Abstract

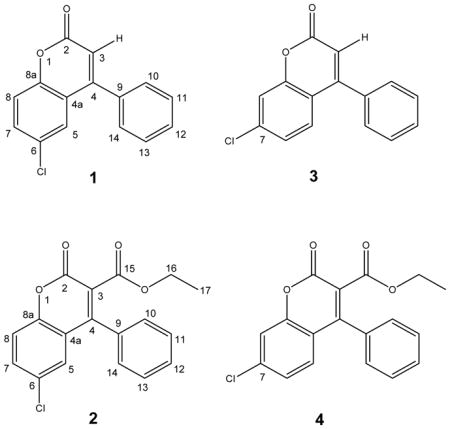
An EtOH extract of the polypore mushroom, Fomitopsis officinalis afforded two new naturally occurring chlorinated coumarins which were identified as the previously synthesized compounds, 6-chloro-4-phenyl-2H-chromen-2-one (1) and ethyl 6-chloro-2-oxo-4-phenyl-2H-chromen-3-carboxylate (2). The structures of the two isolates were deduced ab initio by spectroscopic methods and confirmed by chemical synthesis. In addition, an analogue of each was synthesized as of 7-chloro-4-phenyl-2H-chromen-2-one (3) and ethyl 7-chloro-2-oxo-4-phenyl-2H-chromen-3-carboxylate (4). All four compounds were characterized physicochemically, and their antimicrobial activity profiles revealed a narrow spectrum of activity with lowest MICs against the Mycobacterium tuberculosis complex.
INTRODUCTION
Tuberculosis (TB), along with malaria and HIV, are the leading causes of mortality among infectious diseases.1 Multi-drug resistance and long treatment duration have been the primary obstacles to global TB control.2, 3 Furthermore, emergence of extensive drug resistant TB (XDR-TB) and TB with HIV co-infection have become public health threats throughout the world.4–6 TB is an ancient disease, known for almost 9,000 years, and hence there exists an abundance of ethnomedical information. Such knowledge may facilitate the search for new anti-TB agents. Ethnomedical usage suggests relative safety and efficacy, although the latter may be due partially or entirely to symptomatic relief.7
Fomitopsis officinalis is a wood rotting polypore fungus, found in the old growth forests of Oregon and Washington in the Pacific Northwest United States, and in British Columbia, Canada, as well as in the northern regions of China and Europe. It has been used for the treatment of TB, pneumonia, cough, and asthma.8–11 While F. officinalis is known to be a rich source of unusual triterpenoids, only a few chemical studies have been reported, mostly on the isolation of terpenes.11–15 The present study involved the isolation of two new naturally occurring metabolites from an anti-TB mycelial culture extract (EtOH) of F. officinalis cultivated at Fungi Perfecti, LLC. The structures of the two metabolites were confirmed by chemical synthesis, together with the synthesis of two analogues. For the unambiguous chemical and biological characterization of the four compounds, their 1H NMR fingerprints including all J-couplings and chemical shifts were completely resolved by 1H iterative Full Spin Analysis (HiFSA) to facilitate future dereplication, and their biological activity was extensively profiled, focusing on Mycobacterium tuberculosis susceptibility assays.
