appeared to be the most relevant candidate for functional validation.
Figure 2. Expression pattern of cyp4g16, cyp6n1 and cyp307a1 in adult males.
(A) RT-PCR analysis of cyp4g16, cyp6n1 and cyp307a1 expression pattern in males (T, testes; AG, accessory glands; G, gut and Malpighian tubules; C, carcass). rpL17A is used as a control gene. (B to F) In situ expression pattern of cyp307a1, cyp4g16, cyp6n1 in males (T, testes; AG, accessory glands). (B) cyp307a1 is detected in the anterior part of accessory glands. (C) cyp4g16 is detected at the bottom of testes and in the posterior part of accessory glands. (D) cyp4g16 is detected in the anterior and posterior gut. (E) cyp6n1 is detected at the top of testes. (F) cyp6n1 is expressed in the posterior gut and malpighian tubules. Black arrowheads show expression zones.
CYP307A1 is required for ecdysteroid production in Anopheles gambiae
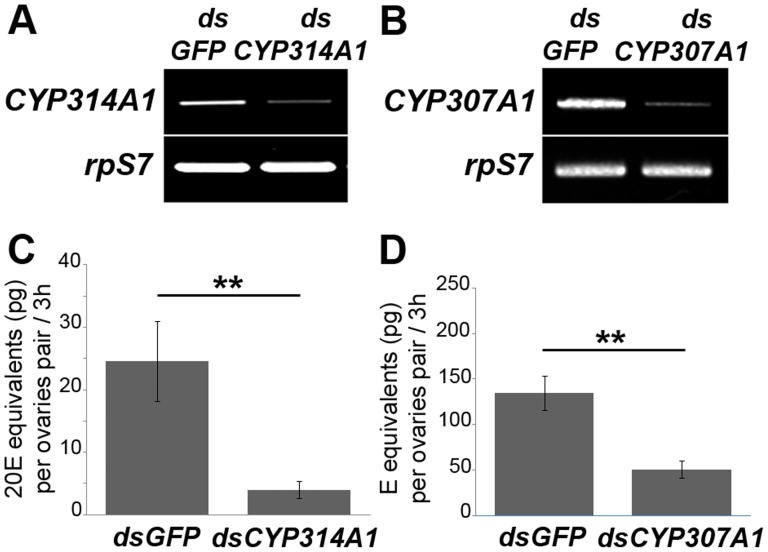
If CYP307A1 is indeed required for ecdysteroid biosynthesis in A. gambiae, knocking down cyp307a1 expression should decrease ecdysteroid production by steroidogenic tissues. To test this hypothesis, we performed transient RNAi on Anopheles females targeting cyp307a1 before measuring in vitro ovarian 20E production 22 h after blood-feeding. As a positive control, we first determined whether knocking-down by transient RNAi a known steroidogenic gene, cyp314a1, would indeed decrease 20E production in ovaries of BF females. As shown in Figure 3A, expression of cyp314a1 was strongly decreased in ovaries from BF females injected with ds-cyp314a1 compared to controls (ds-gfp-injected BF females). The decrease in cyp314a1 RNA led to a significant reduction of ovarian 20E production in ds-cyp314a1-injected BF females compared to controls (Figure 3C). Therefore, transient RNAi targeting a steroidogenic enzyme gene in mosquito female is a powerful method to characterize steroidogenic genes. Injection of ds-cyp307a1 also strongly decreased cyp307a1 expression in ovaries from ds-cyp307a1-injected BF females compared to controls (ds-gfp-injected BF females) (Figure 3B). As depicted in Figure 3D, ovarian ecdysteroid production of ds-cyp307a1-injected females was also significantly decreased compared to controls, demonstrating that cyp307a1 is required for ecdysone biosynthesis in Anopheles.
Figure 3. In vitro ecdysteroid secretion by ovaries of dsRNA injected females.
(A) RT-PCR analysis of cyp314aA1 in ovaries of dsgfp and dscyp314a1 females 22h after blood-feeding. (B) RT-PCR analysis of cyp307a1 in ovaries of dsgfp and dscyp307aA1 females 22h after blood-feeding. (C) In vitro ecdysteroid secretion of ovaries from dsgfp and dscyp314a1 females 22 h after blood-feeding. Results are expressed as mean ± SEM in 20E equivalents (in pg) per ovaries pair. (D) In vitro ecdysteroid secretion of ovaries from dsgfp and dscyp307a1 females 22 h after blood-feeding. Results are expressed as mean ± SEM in E equivalents (in pg) per ovaries pair. Results were subjected to statistical analysis using Mann-Whitney test (**, P<0.01).
