Abstract
When cells encounter substantial DNA damage, critical cell cycle events are halted while DNA repair mechanisms are activated to restore genome integrity. Genomic integrity also depends on proper assembly and function of the bipolar mitotic spindle, which is required for equal chromosome segregation. Failure to execute either of these processes leads to genomic instability, aging, and cancer. Here, we show that following DNA damage in the breast cancer cell line MCF-7, the centrosome protein centrin2 moves from the cytoplasm and accumulates in the nucleus in a xeroderma pigmentosum complementation group C protein (XPC)–dependent manner, reducing the available cytoplasmic pool of this key centriole protein and preventing centrosome amplification. MDA-MB 231 cells do not express XPC and fail to move centrin into the nucleus following DNA damage. Reintroduction of XPC expression in MDA-MB 231 cells rescues nuclear centrin2 sequestration and reestablishes control against centrosome amplification, regardless of mutant p53 status. Importantly, the capacity to repair DNA damage was also dependent on the availability of centrin2 in the nucleus. These observations show that centrin and XPC cooperate in a reciprocal mechanism to coordinate centrosome homeostasis and DNA repair and suggest that this process may provide a tractable target to develop treatments to slow progression of cancer and aging.
Introduction
Each day, endogenous and environmental assaults generate >104 DNA lesions in any given cell, which are corrected by DNA repair pathways responsible for maintenance of DNA integrity (1). Global genome-nucleotide excision repair (GG-NER) is a generic pathway that repairs bulky DNA lesions such as UV-induced thymidine dimers or cisplatin-DNA adducts (2, 3). DNA damage recognition and GG-NER are initiated by the xeroderma pigmentosum complementation group C protein (XPC), which functions as a complex with hRad23B and centrin2 to recognize DNA helix distortions (4–6). The XPC-centrin2 interaction is mediated by binding of the COOH-terminal domain of centrin2 to the W1L4L8 motif of XPC (6–8). Recruitment of the XPC/hRad23B/centrin2 complex is a rate-limiting step in GG-NER and is dependent on XPC abundance and stability (9). On binding to DNA, the XPC/hRad23B/centrin2 complex recruits the DNA helicases XPD and XPB (subunits of TFIIH), which unwind the DNA to allow downstream repair proteins of the pathway to localize to the lesion (9–11). Autosomal recessive defects in this DNA repair process result in xeroderma pigmentosum in which patients carrying mutations in key GG-NER components, including XPC, develop severe UV sensitivity, trichothiodystrophy, neural defects, and an elevated risk of cancer (12, 13).
In addition to repair of DNA lesions, maintenance of genomic integrity is also dependent on exquisite regulation of equal chromosomal segregation during cell division. To form a bipolar microtubule-based mitotic spindle, the centrosome duplicates once in each cell cycle to give rise to the two mitotic spindle poles (14). In many cancers, disruption of this process leads to centrosome amplification characterized by multiple centrosomes, increased accumulation of pericentriolar material (PCM), and/or supernumerary centrioles, which ultimately leads to chromosomal instability and aneuploidy (15, 16). Centrin is a 20-kDa cytosolic calcium-binding regulatory protein required for centrosome homeostasis (17). Humans and mice have four centrin genes. Cetn1 is expressed in male germ cells, certain neurons, and terminally differentiated ciliated cells; Cetn2 and Cetn3 are expressed in all somatic cells; and Cetn4 is expressed in terminally differentiated ciliated cells (18–21). In mammalian cells, centrin2 is distributed diffusely in the cytoplasm with a prominent centriole localization and also sporadically in the nucleus (22, 23). Centrin2 binds diverse centrosome proteins, but the direct binding of centrin2 to the DNA repair protein XPC (5, 6, 8) and the variable nuclear localization of centrin2 lead us to investigate the cellular consequences of the interaction between centrin2 and XPC in more detail.
The studies reported here compare the responses of two different human breast cancer cell lines to DNA damage. One of theses cell lines, MCF-7, represents an early-stage breast cancer phenotype that has retained p53-mediated cell cycle checkpoint controls, whereas the other cell line, MDA-MB 231, represents an advanced breast cancer phenotype with aggressive metastatic properties, loss of p53-mediated checkpoint control, and a high degree of genomic instability (24–26). In addition, we compare the behavior of two primary human fibroblast cell types, one from a normal individual and the second from an individual affected with xeroderma pigmentosum homozygous for an A>C transversion at –2 in intron 5.1 of the XPC gene (IVS5.1-2A>C) resulting in an 83-bp insertion of intron 5.1 with a stop 34 codons downstream (27). DNA damage sufficient to cause cell cycle arrest (e.g., replication fork stalling following nucleotide depletion by hydroxyurea treatment) and damage due to UV-induced thymidine dimer formation share physical and functional cross talk between DNA damage checkpoint controls and proteins involved in the DNA repair pathways (28). Distinctive features of the cellular response to these different DNA damaging agents highlight exceptional aspects of the XPC/centrin2 interaction. We found that in MCF-7 cells following DNA damage, upregulation of the NER protein XPC results in an XPC-dependent wholesale sequestration of the normally cytosolic protein centrin2 in the nucleus. On the other hand, MDA-MB 231 cells are deficient in XPC expression, they fail to sequester centrin2 in the nucleus, and these cells develop centrosome amplification following DNA damage. Reintroduction of XPC expression in MDA-MB 231 cells and fibroblasts null for XPC rescues nuclear centrin2 sequestration and reestablishes control of centriole duplication. Remarkably, we also found that the rate and the extent of repair of UV-induced DNA damage are diminished in cells where centrin2 is depleted by shRNA, implicating a functional role for centrin2 in the NER process itself.
