SUMMARY
Histone modifications play important roles in the regulation of gene expression and chromatin organization. VprBP has been implicated in transcriptionally silent chromatin formation and cell cycle regulation, but the molecular basis underlying such effects remains unclear. Here we report that VprBP possesses an intrinsic protein kinase activity and is capable of phosphorylating histone H2A on threonine 120 (H2AT120p) in a nucleosomal context. VprBP is localized to a large set of tumor suppressor genes and blocks their transcription, in a manner that is dependent on its kinase activity toward H2AT120. The functional significance of VprBP-mediated H2AT120p is further underscored by the fact that RNAi knockdown and small-molecule inhibition of VprBP reactivate growth regulatory genes and impede tumor growth. Our findings establish VprBP as a major kinase responsible for H2AT120p in cancer cells and suggest that VprBP inhibition could be a new strategy for the development of anticancer therapeutics.
INTRODUCTION
The formation of silent chromatin plays important roles in the regulation of gene expression and maintenance of chromosome stability in eukaryotes. Inactive chromatin domains are often associated with distinct histone modifications (Suganuma and Workman, 2011). Like other histone modifications, histone phosphorylation has been linked to various cellular processes such as transcriptional regulation and DNA repair (Banerjee and Chakravarti, 2011). Histone phosphorylation can occur on serine, threonine and tyrosine residues and constitutes a part of the signal to influence chromatin structure and factor recruitment. For example, phosphorylations of H3S10, H3S28 and H2BS32 are linked to the expression of proto-oncogenes such as c-fos, c-jun and c-myc (Choi et al., 2005; Lau et al., 2011; Lau and Cheung, 2011). Phosphorylations of H3S10, H3T11 and H3S28 play a role in combination with H3 acetylation in transcription activation and cell proliferation (Gehani et al., 2010; Lau and Cheung, 2011; Lo et al., 2001; Shimada et al., 2008; Yang et al., 2012). Conversely, H2AS1 phosphorylation inhibits chromatin transcription, and H3 pre-acetylation interferes with this repressive modification (Zhang et al., 2004). In some cases, histone phosphorylation facilitates nucleosome binding by proteins containing phospho-binding modules and restricts their activity as downstream effectors around specific region. While a large number of phosphorylation sites have been identified in core histones, the identification of kinases responsible for these modifications remains areas of intensive investigation.
VprBP is a large nuclear protein that can interact with HIV viral protein R and Cullin 4-DDB1 ubiquitin ligase complex (Li et al., 2010). The cellular function of VprBP has been studied mainly with respect to its role in regulating Cullin 4 E3 ubiquitin ligase activity and cell cycle progression (Hrecka et al., 2007; McCall et al., 2008). However, more recent studies have implicated VprBP in much wider range of cellular processes, as exemplified by its engagement in JNK-mediated apoptosis during cell competition process (Tamori et al., 2010). Another striking example is the demonstration made by us that VprBP acts as an effector that binds histone H3 tails protruding from nucleosomes and establishes chromatin silencing in cancer cells (Kim et al., 2012). These results clearly point to VprBP having a negative regulatory role in transcription, but precisely how VprBP mediates its effects on the formation of repressive chromatin domain is poorly understood.
Here we report that VprBP has an intrinsic kinase activity and phosphorylates histone H2A at threonine 120. Functional studies reveal that H2AT120p by VprBP is sufficient to repress chromatin transcription. RNA interference (RNAi)-mediated knockdown of VprBP impairs H2AT120p, transactivates a large set of tumor suppressor genes, and inhibits cell proliferation. Furthermore, using a highly potent and selective inhibitor for VprBP, we show that downregulation of VprBP-mediated H2AT120p impedes cancer cell proliferation and xenograft tumor progression.
RESULTS
VprBP possesses kinase activity and phosphorylates threonine 120 of histone H2A
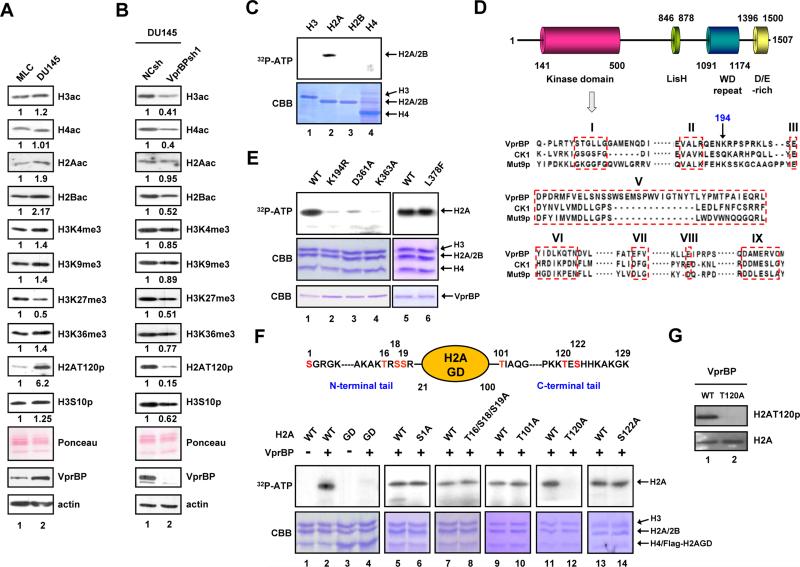
Given that dysregulation of histone modifying activities is linked to human cancers (Chi et al., 2010; Dawson and Kouzarides, 2012), we reasoned that VprBP expression in cancer cells might influence specific histone modifications. As expected, Western blotting of cell lysates confirmed that VprBP is expressed highly in DU145 prostate, LD611 bladder and MDA-MB231 breast cancer cell lines, but minimally in their corresponding normal counterparts (Figures 1A and S1A). In exploring whether any histone modifications are altered in the cancer cell lines, we detected much higher levels of H2AT120p in chromatin fractions. To assess the relationship between VprBP expression and H2AT120p more directly, we examined a possible effect of VprBP depletion. Upon the stable knockdown of VprBP, the abundant H2AT120p found in the cancer cell lines was drastically reduced, but changes in other modifications were much less pronounced or absent (Figures 1B and S1B).
