Abstract
High throughput technologies have been applied to investigate the underlying mechanisms of complex diseases, identify disease-associations and help to improve treatment. However it is challenging to derive biological insight from conventional single gene based analysis of “omics” data from high throughput experiments due to sample and patient heterogeneity. To address these challenges, many novel pathway and network based approaches were developed to integrate various “omics” data, such as gene expression, copy number alteration, Genome Wide Association Studies, and interaction data. This review will cover recent methodological developments in pathway analysis for the detection of dysregulated interactions and disease-associated subnetworks, prioritization of candidate disease genes, and disease classifications. For each application, we will also discuss the associated challenges and potential future directions.
Keywords: Pathway analysis, dysregulated interaction, disease association, Genome Wide Association Studies (GWAS), gene prioritization, disease classification
Introduction
Biomedical research has been revolutionized by advanced high-throughput (HT) technologies for study of genomic, transcriptomic, proteomic and metabolomic “molecular phenotypes” provided by technologies such as microarray, next generation sequencing, RNAi library screening, and high-throughput and high-resolution mass spectrometry[1-3]. However, due to the complexity of diseases, background noise in HT experiments, need for multiple hypothesis testing corrections, and patient heterogeneity, it has been challenging to interpret the direct results from experiments to elucidate biological mechanisms relevant to complex disease[4-6]. Recently, methods targeted on pathway level analyses have been developed and applied to investigate the underlying mechanism of complex diseases[7]. The rationales behind these methods are multiple: genes/proteins do not work alone, but in an intricate network of interactions and pathways. In addition, complex diseases are more likely caused by the dysregulation of multiple targets in connected pathways and/or different genes in the same pathways in different patients. Pathway analysis has statistical advantages in that it can reduce dimensionality of HT-data sets and provides a focused set of targets for biological validation. However, error rate estimation is more likely to be empirical than grounded in theory. Identifying disease-associated pathways can help to understand disease mechanisms and has potential to improve diagnostics and develop efficient treatments.
Pathway analysis has many implementations including: enrichment analyses of gene sets[8, 9] or Gene Ontology (GO) terms[10-12], clustering or module analysis of interaction networks (e.g., protein-protein and/or regulatory interactions)[13-15], kinetic analysis of pathways[16], flux-balance analysis[17], and inference of protein function and novel pathways[18-21]. This review concerns four types of analyses, namely, the detection of dysregulated interactions and disease-associated subnetworks, prioritization of candidate disease genes, and disease classification (Figure 1). We will focus on the latest methodological developments for these analyses, particularly, the methods that integrate multiple “omics” data, such as mRNA expression, genome wide association studies (GWAS), copy number alteration, protein-protein interaction (PPI) interactome and disease-disease association (diseasome). The current challenges and future directions are also discussed.
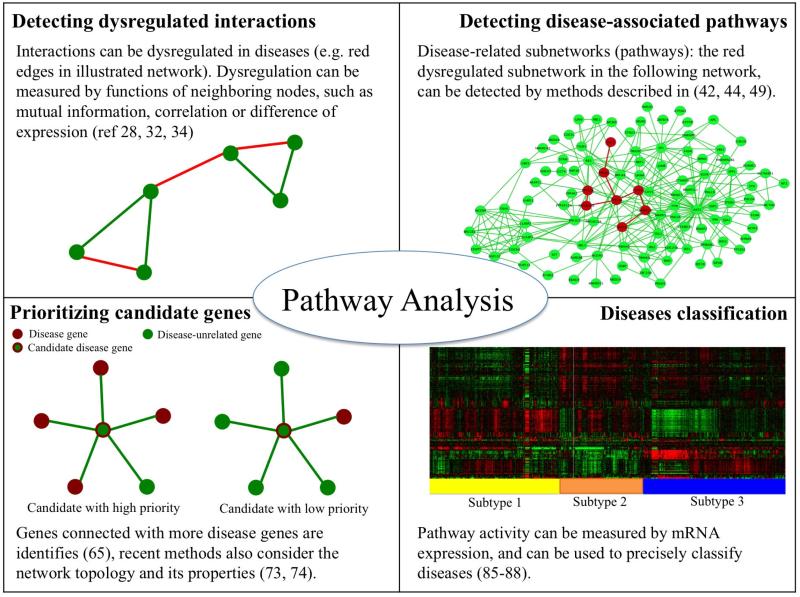
Figure 1.