Materials and Methods
Cell lines
Human breast cancer cell lines MDA-MB 231, MCF-7 (American Type Culture Collection) and primary human fibroblasts GM03377 (normal) and AG10032 (XPC null; Coriell Cell Repositories) were cultured at 37°C, 5% CO2 in EMEM containing 15% fetal bovine serum, 5 mmol/L glutamine, and 1% penicillin/streptomycin. Cultures were resuscitated from stocks frozen at low passage within 6 mo of purchase. 293FT cells (Invitrogen) used for the recombinant lentivirus production were grown in DMEM (Life Technologies, Inc.) for transformed cells and in MEM (Life Technologies) for primary and MDA-MB 231 cells, supplemented with heat-inactivated 10% FCS (Life Technologies) and antibiotics (as above).
Immunofluorescence microscopy
Cells were grown on glass coverslips or glass-bottomed 12-well plates (P12G1.014F, Mattek Corporation) for 48 h followed by specific treatment. Cells were washed twice with PBS, fixed 10 min in –20°C cold methanol, rehydrated for 5 min, and then treated with 0.1% Triton X-100 for 4 min and blocked for 1 h at room temperature in blocking buffer (5% goat serum, 1% glycerol, 0.1% bovine serum albumin, 0.1 fish skin gelatin, 0.04% sodium azide). The cells were incubated with primary antibodies for 60 min at room temperature, washed four times, followed by secondary antibody conjugated to Alexa 488 or Alexa 568 fluorescent dyes (Molecular Probes) at 1:800 dilution. Coverslips were mounted on a microscope slide using antifade containing 4′,6-diamidino-2-phenylindole (DAPI) from Molecular Probes. Microscopy was done with a Zeiss Axiovert 200M microscope, using a 63×/1.4-numerical-aperture Plan-Apo objective, high-resolution Aixiocan digital camera, and AxioVision software. Final figures were adjusted for brightness and color balance to include the full threshold range in Photoshop (CS3 v10) without changing nonlinear (gamma) setting. For DNA damage and repair fluorescence-based assays, cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min, then permeabilized with 0.5% Triton X-100 for 10 min. To denature the DNA, cells were incubated for 30 min in 2 mol/L HCl. Following five washes with PBS, the procedure was identical to that stated above.
Antibodies
Antibodies developed in our laboratory include anti-centrin monoclonal 20H5, which recognizes centrin1, centrin2, and centrin3; monoclonal Cetn-2.4, which is specific for centrin2 (17, 29); and rabbit anti-XPC rabbit IgG MC12183. Antibodies obtained commercially include anti-XPC monoclonal (MS-XPC-26-PX1, GeneTex, Inc.); anti-XPA monoclonal (NeoMarkers); phospho-Ser139-γ-H2AX (Cell Signaling); monoclonal anti–(6-4) DNA photoproduct (clone 64M-2, MBL International); hRAD23 monoclonal (LCG Bioscience); anti–Cu/ZN superoxide dismutase (SOD) rabbit IgG (SOD-100, Stressgen); lamin B1 rabbit IgG (Santa Cruz Biotech); γ-tubulin monoclonal GTU88 rabbit IgG T5192, pericentrin, and β-actin monoclonal AC74 (Sigma-Aldrich).
Western blot analysis
Whole-cell lysates and cell fractions were analyzed by SDS-PAGE and Western blot. Total proteins (10 μg) for whole-cell lysates and for cell fractionation experiment preparations from the equivalent of 20,000 cells were loaded in each lane. Protein was subsequently transferred onto polyvinylidene difluoride membrane (Amersham Pharmacia Biotech), fixed in 0.25% glutaraldehyde, blocked in 5% nonfat dry milk, probed with the specific antibodies indicated in the figure legends, followed by horseradish peroxidase–conjugated secondary antibody, using chemiluminescence (ECL Plus kit from Amersham), and captured with the UVP AutoChemi System. Band densitometry was determined using the NIH ImageJ 1.36b software.
Plasmid construction: expression and knockdown vectors
Knockdown constructs were either based on a plasmid-based system H1 promoter (pCMS4) or a lentivirus system PLL3.7 (U6 promoter). For XPC and XPA expression, we used vectors and methods as described (30). Full-length cDNA sequences encoding HA-XPA and flag-XPC were tagged at the NH2 terminus. Centrin and flag-XPC expression were done using the PSCMV Lentivirus system. To knock down centrin2, we used constructs based on a derivative of pCDNA3-delCMVp plasmid designed with the help of Dr. Billadeau (Mayo Clinic). For shRNA-mediated ablation of centrin2, MCF-7 or human fibroblast cells were transfected with plasmid targeting the 3′ untranslated region of human centrin-2 (AGCTTTGAGCACCTGCCAT and GCAGTCATTCTTGACGGCT). The plasmids were transfected using FuGene6 (Roche) transfection reagent at 3:1 ratio as described in the manufacturer's technical sheet.
Cell cycle and viability analysis by flow cytometry
Cells were trypsinized, washed with cold PBS, fixed in 95% ethanol for 24 h, and stained with propidium iodide for 30 min. For viability assay, unfixed cells were stained with propidium iodide for 5 min. Twenty thousand fluorescence-activated cell sorting events were captured using a FACScan (Becton Dickinson) and analyzed with ModFit (Verity Software House).
DNA damage
Cells were cultured for 48 h before the treatment. Medium was changed daily and 2 mmol/L hydroxyurea or 25 μmol/L cisplatin (Sigma) was added. For UV treatment, cells were washed twice with PBS and excess medium was removed; cells were irradiated with a Mineralight Lamp (model UVGL-25, UVP) emitting predominantly at 254 nm (UVC) at 20 J/m2 delivered at a rate of 0.1 J/m2 for 200 s. Fresh culture medium was added and the cells were incubated under standard culture conditions until harvested or fixed for microscopy.