Figure 1. VprBP phosphorylates histone H2A at T120.
(A) Chromatin was prepared from human prostate cancer (DU145) and normal (MLC) cell lines and subjected to Western blotting with the indicated antibodies. Ponceau S staining and β-actin served as loading controls in all Western blot analyses in this study. Quantifications of the band intensities by densitometry are shown below the Western blots, and similar results were obtained from two additional experiments. ac, acetylation; p, phosphorylation; me3, trimethylation.
(B) DU145 cells were infected with lentiviruses expressing VprBP shRNA (lane 2) or non-specific control shRNA (lane 1), and chromatin fractions were analyzed by Western blotting as in (A).
(C) Individual core histones were incubated with recombinant VprBP in the presence of [γ-32P] ATP. The reactions were resolved by 15% SDS-PAGE and analyzed by autoradiography (upper panel) and Coomassie blue staining (lower panel).
(D) VprBP contains the putative kinase domain in the N-terminal region, the Lis homology motif in the central region, and the WD repeat and D/E-rich motif in the C-terminal region. Numbers denote amino acid positions. Sequence alignment of the putative kinase domain in VprBP with the kinase domains of human CK1 and Mut9p is shown in the lower panel. The boxed regions correspond to the conserved kinase subdomains I-III and V-IX.
(E) Nucleosomes were reconstituted on a 207 bp 601 nucleosome positioning sequence using recombinant histones and incubated with wild type VprBP or the indicated mutants. H2A phosphorylation was detected by autoradiography.
(F) Kinase assays were performed as in (E), but using nucleosomes containing wild type, tailless or mutant H2A. The residues that were mutated are indicated at the top. The H2A mutations did not affect histone octamer and nucleosome formation during reconstitution (data not shown).
(G) Nucleosomes containing H2A wild type or T120A mutant were incubated with VprBP and ATP. H2AT120p was analyzed by Western blotting with anti-H2AT120p antibody. See also Figure S1.
The data above suggest that VprBP may be of particular importance for H2AT120p reactions in cancer cells. To test this possibility, we incubated free individual histones with [γ-32P]-ATP and recombinant VprBP produced in baculovirus-infected insect cells. The integrity and purity of the VprBP protein were confirmed by silver-stained SDS-PAGE (Figure S1C) and mass spectrometry (Figure S1D). Autoradiograph of the kinase reaction products showed a robust phosphorylation of H2A, but not other core histones (Figure 1C). Expectedly, VprBP showed no enzymatic activity in our in vitro HAT and HMT assays (Figure S1E). As the core histones exist within nucleosomes in the cell nucleus, kinase assays were repeated with nucleosomes reconstituted from recombinant histones and the 601 nucleosome positioning sequence (Lowary and Widom, 1998). VprBP generated clear labeling of H2A in the nucleosome after autoradiography (Figure S1F). These results were further corroborated by in vitro kinase assays with nucleosomes immobilized on agarose beads (Figure S1G) and with VprBP immunoaffinity-purified from DU145 cell lysates (Figure S1H). In additional support, the in-gel autophosphorylation assays showed a phosphorylated band at the expected molecular weight of VprBP (Figure S1I).
Consistent with these findings, sequence alignments with CK1 and Mut9p kinases identified 8 out of the 12 protein kinase subdomains (Hanks et al., 1988; Taylor et al., 1992) in the N-terminal region of VprBP (Figure 1D, residues 141-500). The lysine residue in the subdomain II is critical for kinase enzymatic activity (Casas-Mollano et al., 2008; Zhai et al., 1992). VprBP does not have this conserved residue in its subdomain II, but has lysine 194 immediately adjacent to the subdomain II. Notably, mutation of this lysine residue completely abrogated kinase activity (Figure 1E, lanes 1 and 2; Figure S1J, lanes 1 and 2). Mutation at either D361 or K363 that is conserved in the subdomain VI also impaired the catalytic activity (Figure 1E, lanes 3 and 4; Figure S1J, lanes 3 and 4). On the contrary, mutation of L378 lying outside the conserved subdomains did not affect VprBP kinase activity (Figure 1E, lane 6; Figure S1J, lane 5). These results exclude the possibility of H2AT120p by a contaminating kinase activity in the preparation of recombinant VprBP. All VprBP mutants exhibited circular dichroism spectra almost identical to those of the wild type VprBP (Figure S1K), thus ruling out the possibility that the altered structure of the VprBP mutants is responsible for their reduced kinase activity.
