1 Introduction
Long-chain alkane waxes are synthesized by a wide variety of organisms, including plants, animals and microorganisms.1-4 Although chemically alkanes are amongst the most boring of organic molecules, devoid of functionality and thus unreactive, they play essential roles in these organisms’ survival. Plants secrete what are termed “very long chain” alkane waxes (i.e. chain length ~ 30 carbons) onto their leaves and stems.3 These, together with wax esters formed from aliphatic alcohols and carboxylic acids of similar chain length, serve as a waterproof barrier to prevent desiccation. A wide variety of hydrocarbons, including branched and unsaturated molecules, are biosynthesized by insects.2 These are secreted onto the cuticle and play an important role as contact pheromones that mediate all manner of insect:insect recognition and social behaviors; they are also important in preventing desiccation, especially of larval forms.5 Waterfowl secrete alkane-rich oils from their uropygial (preening) glands; these are essential for waterproofing the birds’ feathers, which would otherwise become waterlogged and thus useless for insulation and flight.1 Many algae synthesize large amounts of long-chain alkanes that can comprise up to 30 % of the algae's dry weight.6 These accumulate in the area contained by the inner and outer cell walls known as the trilamellar structure and can be utilized to provide energy when photosynthesis is not possible.7
In recent decades many human beings have become equally dependent on alkanes, in that this class of molecules is the major component of transportation fuels. These fuels are, of course, currently derived from fossil reserves and their combustion is a major source of greenhouse gases. The challenge of producing the next generation of biofuels – those that can effectively function as “drop-in” replacements for gasoline, diesel, and jet fuel – has led to renewed interest, especially in the area of mechanistic enzymology, in how alkanes are biosynthesized.8
The synthesis of completely unfunctionalized molecules presents a challenge for a cell.9 Whereas as a chemist might accomplish this by reducing the corresponding alkene to an alkane using hydrogen and an activated transition metal catalyst, this approach is clearly not biochemically feasible. Historically, an important observation was that naturally-synthesized alkanes invariably comprise an odd number of carbons, suggesting that they are derived from fatty acid biosynthesis through loss of the carboxyl carbon.10 It was subsequently established that long-chain alkanes are synthesized from fatty acids through the intermediacy of the corresponding fatty aldehydes. These are the substrates for a group of enzymes, the aldehyde decarbonylases, which catalyze the removal of the aldehyde carbonyl group to form the alkane. It is the mechanisms of these enzymes that are the focus of this viewpoint. Whereas alkanes might be boring molecules, we hope to show that the mechanisms by which they are formed in Nature, and about which much still remains to be understood, are definitely not!
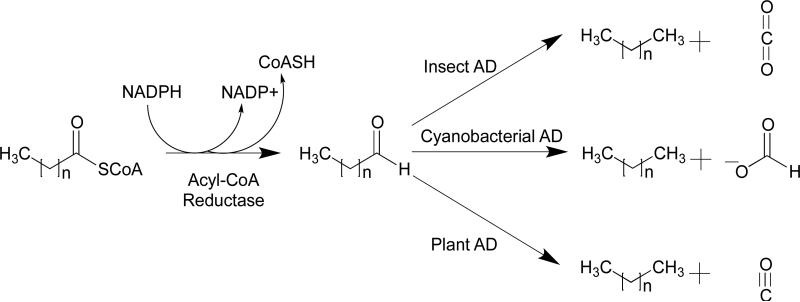
Although all the various organisms so far investigated appear to synthesize alkanes using fatty aldehydes as the precursor, it has become clear that there are several mechanistically distinct classes of decarbonylases, as illustrated in Figure 1. In insects, the enzyme has been shown to be a cytochrome P450 protein,11,12 whereas in cyanobacteria it is a non-heme di-iron enzyme that is structurally related to enzymes such as ferritin and methane monooxygenase.3,13 In plants, the enzyme is an integral membrane protein that has some sequence similarity to the fatty acid hydroxylase superfamily and stearoyl-CoA desaturase.14 On this basis, it too is presumed to be a metalloenzyme, with iron the most likely metal. The different mechanisms of decarbonylation are reflected in the fate of the aldehyde carbon, which is converted to CO2 in insects,12 HCO2H in cyanobacteria,15,16 and CO in plants and algae.6,17
Figure 1.
The biosynthesis of alkanes proceeds in two steps from fatty acyl-CoA esters. The decarbonylation of the intermediate fatty aldehyde proceeds by one of three different mechanisms depending upon the organism. The insect AD is a membrane-bound P450 type enzyme . The cyanobacterial AD is a soluble non-heme di-iron oxygenase. The plant AD is thought to be similar to the non-heme iron membrane proteins represented by stearoyl desaturase and fatty acid hydroxylase.