Discussion
Our microarray analysis revealed a large set of genes encoding CYP overexpressed in ovaries of BF female and in male reproductive tracts. Among these genes and except the CYPs previously known to be involved in 20E biosynthesis, we identified 3 genes encoding P450 enzymes, cyp4g16, cyp6n1 and cyp307a1, that are overexpressed in adult active steroidogenic tissues of both sexes, i.e. reproductive tracts of mature males and ovaries of BF females, compared to non active steroidogenic tissues. In addition, we demonstrated that, among these three genes, only cyp307a1 has a similar expression pattern as other CYP genes involved in ecdysteroid biosynthesis in A. gambiae adults [26]. We further demonstrated that transient loss-of-function of cyp307a1 leads to a decreased E production in A. gambiae, validating the involvement of cyp307a1 in steroidogenesis in this mosquito species.
We found that the previously characterized genes cyp306a1, encoding the 25-hydroxylase, and cyp302a1, encoding the 22-hydroxylase, are up-regulated in ovaries of BF females from 16 h to 22 h PBM, time at which ovaries produce high amounts of steroids compared to ovaries from NBF females. In contrast, cyp315a1 and cyp314a1, which encode respectively the 2- and the 20-hydroxylase, the two final steps leading to the active hormone 20E, are not significantly up-regulated in active steroidogenic ovaries versus non active ones. This is consistent with the fact that these genes are already expressed in ovaries of NBF females and also expressed in some peripheral tissues in A. gambiae and D. melanogaster [4], [14], [15], [26], [36]. The observation that these two final steps 20E biosynthesis, and not only the 20-hydroxylase, are not restricted to steroidogenic tissues compared to the earlier steps could possibly be correlated to the less polar nature of the final steroid compounds. Indeed, 2-deoxyecdysone (2dE) and E are more soluble compounds than earlier intermediates and are likely to easily diffuse from steroidogenic cells to target cells/tissues that would possess the capacity of converting 2dE into the biologically active 20E hormone. A similar situation has also been reported in crustaceans [37]. In MRTs, the strong overexpression of the genes involved in steroidogenesis, except cyp315a1, matches with the huge steroidogenic capacity of the accessory glands of A. gambiae males that exceeds by far that of vitellogenic ovaries [26]. As observed in active steroidogenic ovaries, cyp315a1 is not overexpressed in steroidogenic MRTs. In contrast, MRTs overexpress cyp314a1 (encoding the 20-hydroxylase). While in BF females, ovaries produce a mixture of E and 20E, MAGs, the steroidogenic tissue of MRT, produce the active hormone 20E. MAGs then represent a target tissue-like, which possess a strong 20-hydroxylase activity to ensure the production of large amounts of 20E that is then transferred to female during copulation [26].