RESULTS AND DISCUSSION
Isolation and Structure Elucidation
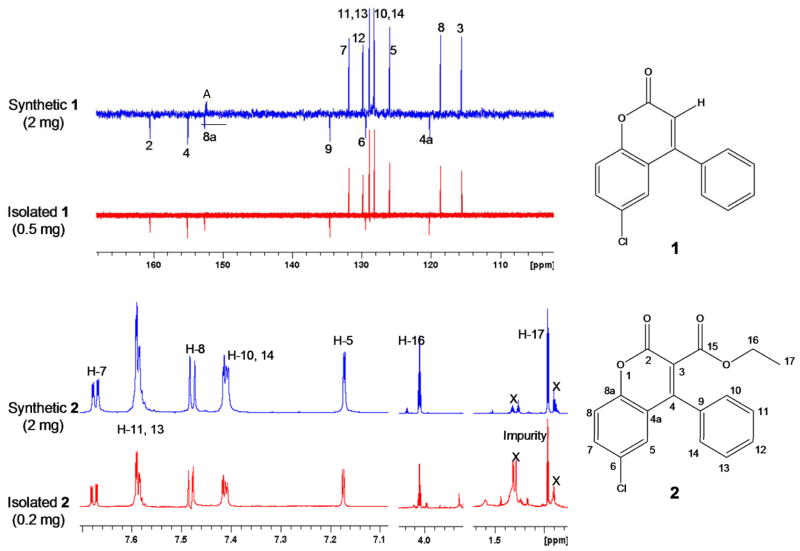
Two new naturally occurring coumarins, 1 (0.5 mg) and 2 (0.2 mg), were isolated from an EtOH extract of the polypore mushroom F. officinalis. For 1, the HRMS measurement indicated the presence of one chlorine in the molecule with [M+H]+ molecular ion peaks for 35Cl and 37Cl isotopes at m/z 257.0372 and 279.0232 respectively, corresponding to a molecular formula of C15H9ClO2. The 1H, 13C DEPTQ, and HSQC NMR (MeOH-d4) spectra at 900/225 MHz confirmed the presence of six quaternary (δ 160.5, 155.1, 152.6, 134.6, 129.4, and 120.2 ppm) and nine methine carbons (δ 131.8, 129.8, two at 128.8, two at 128.2, 125.9, 118.6, and 115.5 ppm). The HRMS measurements and the 13C and DEPTQ NMR were in agreement with the molecular formula of C15H9ClO2. The 1D 1H, 2D 1H-1H COSY, 13C DEPTQ, HSQC, and HMBC NMR spectra for 1 are provided in the Supporting Information (S1–S5). The aromatic region of the 1H NMR spectrum clearly showed the presence of one AMX and one AA′BB′C spin system, indicative of a tri- and a mono-substituted aromatic ring, respectively, which was confirmed in the COSY spectrum (Figure 1). In addition, the IR spectrum exhibited a strong carbonyl absorption band at 1731 cm−1, which was consistent with an ester carbonyl resonance in the DEPTQ spectrum corresponding to the quaternary carbonyl carbon C-2 (δ 160.5). Long-range 1H, 13C couplings from both aromatic protons H-10/H-14 (δ 1.036) on the stated phenyl ring to the quaternary carbon C-4 (δ 155.1) confirmed the phenyl substitution at C-4. HMBC correlations from the olefinic proton H-3 (δ 6.405) to C-2 (δ 160.5), C-4a (δ 120.2), and C-9 (δ 134.6) strengthened the evidence for phenyl substitution at C-4. The presence of an aromatic AMX spin system with a dd splitting pattern for H-7 (8.84, 2.50 Hz) was consistent with ortho-coupling to H-8 and meta-coupling to H-5, respectively, placing the chlorine substitution at C-6 (δ 129.4). Additional HMBC correlations of H-5 (δ 7.433) to C-8a (δ 7.433) and C-4 as well as H-7 to C-8a and C-5 confirmed the remaining coumarin skeleton. Compound 1 turned out to be a new naturally occurring chlorinated coumarin and was identified as the previously synthesized compound,20 6-chloro-4-phenyl-2H-chromen-2-one. Since only sub-milligram quantities could be isolated, the compound was chemically synthesized in order to enable in depth chemical and biological evaluation. The isolated and the synthetic product resulted in identical 13C DEPTQ spectra (Figure 2).
Figure 1.