2 Insect decarbonylases
The biosynthesis of hydrocarbons has been studied in a number of insect species including the domestic house fly, Musca domestica, the fruit fly Drosophila melanogaster, cockroaches Periplaneta americana and Blattella germanica, and the termite Zootermopsis angusticollis.18 Experiments on microsomal preparations of M. domestica demonstrated that molecular oxygen and NADPH were required for the conversion of (Z)-15-tetracosenal to (Z)-9-tricosene and, using 1-14C-material, that the aldehyde carbon was oxidized to CO2 in the process.12 Furthermore, antibodies to either house fly cytochrome P450 reductase or to a cytochrome P450 (CYP6A1) purified from the house fly inhibited (Z)-9-tricosene.19 These initial observations provided support for the involvement of a P450 enzyme in the reaction; this was more recently confirmed with the cloning and heterologous expression of the enzyme, CYP4G1, from Drosophila.11
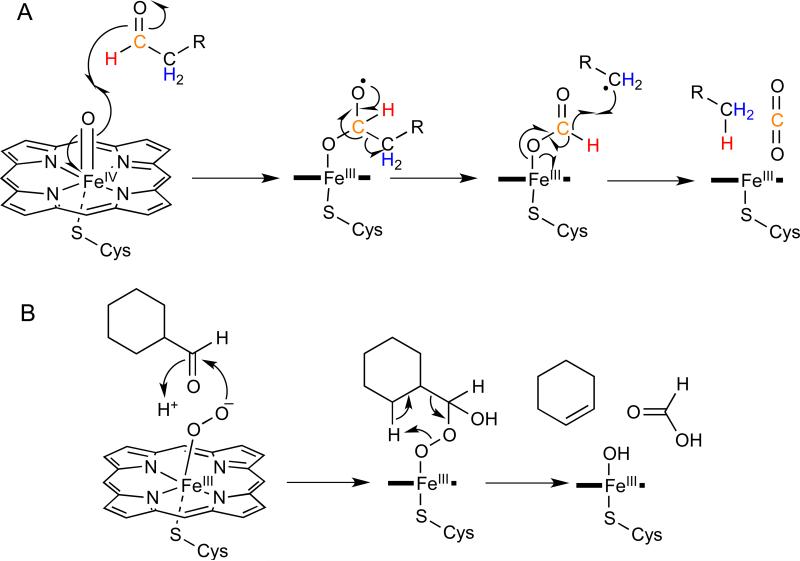
Further clues to the mechanism of hydrocarbon formation came from analyses of n-tricosane formed by M. domestica microsomal preparations incubated with [2,2-2H2,2-13C]-tetracosanoyl-CoA and [3,3-2H2,3-13C]-tetracosanoyl-CoA which demonstrated that the deuterium atoms on the 2- and 3-positions were retained in the hydrocarbon product.19 Analysis of [l-2H]-tetracosenal incubated with microsomal preparations demonstrated that the aldehyde proton on C-l was transferred to the (Z)-9-tricosene product. Alternative oxidizing agents such as hydrogen peroxide, cumene hydroperoxide, and iodosobenzene were shown to substitute for O2 and NADPH in the reaction. To accommodate these observations, a mechanism has been proposed, shown in Figure 2A, in which the high valent iron-oxo species, resulting from heterolytic cleavage of the O-O bond of the iron-peroxy intermediate, abstracts an electron from the carbonyl group of the aldehyde.19 The reduced iron-oxo species then attacks the carbonyl carbon of the aldehyde to form an iron-hemiacetal diradical. This intermediate is proposed to fragment to form an alkyl radical and an iron-bound formyl radical. In the final step, the alkyl radical then abstracts the formyl hydrogen to produce the hydrocarbon and CO2.
Figure 2.
Comparison of the deformylation reactions catalyzed by insect AD (CYP4G1) and CYP2B4. A – Deformylation of fatty aldehydes by the insect AD is proposed to start with the high-valent iron-oxo species and results in the formation of CO2. The color-coding indicates positions in the substrate that have been isotopically-labeled to establish the mechanism. B - Decarbonylation of cyclohexanecarboxaldehyde by CYP2B4 is initiated by the iron peroxide species and results in the formation of cyclohexene and formic acid.
P450 enzymes catalyze a notably diverse range of oxidative transformations on a very wide range of substrates.20 Even so, the decarbonylase reaction stands out as being unusual. In most cases P450 enzymes oxidize aldehydes to carboxylic acids through a well understood mechanism involving hydrogen atom abstraction by the high valent iron-oxo intermediate followed by “rebound” of the iron-bound hydroxyl group to give the hydroxylated substrate.21 In some cases decarbonylation of the aldehyde does occur, notably in the aromatization reaction of androst-4-ene-3,17-dione to estrone catalyzed by human aromatase and the deformylation of cyclohexanal catalyzed by CYP2B4 (Figure 2B).20 However in these reactions, which are believed to involve the iron-peroxide form of P450, the aldehyde carbon is converted to formate, rather than CO2, and a double bond is introduced into the oxidized product.20 An intriguing question is how CYP4G1 controls the highly reactive iron-oxo intermediate to accomplish oxidative decarbonylation rather than simply oxidizing the aldehyde to a carboxylic acid, which, a priori, would appear to be a more likely fate for the substrate.
3 Cyanobacterial decarbonylases
The cyanobacterial pathway for alkane biosynthesis is the most recently discovered pathway,13 and the only one for which, somewhat surprisingly, the acyl-CoA reductase and AD enzymes are soluble proteins. This fact has rendered the cyanobacterial enzyme more amenable to mechanistic analysis than the membrane-bound animal and plant decarbonylases. In the literature the cyanobacterial enzyme is referred to as either aldehyde decarbonylase (cAD) or, more recently, as aldehyde deformylating oxygenase (cADO),22 which more accurately describes the reaction catalyzed by this enzyme.