With the exception of the CYPs previously known to be involved in 20E biosynthesis, only 3 additional CYP genes were found to be overexpressed in steroidogenic tissues of both females and males compared to non steroidogenic tissues: cyp4g16, cyp6n1 and cyp307a1. For two main reasons, only cyp307a1 was further investigated as a candidate gene in the steroid biosynthesis pathway in A. gambiae mosquitoes. First, we show here that only cyp307a1 has the same expression pattern as the previous characterized genes being specifically expressed in the anterior part of the MAGs, the unique steroidogenic tissue in A. gambiae males [26]. In contrast, cyp4g16 and cyp6n1 are expressed mainly in the testes and in the gut, tissues that do not produce steroids. Secondly, the critical physiological function of steroidogenic enzymes has imposed constraints on their selection. As a consequence, cyp genes involved in steroid biosynthesis are well conserved among ecdysteroid producing animals [4], [20], [34]. In contrast, CYP genes involved in detoxification processes present a higher diversification among species, likely due to differences in their ecological niches and adaptive strategies [38]–[39]. Indeed, phylogenetic analyses showed that cyp307a1 possesses a true ortholog in insect genomes, even if, in contrast with genes encoding the terminal hydroxylases which have a single ortholog in any arthropod species investigated so far, cyp307a1 has also two paralogs, cyp307a2 and cyp307b1 [4], [20], [40]. Conversely, cyp4g16 and cyp6n1 do not show such a high degree of conservation in other insect species, suggesting that these two genes are not involved in a highly conserved metabolic pathway like steroid biosynthesis. As cyp4g16 has been associated with insecticide-spraying periods in Anopheles arabiensis (A. gambiae complex) in Cameroon [41] and cyp6n1 has been reported to be overexpressed in A. gambiae after exposure to insecticides [42], it is tempting to speculate that these genes are rather involved in detoxification processes. However, many insect cyp4g have been associated with diverse functions different from detoxification of exogenous compounds. The closest gene of cyp4g16 in D. melanogaster, cyp4g15, which is expressed in the larval brain [43] as well as cyp4c15 in the crayfish Orconectes limosus [32] have been postulated to play a role in ecdysteroid metabolism rather than detoxification. Similarly, another gene, cyp4g1 is highly expressed in the steroidogenic organ in D. melanogaster larvae and might be involved in lipid metabolism, which may indirectly regulate ecdysone biosynthesis [44]–[45]. Cyp4g25 of the silkmoth Antheraea yamamai is expressed in the integument of larvae and seems to be in relation to diapause [46]. Cyp4g16 and its closest homologs in insect species seem therefore to be linked to steroidogenesis, even if this enzyme is probably not a steroidogenic enzyme stricto sensu.
Transient knock-down of cyp307a1 in BF mosquito females leads to a decrease of ovarian ecdysteroid production, further demonstrating that this gene is required for 20E biosynthesis in adult steroidogenic tissues. The involvement of cyp307a1 in steroidogenesis in A. gambiae fits well with the previous identification of cyp307a genes being involved in steroid biosynthesis in D. melanogaster, B. mori and Manduca sexta and more recently in Tribolium castaneum [13], [18]–[19], [29], [31]. Up to now, the precise enzymatic activity of CYP307 proteins has not been elucidated. The evolutionary history of the cyp307 family is quite complex and occurrence and expression pattern during development can vary depending on species [20], [31], [40], [47]. For instance, Drosophila carries two paralogs, cyp307a1, which is expressed only in embryos and in the follicle cells of ovary but not during the larval stages, and cyp307a2, which is expressed only during larval stages within the PG cells [19]. In contrast, in Tribolium, cyp307a1 is expressed in embryos, larvae and adult females while cyp307b1 is only expressed in the male accessory glands [31]. Like Tribolium, A. gambiae possesses cyp307a1 and cyp307b1 paralogs [20]. By RT-PCR, cyp307a1 is detected in larvae, nymphs, adult females and males, but not in embryos, while cyp307b1 is detected at every developmental stage (data not shown). In our microarray analysis, although cyp307b1 was detected in ovaries and MRTs, its expression did not significantly vary in adult steroidogenic organs. The reason, if any, why evolution has allowed flexibility for cyp307 genes is still not clear. The cyp307 paralogs show the highest degree of identity between all steroidogenic CYP proteins and ectopic expression of cyp307a1 rescues Drosophila cyp307a2 mutants [19], [31], [47]. Although subtle catalytic differences may exist between CYP307 enzymes, these conserved paralogs are likely to be functionally redundant products of gene duplications that occupy different spatio-temporal patterns of expression to precisely control ecdysteroid titers during development [20], [40]. In A. gambiae adults, cyp307a1 is highly up regulated in active steroidogenic tissues and this highlights that this gene must encode one of the early steps of steroidogenesis which are known to be more tightly regulated than the last steps leading to 20E [4]. This is consistent with results obtained in D. melanogaster indicating that they could act in the currently uncharacterized black box, from which one or multiple steps are believed to limit the production of ecdysone, in that no stable intermediate has been yet identified [4], [17]. No conversion of C or 7dC was observed in S2 cells transfected with cyp307a1 but since the black box is supposed to contain several oxidative transformations, unless this gene catalyzes the initial reaction, expressing cyp307a1 alone with 7dC would not be expected to produce a product [18]–[19], [48].