Summary of pathway analysis and understanding disease associations. Pathway analysis can be used to detect dysregulated interactions and disease-associated pathways, prioritize candidate disease genes, and classify diseases.
Detecting interaction dysregulation
The majority of pathway analyses can be grouped into three classes: over-representation analysis, functional class scoring and pathway topology-based methods[22]. The over-representation analysis starts from a list of genes and a set of pathways; every pathway is tested for over- or under-representation in the list of input genes using a statistical test based on hypergeometric or chi-square distribution. This approach treats each gene equally, and ignores data associated with each gene, like mRNA expression levels or p-values from GWAS. Many popular methods, such as FatiGO[11] and GoMiner[10], belong to this class. Alternatively, the functional class scoring approach, such as the well-known Gene Set Enrichment Analysis (GSEA), Gene Set Analysis (GSA) and similar methods[8, 9], takes genes and their associated expression values as inputs. A gene-level statistic is computed typically using t-test; then, for each pathway, a single pathway-level statistic is computed by aggregating gene-level statistics; finally, the significance of the pathway-level statistics are evaluated empirically by permutation. The basic steps of pathway topology-based approach are quite similar to functional class scoring, except it takes into account the pathway topology when computing the gene-level statistics[23, 24]. However, almost all methods described above are designed to identify disease-associated pathways by investigating the changes of genes, which are one of the components in pathways. More recently, approaches have been proposed to investigate other components in the pathway, such as interactions.
The physical entities in a pathway, like genes, are only one of the fundamental components in the pathways (in the network model, genes are represented as nodes). Other important components are interactions among them, i.e., gene and protein interactions, and dynamics of those interactions (in the network model, interactions are represented as edges). Both genes and interactions among them are essential and tightly regulated for the proper functioning of the system; perturbation of either of them can lead to dysregulation, i.e., diseases[25, 26]. Studies have showed that cellular networks exhibit systems properties underlying phenotypic variations[5, 27, 28]. Zhong et al. analyzed 50,000 known disease-causative mutations, and proposed two distinct mutations: one type leads to node removal from network due to the destruction of reading frame or destabilization of protein structure; the other type, such as single amino-acid substitution at binding site, may affect the ability to bind/interact with its partners[29]. The latter type was considered as edge-specific (edgetic) perturbations, which confer distinct functional consequences compared to node removal[29]. Identifying and distinguishing both types of mutations will improve our understanding of diseases and help to develop efficient treatments. In this section, we focus on the methods that are designed to detect dysregulated pathways in term of interactions.
Liu et al. proposed the Gene Interaction Enrichment and Network Analysis (GIENA) to identify dysregulated gene interactions and pathways using functions that model the relationship of cooperation, competition, redundancy, and dependency among the expression levels of genes[30]. These functions are defined as follows: the sum of mRNA expression levels, which models cooperation; the difference between mRNA expression levels models competition; the maximum/minimum mRNA expression level models redundancy/dependency between a pair of genes. Moreover, the regulatory logic governing the perturbation in diseases can be constructed based on the detected dysregulated interactions. The proposed framework was applied to identify dysregulated pathways in cancer. The results showed that GIENA can identify pathways that are well-known and biologically meaningful, the results are highly reproducible, and GIENA is efficient in terms of extracting weak signals and identifying pathways that are missed by with gene centered method, such as GSEA/GSA[8, 9]. In other studies, the relative expression of two genes has also been applied to classify two closely related cancers, and identify tightly regulated networks and their changes in diseases[31, 32]. In another study, Taylor et al. defined the difference in the expression of the hub gene with each of its partners as interaction coherence, and the change of interaction coherence was measured between diseases and control samples[33].
Mani et al. developed a method to identify gene pairs showing either a gain of correlation (GoC) or a loss of correlation (LoC) pattern of gene expression in the diseases, compared with the pattern in healthy individuals[34]. A gene set is constructed and its interactions are catalogued, and these interactions are either gained (GoC) or lost (LoC), i.e., dysregulated, in the diseases under investigation. The dysregulated interactions are pooled together to identify genes with significantly high number of dysregulated interactions in its neighborhood. Combining the B-cell interactome with gene expression profiles from three malignant B-cell phenotypes, the authors demonstrated that their method can identify genes and pathways enriched for such gained or lost correlation, which are likely implicated in tumorigenesis, and their method can detect some well-known oncogenes, such as BCL2 and SMAD1, which traditional methods can fail to detect[34]. They also found that the patterns of dysregulated interactions are dramatically different among three malignant B-cell phenotypes, indicating different underlying mechanisms among them. In another study, Zhang et al. proposed a similar method to detect dysregulated interactions and pathways in diseases[35]. In their study, the difference of co-variances or correlations between two genes from healthy and diseases groups represented the interaction among them. Coupled with GSA[9], their method was able to detect pathways with dysregulated interaction enrichments[35].