Results
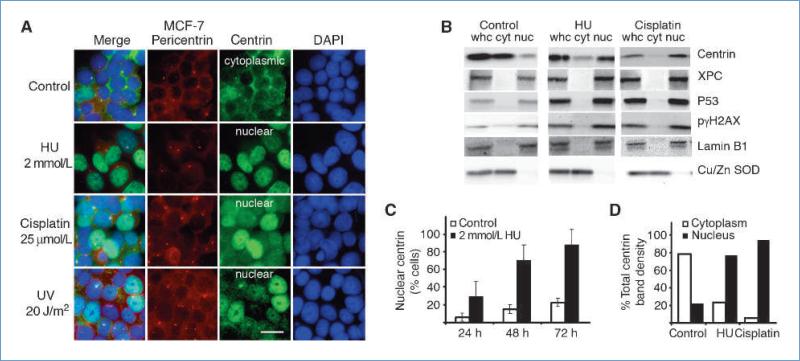
In the absence of exogenous DNA damage, 80% to 95% of control cells showed predominant cytoplasmic centrin2 localization (Fig. 1A). Following DNA damage in MCF-7 cells, centrin2 underwent a striking redistribution where up to 80% of cells showed distinct time-dependent redistribution of centrin2 from the cytoplasm into the nucleus (Fig. 1A and C). Redistribution of centrin2 into the nucleus was observed following treatment with a variety of DNA-damaging agents, including UV irradiation and the genotoxic chemotherapeutic agents hydroxyurea and cisplatin (Fig. 1A). Centrin2 redistribution into the nucleus following DNA damage was also observed in normal human primary fibroblasts (Fig. 2C–E) but not in the human breast cancer cell line MDA-MB 231 (Fig. 3A). Cell fractionation and densitometry of Western band intensity revealed that nuclear centrin2 increased up to 4-fold and cytoplasmic centrin2 was reduced to nominal levels following DNA damage in bulk preparations of MCF-7 (Fig. 1B and D). Validation of the enrichment of cellular fractions in these experiments was established by phase-contrast microscopy (Supplementary Fig. S1) and by the differential partition of the nuclear envelope protein lamin B from the nuclear fraction and of Cu/Zn SOD from the cytosol (Fig. 1B). Because β-actin partitions into both nuclear and cytoplasmic compartments, lamin B and Cu/Zn SOD also served as loading controls for Western blots in these experiments. Cellular response to DNA damage was confirmed by an increase in nuclear levels of XPC, p53, and phospho-γ-histone-2AX (γH2AX; Fig. 1B) and by S-phase arrest (Supplementary Fig. S2). Redistribution into the nucleus was unique for the centrosome protein centrin2 because other centrosome proteins such as γ-tubulin (not shown) and pericentrin remained at the centrosome following DNA damage (Fig. 1A and see later discussion).
Figure 1.
Nuclear sequestration of centrin following DNA damage. A, control MCF-7 cells show centrin largely confined to the cytoplasm, whereas centrin sequestration into the nucleus was seen following DNA damage using 2 mmol/L hydroxyurea (HU) for 48 h, 25 μmol/L cisplatin for 24 h, or 20 J/m2 UV. B, Western blot analysis of whole cells (whc) and cytoplasmic (cyto) and nuclear (nuc) fractions before (Control) and after hydroxyurea- or cisplatin-induced DNA damage. Centrin, XPC, p53, and γH2AX show substantial increase in the nucleus following DNA damage. Lamin B1 and Cu/Zn SOD were used as nuclear and cytoplasmic markers, respectively. C, time course for centrin redistribution into MCF-7 cell nuclei following hydroxyurea treatment. D, densitometry of Western bands of whole-cell lysates and nuclear and cytoplasmic fractions shows that the amount of nuclear centrin increased from ~20% of the total in control cells to ≥80% following DNA damage. Columns, mean from at least three independent experiments (n = 200); bars, SEM.
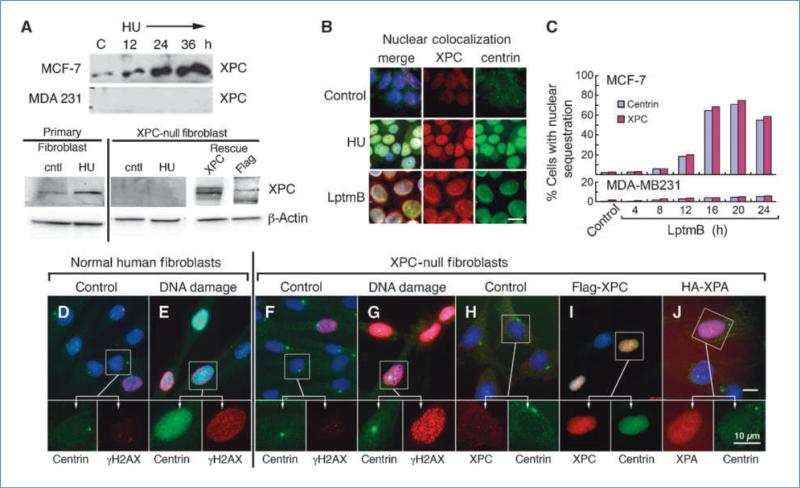
Figure 2.
Nuclear sequestration of centrin is XPC dependent. A, Western blot analysis of XPC expression in MCF-7 and MDA-MB 231 cells before and after treatment with 2 mmol/L hydroxyurea over a 36-h time course and in normal primary human fibroblasts and XPC-null human fibroblasts before and after 48-h hydroxyurea treatment and following rescue with flag-XPC. Centrin and XPC colocalize in MCF-7 nuclei following DNA damage (hydroxyurea, 48 h) or treatment with leptomycin B (LptmB; 12 h). B, nuclear colocalization of centrin and XPC following DNA damage and leptomycin B treatment. C, time course analysis for accumulation of centrin and XPC in nuclei of MCF-7 and MDA-MB 231 cells following treatment with the nuclear export inhibitor leptomycin B. Primary human fibroblasts (D and E) and XPC-null human fibroblasts (F and G) labeled for centrin (green) and γH2AX (red) before and after hydroxyurea-induced DNA damage. Note that in inset for G the centrosome lies over the nucleus. H to J, control XPC-null human fibroblasts (H) and cells transduced with lentivirus flag-XPC or HA-XPA (I and J, respectively) and labeled for centrin (green) and XPC and XPA (I and J, red). Insets in top images are enlarged below. Bar, 10 μm.