In determining VprBP phosphorylation sites in H2A, we found that simultaneous deletion of the N- and C-terminal tails of H2A blocks H2A phosphorylation by VprBP (Figure 1F, lanes 1-4). Moreover, VprBP-mediated phosphorylation is completely abolished by mutation of T120, whereas mutations of six other potential modification sites on the tail domains had little effect (lanes 5-14). Western blot analysis of the kinase reactions using anti-H2AT120p antibody further confirmed that VprBP stimulates the phosphorylation of H2AT120 in the nucleosome (Figure 1G).
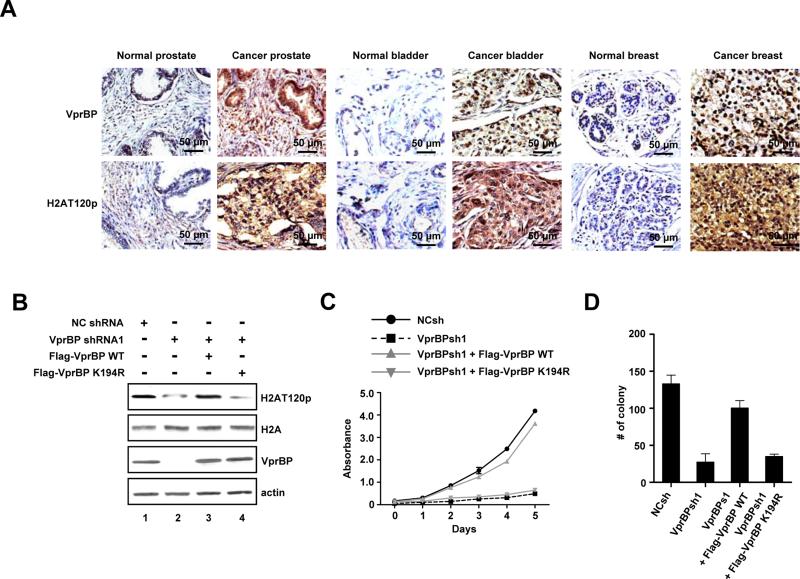
VprBP-mediated H2AT120p is highly abundant in tumors and necessary for cancer cell proliferation
To decipher the clinical significance of VprBP-mediated H2AT120p, we next analyzed the levels of VprBP and H2AT120p in multiple patient-matched normal and tumor tissue microarray (Figure 2A and Table S1). Immunohistochemical analysis on 16 types of organ cancer with matched adjacent normal tissue demonstrated a clear link between elevated expression of VprBP and increased levels of H2AT120p in more than 70% of the tumor samples. This trend was more evident in bladder, breast and prostate tumor samples. In cases where there was no change in VprBP expression, the same trend was observed for H2AT120p. These findings validate the results from cell lines, and support the conjecture that VprBP possesses oncogenic properties and its kinase activity contributes to the observed changes. To address this issue, we tested the effects of VprBP depletion on the proliferation of DU145 cancer cells. Expectedly, much lower levels of VprBP were detected in VprBP-depleted cells compared to mock-depleted cells, and the observed reduction of VprBP correlated well with decreased H2AT120p (Figures 2B and S2A). MTT assays over a 5-day time course also revealed that VprBP depletion gradually decreased the viability of cancer cells and that the expression of VprBP wild type, but not VprBP K194R kinase-dead mutant, restored H2AT120p and cell proliferation rates (Figures 2C and S2B). Analogously, VprBP depletion interfered with cell proliferation and thus reduced the number of colony-forming cells; colony numbers increased to about 75% of undepleted cells after the expression of wild type but not K194R-mutated VprBP in the depleted cells (Figures 2D and S2C). Consistent with these observations, VprBP overexpression in MLC cells containing low levels of VprBP increased H2AT120p, thereby facilitating cell proliferation and colony formation (Figures S2D-S2F).
Figure 2. VprBP is overexpressed in tumors and required for cell proliferation.
(A) Tissue microarrays containing primary tumor and adjacent normal samples from cancer patients were subjected to immunohistochemistry with VprBP and H2AT120p antibodies. High power magnifications are shown for six representative samples. Scale bars correspond to 50 μm. See also Table S1.
(B) DU145 cells were depleted of VprBP and infected with lentiviruses expressing the VprBP wild type (WT) or VprBP kinase-dead mutant K194R (KD). The levels of VprBP and H2AT120p were determined by Western blotting.