Watkinson et al. utilized a synergy concept from information theory to define types of gene interactions[36]. The synergy of two genes is defined as a function of mutual information (MI) between gene expression profiles (gene1 and gene2) and phenotype status (phenotype): Synergy (gene1, gene2) = MI(gene1, gene2; phenotype) - [MI(gene1; phenotype) + MI(gene2; phenotype)]. Positive synergy indicates gene interactions, and a synergy network can be constructed based on detected interactions. Using gene expression data from prostate cancer and healthy individuals, the authors found strong synergies between many gene pairs, which can predict prostate cancer much better than the simple additive individual genes. RBP1 appears most frequently in high-synergy gene pairs. RBP1 inhibits the PI3K/Akt survival pathway, indicating PI3K/Akt is associated with prostate tumorigenesis. In another study, mutual information also has been used to measure the activity of a network, dysregulated subnetworks were identified in diseases or different development stages using a heuristic search algorithm[37].
Although the methods described above can detect dysregulated interactions in diseases, this field is still in its early stage of development. Several important questions need to be addressed before they are widely applied, e.g., which method performs better, how to validate the detected interactions, and what is the nature of the interactions. Furthermore, the gene-based and interaction-based methods are complementary; thus, it is desirable to integrate both approaches to provide a comprehensive understanding of complex diseases.
Pathway-based methods to detect disease-association
Pathway-based analysis was first developed for the analysis of gene expression profiling from microarray experiments to identify pathways that have modest, but consistent expression changes in diseases[22]. In last 5 years, over 1000 GWAS have been conducted searching for genetic association of common diseases, and pathway analyses of GWAS data have been extended to understand the underlying disease mechanisms[38, 39]. More recently, integrative approaches have been developed to combine GWAS data with multiple “omics” data, such as mRNA expression, copy number alteration and the interaction network data (PPI and gene regulatory networks). The integration of interaction networks is expected to extend and improve our current pathway knowledge since our pathway knowledge is far from complete, and strong evident suggesting that disease-associated proteins tend to interact with each other[28, 40-43]. In this section, we will focus on the latest methodological development to pathway-based detection of disease association, especially, methods integrating GWAS with other “omics” data.
Many studies have demonstrated that integrating GWAS data with other “omics” can provide additional information and biological insight to conventional GWAS analysis, e.g., the underlying disease pathways that conventional method failed to identify. Jia et al. integrated both GWAS and PPI network data to identify disease-associated subnetworks[44]. The method first mapped all SNPs and their p-values in a GWAS dataset to genes based on the SNP-gene association (the most significant p-value among SNPs of each gene, was considered to represent the p-value of the gene); then, genes and their p-values were loaded onto a human PPI network; finally, dense module searching (DMS) previously developed for gene expression datasets was used to search for subnetworks that locally maximize the proportion of low-P-value genes in the GWAS dataset. The method was applied to two GWAS datasets for breast cancer and pancreatic cancer; identified gene sets and the connections among these genes (subnetworks) in the context of PPI networks, and further analyses showed that several cancer-related pathways were enriched in both gene sets[44].
To detect the disease-associated subnetworks from GWAS data and reduce the burden for multiple hypothesis testing problem, Pan introduced a network-based approach to give higher weight to subnetworks that contain known diseases genes or their partners[45]. Two weighting schemes are proposed based on exponential and inverse probabilities. Compared with exhaustive search, this approach significantly decreases the search space. Using a human PPI network and 23 known ataxia-causing genes, the author demonstrated that ataxia-genes are clustered in the network and subnetworks containing both disease genes and novel genes are detected[45]. Taking advantage of previous knowledge about disease associated genes, PPI networks and pathways, and eSNPs, Liu et al. proposed four frameworks to discover diseases associated interactions from GWAS data[46]. Four types of SNP sets were constructed first based on prior knowledge (e.g., all SNPs associated with genes in a single pathway, or SNPs in genes in a diseases-associated PPI network), and then exhaustive SNP-SNP interactions within each set were tested for diseases association using a logistic regression model. These approaches significantly decreased the search space and reduced hypothesis testing, and were applied to detect interactions in a GWAS dataset for type 2 diabetes (T2D). Interestingly, SNP interactions detected from four frameworks partially overlapped, and a connected network could be constructed[46]. More importantly, diseases associations of some SNP pairs were not tested because they are never present in same pathway or network; additional testing revealed two interactions that were significantly associated with T2D, which gives additional support for the association between the network with T2D[46].