In conclusion, our approach has led to the identification of cyp307a1 as playing a role in steroid biosynthesis in the malaria mosquito A. gambiae. Our study did not reveal any other CYP gene except cyp307a1 that could be involved in this metabolic pathway, provided that the early steps are regulated at the transcriptional level. Although several studies strongly implicated cyp307 genes in ecdysteroid biosynthesis and more particularly in the black box, additional experiments are necessary to clarify their precise biochemical activity. The recent identification of Sro as also playing a role in the black box [22] plus the availability of new ecdysteroid intermediates [17] should facilitate the characterization of these mysterious steps in the near future.
Materials and Methods
Mosquito strains
Two different A. gambiae strains were used for the microarray experiments: the Kisumu strain (molecular S form, from Kisumu, Western Kenya) and the Yaoundé strain (molecular M form, from Yaoundé, Cameroon). Based on population genomic evidences, it has been recently proposed to assign distinct species names to A. gambiae M and S forms [49]. The S form should conserve the A. gambiae s.s. name while the M form should now be A. coluzzii.
For all other experiments, only the Yaoundé strain was used. Mosquitoes were reared at 27°C under standardized conditions of 70% relative humidity and 12/12 h light/dark cycle, on 10% w/v sucrose solution.
Microarray experimental design and sample collection
The microarray used in this study contains probes for 103 P450s, 31 COEs, 35 GSTs, 41 Red/Ox genes, 5 ATP-binding-cassette transporters, tissue-specific genes and housekeeping genes of the Kisumu strain of A. gambiae [33]. This array was used with different species of the Anopheles gambiae complex and exhibited similar performance between species [41]. Thus, we were confident that hybridizations with the Yaoundé strain of A. gambiae would be similar to the ones with the Kisumu strain. At most, the number of candidate genes in female experiments would be underestimated. To identify steroidogenic genes, we compared transcription levels of genes encoding CYP between steroidogenic tissues (ovaries from blood-fed females and male reproductive tracts, MRTs) and non steroidogenic tissues (ovaries from non blood-fed females). Each set of microarray experiment consisted of four hybridizations comprising two biological replicates (ovaries from BF females or MRTs) compared to a unique reference with dye swap of Cy3 and Cy5 fluorophores. The reference is a pool of ovaries from 3 independent cohorts of 3 days-old non-blood-fed females (n = 30 per cohort) either Yaoundé (female experiments) or Kisumu (MRTs experiments). For each biological replicate, about 300 adult mosquitoes synchronized at emergence were reared simultaneously. Each biological replicate consisted of mosquitoes (n = 30) from distinct generations to take into account stochastic variations.
For female experiments, ovaries from NBF females of the Yaoundé strain (reference in female experiments) were compared to ovaries of blood-fed (BF) females of the Yaoundé strain at 5 h, 16 h and 22 h post-blood-meal (PBM). 3 days-old females were allowed to feed on blood for 20 minutes. Partially or unfed females were discarded. For MRTs experiments, ovaries from NBF females of the Kisumu strain (reference in MRTs experiments) were compared to MRTs from 3 days-old males of the Kisumu strain. Ovaries from NBF females, non steroidogenic, were also used as the reference as MRTs have been shown to be steroidogenic during all the life of adult males [26]. Ovaries and MRTs were dissected in PBS (0.22 µm filtered) and stored in RNAlater (Applied Biosystems) at 4°C until RNA extraction.
Target preparation and microarray hybridizations
RNA extractions, antisense RNA (aRNA) synthesis, and labelling reactions were performed independently for each replicate to take into account technical variation. Total RNA was extracted from batches of 40 to 60 ovaries/MRTs using the Picopure RNA isolation kit (Arcturus) with a DNAse treatment according to manufacturer's instructions. A batch contained mosquitoes from the same generation, collected on the same day. Total RNA quantity and quality were assessed by using a Nanodrop spectrophotometer (Nanodrop Technologies, Oxfordshire, U.K.) and agarose gel electrophoresis. From 1 (MRTs) to 5.5 µg (ovaries) of total RNA from each batch were amplified in one amplification round using the Riboamp RNA Amplification Kit (Arcturus) to generate purified aRNA. aRNA quantity and quality were further assessed by a Nanodrop spectrophotometrer and agarose gel electrophoresis. Final target preparation (aRNA fluorescent labelling and purification), hybridizations and microarray scanning were performed as previously described [50].