(C) VprBP-depleted DU145 cells were complemented with VprBP WT or KD, and cell proliferation was measured by MTT assay. Results represent the means ± S.D. of three experiments performed in triplicate.
(D) VprBP-depleted DU145 cells were infected with VprBP WT or KD as in (C), and the colonies grown up in soft agar were stained and counted. The Y axis indicates the number of colonies with a diameter of > 0.05 mm per view. Three independent experiments in triplicate wells were performed. Data represent the means ± S.D. of three independent experiments. See also Figure S2.
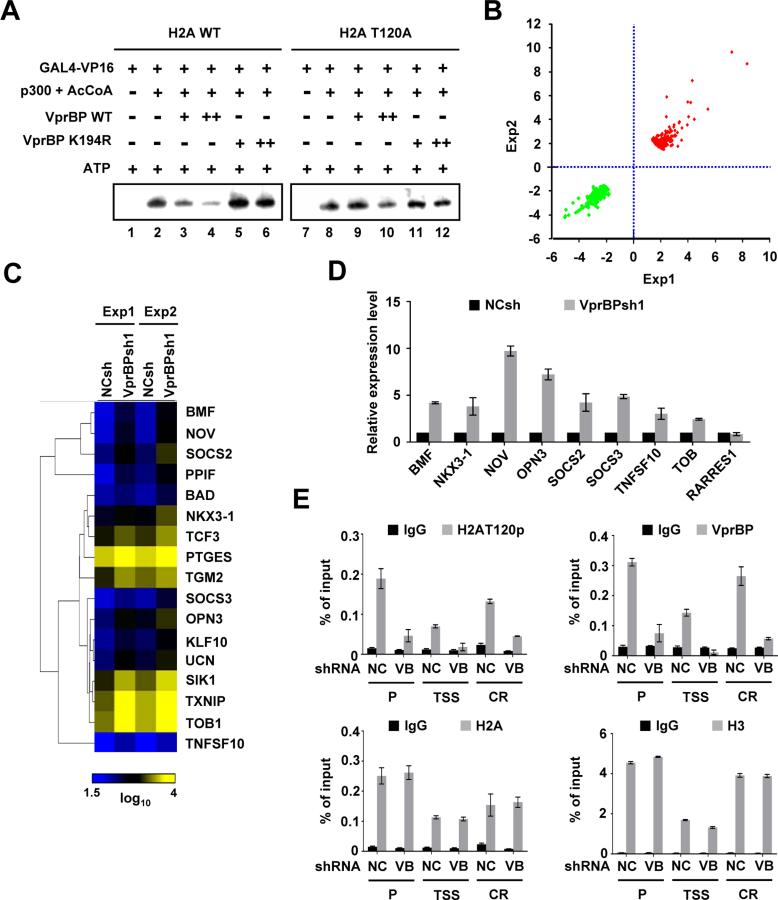
VprBP-mediated H2AT120p inactivates cell growth regulatory genes
As VprBP has been reported to act as a negative regulator of chromatin transcription (Kim et al., 2012), we sought to determine whether H2AT120p is required for VprBP function. In the absence of VprBP, high levels of transcription from chromatin reconstituted from G5ML-601 array DNA and recombinant histones were achieved by Gal4-VP16 and p300 (Figure 3A, lanes 1 and 2). When chromatin was phosphorylated by VprBP, significant repression of transcription was evident (lanes 3 and 4). Intriguingly, however, the ability of VprBP to block transcription was compromised upon mutation of H2AT120 in chromatin (lanes 9 and 10) or omission of ATP from the reaction (Figure S3A). Furthermore, addition of VprBP kinase dead mutant to transcription reactions had no detectable effect on transcription (Figure 3A, lanes 5, 6, 11, and 12), strongly arguing that H2AT120p is the cause of the observed repression.
Figure 3. Functional analysis of VprBP-mediated H2AT120p.
(A) Chromatin templates containing wild type or T120-mutated H2A were transcribed in the presence of Gal4-VP16, p300 + AcCoA and/or VprBP as indicated above the panel. VprBP was added to the reaction before p300. The results shown are representative of three independent experiments.
(B) Shown are scatter plots of the global gene expression patterns comparing VprBP-depleted DU145 cells with mock-depleted cells. Dots represent expression values for the genes with a change > 1.7-fold in either of two independent experiments.
(C) Clustering and heat map representation of the genes upregulated upon VprBP depletion and related to cell death and proliferation. Yellow and blue indicate high and low expression, respectively. See also Tables S2 and S3.
(D) RNA was isolated from VprBP-depleted DU145 cells as in (B) and subjected to real-time qRT-PCR using primers specific for the indicated genes and listed in Supplemental Experimatal Procedures. Expression levels were normalized to β-actin level and shown relative to those of mock-depleted cells, and were arbitrarily assigned a value of 1. Data represent the means ± S.D. of three independent experiments.