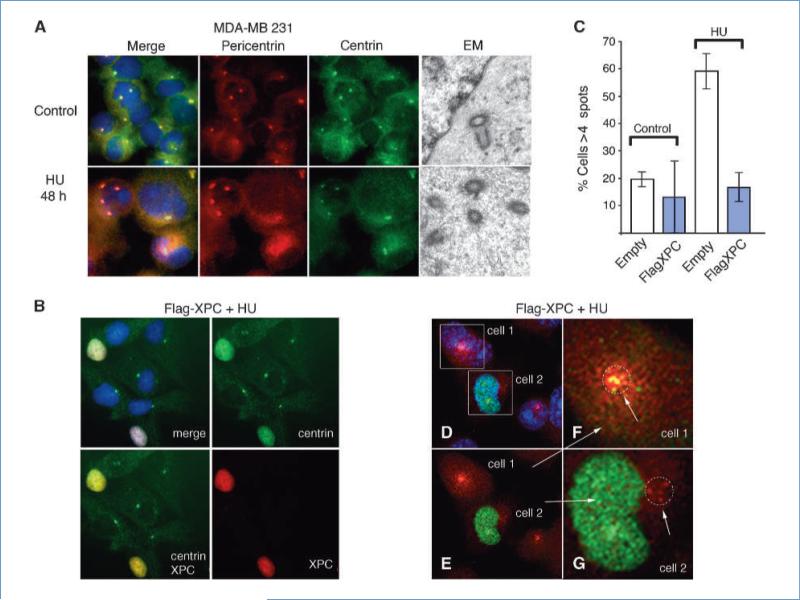
Figure 3.
Centrin nuclear sequestration prevents DNA damage–induced centriole overduplication and centrosome amplification. A, MDA-MB 231 cells fail to sequester centrin in the nucleus following DNA damage. Fluorescence microscopy of cells stained for centrin (green), pericentrin (red), and DNA (blue) before (control) and after DNA damage (2 mmol/L hydroxyurea treatment for 48 h) and electron microscopy showing a pair of centrioles before and multiple centrioles after hydroxyurea treatment. B, MDA-MB 231 cells stained for centrin (green), XPC (red), and DNA (blue) following hydroxyurea treatment and rescue of XPC expression using flag-XPC. Note that only those cells that express flag-XPC also show translocation of centrin into the nucleus. C, analysis of cells with more than four centrin-staining spots before and after hydroxyurea treatment and with control vector (open column) or recombinant flag-XPC expression (blue column). Columns, mean from at least three independent experiments; bars, SEM. D to G, adjacent nontransduced (cell 1) and flag-XPC lentiviral–transduced (cell 2) cells stained for centrin (green), γ-tubulin (red), and DNA (blue). E, the same image as in D but with the DAPI channel removed. F and G, enlarged image of nontransduced (cell 1) and flag-XPC–transduced (cell 2) cells. The centrosome region of each cell (1 and 2) is indicated by the red spots (pericentrin), arrows and circle.
We next determined whether XPC plays a direct role in the nuclear sequestration of centrin2. To do this, we used four cell lines, two of which express XPC and the other two do not. Before DNA damage, MCF-7 cells and normal primary human fibroblasts express low but detectable levels of XPC (Fig. 2A). Following DNA damage by hydroxyurea treatment, XPC abundance increased appreciably over 36 hours in MCF-7 cells (Fig. 2A). Importantly, double-label immunofluorescence microscopy confirmed an increase in abundance of XPC and centrin2 and revealed their colocalization in the nucleus following DNA damage (Fig. 2B, HU). In contrast, the breast cancer cell line MDA-MB 231 and XPC-null human fibroblasts derived from a xeroderma pigmentosum patient showed no detectable XPC expression before or following hydroxyurea treatment (Fig. 2A and H).
To test whether XPC was required for centrin2 redistribution into the nucleus, we next compared centrin2 localization in normal fibroblasts and XPC-null fibroblasts before and after DNA damage (Fig. 2D–G). Before DNA damage, both cell types showed cytoplasmic centrin2 localization with predominant centrosome staining (Fig. 2D and F). Control cells also showed low-level staining with antibodies specific for γH2AX, indicating nominal chromatin remodeling characteristic of basal DNA repair activity. Importantly, following DNA damage, all of the normal fibroblasts that showed increased nuclear γH2AX staining (~87%, N = 200) also showed dramatic sequestration of centrin2 into the nucleus (Fig. 2E). In contrast, none of the XPC-null fibroblasts that showed γH2AX nuclear staining showed increased nuclear centrin2 (0 of 200), even after 72 hours of hydroxyurea treatment (Fig. 2G). Western blot analysis showed an increase in XPC (~2.2-fold) in normal fibroblasts following DNA damage, whereas XPC-null cells showed no detectable XPC (Fig. 2A). In contrast, when XPC-null fibroblasts were rescued with a lentiviral expression construct of flag-tagged XPC (Fig. 2A), fluorescence microscopy showed nuclear sequestration of centrin2 only in XPC-null cells rescued by expression of recombinant flag-XPC (Fig. 2I). The rescued cells also showed greatly diminished centrin2 localization at the centrosome; this observation is addressed more thoroughly in experiments below (Fig. 3). Importantly, XPC overexpression alone was sufficient for nuclear sequestration of centrin2 even in the absence of DNA damage. To test if other downstream GG-NER proteins could cause centrin2 sequestration into the nucleus, we expressed HA-XPA in the XPC-null cell line (Fig. 2J). All of the XPA-expressing cells showed centrin2 at the centrosome and none showed nuclear sequestration of centrin2 (0 of 200). To further investigate the mechanism for nuclear sequestration, we first analyzed centrin2 and XPC protein sequences for predicted nuclear localization import (NLS) and export (NES) signals (31, 32). Whereas the XPC sequence had readily identifiable NLS and NES motifs, the centrin2 sequence was devoid of any obvious nuclear transport sequences (Supplementary Fig. S3). We then treated cells with a potent inhibitor of CRM1-mediated nuclear export, leptomycin B (33), and monitored nuclear localization of centrin2 and XPC (Fig. 2B and C). Centrin2 and XPC accumulated in the nucleus with similar kinetics over a 24-hour time course in leptomycin B–treated MCF-7 cells, whereas MDA-MB 231 cells failed to sequester centrin2 in the nucleus. Thus, cells that fail to make XPC also fail to retain centrin2 in the nucleus. Taken together, the experiments shown above show that following DNA damage, nuclear sequestration of centrin2 is XPC dependent. These observations led us to further investigate the functional cellular consequences of centrin2 sequestration into the nucleus.