Microarray data analysis
Data analysis was performed as described in David et al. [33] except that genes showing a t test P value<0.05 and an expression ratio >1.5-fold in either direction were considered differentially transcribed. In our screen, only genes overexpressed in steroidogenic tissues with an expression ratio >1.5-fold were further considered.
As a control, a calibration experiment was performed in which two aliquots of labelled aRNA derived from the same sample were co-hybridized to two arrays with dye-swap. As expected, none of the gene probes came out significantly differentially transcribed, supporting the statistical approach described above (data not shown).
All microarray data have been deposited at ArrayExpress (E-MTAB-1697).
mRNA expression analysis by RT-PCR
4-day-old male tissues were carefully dissected in ice-cold, RNase-free phosphate buffered saline (100 mM, pH 7.4), containing 0.1% Tween (PBT). Total RNA was then extracted with SV Total RNA Isolation System (Promega) and quantified by spectrometry at 260 nm. cDNAs were generated using M-MLV reverse transcriptase from 100 ng of total RNA. rpL17A, coding for the ribosomal protein rpL17A, a domestic gene, was used as internal control.
For mRNA ovarian expression analysis after transient RNAi, ovaries from 10 BF females were subjected to RNA extraction. cDNAs were then generated from 500 ng of total RNA. Sense and antisense primers used for PCR analysis are located inside and outside the dsRNA sequence respectively, to avoid any amplification of dsRNA. rpS7, coding for the ribosomal protein S7, was used as internal control. Sequences of all primers used are given in Table S1.
Gene cloning
Total RNA was isolated with Trizol reagent (Invitrogen) from vitellogenic ovaries and reverse transcribed with M-MLV reverse transcriptase (Promega). The A. gambiae genome is sequenced and genomic data are available on the website http://www.Ensembl.org/. Full length cDNA sequences of Agcyp314a1, Agcyp307a1, Agcyp6n1, Agcyp4g16 were amplified from total cDNAs by PCR with specific primers (see Table S1). cDNAs were gel purified, cloned into pIB/V5-His (TA cloning, Invitrogen) and insert sequences verified (Genome Express; GenBank Accession numbers KF656700, KF656701, KF656702). Egfp (described in [51]) was cloned into pGEM-T Easy vector (pGEM-GFP) with specific primers (Table S1).
In situ hybridization
MRTs were carefully dissected in PBT and fixed with 4% paraformaldehyde. RNA probes and in situ hybridization on MRTs from 4-day-old males were performed according to the method described in Parvy et al. [27]. Probes were synthesized from Agcyp307a1, Agcyp6n1 and Agcyp4g16 full-length cDNA cloned into pGEMT-easy.
Preparation of dsRNA, injection procedure, and sample preparation
cDNA fragments corresponding to the C-terminal sequence of cyp307a1 (783 bp), cyp314a1 (788 bp) and to Egfp were produced by RT-PCR using pIB/V5-cyp307a1, pIB/V5-cyp314a1 and pGEM-GFP respectively as a template and gene-specific primers extended with a T7-promoter sequence containing a purine tail (Table S1). Those amplicons were then used as template to generate dsRNA by in vitro transcription (MEGAscript RNAi Kit, Ambion). dsRNA concentration and quality were estimated by spectrometry at 260 nm and electrophoresis on an ethidium bromide containing agarose gel. dsRNA were injected into one-day-old cold-anesthetized virgin females using a nanoject micro-injector (Drummond Scientific). 800 ng of dsRNA in 120 nl of water were injected per mosquito. On day 4 post-injection, injected virgin females were allowed to feed on mouse blood for 30 minutes; unfed females were discarded just after the blood meal. Ovaries from females were then carefully dissected 22 h after the blood meal and were subjected to RNA extraction/RT-PCR or in vitro incubation for ecdysteroid quantification. Experiments were performed on 2 independent cohorts of mosquitoes.
In vitro culture and ecdysteroid quantification
Cultures were performed according to the method described in Pondeville et al. [26] except that ovaries were incubated for 3 h at 25°C. After incubation, culture medium was collected and stored at −20°C until ecdysteroid quantification.