(E) The levels of H2AT120p, H2A, VprBP and H3 at the OPN3 gene were assessed in mock- and VprBP-depleted DU145 cells by ChIP analysis. Precipitation efficiencies were determined for promoter (P), transcription stat site (TSS) and coding region (CR) by quantitative PCR (qPCR) with primers listed in Supplemental Experimental Procedures. Quantitative results were averaged from three separate determinations. Results represent the means ± S.D. of three independent experiments. See also Figure S3.
Next, we performed comprehensive microarray analysis with total RNA isolated from mock- or VprBP-depleted DU145 cancer cells. With a fold-change cutoff of > 1.7 and stringent P < 0.005, the gene expression profiling showed that 292 genes were activated and 208 genes were repressed (Figure 3B; Tables S2 and S3) in response to VprBP knockdown. Many of the genes up-regulated upon VprBP depletion encode cell proliferation and growth regulators (Figure S3B), including those known to be key regulatory components for cancer initiation and progression (Figure 3C). The transcriptional changes detected by microarray were validated by qRT-PCR of eight genes whose expression was increased upon VprBP depletion and one unaffected control gene (Figure 3D). To check whether the candidate target genes harbor VprBP and H2AT120p, we conducted ChIP assays. In mock-depleted cells, VprBP occupied the promoter and coding regions of the target genes, and H2AT120p showed similar distribution across the loci (Figures 3E and S3C). Consistent with previous studies (Schones et al., 2008), nucleosomes are depleted in the vicinity of transcription start site (TSS), as indicated by the low levels of H2A and H3. For this reason, ChIP analysis exhibited low levels of VprBP and H2AT120p over the TSS of the target genes. Importantly, VprBP depletion resulted in greatly reduced levels of VprBP and concomitant loss of H2AT120p at the candidate target genes, reinforcing the conclusion that H2AT120p observed in these genes is dependent of VprBP. In the case of RARRES1 gene which is not affected by VprBP knockdown (Figure 3D), the H2AT120p levels were low and remained unchanged under control and VprBP knockdown conditions (Figure S3C).
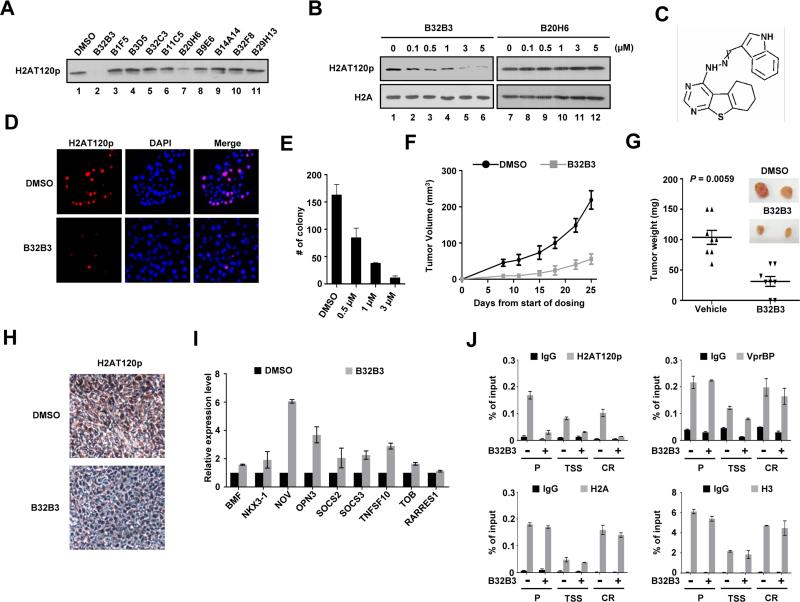
B32B3 is a potent and selective inhibitor of VprBP and suppresses tumor growth
The fact that VprBP knockdown abrogates H2AT120p and slows cancer cell growth prompted us to look for highly potent and selective inhibitors for VprBP. To this end, we screened our in-house small-molecule library of 5000 compounds. When the inhibitory potential of the compounds was assessed by in vitro kinase assays, two of them (0.002% hit rate), designated as B32B3 and B20H6, inhibited VprBP and decreased H2AT120p at a concentration of 5 μM (Figure 4A). To evaluate their cellular effects, DU145 cells were treated with the compounds in the concentration range of 0-5 μM for 24 h. B32B3 potently inhibited H2AT120p with a half-maximal inhibitory concentration (IC50) value of 0.5 μM, as evaluated by Western blotting and immunostaining (Figures 4B, 4D and S4A). The observed reduction in H2AT120p was paralleled by inhibition of cell proliferation (Figures 4E and S4B). By comparison, B20H6 failed to produce any detectable changes in H2AT120p and cell growth after treatment (Figure 4B, lanes 7-12 and Figure S4B), suggesting that this compound might be relatively unstable with poor cellular uptake. Importantly, the knockdown of VprBP sensitized DU145 cells to B32B3 with a circa two-fold decrease in the IC50, whereas B32B3 was considerably less potent in DU145 cells overexpressing VprBP (Figure S4A). Furthermore, B32B3 treatment at concentrations up to 5 μM minimally antagonized the proliferation of MLC cells lacking VprBP-mediated H2AT120p (Figures S4C and S4D). These results indicate that B32B3 preferentially targets cancer cells exhibiting high levels of VprBP and H2AT120p. That the IC50 value of B32B3 was increased in the presence of incremental concentrations of ATP argues strongly that B32B3 competes with ATP and may bind to the kinase active site (Figure S4E). Additionally, when tested against a panel of 33 human kinases, B32B3 showed greater than 100-fold selectivity for VprBP over 33 other kinases with an IC50 of 0.6 μM (Table S4). Thus, although we cannot exclude the possibility that other kinases that were not included in the selectivity screen might be affected, B32B3 can be defined as a highly specific VprBP inhibitor at present.