Next, we tested whether XPC-dependent nuclear sequestration of centrin2 was sufficient to prevent inappropriate centrosome amplification following DNA damage. We chose the human breast cancer cell line MDA-MB 231 because we previously showed that these cells develop striking centrosome amplification characterized by supernumerary centrioles and increased PCM volume following DNA damage (34). MDA-MB 231 cells lack detectable XPC expression and also fail to sequester centrin2 into the nucleus following DNA damage (Figs. 2A and 3A). Furthermore, as for XPC-null fibroblasts, expression of recombinant flag-XPC rescued the ability of MDA-MB 231 cells to sequester centrin2 into the nucleus (Fig. 3B–G). First, we confirmed that DNA damage leads to centrosome amplification in MDA-MB 231 by counting the number of centrin2-stained spots, by assessing PCM volume by pericentrin staining, and by electron microscopy (Fig. 3A and C). Cells with amplified centrosomes characterized by more than four centrin-staining spots increased from 20% in controls to ~60% following DNA damage by hydroxyurea treatment (Fig. 3C). This increase in centrin-staining spots corresponded to an increase in centriole number as determined by the presence of multiple centrioles observed in electron microscopy of thin-sectioned cells (Fig. 3A). We found that cells rescued using recombinant flag-XPC regained the ability to move centrin2 into the nucleus following hydroxyurea treatment (Fig. 3B and D–G). Centrin2 translocation into the nucleus is seen only in cells that express flag-XPC and not in untransfected cells (Fig. 3B). This image shows a transient transfection of MDA-MB 231 with a plasmid expression construct for flag-XPC and immunofluorescence for centrin (green) and XPC (red). All cells that express XPC also show nuclear centrin. All untransfected cells, those that fail to express XPC, show cytoplasmic centrin. Therefore, when using MDA-MB 231, following transient transfection with flag-XPC, any cell that shows nuclear centrin is a transfected cell.
Remarkably, the flag-XPC transfected cells also showed a dramatic reduction of pericentrin staining and a near-complete loss of centrin staining at the centrosome following DNA damage (Fig. 3B and C–G). This was clearly evident when comparing cells in low-magnification fields (Fig. 3B) and when adjacent recombinant XPC-transduced and non-transduced cells were examined closely at higher magnification (Fig. 3D–G). Those cells that showed significant centrin2 accumulation in the nucleus also showed nominal pericentrin and centrin2 staining (Fig. 3D and E–G, cell #2). In contrast, those cells that failed to sequester centrin2 into the nucleus showed centrosome amplification (more than four centrin spots and pervasive pericentrin staining; Fig. 3D and E–F, cell #1). These observations show that centrosome amplification is effectively prevented in cells that show XPC-dependent sequestration of centrin2 into the nucleus and that expression of XPC alone is sufficient to arrest centrosome amplification in cells that have experienced DNA damage sufficient to cause cell cycle arrest.