Ecdysteroids were quantified by EIA, with 20-hydroxyecdysone-2-succinate coupled to peroxidase as a tracer (dilution 1∶80,000) and either the L2 antiserum (a generous gift from Dr. M. De Reggi, dilution 1∶40,000) or the EC19 antiserum (a generous gift from Dr. J.-P. Delbecque, dilution 1∶10,000). The L2 antibody recognizes both E and 20E, as calculated from the comparison of reference standard curves (data not shown). The EC19 antibody recognizes only 20E. Calibration curves were generated with E or 20E (3.6 to 500 pg/tube) diluted in Schneider's medium and the in vitro production was expressed in E or 20E equivalents. Under these conditions, detection limits are 7 pg E equivalents for the L2 antibody and 5 pg 20E equivalents for the EC19 antibody. Ecdysteroids secreted by tissues were measured directly on incubation media. For each sample, measurements were performed in duplicate and the results are expressed as mean values ± S.E.M. of several (n = 20) independent ovary pairs. All experiments have been repeated on 2 independent cohorts of mosquitoes. Data were subjected to statistical analysis using Mann-Whitney test.
Supporting Information
Primers used in the study.
(XLS)
Acknowledgments
We thank Marie-Thérèse Lecoq (CEPIA) for A. gambiae rearing. We address many thanks to the Vector group of LSTM for mosquito rearing, helpful technical advices and discussions. We also thank Dr. Jean-Philippe Parvy for helpful discussions and comments on the manuscript.
Funding Statement
This work was supported by the Université Pierre et Marie Curie and the Ministère de la Recherche Scientifique (EP, EG, AM, and CDV), by the Fondation pour la Recherche Médicale and a Fondation des Treilles award (to EP), by the Institut Pasteur (to EP, JCJ and CB), and by the Wellcome Trust (Grant number 072833/Z/03/Z, to JPD and HR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Thummel CS (2001) Molecular mechanisms of developmental timing in C. elegans and Drosophila . Dev Cell 1 (4) 453–465. [DOI] [PubMed] [Google Scholar]
- 2.Raikhel AS, Brown MR, Belles X (2005) Hormonal control of reproductive processes. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 1. pp. 433–491.
- 3. Spindler KD, Hönl C, Tremmel Ch, Braun S, Ruff H, et al. (2009) Ecdysteroid hormone action. Cell Mol Life Sci 66 (24) 3837–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafont R, Dauphin-Villemant C, Warren J, Rees HH (2012) Ecdysteroid chemistry and biochemistry. In : Gilbert LI, editor. Insect Endocrinology. pp. 106–176.
- 5. Yamanaka N, Rewitz KF, O'Connor MB (2013) Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol 58: 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilbert LI, Rybczynski R, Warren JT (2002) Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol 47: 883–916. [DOI] [PubMed] [Google Scholar]
- 7. Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, et al. (2006) Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell 10 (4) 473–482. [DOI] [PubMed] [Google Scholar]
- 8. Yoshiyama T, Namiki T, Mita K, Kataoka H, Niwa R (2006) Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133 (13) 2565–2574. [DOI] [PubMed] [Google Scholar]
- 9. Yoshiyama-Yanagawa T, Enya S, Shimada-Niwa Y, Yaguchi S, Haramoto Y, et al. (2011) The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J Biol Chem 286 (29) 25756–25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lang M, Murat S, Clark AG, Gouppil G, Blais C, et al. (2012) Mutations in the neverland gene turned Drosophila pachea into an obligate specialist species. Science 337 (6102) 1658–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, et al. (2004) CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila . J Biol Chem 279 (34) 35942–35949. [DOI] [PubMed] [Google Scholar]
- 12. Warren JT, Petryk A, Marqués G, Parvy JP, Shinoda T, et al. (2004) Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem Mol Biol 34 (9) 991–1010. [DOI] [PubMed] [Google Scholar]
- 13. Chávez VM, Marqués G, Delbecque JP, Kobayashi K, Hollingsworth M, et al. (2000) The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 127 (19) 4115–4126. [DOI] [PubMed] [Google Scholar]
- 14. Warren JT, Petryk A, Marques G, Jarcho M, Parvy JP, et al. (2002) Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster . Proc Natl Acad Sci U S A 99 (17) 11043–11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petryk A, Warren JT, Marqués G, Jarcho MP, Gilbert LI, et al. (2003) Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Natl Acad Sci U S A 100 (24) 13773–13778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gilbert LI, Warren JT (2005) A molecular genetic approach to the biosynthesis of the insect steroid molting hormone. Vitam Horm 73: 31–57. [DOI] [PubMed] [Google Scholar]