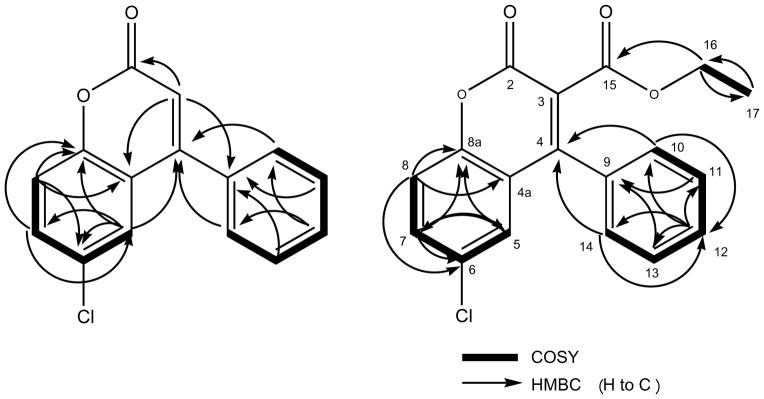
Key 2D correlations used for the structure determination of 1 and 2.
Figure 2.
Demonstration of congruence between the synthetic and isolated compounds 1 and 2 by 13C and 1H NMR (225/900 MHz, MeOH-d4; A = image artifact from the solvent)
In order to unambiguously differentiate the structure of 1 from a possible isomer with chlorine substitution at C-7, the compound 7-chloro-4-phenyl-2H-chromen-2-one, 3, was synthesized as well. The 1H and 13C DEPTQ NMR spectra are available in the Supporting Information (S15 and S16). In addition, a 1H iterative Full Spin Analysis (HiFSA) was carried out for both synthetic compounds, 1 and 3, using the PERCH software tool and in approach described recently,16–19 and this resulted in the unambiguous assignment of all 1H resonances including their multiplicities, as shown in Table 2 and the Supporting Information (S19 and S21).
Table 2.
Anti-TB activity profiles of the coumarins, 1 – 4, in comparison with standard TB drugs and cytotoxicity.
| Compounds | MIC90 (μg/mL) | IC50 (μg/mL) | SId | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| H37Rva | H37Rvb | rRMPc | rINHc | rSMc | rKANc | rCSc | Vero | ||
| 1 | > 100 | > 100 | > 100 | > 100 | > 100 | > 100 | > 100 | >100 | - |
| 2 | 44.7 | 37.1 | 46.6 | 44.7 | 49.5 | > 100 | > 100 | 36.7 | 0.82 |
| 3 | 23.9 | 21.9 | 45.4 | 76.5 | 46.9 | 47.4 | 47.3 | 30.4 | 1.27 |
| 4 | 35.9 | 23.2 | 44.7 | 31.7 | 46.9 | 43.9 | 47.3 | 39.4 | 1.10 |
| rifampin | 0.08 | <0.05 | >3.29 | 0.04 | 0.09 | 0.02 | 0.02 | >100 | >1250 |
| isoniazid | 0.03 | - | 0.03 | >2.19 | 0.03 | 0.07 | 0.04 | >100e | >3333 |
| streptomycin | - | 1.07 | 0.47 | 0.56 | >9.31 | 1.48 | 0.53 | - | - |
| PA824 | 0.02 | 0.25 | 0.07 | 0.05 | 0.17 | 0.34 | 0.18 | >100e | >5000 |
| kanamycin | - | - | 0.73 | 0.72 | 0.76 | > 25 | 1.12 | - | - |
| cycloserine | - | - | 4.8 | 5.0 | 4.9 | 4.9 | >10.2 | - | - |
Determined by the microplate Alamar blue assay (MABA) for replicating M. tuberculosis.
Determined by the low oxygen recovery assay LORA) for non-replicating M. tuberculosis.
M. tuberculosis strains resistant to rifampin (rRMP), isoniazid (rINH), streptomycin (rSM), kanamycin (rKAN), and cycloserine (rCS).
Selectivity Index = IC50/ MABA MIC.
Values observed in separate experiments over time in our laboratory.