Although the biological functions of the hydrocarbon waxes produced by plants and animals are well understood, it is unclear why cyanobacteria biosynthesize these molecules, and indeed not all strains of cyanobacteria do. It was this latter observation that allowed the genes for alkane biosynthesis to be identified using a genomic subtraction approach to identify genes that were present in producing strains but absent in non-producers.13
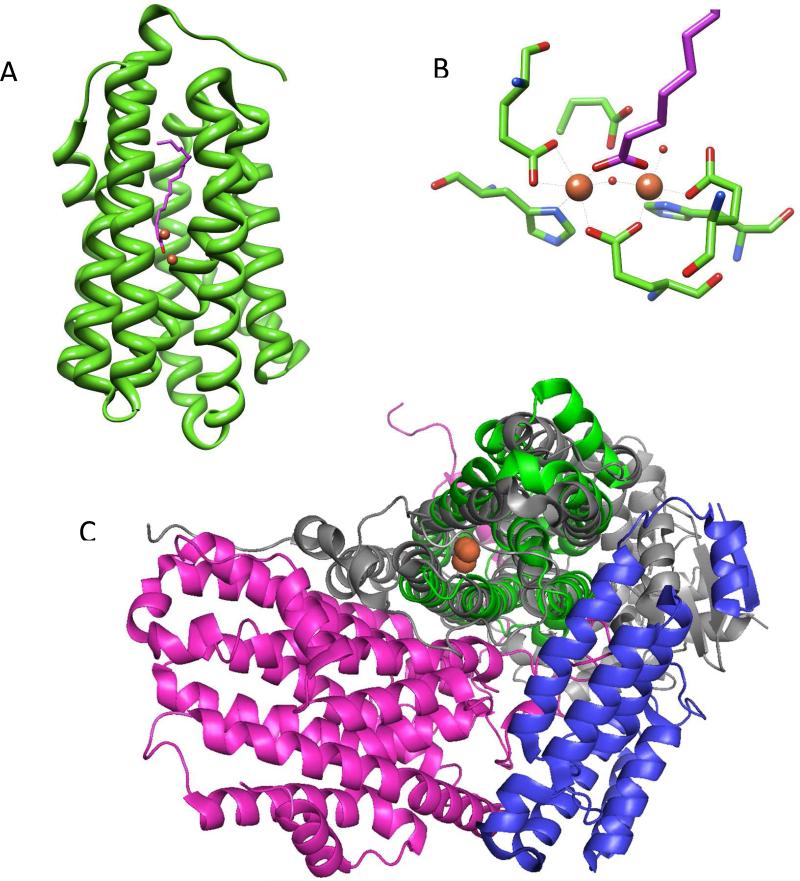
The cADO is the only decarbonylase for which an x-ray structure, shown in Figure 3A, is available.23 The structure reveals it to be a member of the non-heme di-iron family of oxygenases24,25 exemplified by enzymes such as methane monooxygenase (MMO), class I ribonucleotide reductase, and fatty-acyl-ACP desaturase. The active site of the enzyme is housed within an antiparallel 4-helix bundle in which the two iron atoms are each coordinated by a histidine and two carboxylate ligands from the protein, shown in Figure 3B. A further four α-helixes pack against the 4-helix bundle. The substrate-binding site comprises a long hydrophobic channel that terminates at the di-iron center. Compared with other enzymes of this family, cADO presents a rather “minimalist” structure and the enzyme is much smaller (29 kDa monomer) than most other enzymes in this family. For example, the core structure of MMO, compared in Figure 3C, is significantly larger (251 kDa α2β2γ2 homodimer) and the enzyme includes additional β and γ subunits that interact with the obligate reductase system (MMOR) as well as a regulatory component B (MMOB).24,26 As discussed below, cADO similarly requires molecular oxygen and an external reducing system for activity, but proteins analogous to MMOR and MMOB, if they exist, have not yet been identified for this system.27
Figure 3.
A Crystal structure of cADO (pdb 2OC5) from P. marinus. The iron atoms of the di-iron center are shown in orange and the co-crystallized fatty acid is shown in purple. B – Detail of the active site of cADO showing the ligands to iron. C – Comparison of the structures of cADO and MMOH (pdb 1XVC). The structure of cADO, shown in green, is overlaid on the α subunit of MMOH, shown in gray. The additional β and γ subunits of MMOH are shown in pink and blue; the di-iron centers of both enzymes closely overlay and are shown in orange. Figures 3A and 3B are based on reference 30.
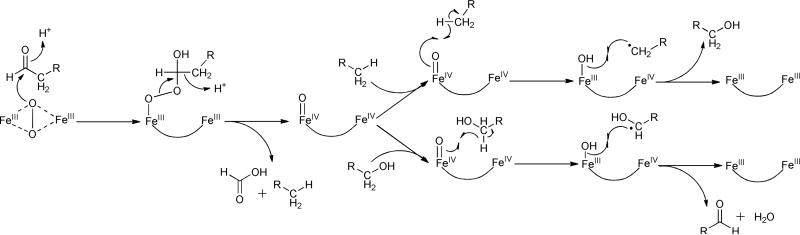
Our current understanding of the mechanism of cADO is drawn from a combination of isotopic labeling studies, reactions with mechanism-based inhibitors, spectroscopic characterization by Mössbauer and EPR spectroscopy and inferences based on its structural similarity to the MMO system. Initial studies established the conversion of aldehydes to alkanes required an external reducing system – either reduced ferredoxin or a chemical reducing system such as phenazine methosulfate/NADH – to support activity.16,27 In contrast to the insect or plant enzymes, the aldehyde carbon was shown to be converted to formate, rather than CO2 or CO, in the reaction.15,16 Deuterium labeling studies established that the aldehyde hydrogen was retained in formate, whereas the proton in the alkane derives from the solvent.15,16 A key observation, based on 18O2-labelling studies was that one of the oxygen atoms in formate derives from molecular oxygen.27 However, to accommodate the overall stoichiometry of the reaction it is necessary to invoke complete reduction of the oxygen consumed during turnover to give the equivalent of two molecules of water. In this respect cADO is unique among iron oxygenases and the reaction has been referred to as a “cryptic” oxidation because the overall conversion of aldehyde to alkane + formate is redox neutral.