- 17. Warren JT, O'Connor MB, Gilbert LI (2009) Studies on the Black Box: incorporation of 3-oxo-7-dehydrocholesterol into ecdysteroids by Drosophila melanogaster and Manduca sexta . Insect Biochem Mol Biol 39 (10) 677–687. [DOI] [PubMed] [Google Scholar]
- 18. Namiki T, Niwa R, Sakudoh T, Shirai K, Takeuchi H, et al. (2005) Cytochrome P450 CYP307A1/Spook: a regulator for ecdysone synthesis in insects. Biochem Biophys Res Commun 337 (1) 367–374. [DOI] [PubMed] [Google Scholar]
- 19. Ono H, Rewitz KF, Shinoda T, Itoyama K, Petryk A, et al. (2006) Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev Biol 298 (2) 555–570. [DOI] [PubMed] [Google Scholar]
- 20. Rewitz KF, O'Connor MB, Gilbert LI (2007) Molecular evolution of the insect Halloween family of cytochrome P450s: phylogeny, gene organization and functional conservation. Insect Biochem Mol Biol 37 (8) 741–753. [DOI] [PubMed] [Google Scholar]
- 21. Rewitz KF, Larsen MR, Lobner-Olesen A, Rybczynski R, O'Connor MB, et al. (2009) A phosphoproteomics approach to elucidate neuropeptide signal transduction controlling insect metamorphosis. Insect Biochem Mol Biol 39 (7) 475–483. [DOI] [PubMed] [Google Scholar]
- 22. Niwa R, Namiki T, Ito K, Shimada-Niwa Y, Kiuchi M, et al. (2010) Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the ‘Black Box’ of the ecdysteroid biosynthesis pathway. Development 137 (12) 1991–1999. [DOI] [PubMed] [Google Scholar]
- 23. Ou Q, Magico A, King-Jones K (2011) Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol 9 (9) e1001160 10.1371/journal.pbio.1001160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rewitz KF, O'Connor MB (2011) Timing is Everything: PTTH Mediated DHR4 Nucleocytoplasmic Trafficking Sets the Tempo of Drosophila Steroid Production. Front Endocrinol 2: 108 10.3389/fendo.2011.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swevers L, Raikhel AS, Sappington TW, Shirk P, Iatrou K (2005) Vitellogenesis and post-vitellogenic maturation of the insect ovarian follicle. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 1. pp. 87–155.
- 26. Pondeville E, Maria A, Jacques JC, Bourgouin C, Dauphin-Villemant C (2008) Anopheles gambiae males produce and transfer the vitellogenic steroid hormone 20-hydroxyecdysone to females during mating. Proc Natl Acad Sci U S A 105 (50) 19631–6 10.1073/pnas.0809264105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parvy JP, Blais C, Bernard F, Warren JT, Petryk A, et al. (2005) A role for betaFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster . Dev Biol 282 (1) 84–94. [DOI] [PubMed] [Google Scholar]
- 28. Sieglaff DH, Duncan KA, Brown MR (2005) Expression of genes encoding proteins involved in ecdysteroidogenesis in the female mosquito, Aedes aegypti . Insect Biochem Mol Biol 35 (5) 471–490. [DOI] [PubMed] [Google Scholar]
- 29. Rewitz KF, Rybczynski R, Warren JT, Gilbert LI (2006) Identification, characterization and developmental expression of Halloween genes encoding P450 enzymes mediating ecdysone biosynthesis in the tobacco hornworm, Manduca sexta . Insect Biochem Mol Biol 36 (3) 188–199. [DOI] [PubMed] [Google Scholar]
- 30. Iga M, Smagghe G (2009) Identification and expression profile of Halloween genes involved in ecdysteroid biosynthesis in Spodoptera littoralis . Peptides 31 (3) 456–467. [DOI] [PubMed] [Google Scholar]
- 31. Hentze JL, Moeller ME, Jørgensen AF, Bengtsson MS, Bordoy AM, et al. (2013) Accessory gland as a site for prothoracicotropic hormone controlled ecdysone synthesis in adult male insects. PLoS One 8 (2) e55131 10.1371/journal.pone.0055131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aragon S, Claudinot S, Blais C, Maïbèche M, Dauphin-Villemant C (2002) Molting cycle-dependent expression of CYP4C15, a cytochrome P450 enzyme putatively involved in ecdysteroidogenesis in the crayfish, Orconectes limosus . Insect Biochem Mol Biol 32 (2) 153–159. [DOI] [PubMed] [Google Scholar]
- 33. David JP, Strode C, Vontas J, Nikou D, Vaughan A, et al. (2005) The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Natl Acad Sci U S A (11) 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feyereisen R (2005) Insect Cytochrome P450. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Vol. 4. pp. 1–77.