Compound 2, ethyl 6-chloro-2-oxo-4-phenyl-2H-chromen-3-carboxylate, displayed a very similar 1H and 13C NMR peak patterns compared to that of 1, including the presence of AMX and AA′BB′C spin systems. The HRMS measurement also indicated the presence of one chlorine atom in the molecule by affording a molecular ion peak at m/z 329.0614 [M+H]+ for the 35Cl isotope, representing a 72 amu difference from that of 1. Although the 13C DEPTQ, HSQC, and HMBC spectra were obtained at high-field with a 5mm inverse cryoprobe (225/900 MHz, 3 mm and 1.7 mm samples, in MeOH-d4,) and even with the long accumulation time (over 24 hours), they showed relatively low signal-to-noise. The 1D 1H, 2D 1H-1H COSY, 1D 13C DEPTQ, HSQC, and HMBC NMR spectra for the isolated compound 2 are provided in the Supporting Information (S6–S10). Despite these limitations, the data still confirmed that 2 possesses a coumarin skeleton with a chlorine at C-6 (δ 131.4) and phenyl substitution at C-4 (δ 153.7). In addition, 2 clearly did not have an olefinic proton at C-3, but instead contained an ethoxycarbonyl moiety with H-17 methylene (2H, δ 4.052) and H-17 methyl protons (3H, δ 0.966). The 1H-1H COSY confirmed the correlation between H2-16 and H3-17. Deshielding of both the carbon and the proton of C-16/H-16 (δ 61.5/δ 4.052) indicated close proximity to an electronegative oxygen atom. Long-range couplings from H-17 (δ 4.052) to a quaternary carbon C-15 (δ 165.3) in the HMBC confirmed the presence of the ethyl ester moiety, which also accounted for the 72 amu difference. Therefore, 2 was assigned to contain an ethyl ester moiety attached to C-3 (δ 121.8) of 1. Overall, 2 was deduced as another new naturally occurring chlorinated coumarin, ethyl 6-chloro-2-oxo-4-phynyl-2H-chromen-3-carboxylate, which also had previously been synthesized.21 In order to confirm the NMR-based structural assignment, 2 was synthesized and revealed identical 1H NMR spectra, as shown in Figure 2. The detailed synthetic schemes are provided in the Supporting Information (S24).
In addition to the naturally occurring compounds, the analogue with chlorine substitution at C-7, ethyl 7-chloro-2-oxo-4-phenyl-2H-chromen-3-carboxylate (4), was synthesized. Considering the differences in their spectroscopic data, this also ruled out the positional isomer 4 as a potential structure of 3. The 1H and 13C DEPTQ NMR spectra for the 4 are available in the Supporting Information (S17 and S18). In addition, HiFSA was carried out for the 900 MHz spectra of both 2 and 4, which resulted in the full assignment of the 1H resonances and multiplicities. The results are summarized in Table 2 and in the Supporting Information (S20 and S22).
Biological Evaluation
The anti-TB activities of the four coumarins, 1–4, were assessed by the microplate Alamar blue assay (MABA) and the low oxygen recovery assay (LORA) (Table 2). MABA is used to measure the activity against replicating M. tuberculosis, while LORA is designed to assess the activity against non-replicating M. tuberculosis. MABA and LORA MICs indicated similar anti-TB activities with MICs ranging from 20 to 50 μg/mL, except for compound 1 (>100 μg/mL). The 7-chloro congener 4 containing an ethyl ester had superior anti-TB activity. The olefinic proton substitution (in 3) of the two 7-chloro variations (in 3 and 4) seemed to have slightly greater potency than the ethyl ester variation, but the difference in the MIC values was insignificant (less than two-fold). On the other hand, in the 6-chloro variation (in 1 and 2), the ethyl ester substitution (in 2) caused significantly stronger anti-TB activity. Based on these observations, it can be concluded that the structural variation containing an ethyl ester has superior anti-TB activity if it contains chlorine in position 6, while coumarins with a 7-chlorine substitution are less active. In summary, of the four structural variations in 1–4, compound 3 was the most active against both replicating and non-replicating M. tuberculosis with MICs of 23.9 and 21.9 μg/mL, respectively. Furthermore, activity in the LORA indicated that these coumarins might have the potential for shortening the duration of therapy via efficient killing of the non-replicating persistor cells.
Activity against a panel of mono drug-resistant isolates was determined by the MABA and resulted in a similar susceptibility profile to those obtained against wild type M. tuberculosis. This indicated that there is no significant difference in the anti-TB activity for an ethyl ester with 7-chloro substitution, while better activity was observed with the 6-chloro analogues (Table 2). Activity against mono drug-resistant isolates suggests a molecular target distinct from those for the corresponding existing clinical drugs.