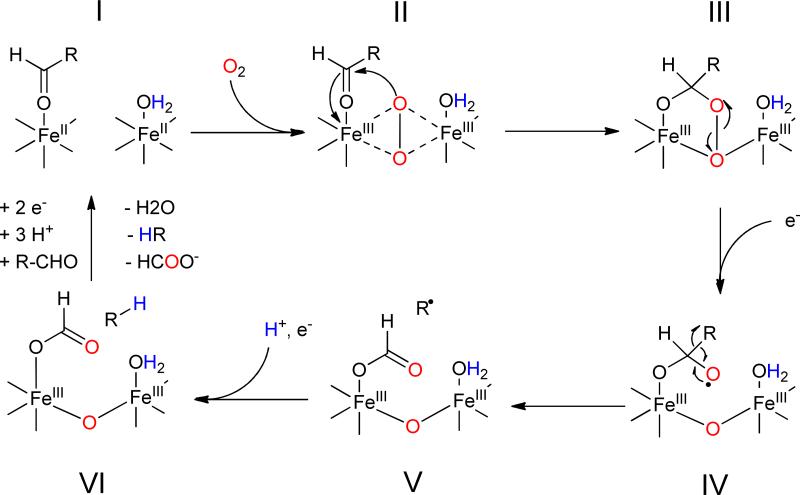
Based on these observations, the mechanism shown in Figure 4 has been proposed that extrapolates from other, better-understood non-heme di-iron oxygenases.27 The reaction starts with the reduction of the di-ferric resting state of the enzyme to the active di-ferrous form that is able to bind molecular oxygen. It is hypothesized that oxygen forms an intermediate peroxo-bridged di-iron core (P-type species), as has been observed in the MMO system. In the next step, the peroxo species undergoes nucleophilic addition to the aldehyde, forming a peroxy-hemiacetal intermediate. At this point it is necessary to invoke injection of a further electron into the active site that results in breakdown of the peroxide and transiently generates a formyl radical. Fragmentation of this species results in homolytic cleavage of the carbon-carbon bond to produce a primary alkyl radical and formate as the co-product. Lastly, further reduction of the alkyl radical, possibly as a proton-coupled electron transfer reaction, results in formation of the alkane.
Figure 4.
Proposed mechanism for deformylation of aldehydes by cADO. The color-coding indicates the origins of oxygen atoms and protons in the products established by isotope-labeling.
Support for a peroxy-hemiacetal species (intermediate III in Figure 4) comes from recent U.V.-Visible stopped-flow measurements and Mössbauer spectroscopy28. The formation of a Fe2III/III bridged peroxide or peroxy-hemiacetal species was inferred from the appearance of a transient and characteristic band at 450 nm, whose appearance was dependent upon the presence of substrate. The freeze-quenched Mössbauer spectrum of this intermediate exhibited isomer shifts characteristic of a diferric core and consistent with the formation of a peroxy or peroxy-hemiacetal species. In the absence of external reductant the species was relatively stable, t1/2 ~400 s; however upon addition of a stoichiometric amount of reduced MeOPMS the intermediate rapidly decayed and resulted in ~0.50 equivalents of formate being produced. Reduction by MeOPMS resulted in the formation of an additional Fe2III/III state distinct, as characterized by Mössbauer spectroscopy, from either the peroxy intermediate or the resting diferric enzyme, which likely reflects the presence of bound products.
Support for a radical mechanism for C-C bond scission (conversion of IV to V in Figure 4) comes from the reaction of cADO with an aldehyde substrate containing a strategically placed cyclopropyl group that could act as a “radical clock”.29 Cyclopropylcarbinyl radicals, formed when radicals are generated adjacent to the cyclopropyl ring, undergo rapid and very well characterized ring-opening reactions and have been employed to investigate the reactions of many enzymes.30 When this substrate was reacted with cADO only the rearranged alkene was observed as a product, indicating that an alkyl radical with a relatively long lifetime ( > 10 ns) was formed as an intermediate. Interestingly, the cyclopropyl aldehyde was found to partition approximately equally between turnover and acting as a mechanism-based inhibitor. Inactivation resulted from the covalent attachment of the alkyl chain to the enzyme through a phenylalanine residue that lines the substrate-binding channel. The modification was presumably a consequence of the cyclopropyl ring opening up, with the result that the repositioned alkyl radical could react with the protein.
Most interestingly, it has recently been shown that cADO is capable of catalyzing other oxidative reactions.31 The reactions of nonanal and decanal, which are very slow substrates, results in the formation of n-1 alcohols and aldehydes in addition to n-1 alkanes. (These products may have been missed in previous studies because longer chain aldehydes typically used to assay the enzyme don't produce significant amounts of these side products.) The oxygen in the alcohol was shown to derive from O2 , whereas the only C1 product formed was formate. Intriguingly, simply incubating reduced cADO with alkanes or alcohols, in the absence of aldehydes, did not result in oxidation products. But when 13C-labeled nonanol was incubated with the enzyme in the presence of unlabeled decanal 13C-labeled nonanal was produced. These observations suggest the formation of a diffusible intermediate in the reaction, and to accommodate them a quite different mechanism for deformylation has been proposed (Figure 5).
Figure 5.
Alternative carbanionic mechanism proposed for deformylation of aldehydes by cADO resulting in a reactive FeIV superoxo species capable of additional reactions producing n-1 alcohols and aldehydes.