Methods have been developed to combine expression data with GWAS data to identify disease-associated pathways[47, 48]. Xiong et al. developed Gene Set Association Analysis (GSAA), which simultaneously takes into account the SNP and gene expression variation to identify diseases associated pathways that enriched for differential expression and/or trait-associated SNPs[47]. In another study, pathways enriched for SNPs that associated with expression of genes (eSNPs) are targeted [48]. Zhong et al. identified eSNPs that associated with the expression of genes in liver, subcutaneous adipose, and omental adipose[48, 49]. Each eSNPs was tested for the association with diseases, generating a p-value; the p-value is assigned to the gene whose expression is associated with the eSNP. A previous method based on GSEA is used to detect pathways enriched for eSNPs[50]. This approach was applied to identify pathways associated with T2D, and many of the pathways identified have been proposed as important candidate pathways for T2D, and novel associated pathways, including the tight junction, complement and coagulation pathway, and antigen processing and presentation pathway[48].
Based on the observation that some genomic events (somatic mutations or copy number alterations) within oncogenic pathways exhibit a statistically significant level of mutual exclusivity, it was proposed that mutation or alteration of two or more genes within the same oncogenic pathway doesn’t offer selective advantage for tumor cells[51]. Ciriello et al. designed a novel method, Mutual Exclusivity Modules in cancer (MEMo) to identify network modules in which oncogenic mutations are mutually exclusive, by integrating somatic mutations, copy number alteration, mRNA expression and PPI network data and using correlation analysis[51]. The application of this method to glioblastoma identified multiple gene pairs in PI3K, p53 and Rb pathways that show significant mutual exclusivity of mutation or genomic alterations[51]. The authors suggested that the mutual exclusivity of mutations from two genes is due to the fact that the alteration to a second gene within the same pathway offers no further selective advantage[51]. Similar network-based integrative methods have been proposed to identify pathways that drive cancer subtypes and cooperative genetic alterations in brain tumors, and infer the patient-specific pathway activities and driver genes[52-54].
Kim et al. developed another approach to identify disease-causal genes and associated dysregulated pathways by integrating gene expression, copy number alterations, and interaction networks (including interaction data such as PPI, phosphorylation events, and protein-transcription factor interactions)[55]. An expression Quantitative Trait Loci (eQTL) analysis was applied to determine the causal loci of each differentially expressed gene (target genes) by using a linear regression model on the differentially expressed genes and copy number alterations of 911 selected loci. To filter the false positive associations and determine the pathways associated with causal and target genes, a circuit flow algorithm was adopted to search the path from one causal to the target genes in the PPI, protein-DNA networks and phosphorylation events. The results were further filtered by accounting for multiple hypothesis testing corrections or selecting the set of genes that best explained most disease cases.
The challenges in detection of disease-associated pathways include lack of a comprehensive and accurate human interactome, poor understanding of the biological functions and role of intergenic regions of the human genome, and lack of comprehensive epigenetic data sets. PPI networks have been commonly integrated with mRNA expression, GWAS and other “omics” data to identify diseases-associated subnetworks. Although this approach can provide many novel insights for the underlying disease mechanisms, we should keep in mind problems like: the poor correlation between expression of mRNA and protein expression [56], PPI networks are likely tissue specific and dynamic[57], and the existence of other important interactions, such as transcription factor binding to DNA, microRNA interactions with mRNA[58], and other potential genetic interactions [59]. As many SNPs identified by GWAS are located in intergenic regions and their functional connections are unknown, it is currently challenging to include them appropriately in pathway analysis. Those SNPs might have strong effects on expression of distant genes by altering regulation or amplification status, i.e., as enhancers. Recent studies provided evidence that SNPs in “gene deserts” can physically interact with the promoter via transcription factor binding and act in an allele specific manner to regulate oncogene expression[60]. Epigenetic events, such as DNA methylation and histone modification, are another layer of regulation of gene expression[61] and post-translational modifications of proteins are an obvious new area of interest and importance. Many studies showed that all these types of alterations are associated with cancer and other diseases[62, 63], but it is challenging to integrate with other data due to the lack of data and poor understanding of the functional mechanisms of regulation.