- 35. David JP, Ismail HM, Chandor-Proust A, Paine MJ (2013) Role of cytochrome P450s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos Trans R Soc Lond B Biol Sci 368 (1612) 20120429 10.1098/rstb.2012.0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clements AN (1992) The Biology of Mosquitoes: Development, nutrition, and reproduction. Chapman & Hall. 509 p.
- 37. Mykles DL (2011) Ecdysteroid metabolism in crustaceans. J Steroid Biochem Mol Biol 127 (3–5) 196–203. [DOI] [PubMed] [Google Scholar]
- 38. Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, et al. (2002) Evolution of supergene families associated with insecticide resistance. Science 298 (5591) 179–181. [DOI] [PubMed] [Google Scholar]
- 39. Feyereisen R (2006) Evolution of insect P450. Biochem Soc Trans 34 (6) 1252–1255. [DOI] [PubMed] [Google Scholar]
- 40. Sztal T, Chung H, Gramzow L, Daborn PJ, Batterham P, et al. (2007) Two independent duplications forming the Cyp307a genes in Drosophila . Insect Biochem Mol Biol 37 (10) 1044–1053. [DOI] [PubMed] [Google Scholar]
- 41. Müller P, Chouaïbou M, Pignatelli P, Etang J, Walker ED, et al. (2008) Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol Ecol 17 (4) 1145–1155. [DOI] [PubMed] [Google Scholar]
- 42. Djouaka RF, Bakare AA, Coulibaly ON, Akogbeto MC, Ranson H, et al. (2008) Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics 10.1186/1471-2164-9-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maïbèche-Coisne M, Monti-Dedieu L, Aragon S, Dauphin-Villemant C (2000) A new cytochrome P450 from Drosophila melanogaster, CYP4G15, expressed in the nervous system. Biochem Biophys Res Commun 273 (3) 1132–1137. [DOI] [PubMed] [Google Scholar]
- 44. Gutierrez E, Wiggins D, Fielding B, Gould AP (2007) Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445 (7125) 275–280. [DOI] [PubMed] [Google Scholar]
- 45. Niwa R, Sakudoh T, Matsuya T, Namiki T, Kasai S, et al. (2011) Expressions of the cytochrome P450 monooxygenase gene Cyp4g1 and its homolog in the prothoracic glands of the fruit fly Drosophila melanogaster (Diptera: Drosophilidae) and the silkworm Bombyx mori (Lepidoptera: Bombycidae). Appl Entomol Zool 46 (4) 533–543(11). [Google Scholar]
- 46. Yang P, Tanaka H, Kuwano E, Suzuki K (2008) A novel cytochrome P450 gene (CYP4G25) of the silkmoth Antheraea yamamai: cloning and expression pattern in pharate first instar larvae in relation to diapause. J Insect Physiol 54 (3) 636–43. [DOI] [PubMed] [Google Scholar]
- 47.Feyereisen R (2012) Insect CYP genes and P450 enzymes. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. pp. 236–316.
- 48. Ono H, Morita S, Asakura I, Nishida R (2012) Conversion of 3-oxo steroids into ecdysteroids triggers molting and expression of 20E-inducible genes in Drosophila melanogaster . Biochem Biophys Res Commun 421 (3) 561–566. [DOI] [PubMed] [Google Scholar]
- 49. Coetzee M, Hunt R, Wilkerson R, Della Torre A, Coulibaly M, et al. (2013) Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619 (3) 246–274. [PubMed] [Google Scholar]
- 50. Müller P, Donnelly MJ, Ranson H (2007) Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics 8: 36 10.1186/1471-2164-8-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boisson B, Jacques J-C, Choumet V, Martin E, Xu J, et al. (2006) Gene silencing in mosquito salivary glands by RNAi. FEBS Lett 580 (8) 1988–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in the study.
(XLS)