The mechanism invokes heterolytic C-C bond cleavage to produce formate and a carbanion, which would be rapidly protonated, and a FeIV superoxo species. At this point the resulting alkane could undergo hydroxylation to give the n-1 alcohol, or alternatively diffuse from the enzyme. This would allow other alkanes or alcohols to enter the active site and be oxidized by the FeIV superoxo species. Although this mechanism accounts for the oxidation products observed with medium chain length aldehydes, it appears at odds with the reductive homolytic bond cleavage previously proposed that is supported by the experiments discussed above. Further experiments are clearly needed to resolve this mechanistic paradox!
The reaction catalyzed by cADO raises some intriguing mechanistic questions that are similar to those posed by the insect P450 AD, as the high-valent iron-oxo species formed by the non-heme di-iron oxygenases are comparable in reactivity to those in P450 oxygenases. Thus, one likely fate of the aldehyde would be oxidation to a carboxylic acid through a mechanism analogous to alkane hydroxylation by MMOH. A further possibility would be oxidative deformylation to give formate and an alkene, as occurs in the aromatase reaction. Lastly, oxidative decarboxylation to give CO2 and an alkane would also appear reasonable, given that this is the reaction catalyzed by the insect AD. Indeed, given the feasibility of this last reaction, it is all the more puzzling why the “cryptic” oxidation of aldehydes to alkanes would have evolved. How cADO discriminates against the many other potential oxidative pathways open to its substrates is one of the major challenges for understanding this enzyme.
cADO is the only decarbonylase for which the kinetics of the reaction have been investigated.22,32,33 Kinetic analysis of the reaction is far from straightforward and is complicated by the very poor solubility of longer-chain aldehydes, and the requirement for an external reducing system. Nevertheless, it is clear that, in comparison with most enzymes, the reaction is extremely slow. Different laboratories have assayed the enzyme using different substrates, reducing agents and oxygen concentrations, making direct comparisons difficult, but the highest steady state turnover numbers reported are only about 1 min−1. Given the interest in engineering this enzyme for use in biofuels biosynthesis,34,35 the reason why the reaction is so sluggish is of more than academic interest.
We note that the extent to which the low activity measured in vitro represents the physiological activity of the enzyme in vivo is currently unclear. Cyanobacteria produce only low levels of hydrocarbons (the benefit to the organism, if any, is unknown) and the level at which cADO is expressed in cyanobacteria has not been reported, so it is quite possible that cADO may not be significantly more active in vivo. On the other hand, technical difficulties inherent to the assay, as discussed below, most likely contribute somewhat to the slow rates of turnover that have been reported.
In air-saturated buffer a significant amount of O2 reacts non-enzymatically with the reducing system (both reduced ferredoxin and PMS are effective oxygen scavengers). This side reaction rapidly depletes both the reducing system and O2 causing the enzyme-catalyzed reaction to cease after only a few turnovers. The total number of turnovers can be significantly increased by conducting the reaction at low oxygen concentrations, although under oxygen-limiting conditions the rate of reaction is several-fold slower.32 An additional complication is that hydrogen peroxide generated by reaction of O2 with the reducing system is an effective inhibitor of the enzyme (Ki ~ 16 μM), further depressing turnover.33 Addition of catalase was found to prolong the activity of cADO in the reaction, but it did not significantly increase the enzyme's specific activity.
Mechanistically, a more interesting reason for the enzyme's low activity is that additional protein cofactors may be necessary to activate it. One reason to suggest this is that in some mechanistically related enzymes, such as MMO, toluene monooxygenase, and alkene monooxygenase, several additional subunits are required for activity.26 The hydroxylase component of MMO (MMOH), for example, comprises an α2β2γ2-homodimer of Mr ~ 251 kDa. The β and γ subunits provide supporting protein architecture (Figure 3C) that surrounds the catalytic α subunit and mediates interactions with the 39 kDa reductase component (MMOR) and an additional 16 kDa regulatory subunit, MMOB. This regulatory subunit, which is common to other enzymes in this group of oxygenases, dramatically alters the activity of the system by effecting a conformational change in the hydroxylase subunit.36 MMOB binding results in up to a 1000-fold increase in activity.24,26 The changes in structure are subtle and affect both the reduction potential of the iron center as well as the relative affinity of the enzyme for inhibitors and substrates.36 It seems unlikely that cADO possesses additional tightly bound subunits analogous to the β and γ subunits of MMOH; if it did, the enzyme would probably not fold properly in their absence. However, a freely dissociable activating subunit, analogous to MMOB, seems at least a reasonable possibility.
Another possibility is that there is a specific, yet to be identified, reductase for the enzyme. Although ferredoxin supports activity, it interacts very weakly with cADO in comparison with most enzymes that use ferredoxin as a reductant. Moreover, cADO is unique in requiring additional electrons during the catalytic cycle, whereas in iron-oxygenase enzymes electrons are required only for initial reduction of the di-ferric resting state. The introduction of these additional electrons at the correct point in the catalytic cycle is crucial to the mechanism of deformylation as even pre-reduced enzyme will not turn over without the presence of the reducing system.16 Thus a cADO-specific auxiliary redox protein that could supply electrons at the right point in the catalytic cycle would be an appealing solution to this problem.
Lastly, there remains a distinct possibility that the deformylating activity of cADO may be an adventitious side reaction, and that the physiological role of the enzyme may involve a more conventional oxidation of an unknown substrate. In this context, the recent observation, albeit at very low activity, of n-1 alcohols and aldehydes derived from the reaction of C9 and C10 aldehydes is certainly intriguing31.