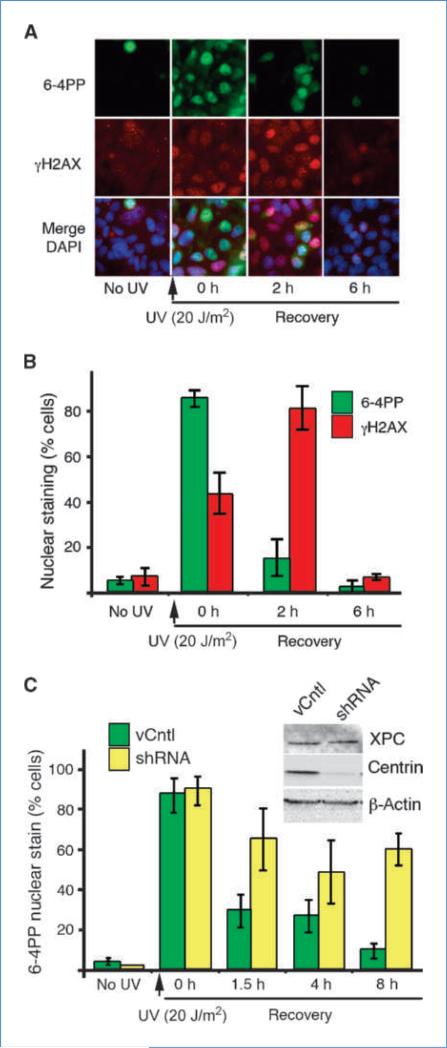
We then assessed whether or not centrin2 plays a role in the process of DNA repair. To do this, we established conditions for monitoring UV-induced DNA damage and repair in MCF-7 cells by following individual cells stained for thymidine dimer 6-4 photoproducts (6-4PP) and γH2AX. MCF-7 cells showed a time-dependent recovery following UV exposure (Fig. 4A and B). Both gain and loss of γH2AX staining were delayed relative to that of 6-4PP, indicating that chromatin remodeling occurred after UV damage and was completed following DNA repair (Fig. 4B). We then knocked down centrin2 using a plasmid-based shRNA construct (Fig. 4C, inset) and monitored UV-induced DNA damage and repair using these same assays. DNA repair proceeded normally in control cells where recovery to near basal level of DNA damage was seen by 8 hours after UV exposure. Conversely, in cells where centrin2 had been depleted by shRNA, both the rate and the extent of DNA repair were significantly reduced. By 8 hours after UV treatment, ~90 of control cells had lost 6-4PP staining while only ~40% of the cells treated with centrin-directed shRNA had recovered by this time (Fig. 4C). Because detection of thymidine dimer 6-4PP required acid treatment of the specimen for epitope accessibility and this treatment eliminated centrin staining in controls, we were unable to colabel for 6-4PP and centrin in the same cells. Nonetheless, we note that the depressed level of DNA repair roughly paralleled the level of centrin2 knockdown, whereas XPC levels remained unchanged (Fig. 4C). Because centrin2 knockdown alone resulted in loss of cell viability by 72 hours after transfection (17), survival curves following UV treatment +/– centrin2 knockdown were not informative. Taken together with earlier in vitro assays by others (5, 35), these results suggest that centrin2 plays a functional role in the control of GG-NER through its interaction with XPC. Because centrin2 does not directly bind DNA, its effect on GG-NER is likely to be either through stabilization of XPC, preventing its degradation (36), or as an allosteric regulator of XPC (5, 6, 37).
Figure 4.
Centrin knockdown leads to impaired DNA repair. A, DNA damage and repair kinetics in MCF-7 cells as determined by immunofluorescence for 6-4PP (green; top row), γH2AX (red; middle row), and images merged with Hoechst-stained DNA (blue; bottom row). Cells were exposed to 20 J/m2 UVC and allowed to recover for up to 6 h. B, graph showing repair kinetics (percent cells with distinct nuclear staining versus time). Green columns, 6-4PP; red columns, pγH2AX; bars, SD. C, kinetics of DNA repair (6-4PP removal) with (yellow columns) and without (green columns) shRNA knockdown of centrin2. Inset, Western blot showing centrin knockdown and XPC level. Columns, mean from at least three independent experiments; bars, SEM.
Discussion
Although evidence for a role of XPC and centrin2 in DNA repair has been recognized for some time now (5–8, 23, 35, 36), a potential role for the complex in coordination of DNA repair and centrosome dynamics has not been addressed experimentally. Here, we present evidence for a mechanism involving the XPC-dependent nuclear sequestration of centrin not only in the function of the GG-NER pathway but also to coordinate GG-NER with centrosome homeostasis. Our studies also show that the human breast cancer cell line MCF-7 shows an operational coordination of DNA repair and centrosome behavior, whereas MDA-MB 231 cells are defective in this process.
GG-NER and centrosome dynamics are linked through wide-ranging cell cycle and DNA-damage checkpoint controls involving the tumor suppressor p53, whose abundance is itself controlled in response to DNA damage. Efficient DNA damage–induced transcriptional control of XPC gene expression operates through a p53 response element 5′ of the XPC coding sequence (ref. 38; for conflicting observations, see also ref. 39). Conversely, XPC may also modulate p53 function because XPC-null cells show reduced p53-mediated response to cisplatin-induced DNA adducts, including reduction in expression of the cell cycle cyclin-dependent kinase (cdk) inhibitor p21 and decrease in activation of caspase-3 and apoptosis (40, 41). XPC abundance is also determined through control of XPC degradation by ubiquitin-mediated proteasome degradation (9, 36, 42, 43). Importantly, p53 has been implicated in the control of centrosome homeostasis involving distinct roles for cdk2, microtubules, dynein, Hsp90, and transcriptional activation of p21 (26, 34, 44–46). Several recent studies implicate a role for centrin in XPC stability and nuclear transport. Saccharomyces cerevisiae expressing a Cdc31 (yeast centrin) mutant that forms a weak interaction with Rad4 (yeast XPC) showed greater sensitivity to UV radiation and conferred proteolytic defects on XPC degradation (36). Similarly, a role for Cdc31 in protein degradation was also implicated by its interaction with Dsk2 (47), a protein that functions in the ubiquitin/proteasome pathway (48–50). Klein and Nigg (23) recently identified a small pool of SUMO-modified centrin2 and they showed that interference with the SUMOylation machinery leads to exclusion of centrin from the nucleus. Studies by Prosser and coworkers (46) also suggest that, in p53-deficient cells, nuclear export of centrin is required for centriolar satellite formation and centrosome overduplication. In this regard, genetic studies in yeast and structural studies in Xenopus and mammalian cells have implicated a role for centrin in transport across the nuclear pore itself (45, 51, 52). Taken together, with in vitro studies of Araki and coworkers (5), these observations suggest that p53, centrin2, and SUMOylation may function to augment GG-NER through regulation of XPC expression, nuclear transport, and stability and may cooperate in the control of cell cycle progression, centrosome homeostasis, and GG-NER following DNA damage.