In order to better understand the spectrum of antimicrobial activity, MICs of these chlorinated coumarins were determined for a panel of 8 species: S. aureus, E. coli, C. albicans, E. faecalis, P. aeruginosa, A. baumanii, S. pneumoniae, and M. smegmatis. All four compounds were inactive at the highest test concentration, 100 μg/mL. In addition, activity against other non-tuberculous mycobacteria (NTM) was assessed with M. chelonae, M. abscessus, M. marinum, M. kansasii, M. avium, and M. bovis. In this series, M. bovis has the closest gene homology to M. tuberculosis, and not surprisingly showed the most similar susceptibility profile to that of M. tuberculosis. Compounds 2 against M. marinum and 3 against M. kansasii exhibited MICs of 97.1 and 49.3 μg/mL, respectively. Otherwise, no activity was observed at 100 μg/mL. The activities against the 8 non-mycobacterial species and NTM are documented in S23 of the Supporting Information.
Mammalian cell cytotoxicity using Vero cells for 1–4 was evaluated and described as the 50% growth Inhibitory Concentration (IC50), resulting in values of >100, 36.7, 30.4, and 39.4 μg/mL, respectively. This translates into Selectivity Indices (SIs; IC50/MIC) around unity for the active coumarins 2–4 (Table 2). Cytotoxicity against Vero cells was observed at a similar concentration as that causing significant inhibition against M. tuberculosis and M. bovis, whereas activities against other microorganisms are essentially lacking (S23, Supporting Information). However, it is not clear at this point whether the chlorinated coumarins are hitting similar targets in the M. tuberculosis complex species and VERO cells. As SIs greater than unity are generally regarded as requirement for progression as TB drug leads, analogues of these coumarins with significantly higher SIs will need to be found or synthesized in order to provide a useful novel anti-TB pharmacophore. Although these isolated coumarins, based on the yield and the concentration within the crude extract and the activity of the compounds in question, do not explain the claimed vitro anti-TB activity of the crude extract with respect to its ethnomedical use, the present study led to the identification of a new compound class for this organism and revealed structural entities that show anti-TB potency and could represent a possible link to the traditional use of F. officinalis as an anti-TB treatment.
EXPERIMENTAL SECTION
General Experimental Procedures
The UV-vis spectra were obtained with a SpectraMax Plus 384 at 25 °C. Optical rotations [α]D were measured on a Perkin-Elmer 242 polarimeter at 25 °C. IR spectra were measured on a Thermo Nicolet 6700 FT-IR spectrometer. All NMR experiments were obtained at either 600 or 900 MHz and performed on Bruker AVANCE-600 or AVANCE II-900 instruments, each equipped with a cryogenic sensitivity-enhanced triple-resonance 5mm inverse TCI cryoprobe. The samples were dissolved in 200 μL of MeOH-d4 and 150 μL transferred to 3 mm (or 50 μL of MeOH-d4 and 40 μL transferred to 1.7 mm NMR tubes). All NMR experiments were performed using standard Bruker pulse sequences and the temperature was maintained at 25 °C (298 K). High-resolution ESI mass spectra were obtained using a Shimadzu IT-TOF LC mass spectrometer. The 1H iterative Full Spin Analysis was performed with the PERCH NMR software (v.2010.1, PERCH Solutions Ltd., Kuopio, Finland). The 1H NMR spectra were processed with NUTS (Acorn NMR Inc.), imported into PERCH as JCAMP-DX files, and subjected to baseline correction, peak picking, and integration. The 1H NMR parameters in MeOH-d4 were predicted using the PERCH Molecular Modeling System (MMS). After a manual examination of the 1H assignments, the calculated 1H chemical shifts, signal line widths, and J-couplings were refined by using the integral-transform (D) and total-line-fitting (T) modes until an excellent agreement between the observed and simulated spectra was attained (total RMS ≤0.5%). Cellular viability was assessed by the measurement of fluorescence, luminescence, or absorbance with the Victor3 multilabel reader (PerkinElmer Life Sciences).