Figure 4. Discovery and characterization of a small-molecule VprBP inhibitor.
(A) In vitro kinase assays were performed with recombinant H2A and VprBP in the presence of the indicated compounds (5 μM). The effects of the compounds were evaluated by Western blotting with H2AT120p antibody.
(B) DU145 cells were grown in the presence of the indicated concentrations of either B32B3 or B20H6 for 24 h, and immunoblotted with H2A and H2AT120p antibodies.
(C) Molecular structure of B32B3.
(D) DU145 cells were treated with DMSO or 0.5 μM B32B3 for 24 h. The cellular levels of H2AT120p were assessed by immunostaining.
(E) DU145 cells were treated with increasing concentrations of B32B3 for 24 h, and the colonies were counted 3 weeks after seeding the cells on soft agar. The data are the means of three independent experiments ± S.D.
(F) Nude mice were implanted with 1 ×107 DU145 cells on the left flank. Five days after implantation, mice bearing established tumors were randomized into groups and treated with twice-weekly i.p. injections of either DMSO or B32B3 at a dose of 5 mg/kg (n = 8 per group). Tumor volumes (mm3) were measured at the indicated time points and shown as mean tumor volumes ± S.E.M.
(G) Mice were killed at day 25 of the tumor growth, and the tumors were dissected and weighed. Mean tumor weights ± S.E.M. are shown, and the P value was calculated by unpaired Student's t test.
(H) DU145 xenografts were excised from DMSO-treated and B32B3-treated mice, and were analyzed by immunohistochemistry. Representative view was photographed
(I) Relative mRNA levels of the VprBP target genes in DMSO-treated (black bars) and B32B3-treated (1 μM, gray bars) DU145 cells were determined by qRT-PCR. Data represent the means ± S.D. of three independent experiments.
(J) DU145 cells exposed to DMSO or 1 μM B32B3 were subject to ChIP analysis using the indicated antibodies. The data are the means of three independent experiments ± S.D. See also Figure S4.
A key question that arises from our findings is whether B32B3 exhibits antitumor efficacy through its VprBP inhibitory activity. To address this question, we inoculated 1 × 107 DU145 cancer cells into nude mice and treated tumor xenografts with intraperitoneal injections of B32B3 at a dose of 5 mg/kg twice a week over 3 weeks. Tumor growth was inhibited, as calculated the day after the last treatment of B32B3, by 70-75% (Figures 4F and 4G). At these doses, B32B3 appeared to be well tolerated in mice, and it did not cause any significant weight loss during treatment (Figure S4F). In evaluating the pharmacokinetic properties of B32B3, we found that B32B3 has a half-life of approximately 7 h in mouse plasma and a Cmax of 1 μM at a dose of 5 mg/kg in mice (Figure S4G and S4H). To correlate B32B3 antitumor activity with VprBP inhibition, DU145 xenograft tumors explanted from DMSO- or B32B3-treated mice were analyzed by immunohistochemistry. The levels of H2AT120p were greatly decreased in the tumors of B32B3-treated mice, compared to DMSO-treated controls (Figure 4H). To elucidate the mechanistic basis of the B32B3 effects, we tested if the compound could rescue the transcriptional inhibition caused by VprBP. As summarized in Figure 4I, treatment of DU145 cells with B32B3 (1 μM) resulted in, albeit to a varying extent, higher expression of VprBP target genes. Because H2AT120p is essential for VprBP transrepression, we also examined the effects of B32B3 on H2AT120p at the target genes. VprBP was present at high levels at both promoter and coding regions, which did not alter upon B32B3 treatment. However, H2AT120p at the regions was reduced by 70%, following B32B3 treatment at the same dose (Figures 4J and Figure S4I). B32B3 thus display anticancer properties at least in part, by interfering with VprBP-mediated H2AT120p at the target genes.