Prioritizing candidate disease genes using network knowledge
Gene prioritization aims to rank a list of candidate genes based on their likelihood to be disease-associated for further validation through integrative analyses of available data, such as literature, function annotation, sequence similarity, linkage and association data and gene expression profiling[64-67]. Recently, network knowledge, like disease networks, and PPI or functional linkage networks have been integrated to prioritize candidates. Most of the early methods made the assumption that genes closer to each other in the network likely associate with similar diseases (guilt by association assumption)[68]. For example, Wu el at. constructed an integrated network by combining disease networks and PPI networks using disease-gene associations[69]. A score is calculated to measure the concordance between the phenotype similarities and the functional genetic relatedness of genes. The candidate genes are ranked based on their score. It has been shown that in 709 out of 1444 cases, this method successfully ranks disease genes at the top[69]. Linghu et al. and others constructed functional linkage networks by integrating multiple “omics” data (PPI, coexpression, functional annotation, co-occurrence in literature, etc.), and applied it to prioritize candidate genes[70-72]. Goncalves et al. compared the performance of the gene prioritization methods using PPI network alone and network integrating heterogeneous resources, and found integrative network consistently perform better over single PPI network in most cases[73].
Methods based on guilt by association have been questioned because of concern of statistical artifacts that results from node degree effects or exceptional edges[74]. Kohler et al. developed a method that takes into account the indirect interactions between candidate and disease genes[75]. This method gave more weight to candidate genes that share more interacting partners with disease genes. More recently, methods using global network properties have been developed. Proteins with different functions are connected in interacting networks to reveal signaling or metabolic functions so that PPI networks are organized into recurrent schemas[76]. Based on these observations, Erten et al. proposed that disease genes likely exhibit topological profile similarity, and topological profiles of candidate genes can be measured and compared with diseases genes, and used to prioritize potential candidates[77]. The topological profile of a protein is represented by effective conductance, a concept from electrical circuit, which can be efficiently computed using random walks. If the protein products of candidate genes are topologically similar to the products of disease genes (i.e., the effective conductance of candidates and diseases are significantly correlated), then the candidate genes are likely associated with the diseases. Thus, the correlation of effective conductance is used to prioritize the candidate genes[77]. Similar methods considering the network properties have also been proposed[73, 78]. Results show that these methods significantly outperformed those based on guilt by association assumptions[43, 73, 75, 77, 78]. Machine learning approaches coupled with statistical procedures have also been applied to filter background SNPs, construct networks and rank SNPs. McKinney and colleagues developed evaporative cooling (EC) to filter SNPs and detect the disease-associated networks from GWAS data[79-81]. This approach has been applied to GWAS data for bipolar disorder, and identified top ranked SNPs in ANK3 and DGKH, which were associated with bipolar diseases previously[79].
Although a few “top ranking genes” from prioritization methods have been experimentally validated [82], the order or ranks of candidate genes are almost impossible to confirm and hard to biologically interpret, which makes it difficult to evaluate the overall performance of the prioritization methods. Moreover, a network of several genes with small effects may have stronger effect than the top-ranking gene. Thus, results from prioritization should be interpreted carefully.
Pathway-based diseases classification
Accurate classification of diseases and disease stages is important for understanding of the underlying mechanism and design of efficient treatment. Gene expression profiling has been applied to identify cancer subtypes and predict treatment outcomes for over a decade [83-87]. In those early studies, genes are typically selected by their power to discriminate between different classes of disease without acknowledging the fact that genes are functioning by coordinately interacting with each other. The performance of those methods was not satisfactory, and the selected gene sets from different studies have limited overlap, even for the same cancer [84, 86], which is likely due to the genetic heterogeneity across patients and dysregulation at the pathway level instead of the gene level. Pathway and network-based methods have been developed to improve the classification and cope with these issues.
Nevis and his colleagues developed pathway-based methods to detect cancer subtypes[88-90]. Their approach identified gene expression signatures that reflect the activation status of several oncogenic pathways, and detected cancer subtypes using these signatures. To identify the expression signature, first, human mammary epithelial cells (HMEC) were infected with adenovirus expressing a specific oncogene, such as Myc, Ras, or Src. Then, the activation status for each oncogenic pathway was measured, and gene expression signatures that reflect the activities of a given pathway were selected. Finally, the signatures were used to detect cancer subtypes. Results showed that the identified patients in the same subtypes share similar clinical and biological properties[89].