Organism Collection, Identification, Culture, and Extraction
Fomitopsis officinalis was collected from Morton, WA in September of 2001. The tissue culture and stock cultures are maintained at Fungi Perfecti Research Laboratories in Shelton, WA. Partial sequence of 18S and 28S ribosomal RNA genes established the identification of Fomitopsis officinalis. Sequence data is available on Genebank (EU854436.1).
Mycelial cultures were grown in sterile Petri dishes containing sterilized antibiotic malt extract yeast agar. After three weeks of colonization in a clean room laboratory at temperatures between 21 – 24 °C, the cultures were aseptically transferred into a 1000 mL Eberbach™ stirrer containing 800 mL sterilized water. The Eberbach™ container was activated using a Waring™ blender base and the mycelium was fragmented in a process known as liquid fragmentation (the dissociated fragmented mycelial mass allows for a multiple loci inoculation, resulting in accelerated colonization). Approximately 50 – 100 mL myceliated broth was then transferred into a polypropylene incubation bag containing approximately 3 kg of moistened sterilized rice (approximately 45–50% moisture content). These bags of freshly inoculated rice were then incubated for 60 – 120 days in a class 100 clean room. Once colonization was determined to be sufficient, the mycelium-colonized rice was transferred to glass containers for extraction. The mycelium being delicate in nature was handled with utmost gentle care, so as to not cause cell damage during transfer, and immediately covered with an equal weight of 95% EtOH. The mixture was agitated and then allowed to macerate at room temperature.
Isolation
The Fomitopsis officinalis mycelium culture extract (1.75 L, 95% EtOH) was evaporated under vacuum to give 17 g of dried crude extract. The extract was re-dissolved in 1 L of 75% EtOH and partitioned with petroleum ether, hexane, CHCl3, EtOAc, and n-BuOH (1:1, v/v) to give 474, 30, 4270, 48, and 8569 mg of dried extract, respectively. The CHCl3 partition fraction was further separated on an Isolera™ Flash purification system (SNAP 100 g, 30 mL/min) with a linear solvent gradient of CHCl3-MeOH (100:0, v/v) to 100% MeOH over 100 min, to afford 135 fractions (25mL/fraction). The recombination into 22 fractions was based on TLC (silica; CHCl3-MeOH, 85:15) analysis. From the 22 fractions, fractions 11, 12, and 13 were again recombined (300 mg) for further separation on an Isolera™ Flash purification system (SNAP 10 g, 9 mL/min) with a linear solvent gradient of CHCl3-MeOH (90:10, v/v) to 15% MeOH over 28 min, affording 85 sub-fractions (3mL/fraction). These were recombined to 7 fractions based on TLC analysis and dried to yield 17, 9, 6, 24, 31, 37, and 17 mg, respectively. The first combined fraction (17 mg) was further fractionated using reverse-phase HPLC on a Waters Delta 600 system with a Waters 996 photodiode array detector using a semi-preparative column (Waters, C18, 5 um, 250 × 10 mm, 3 mL/min). Five separated injections with a linear solvent gradient of MeOH-H2O (80:20, v/v) to 100% MeOH over 25 min afforded 0.2 mg of 2 at 15 min and 0.5 mg of 1 at 17 min. General fraction monitoring for chromatographic separation was done by TLC analysis with precoated Alugram SIL G/UV plates (Macherey-Nagel, Düren, Germany).
6-Chloro-4-phenyl-2H-chromen-2-one (1).20
An amorphous white solid: [α]25D 0 (c 0.001, MeOH); UV/Vis (MeOH) λmax (log ε) 257 (2.87), 264 (3.09), 288 nm (3.42); IR (neat) ν max 2359, 2341, 1731 cm−1; 1H/DEPTQ 13C NMR (900/225 MHz, MeOH-d4), see Table 1; HRMS m/z 256.0274 [M+H]+ (calculated for C15H9ClO2, 256.0291). Synthetic methodology is available in Supporting Information (S24).
Table 1.