DISCUSSION
This work describes the systematic biochemical and cellular analysis of VprBP and unveils a surprising mechanism underlying the formation of repressive chromatin by VprBP. A key finding is that the N-terminal region of VprBP possesses a previously unrecognized kinase activity for H2AT120. VprBP contains 8 out of the 12 conserved protein kinase subdomains, and mutations of these subdomains abolish the catalytic activity, further confirming that VprBP is a bona fide protein kinase. Interestingly, while most histone kinases identified so far are incapable of phosphorylating nucleosomal histones, VprBP displays an unusual additional activity as an effective kinase of nucleosomes. In this respect, VprBP resembles Drosophila NHK-1, which catalyzes phosphorylation of H2AT119 (equivalent to human H2AT120) in the nucleosomal context (Aihara et al., 2004). It has been reported that Bub1 acts as a centromere-specific kinase for H2AS121 (H2AT120 in human) during prometa- and metaphase in fission yeast (Kawashima et al., 2010). Our attempts to detect any significant changes in H2AT120p in Bub1-depleted DU145 cells have been unsuccessful (Figure S1L), probably because we have used unsynchronized interphase cells. We also observed that Bub1 is expressed at similar levels in MLC normal and DU145 cancer cells (Figure S1M). It thus appears that the role of VprBP-mediated H2A phosphorylation is distinct from that of Bub1-mediated H2A phosphorylation. Further structural and biological analyses of VprBP and Bub1 would be helpful for understanding the functional differences of these two kinases.
There has been no demonstration that H2AT120p is involved in the regulation of gene expression, although recently this possibility has been discussed. Our well-defined in vitro assay system allows us to provide the first direct connection between H2AT120p and transcriptional repression. Importantly, blocking VprBP-mediated H2AT120p by point mutation, we have been able to verify that H2AT120p is critical determinant of repressive action of VprBP. In accord with these in vitro data, gene expression profiling demonstrated that VprBP downregulates 292 genes, many of which are involved in cell proliferation and programmed cell death. An intriguing question raised by these results is how H2AT120p by VprBP modulates chromatin transcription. One possible mechanism is that H2AT120p can affect the occurrence of other histone modifications on the same or different histone tails. Recent work from our lab has shown that VprBP interacts with HDAC1, thereby inhibiting H3 acetylation at p53 target genes (Kim et al., 2012). This suggests that H2AT120p at target genes may influence HDAC1 activity required for gene repression. Another possibility is that H2AT120p could serve as an integrating platform for repressor proteins. Considering the fact that the centromere cohesion protector shugoshin recognizes H2AT120p and recruits heterochromatin protein Swi6/HP1 at centromeres in fission yeast (Kawashima et al., 2010; Yamagishi et al., 2008), the recruitments of factors to specific chromatin domains are likely to be part of the mechanisms for VprBP-induced gene silencing. Another question unsolved in our study is how VprBP is initially localized at target genes. A likely model is that VprBP physically associates with gene specific factors to influence the transcription of target genes, as supported by our recent finding that VprBP-p53 interaction is a key event in VprBP action on p53 target genes (Kim et al., 2012). Thus, more extensive studies of VprBP interaction with DNA-binding factors and other coregulators would provide a molecular explanation to gene-specific function of VprBP.
VprBP expression is significantly higher in breast, bladder and prostate cancer tissues, compared to their benign counterparts. The observation that VprBP knockdown significantly decreased H2AT120p and cancer cell growth indicates that VprBP could be an ideal target for cancer therapy. As the first step toward checking this possibility, we screened large numbers of compounds in a high-throughput manner and identified B32B3 as a selective inhibitor of VprBP. B32B3 is thought to inhibit VprBP kinase activity by competing with ATP. Importantly, B32B3 recapitulates the most molecular phenotypes that are arisen from VprBP knockdown; (1) the reduction of H2AT120p at target genes, (2) higher expression of VprBP target genes, and (3) the impairment of cancer cell growth. Thus, B32B3 represents a unique tool to investigate the regulatory pathways governing H2AT120p in physiological and tumorigenic conditions. Moreover, the selectivity for cancer cells in culture, and our ability to demonstrate efficacy in a mouse model of VprBP at doses that were well tolerated suggest that inhibition of VprBP by B32B3 may provide a pharmacological basis for therapeutic intervention against cancers.
EXPERIMENTAL PROCEDURES
In vitro kinase and transcription assays
Recombinant mononucleosomes and nucleosome arrays were reconstituted using recombinant histone octamers as recently described (Jaskelioff et al., 2000; Robinson et al., 2008). For kinase assays, recombinant VprBP was incubated with free histones (1 μg) or reconstituted nucleosomes (2 μg) in kinase buffer (50 mM Tris–HCl, pH 7.5, 20 mM EGTA, 10 mM MgCl2, 1 mM DTT, and 1 mM β-glycerophosphate) containing 10 μCi of [γ-32P] ATP and 4 mM ATP for 30 min at 30°C. Proteins from each reaction were separated by SDS-PAGE, Coomassie blue stained, dried, and visualized by autoradiography. To create VprBP inhibitors, a collection of 5000 compounds were screened in the same kinase assays at a final concentration of 5 μM. The selectivity of B32B3 toward VprBP kinase was assessed in a panel of 33 kinases listed in Table S4. In vitro transcription assays were as described (Kim et al., 2012) except that G5ML-601 nucleosome arrays (100 ng) and Gal4-VP16 (15ng) were used for the reactions. Recombinant VprBP (25 or 50 ng) and ATP (10 mM) were added before p300 (20 ng) and AcCoA (10 μM).