Ideker and his colleagues proposed a method to identify subnetworks that correlated with cancer metastasis[37, 91]. Their method integrated PPI networks with gene expression profiling from metastatic or non-metastatic cancer cells. For one given subnetwork, MI was calculated to detect the correlation between expression profiling and metastasis. The subnetwork with optimal MI was searched using a greedy algorithm. Permutation was used to test the statistical significance of the subnetwork. Results showed that network based methods achieve higher accuracy and are more reproducible than alternative approaches. This approach has been extended to integrate the proteins that were differentially expressed in colon cancer from proteomics experiments[92].
Conclusion
Many novel methods for pathway analysis have been developed and applied to many aspects of biomedical research to understand the underlying mechanism of diseases. The pathway-based approach outperforms previous methods because it is based on the activity of biologically connected and validated gene sets rather than on the expression levels of individual genes. The methods described above that integrate genome wide expression or GWAS data with pathways and networks are very promising, but they can be improved by taking into account other information, such as epigenetics. However, the field is still far from maturity due to incomplete pathway knowledge. Furthermore, pathway analysis is currently coding gene-centered; non-protein coding elements (noncoding RNA, non-transcribed regions and epigenetic marks) have not been sufficiently integrated in the analysis. Recent studies demonstrated that 80% of human genome might be functional[93] and epigenetics plays important role to maintain proper cellular functions[62, 94-96]. As the cost for HT-data acquisition keeps decreasing dramatically, genomic, epigenomic, and ultimately proteomics data from biomedical research will be accumulated even more rapidly. This will accelerate the integration of information form coding and non-coding regions to significantly improve pathway analysis.
Acknowledgements
This publication was made possible in part by the Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research and in part through support from the National Cancer Institute (P30-CA-043703), and the National Institute for Allergy and Infectious Diseases (P30-AI-036219).
Footnotes
Conflict of Interest
Y Liu declares no conflicts of interest.
MR Chance declares no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Reference
Papers of particular interest, published recently, have been highlighted as:
• Of importance;
•• Of major importance
- 1.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 4.Friend SH, Ideker T. POINT: Are We Prepared For the Future Doctor Visit? Nat Biotechnol. 2011;29(3):215–218. doi: 10.1038/nbt.1794. [DOI] [PubMed] [Google Scholar]
- 5••.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. This article presnts an excellent review about how networks can be used to study human diseases.
- 6.Fernald GH, Capriotti E, Daneshjou R, et al. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27(13):1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annual review of cell and developmental biology. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efron B, Tibshirani R. On Testing the Significance of Sets of Genes. Ann Appl Stat. 2007;1(1):107–129. [Google Scholar]
- 10.Zeeberg BR, Feng WM, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4(4) doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20(4):578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 12.Eden E, Navon R, Steinfeld I, et al. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ideker T, Ozier O, Schwikowski B, et al. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 14.Song JM, Singh M. How and when should interactome-derived clusters be used to predict functional modules and protein function? Bioinformatics. 2009;25(23):3143–3150. doi: 10.1093/bioinformatics/btp551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M, Liberzon A, Kong SW, et al. Network-based analysis of affected biological processes in type 2 diabetes models. Plos Genet. 2007;3(6):958–972. doi: 10.1371/journal.pgen.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendes P, Kell DB. Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics. 1998;14(10):869–883. doi: 10.1093/bioinformatics/14.10.869. [DOI] [PubMed] [Google Scholar]
- 17.Kauffman KJ, Prakash P, Edwards JS. Advances in flux balance analysis. Curr Opin Biotech. 2003;14(5):491–496. doi: 10.1016/j.copbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Ourfali O, Shlomi T, Ideker T, et al. SPINE: a framework for signaling-regulatory pathway inference from cause-effect experiments. Bioinformatics. 2007;23(13):I359–I366. doi: 10.1093/bioinformatics/btm170. [DOI] [PubMed] [Google Scholar]
- 19.Dutkowski J, Kramer M, Surma MA, et al. A gene ontology inferred from molecular networks. Nat Biotechnol. 2013;31(1):38–+. doi: 10.1038/nbt.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007:3. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McShan DC, Rao S, Shah I. PathMiner: predicting metabolic pathways by heuristic search. Bioinformatics. 2003;19(13):1692–1698. doi: 10.1093/bioinformatics/btg217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS computational biology. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. This article reviews latest approaches for pathway analysis and challenges.