Since the early days of cancer cell biology, there has been speculation that some form of signaling activity functions to coordinate the behavior of the cell genome and the cell center or centrosome. Disregulation of this proposed pathway could account for the genomic instability common to most cancers (16). Here, we show that following DNA damage, centrin2 moves from the cytoplasm and accumulates in the nucleus in an XPC-dependent manner in MCF-7 cells. Nuclear sequestration of centrin2 reduced the available cytoplasmic pool of this key centriole protein, thereby preventing centrosome amplification. Conversely, in XPC-deficient MDA-MB 231 and XPC-null (XPC–/–) fibroblast cells, centrin2 did not accumulate in the nucleus, and centrosome amplification occurred following DNA damage. Importantly, rescue of XPC-deficient cells by recombinant flag-XPC expression alone restored control, preventing centrosome amplification following DNA damage in primary XPC-null (XPC–/–) human fibroblasts as well as in MDA-MB 231 cells that have abrogated p53. Finally, we found that the capacity to repair UV-induced DNA damage in vivo was dependent on the avail ability of centrin2 in the nucleus. Taken together, our observations suggest that cellular response to severe DNA damage includes the integration of DNA damage repair and centro-some homeostasis involving the proteins centrin2 and XPC, and that the efficiency of this response differs among established breast tumor cell lines. Further, our observations suggest a direct mechanistic role for centrin and XPC in coordinating GG-NER and centrosome behavior that operates apart from that of p53 and provides a direct mechanistic link coordinating these processes to ensure genomic integrity at the nucleotide level through GG-NER and at the chromosomal level through control of centrosome number. A scenario for co-ordination of centrosome homeostasis and genomic integrity through centrin and XPC is presented in the diagram in Fig. 5. Our observations raise the intriguing possibility that centrin2 may operate more generally to coordinate cell cycle progression with other DNA damage repair pathways such as those induced by γ-radiation or oxidative stress (42, 53). Finally, the reciprocal link between centrosome and genomic integrity through the interaction of centrin2 and XPC provides a fresh rationale to augment this process and to develop treatments to slow progression of cancer and aging, diseases that involve both DNA damage and chromosomal instability.
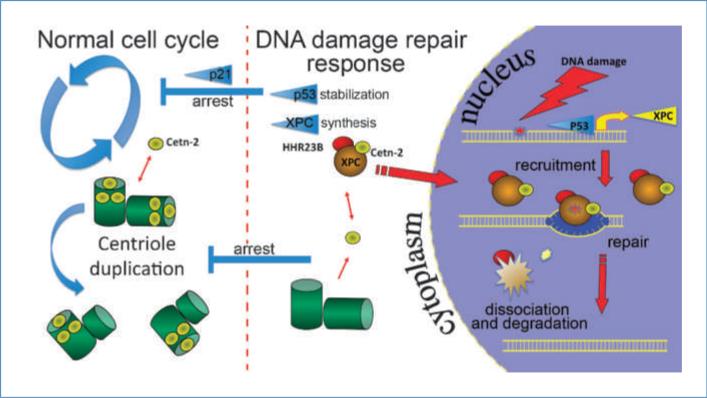
Figure 5.
Scheme for coordination of genomic integrity and centrosome homeostasis through centrin and XPC. Normal cell cycle progression and centrosome duplication are interrupted on excess DNA damage by the stabilization of p53 and its DNA damage response. Subsequent elevation of XPC expression, XPC binding to centrin2, and their nuclear sequestration lead to DNA damage repair and arrest of the centrosome duplication cycle due to depletion of cytoplasmic centrin. Other cell cycle events are simultaneously halted through p53-mediated elevation of the cyclin inhibitors such as p21. In the absence of XPC expression, centrosome homeostasis is disrupted.
Acknowledgments
We thank members of the laboratory and Drs. D. Billadeau, D. Tindall, E. Trushin, and J. van Deursen for comments on the manuscript.
Grant Support
NIH grant CA72836 (J.L. Salisbury), Mayo Clinic Breast Cancer Specialized Program of Research Excellence NIH grant CA116201, and the Mayo Foundation.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
I.D. Acu, T. Liu, and K. Suino-Powell contributed equally to this work.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 3.Cline SD, Hanawalt PC. Who's on first in the cellular response to DNA damage? Nat Rev Mol Cell Biol. 2003;4:361–72. doi: 10.1038/nrm1101. [DOI] [PubMed] [Google Scholar]
- 4.Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. 2000;300:275–90. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 5.Araki M, Masutani C, Takemura M, et al. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision Repair. J Biol Chem. 2001;276:18665–72. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 6.Popescu A, Miron S, Blouquit Y, Duchambon P, Christova P, Craescu CT. Xeroderma pigmentosum group C protein possesses a high affinity binding site to human centrin 2 and calmodulin. J Biol Chem. 2003;278:40252–61. doi: 10.1074/jbc.M302546200. [DOI] [PubMed] [Google Scholar]
- 7.Charbonnier JB, Renaud E, Miron S, et al. Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein. J Mol Biol. 2007;373:1032–46. doi: 10.1016/j.jmb.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 8.Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J Biol Chem. 2006;281:18746–52. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- 9.Ng JM, Vermeulen W, van der Horst GT, et al. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–45. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch ME, Cross IV, Ford JM. p53 responsive nucleotide excision repair gene products p48 and XPC, but not p53, localize to sites of UV-irradiation-induced DNA damage, in vivo. Carcinogenesis. 2003;24:843–50. doi: 10.1093/carcin/bgg031. [DOI] [PubMed] [Google Scholar]
- 11.van der Spek PJ, Eker A, Rademakers S, et al. XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 1996;24:2551–9. doi: 10.1093/nar/24.13.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francisco G, Menezes PR, Eluf-Neto J, Chammas R. XPC polymorphisms play a role in tissue-specific carcinogenesis: a meta-analysis. Eur J Hum Genet. 2008;16:724–34. doi: 10.1038/ejhg.2008.6. [DOI] [PubMed] [Google Scholar]
- 13.Shore RE, Zeleniuch-Jacquotte A, Currie D, et al. Polymorphisms in XPC and ERCC2 genes, smoking and breast cancer risk. Int J Cancer. 2008;122:2101–5. doi: 10.1002/ijc.23361. [DOI] [PubMed] [Google Scholar]
- 14.Sluder G. Centrosome duplication and its regulation in higher animal cells. In: Nigg EA, editor. Centrosomes in Development and Disease. Wiley-VCH; Weinheim: 2004. pp. 167–90. [Google Scholar]