4 Plant and algal decarbonylases
Although alkane biosynthesis was first investigated in plants and green algae, these decarbonylases remain the least well-understood class, in large part because they are integral membrane proteins that are not highly produced and are hard to over-express and purify. Early experiments on crude microsomal preparations of pea leaves, Pisum sativum, and the green algae, Botryococcus braunii, demonstrated that these extracts were capable of converting octadecanal to heptadecane.6,17 A key finding was the demonstration, in experiments utilizing [1-3H,1-14C]octadecanal, that the carbonyl group was released as CO, which could be trapped by RhClPh3.17 The aldehyde hydrogen was, at least partly, transferred to the hydrocarbon as shown by the appearance of tritium in the heptadecane. The reaction required no additional cofactors and was inhibited by metal chelators, suggesting the enzymes were metallo-proteins.
Further purification of the decarbonylase from B. braunii (a technically challenging undertaking) yielded very small quantities of partially purified protein (a few tens of g) but allowed some limited characterization of the algal enzyme.37 The enzyme appeared to be membrane-associated with Mr ~ 66,000 Da, and a rather low specific activity: 3.5 nmol/min/mg. It was suggested on the basis of its UV-Visible spectrum, together with metal analysis, that it might contain a cobalt-porphyrin complex.37 However, in the light of later genetic analyses discussed below, this appears most unlikely to be the case.
Genetic analysis has provided further insights into the plant decarbonylases. In land plants, very-long-chain alkanes are a major component of the waxy surface, known as the eceriferum, that cover the aerial organs and protect the plant against desiccation. Mutations or deletions in the eceriferum (or cer) genes result in a range of phenotypes including reduced hardiness to drought and reduced fertility, and have thus been studied quite extensively.3,38 The aldehyde decarbonylase gene was identified as cer1 through genetic studies in the model organism, A. thaliana.14 The gene encodes a protein of 630 residues that is predicted to be an integral membrane protein and contains the “eight histidine” motif common to stearoyl desaturases and fatty acid hydroxylases.39 These latter enzymes are all integral-membrane, non-heme iron proteins, suggesting that Cer1 is likely also iron-dependent; however this has not been verified experimentally.
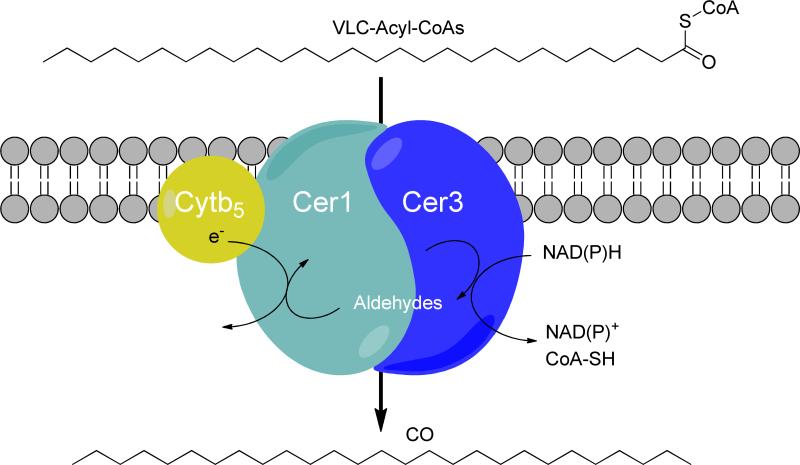
Confirmation that cer1 actually encodes a decarbonylase has only recently been obtained, with the demonstration that heterologous expression of the gene in yeast results in the synthesis of small amounts of very long-chain alkanes, primarily 29 carbons in length.40 However, to achieve this it was also necessary to introduce both the cognate acyl-CoA reductase, encoded by the cer3 gene, and to introduce a mutated version of the yeast fatty acyl-CoA elongase component, Sur4, which allowed production of very long chain fatty acids (C28 and C30) that are not otherwise synthesized by yeast. In vivo protein:protein interaction screening indicated that Cer1 and Cer3 function as a complex in the membrane (Figure 6). The requirement for the mutant elongase component further suggests that this complex is specific for very long chain acyl-CoA esters. Interestingly, the interaction screen also revealed an association between an endoplasmic reticulum-localized cytochrome b5 isoform. The introduction of this cytochrome b5 isoform into yeast enhanced Cer1/Cer3 alkane production, suggesting that cytochrome b5 may be a cofactor for Cer1. Lastly, site-directed mutagenesis of the putative metal-binding histidine residues in Cer1 abolished alkane biosynthesis, demonstrating that they are essential for alkane synthesis.40
Figure 6.
The multi-enzyme complex comprising Cer1/Cer3/Cytb5 that has been proposed to carry out the conversion of very long-chain acyl-CoA esters to alkanes and carbon monoxide in plants. Figure based on reference 38.
The reaction catalyzed by the plant AD enzymes remains the most enigmatic. With so little biochemical data on these enzymes, the mechanism of the reaction is hard to even guess at. Carbon monoxide-producing reactions are extremely rare in biology, and thus it is difficult to draw mechanistic comparisons with other enzymes. The observation that Cer1 is homolgous to stearoyl-CoA desaturase, together with its stimulation by cytochrome b5, hints at the enzyme being iron and O2-dependent, which would provide a common link to the insect and cyanobacterial enzymes. This raises the intriguing possibility that the plant decarbonylases may also employ a cryptic oxidation mechanism analogous to the cyanobacterial enzymes. It is, however, difficult to conceive of an oxygen-involving mechanism that would result in the formation of CO.