- 23.Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome research. 2007;17(10):1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shojaie A, Michailidis G. Analysis of gene sets based on the underlying regulatory network. Journal of computational biology: a journal of computational molecular cell biology. 2009;16(3):407–426. doi: 10.1089/cmb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37(4):413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 26.Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goh KI, Cusick ME, Valle D, et al. The human disease network. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews Genetics. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Q, Simonis N, Li QR, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30• •.Liu Y, Koyuturk M, Barnholtz-Sloan JS, Chance MR. Gene interaction enrichment and network analysis to identify dysregulated pathways and their interactions in complex diseases. BMC systems biology. 2012:6. doi: 10.1186/1752-0509-6-65. This study introduces mathematic measures for dysregulated interactions and methods to identify them.
- 31.Eddy JA, Hood L, Price ND, Geman D. Identifying Tightly Regulated and Variably Expressed Networks by Differential Rank Conservation (DIRAC) PLoS computational biology. 2010;6(5) doi: 10.1371/journal.pcbi.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price ND, Trent J, El-Naggar AK, et al. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(9):3414–3419. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor IW, Linding R, Warde-Farley D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27(2):199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 34.Mani KM, Lefebvre C, Wang K, et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Li J, Deng HW. Identifying gene interaction enrichment for gene expression data. PloS one. 2009;4(11):e8064. doi: 10.1371/journal.pone.0008064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watkinson J, Wang XD, Zheng T, Anastassiou D. Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC systems biology. 2008:2. doi: 10.1186/1752-0509-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends in genetics: TIG. 2012;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Wang K, Li MY, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nature Reviews Genetics. 2010;11(12):843–854. doi: 10.1038/nrg2884. This article presents a review of pathway analysis of GWAS data.
- 40.Gandhi TK, Zhong J, Mathivanan S, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38(3):285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 41.Furlong LI. Human diseases through the lens of network biology. Trends in genetics: TIG. 2013;29(3):150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 42•.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44(8):841–847. doi: 10.1038/ng.2355. This article presents some examples for integrating of network and other “omics” data for disease association study.
- 43.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26(8):1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Jia PL, Zheng SY, Long JR, Zheng W, Zhao ZM. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27(1):95–102. doi: 10.1093/bioinformatics/btq615. This study was among the first to integrate network and GWAS data.
- 45.Pan W. Network-based model weighting to detect multiple loci influencing complex diseases. Hum Genet. 2008;124(3):225–234. doi: 10.1007/s00439-008-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46• •.Liu Y, Maxwell S, Feng T, et al. Gene, pathway and network frameworks to identify epistatic interactions of single nucleotide polymorphisms derived from GWAS data. BMC systems biology. 2012:6. doi: 10.1186/1752-0509-6-S3-S15. This study presents four frameworks for efficiently identifying interactions among SNPs associated with diseases.
- 47.Xiong Q, Ancona N, Hauser ER, Mukherjee S, Furey TS. Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome research. 2012;22(2):386–397. doi: 10.1101/gr.124370.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating Pathway Analysis and Genetics of Gene Expression for Genome-wide Association Studies. Am J Hum Genet. 2010;86(4):581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS biology. 2008;6(5):e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51• •.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome research. 2012;22(2):398–406. doi: 10.1101/gr.125567.111. This study presents a novel method to detect network modules associated with tumorigenesis.
- 52.Dutta B, Pusztai L, Qi Y, et al. A network-based, integrative study to identify core biological pathways that drive breast cancer clinical subtypes. British journal of cancer. 2012;106(6):1107–1116. doi: 10.1038/bjc.2011.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated Network Analysis Identifies Core Pathways in Glioblastoma. PloS one. 2010;5(2) doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaske CJ, Benz SC, Sanborn JZ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YA, Wuchty S, Przytycka TM. Identifying causal genes and dysregulated pathways in complex diseases. PLoS computational biology. 2011;7(3):e1001095. doi: 10.1371/journal.pcbi.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gry M, Rimini R, Stromberg S, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC genomics. 2009:10. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nature Reviews Genetics. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sotelo J, Esposito D, Duhagon MA, et al. Long-range enhancers on 8q24 regulate c-Myc. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 62•.Shen H, Laird PW. Interplay between the Cancer Genome and Epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. This article presents a review for latest development of cancer genomics and epigenomics.
- 63•.Akhtar-Zaidi B, Cowper-Sal-lari R, et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science. 2012;336(6082):736–739. doi: 10.1126/science.1217277. This study shows the significance of epigenomics for tumorigenesis.