- 15.Lingle WL, Barrett SL, Negron VC, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–83. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–92. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 18.Errabolu R, Sanders MA, Salisbury JL. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- 19.Hart PE, Wolniak SM. Molecular cloning of a centrin homolog from Marsilea vestita and evidence for its translational control during spermiogenesis. Biochem Cell Biol. 1999;77:101–8. doi: 10.1139/o99-013. [DOI] [PubMed] [Google Scholar]
- 20.Wolfrum U, Salisbury JL. Expression of centrin isoforms in the mammalian retina. Exp Cell Res. 1998;242:10–7. doi: 10.1006/excr.1998.4038. [DOI] [PubMed] [Google Scholar]
- 21.Manandhar G, Feng D, Yi YJ, et al. Centrosomal protein centrin is not detectable during early pre-implantation development but reappears during late blastocyst stage in porcine embryos. Reproduction. 2006;132:423–34. doi: 10.1530/rep.1.00983. [DOI] [PubMed] [Google Scholar]
- 22.Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- 23.Klein UR, Nigg EA. SUMO-dependent regulation of centrin-2. J Cell Sci. 2009;122:3312–21. doi: 10.1242/jcs.050245. [DOI] [PubMed] [Google Scholar]
- 24.Price JE, Polyzos A, Zhang RD, Daniels LM. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–21. [PubMed] [Google Scholar]
- 25.Levenson AS, Jordan VC. MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 1997;57:3071–8. [PubMed] [Google Scholar]
- 26.D'Assoro AB, Busby R, Acu ID, et al. Impaired p53 function leads to centrosome amplification, acquired ERα phenotypic heterogeneity and distant metastases in breast cancer MCF-7 xenografts. Oncogene. 2008;27:3901–11. doi: 10.1038/onc.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan SG, Metin A, Gozukara E, et al. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum Mol Genet. 2004;13:343–52. doi: 10.1093/hmg/ddh026. [DOI] [PubMed] [Google Scholar]
- 28.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzawa M, Grams J, Madden B, Toft D, Salisbury JL. Identification of a complex between centrin and heat shock proteins in CSF-arrested Xenopus oocytes and dissociation of the complex following oocyte activation. Dev Biol. 1995;171:51–9. doi: 10.1006/dbio.1995.1259. [DOI] [PubMed] [Google Scholar]
- 30.Marchetto MC, Correa RG, Menck CF, Muotri AR. Functional lentiviral vectors for xeroderma pigmentosum gene therapy. J Biotechnol. 2006;126:424–30. doi: 10.1016/j.jbiotec.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 31.la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–36. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 32.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–5. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–4. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 34.D'Assoro AB, Busby R, Suino K, et al. Genotoxic stress leads to centrosome amplification in breast cancer cell lines that have an inactive G1-S cell cycle checkpoint. Oncogene. 2004;23:4068–75. doi: 10.1038/sj.onc.1207568. [DOI] [PubMed] [Google Scholar]
- 35.Sugasawa K. XPC: its product and biological roles. Adv Exp Med Biol. 2008;637:47–56. doi: 10.1007/978-0-387-09599-8_6. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Madura K. Centrin/Cdc31 is a novel regulator of protein degradationg. Mol Cell Biol. 2008;28:1829–40. doi: 10.1128/MCB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishi R, Okuda Y, Watanabe E, et al. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol Cell Biol. 2005;25:5664–74. doi: 10.1128/MCB.25.13.5664-5674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adimoolam S, Ford JM. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci U S A. 2002;99:12985–90. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Zhu Q, Wani MA, Wani G, Chen J, Wani AA. Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. DNA Repair (Amst) 2003;2:483–99. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 40.Wang G, Dombkowski A, Chuang L, Xu XX. The involvement of XPC protein in the cisplatin DNA damaging treatment-mediated cellular response. Cell Res. 2004;14:303–14. doi: 10.1038/sj.cr.7290375. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Chuang L, Zhang X, et al. The initiative role of XPC protein in cisplatin DNA damaging treatment-mediated cell cycle regulation. Nucleic Acids Res. 2004;32:2231–40. doi: 10.1093/nar/gkh541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernardes de Jesus BM, Bjoras M, Coin F, Egly JM. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol Cell Biol. 2008;28:7225–35. doi: 10.1128/MCB.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–34. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene. 2001;20:3173–84. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- 45.Resendes KK, Rasala BA, Forbes DJ. Centrin 2 localizes to the vertebrate nuclear pore and plays a role in mRNA and protein export. Mol Cell Biol. 2008;28:1755–69. doi: 10.1128/MCB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29:1760–73. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–46. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funakoshi M, Geley S, Hunt T, Nishimoto T, Kobayashi H. Identification of XDRP1;a Xenopus protein related to yeast Dsk2p binds to the N-terminus of cyclin A and inhibits its degradation. EMBO J. 1999;18:5009–18. doi: 10.1093/emboj/18.18.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci U S A. 2002;99:745–50. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–5. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 51.Jani D, Lutz S, Marshall NJ, et al. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell. 2009;33:727–37. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6:840–8. doi: 10.1038/ncb1163. [DOI] [PubMed] [Google Scholar]
- 53.Molinier J, Ramos C, Fritsch O, Hohn B. CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell. 2004;16:1633–43. doi: 10.1105/tpc.021378. [DOI] [PMC free article] [PubMed] [Google Scholar]