The non-enzymatic decarbonylation of aldehydes is catalyzed by a variety of transition metal complexes.41 In particular, the decarbonylation of aldehydes by bi-dentate phosphine-ligated rhodium (I) complexes42-44 has been studied in some detail. The reaction most likely occurs through the oxidative addition of the aldehyde to the metal to form a rhodium-formyl-hydride complex, followed by extrusion of CO, to yield a rhodium-alkyl-hydride complex, and lastly reductive elimination of the alkane.43 The only well-studied CO utilizing enzymes are carbon monoxide dehydrogenase (CODH) and carbon monoxide dehydrogenase–acetyl-CoA synthase (CODH-ACS). These enzymes are extremely oxygen sensitive (unlike aldehyde decarbonylases) and the coenzymes employed in these reactions are quite unlike the di-iron center proposed for Cer1. The enzymes contain unique and complex metal clusters, in which CO is bound to either nickel ions that are part of modified iron-sulfur clusters, or a copper ion that is bound to molybdopterin though a bridging sulfide.45 Whereas it seems highly unlikely that similar cofactors are contained within Cer1, it has been suggested that a di-nuclear nickel-iron center might function instead to catalyze decarbonylation chemistry in a manner analogous to that of the rhodium complex.25
5 Concluding Remarks
The biosynthesis of aliphatic hydrocarbons appears to have evolved several times in Nature, resulting in three different solutions to the problem of converting ubiquitous fatty acids to unfunctionalized alkanes and alkenes. The common mechanistic link between the decarbonylases appears to be iron-mediated oxygenation reactions, although this remains rather speculative in the case of the plant decarbonylases. In each case decarbonylation involves a novel biochemical reaction that represents a new variation on the oxidative chemistry catalyzed by other members of the respective enzyme families.
The difficulties associated with expressing and purifying eukaryotic membrane proteins have hindered the mechanistic analysis of the insect and plant enzymes, although the recent reports of the cloning and recombinant expression of the Drosophila P450 enzyme in sf9 (insect gut) cell culture46 and the Arabidopis enzyme (Cer 1) in yeast40 hold out the prospect of being able to better characterize these enzymes in future. The cyanobacterial enzyme being small, soluble, easily expressed and with the benefit of an x-ray structure, has so far been the subject of the most intense mechanistic interest, although, as discussed above, much remains to be understood about the reaction.
Lastly, we note that several studies have sought to use cADO in metabolic engineering of hydrocarbon biosynthesis pathways.13,23,34,35 So far the titres of alkanes produced have been modest, and it seems likely that this is at least in part due to the low actvity of cADO. The kinetics of the plant and insect enzymes have not been investigated, although the reported specific activity of the enzyme from algal preparations indicates that it is similarly slow.37 Whether the activity of cADO (or the other types of decarbonylases) can be increased by the identification of putative activating and/or reductase components, or through directed evolution and protein engineering remains an interesting challenge.
Acknowledgements
We thank Debasis Das, Ben Buer and Ben Ellington for helpful discussions in the preparation of this manuscript. Research in the authors’ laboratory on enzymes involved in hydrocarbon biosynthesis is supported by grants from the National Science Foundation, CHE 1152055, CBET 1336636, the National Institutes of Health, GM 093088, and the European Union, FP17 256808.
References
- 1.Cheesbrough TM, Kolattukudy PE. J. Biol. Chem. 1988;263:2738–2743. [PubMed] [Google Scholar]
- 2.Howard RW, Blomquist GJ. In Annu. Rev. Entymol. 2005;50:371–393. doi: 10.1146/annurev.ento.50.071803.130359. [DOI] [PubMed] [Google Scholar]
- 3.Bernard A, Joubes J. Prog. Lipid Res. 2013;52:110–129. doi: 10.1016/j.plipres.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Ladygina N, Dedyukhina EG, Vainshtein MB. Process Biochem. 2006;41:1001–1014. [Google Scholar]
- 5.Yoder JA, Denlinger DL, Dennis MW, Kolattukudy PE. Insect Biochem. Mol. Biol. 1992;22:237–243. [Google Scholar]
- 6.Dennis MW, Kolattukudy PE. Arch. Biochem. Biophys. 1991;287:268–275. doi: 10.1016/0003-9861(91)90478-2. [DOI] [PubMed] [Google Scholar]
- 7.Wolf FR. Appl. Biochem. Biotechnol. 1983;8:249–260. [Google Scholar]
- 8.Ghim CM, Kim T, Mitchell RJ, Lee SK. Biotechnol. Bioprocess Eng. 2010;15:11–21. [Google Scholar]
- 9.Buist PH. Nat. Prod. Rep. 2007;24:1110–1127. doi: 10.1039/b508584p. [DOI] [PubMed] [Google Scholar]
- 10.Kolattukudy PE. Lipids. 1970;5:259–275. [Google Scholar]