- 64.Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nature reviews Genetics. 2012;13(8):523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- 65.Tranchevent LC, Capdevila FB, Nitsch D, et al. A guide to web tools to prioritize candidate genes. Briefings in bioinformatics. 2011;12(1):22–32. doi: 10.1093/bib/bbq007. [DOI] [PubMed] [Google Scholar]
- 66.Oti M, Ballouz S, Wouters MA. Web tools for the prioritization of candidate disease genes. Methods Mol Biol. 2011;760:189–206. doi: 10.1007/978-1-61779-176-5_12. [DOI] [PubMed] [Google Scholar]
- 67.Piro RM, Di Cunto F. Computational approaches to disease-gene prediction: rationale, classification and successes. The FEBS journal. 2012;279(5):678–696. doi: 10.1111/j.1742-4658.2012.08471.x. [DOI] [PubMed] [Google Scholar]
- 68.Oti M, Brunner HG. The modular nature of genetic diseases. Clinical genetics. 2007;71(1):1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 69.Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Linghu B, Snitkin ES, Hu Z, Xia Y, Delisi C. Genome-wide prioritization of disease genes and identification of disease-disease associations from an integrated human functional linkage network. Genome Biol. 2009;10(9):R91. doi: 10.1186/gb-2009-10-9-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franke L, van Bakel H, Fokkens L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet. 2006;78(6):1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome research. 2011;21(7):1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goncalves JP, Francisco AP, Moreau Y, Madeira SC. Interactogeneous: disease gene prioritization using heterogeneous networks and full topology scores. PloS one. 2012;7(11):e49634. doi: 10.1371/journal.pone.0049634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Gillis J, Pavlidis P. “Guilt by Association” Is the Exception Rather Than the Rule in Gene Networks. PLoS computational biology. 2012;8(3) doi: 10.1371/journal.pcbi.1002444. This study shows that functional information within networks is typically concentrated in only a small region of network, and “guilt by association” can not be applied across whole network.
- 75.Kohler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82(4):949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pandey J, Koyuturk M, Kim Y, et al. Functional annotation of regulatory pathways. Bioinformatics. 2007;23(13):I377–I386. doi: 10.1093/bioinformatics/btm203. [DOI] [PubMed] [Google Scholar]
- 77• •.Erten S, Bebek G, Koyuturk M. VAVIEN: An Algorithm for Prioritizing Candidate Disease Genes Based on Topological Similarity of Proteins in Interaction Networks. Journal of Computational Biology. 2011;18(11):1561–1574. doi: 10.1089/cmb.2011.0154. This study presents method to prioritize genes based on topoligical property instead of “guilt by association”.
- 78.Guney E, Oliva B. Exploiting protein-protein interaction networks for genome-wide disease-gene prioritization. PloS one. 2012;7(9):e43557. doi: 10.1371/journal.pone.0043557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandey A, Davis NA, White BC, et al. Epistasis network centrality analysis yields pathway replication across two GWAS cohorts for bipolar disorder. Translational psychiatry. 2012;2:e154. doi: 10.1038/tp.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davis NA, Crowe JE, Jr., Pajewski NM, McKinney BA. Surfing a genetic association interaction network to identify modulators of antibody response to smallpox vaccine. Genes and immunity. 2010;11(8):630–636. doi: 10.1038/gene.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McKinney BA, Crowe JE, Guo J, Tian D. Capturing the spectrum of interaction effects in genetic association studies by simulated evaporative cooling network analysis. Plos Genet. 2009;5(3):e1000432. doi: 10.1371/journal.pgen.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Erlich Y, Edvardson S, Hodges E, et al. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome research. 2011;21(5):658–664. doi: 10.1101/gr.117143.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 84.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 85.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 87.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 88.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 89• •.Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(15):6994–6999. doi: 10.1073/pnas.0912708107. This study presents method to measure activities of some oncogenic pathways and use them to classify breast cancer.
- 90.Nevins JR. Pathway-based classification of lung cancer: a strategy to guide therapeutic selection. Proceedings of the American Thoracic Society. 2011;8(2):180–182. doi: 10.1513/pats.201006-040MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chuang FY, Rassenti LZ, Salcedo M, et al. Subnetwork-Based Analysis of Chronic Lymphocytic Leukemia Identifies Pathways That Associate with Disease Progression. Blood. 2011;118(21):1521–1522. doi: 10.1182/blood-2012-03-416461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and Scoring of Protein Interaction Subnetworks Discriminative of Late Stage Human Colon Cancer. Molecular & Cellular Proteomics. 2009;8(4):827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou X, Maricque B, Xie MC, et al. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8(12):989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]