Mice xenografts
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee. Tumor xenografts were established by subcutaneous injection of 1 × 107 DU145 cells into 8-week-old female nude mice (n=8). At day 5 after injection, the mice bearing DU145 tumor xenografts were treated with twice-weekly i.p. injections of either DMSO or B32B3 at a dose of 5 mg/kg throughout the duration of the experiment. Tumor dimension was measured by calipers twice a week and tumor mass was calculated as described (Heo et al., 2012). The mice were killed by asphyxiation with CO2, and tumors were excised and weighed 25 day after the cell injection. To analyze the H2AT120p, formalin-fixed and paraffin-embedded sections (5 μm) from DU145 tumor xenografts were subject to immunohistochemistry.
Supplementary Material
HIGHLIGHTS.
VprBP possesses an inherent kinase activity and phosphorylates H2AT120
VprBP overexpression stimulates cell growth by increasing H2AT120p
H2AT120p is required for VprBP transrepression of tumor suppressor genes
B32B3 is a potent inhibotor of VprBP
ACKNOWLEDGEMENTS
We are grateful to Hongtao Yu for bub1 baculovirus. We acknowledge the valuable technical assistance of Mark Ambroso and Ralph Langen in circular dichroism analysis. This work was supported by NIH Grant GM84209 and ACS Research Scholar Grant DMC-1005001 awarded to W.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Details for other experimental procedures can be found in Supplemental Information.
ACCESSION NUMBERS
The NCBI GEO accession number for microarray data reported in this paper is GSE50414.
REFERENCES
- Aihara H, Nakagawa T, Yasui K, Ohta T, Hirose S, Dhomae N, Takio K, Kaneko M, Takeshima Y, Muramatsu M, et al. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 2004;18:877–888. doi: 10.1101/gad.1184604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Chakravarti D. A peek into the complex realm of histone phosphorylation. Mol Cell Biol. 2011;31:4858–4873. doi: 10.1128/MCB.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, Jeong BR, Xu J, Moriyama H, Cerutti H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc Natl Acad Sci U S A. 2008;105:6486–6491. doi: 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Choi BY, Cho YY, Mizuno H, Kang BS, Bode AM, Dong Z. Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer Res. 2005;65:5818–5827. doi: 10.1158/0008-5472.CAN-05-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Heo K, Kim JS, Kim K, Kim H, Choi J, Yang K, An W. Cell-penetrating H4 tail peptides potentiate p53-mediated transactivation via inhibition of G9a and HDAC1. Oncogene. 2012 doi: 10.1038/onc.2012.273. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, Skowronski J. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc Natl Acad Sci U S A. 2007;104:11778–11783. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskelioff M, Gavin IM, Peterson CL, Logie C. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol. 2000;20:3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- Kim K, Heo K, Choi J, Jackson S, Kim H, Xiong Y, An W. Vpr-binding protein antagonizes p53-mediated transcription via direct interaction with H3 tail. Mol Cell Biol. 2012;32:783–796. doi: 10.1128/MCB.06037-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AT, Lee SY, Xu YM, Zheng D, Cho YY, Zhu F, Kim HG, Li SQ, Zhang Z, Bode AM, et al. Phosphorylation of histone H2B serine 32 is linked to cell transformation. J Biol Chem. 2011;286:26628–26637. doi: 10.1074/jbc.M110.215590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PN, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You L, Cooper J, Schiavon G, Pepe-Caprio A, Zhou L, Ishii R, Giovannini M, Hanemann CO, Long SB, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- McCall CM, Miliani de Marval PL, Chastain PD, 2nd, Jackson SC, He YJ, Kotake Y, Cook JG, Xiong Y. Human immunodeficiency virus type 1 Vpr-binding protein VprBP, a WD40 protein associated with the DDB1-CUL4 E3 ubiquitin ligase, is essential for DNA replication and embryonic development. Mol Cell Biol. 2008;28:5621–5633. doi: 10.1128/MCB.00232-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–232. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Tamori Y, Bialucha CU, Tian AG, Kajita M, Huang YC, Norman M, Harrison N, Poulton J, Ivanovitch K, Disch L, et al. Involvement of Lgl and Mahjong/VprBP in cell competition. PLoS Biol. 2010;8:e1000422. doi: 10.1371/journal.pbio.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Knighton DR, Zheng J, Ten Eyck LF, Sowadski JM. Structural framework for the protein kinase family. Annu Rev Cell Biol. 1992;8:429–462. doi: 10.1146/annurev.cb.08.110192.002241. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Graves PR, Longenecker KL, DePaoli-Roach AA, Roach PJ. Recombinant rabbit muscle casein kinase I alpha is inhibited by heparin and activated by polylysine. Biochem Biophys Res Commun. 1992;189:944–949. doi: 10.1016/0006-291x(92)92295-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Griffin K, Mondal N, Parvin JD. Phosphorylation of histone H2A inhibits transcription on chromatin templates. J Biol Chem. 2004;279:21866–21872. doi: 10.1074/jbc.M400099200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.