- 11.Qiu Y, Tittiger C, Wicker-Thomas C, Le Goff G, Young S, Wajnberg E, Fricaux T, Taquet N, Blomquist GJ, Feyereisen R. Proc. Natl. Acad. Sci. USA. 2012;109:14858–14863. doi: 10.1073/pnas.1208650109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed JR, Vanderwel D, Choi SW, Pomonis JG, Reitz RC, Blomquist GJ. Proc. Natl. Acad. Sci. USA. 1994;91:10000–10004. doi: 10.1073/pnas.91.21.10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schirmer A, Rude MA, Li XZ, Popova E, del Cardayre SB. Science. 2010;329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 14.Aarts MGM, Keijzer CJ, Stiekema WJ, Pereira A. Plant Cell. 1995;7:2115–2127. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warui DM, Li N, Norgaard H, Krebs C, Bollinger JM, Booker SJ. J. Am. Chem. Soc. 2011;133:3316–3319. doi: 10.1021/ja111607x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das D, Eser BE, Han J, Sciore A, Marsh ENG. Angew. Chem. 2011;50:7148–7152. doi: 10.1002/anie.201101552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheesbrough TM, Kolattukudy PE. Proc. Natl. Acad. Sci. Biol. 1984;81:6613–6617. doi: 10.1073/pnas.81.21.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mpuru S, Reed JR, Reitz RC, Blomquist GJ. Insect Biochem. Mol. Biol. 1996;26:203–208. [Google Scholar]
- 19.Reed JR, Quilici DR, Blomquist GJ, Reitz RC. Biochemistry. 1995;34:16221–16227. doi: 10.1021/bi00049a038. [DOI] [PubMed] [Google Scholar]
- 20.Meunier B, de Visser SP, Shaik S. Chem. Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 21.Newcomb M, Hollenberg PF, Coon MJ. Arch. Biochem. Biophys. 2003;409:72–79. doi: 10.1016/s0003-9861(02)00445-9. [DOI] [PubMed] [Google Scholar]
- 22.Li N, Chang W-C, Warui DM, Booker SJ, Krebs C, Bollinger JM. Biochemistry. 2012;51:7908–7916. doi: 10.1021/bi300912n. [DOI] [PubMed] [Google Scholar]
- 23.Khara B, Menon N, Levy C, Mansell D, Das D, Marsh ENG, Leys D, Scrutton NS. ChemBioChem. 2013;14:1204–1208. doi: 10.1002/cbic.201300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallar BJ, Lipscomb JD. Chem. Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 25.Krebs C, Bollinger JM, Jr., Booker SJ. Curr. Opin. Chem. Biol. 2011;15:291–303. doi: 10.1016/j.cbpa.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Muller J, Lippard SJ. Angew. Chem. Int. Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Norgaard H, Warui DM, Booker SJ, Krebs C, Bollinger JM. J. Am. Chem. Soc. 2011;133:7148–7152. doi: 10.1021/ja111607x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandelia ME, Li N, Norgaard H, Warui DM, Rajakovich LJ, Chang WC, Booker SJ, Krebs C, Bollinger JM. J. Am. Chem. Soc. ASAP; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul B, Das D, Ellington B, Marsh ENG. J. Am. Chem. Soc. 2013;135:5234–5237. doi: 10.1021/ja3115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb M, Toy PH. Acc. Chem. Res. 2000;33:449–455. doi: 10.1021/ar960058b. [DOI] [PubMed] [Google Scholar]
- 31.Aukema KG, Makris TM, Stoian SA, Richman JE, Münck E, Lipscomb JD, Wackett LP. ACS Catal. ASAP; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eser BE, Das D, Han J, Jones PR, Marsh ENG. Biochemistry. 2011;50:10743–10750. doi: 10.1021/bi2012417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andre C, Kim SW, Yu X-H, Shanklin J. Proc. Natl. Acad. Sci. USA. 2013;110:3191–3196. doi: 10.1073/pnas.1218769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akhtar MK, Turner NJ, Jones PR. Proc. Natl. Acad. Sci. USA. 2013;110:87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howard TP, Middelhaufe S, Moore K, Edner C, Kolak DM, Taylor GN, Parker DA, Lee R, Smirnoff N, Aves SJ, Love J. Proc. Natl. Acad. Sci. USA. 2013;110:7636–7641. doi: 10.1073/pnas.1215966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SJ, McCormick MS, Lippard SJ, Cho US. Nature. 2013;494:380–384. doi: 10.1038/nature11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis M, Kolattukudy PE. Proc. Natl. Acad. Sci. USA. 1992;89:5306–5310. doi: 10.1073/pnas.89.12.5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunst L, Samuels AL. Prog. Lipid Res. 2003;42:51–80. doi: 10.1016/s0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 39.Shanklin J, Whittle E, Fox BG. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 40.Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure J-D, Haslam RP, Napier JA, Lessire R, Joubes J. Plant Cell. 2012;24:3106–3118. doi: 10.1105/tpc.112.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modak A, Deb A, Patra T, Rana S, Maity S, Maiti D. Chem. Commun. 2012;48:4253–4255. doi: 10.1039/c2cc31144e. [DOI] [PubMed] [Google Scholar]
- 42.Doughty DH, Pignolet LH. J. Am. Chem. Soc. 1978:100. [Google Scholar]
- 43.Fristrup P, Kreis M, Palmelund A, Norrby P-O, Madsen R. J. Am. Chem. Soc. 2008;130:5206–5215. doi: 10.1021/ja710270j. [DOI] [PubMed] [Google Scholar]
- 44.Patra T, Manna S, Maiti D. Angew. Chem. Int. Ed. 2011;50:12140–12142. doi: 10.1002/anie.201103860. [DOI] [PubMed] [Google Scholar]
- 45.Ragsdale SW. Chem. Rev. 2006;106:2217–3337. doi: 10.1021/cr0503153. [DOI] [PubMed] [Google Scholar]
- 46.Qui Y, Tittiger C, Wicker-Thomas C, Le Goff G, Young S, Wajnberg E, Fricaux T, Taquet N, Blomquist GJ, Feyereisen R. Proc. Natl. Acad. Sci. (USA) 2012;109:14858–14863. doi: 10.1073/pnas.1208650109. [DOI] [PMC free article] [PubMed] [Google Scholar]