1H and 13C NMR Data of compounds 1 – 4.
| 1b | 2b | 3b | 4b | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| position | δ Cc | multi | δH | multi (J in Hz)e | δ Cc | multi | δH | multi (J in Hz)e | δCc | multi | δH | multi (J in Hz)e | δCc | multi | δH | multi (J in Hz)e |
| 2 | 160.5 | C | - | - | 159.1 | C | - | - | 161.1 | C | - | - | 158.9 | C | - | - |
| 3 | 115.5 | CH | 6.405 | s | 121.8 | C | - | - | 115.1 | CH | 6.382 | s | 119.2 | C | - | - |
| 4 | 155.1 | C | - | - | 153.7 | C | - | - | 156.3 | C | - | - | 155.1 | C | - | - |
| 4a | 120.2 | C | - | - | 123.5 | C | - | - | 118.4 | C | - | - | 122.3 | C | - | - |
| 5 | 125.9 | CH | 7.433 | dd (2.50, 0.41) | 128.4 | CH | 7.176 | dd (2.50, 0.44) | 125.3 | CH | 7.494 | dd (8.58, 0.76) | 126.5 | CH | 7.250 | dd (8.61, 0.22) |
| 6 | 129.4 | C | - | - | 131.4 | C | - | - | 128.8 | CH | 7.322 | dd (8.58, 1.98) | 131.0 | CH | 7.326 | dd (8.61, 2.05) |
| 7 | 131.8 | CH | 7.632 | dd (8.84, 2.50) | 134.3 | C | 7.677 | dd (8.87, 2.50) | 135.5 | C | - | - | 134.0 | CH | - | - |
| 8 | 118.6 | CH | 7.455 | dd (8.84, 0.41) | 120.1 | CH | 7.495 | dd (8.87, 0.44) | 117.7 | CH | 7.504 | dd (1.98, 0.76) | 118.3 | CH | 7.548 | dd (2.05, 0.22) |
| 8a | 152.6 | C | - | - | 153.5 | C | - | - | 155.1 | C | - | - | 154.2 | C | - | - |
| 9 | 134.6 | C | - | - | 133.8 | C | - | - | 138.3 | C | - | - | 140.0 | C | - | - |
| 10 | 128.2 | CH | 7.527 | dddd (2.04, 7.66, 1.25, 0.64) | 130.0 | CH | 7.422 | dddd (1.92, 7.67, 1.25, 0.62) | 128.9 | CH | 7.512 | dddd (2.24, 7.67, 0.33, 1.30) | 129.5 | CH | 7.395 | dddd (1.63, 7.68, 0.63, 1.06) |
| 11 | 128.8 | CH | 7.599 | dddd (7.66, 0.64, 1.25, 7.55) | 130.2 | CH | 7.591 | dddd (1.25, 7.57, 0.72, 7.67) | 129.4 | CH | 7.570 | dddd (7.76, 0.33, 1.33, 7.54) | 130.0 | CH | 7.559 | dddd (7.68, 0.63, 1.28, 7.49) |
| 12 | 129.8 | CH | 7.592 | dd (1.25, 7.55) | 131.2 | CH | 7.588 | dd (7.57, 1.25) | 130.4 | CH | 7.566 | dd (1.30, 7.54) | 130.5 | CH | 7.565 | dd (7.49, 1.06) |
| 13 | 128.8 | CH | 7.599 | dddd (7.66, 0.64, 1.25, 7.55) | 130.2 | CH | 7.591 | dddd (1.25, 7.57, 0.72, 7.67) | 129.4 | CH | 7.570 | dddd (7.76, 0.33, 1.33, 7.54) | 130.0 | CH | 7.559 | dddd (7.68, 0.63, 1.28, 7.49) |
| 14 | 128.2 | CH | 7.527 | dddd (2.04, 7.66, 1.25, 0.64) | 130.0 | CH | 7.422 | dddd (1.92, 7.67, 1.25, 0.62) | 128.9 | CH | 7.512 | dddd (2.24, 7.67, 0.33, 1.30) | 129.5 | CH | 7.395 | dddd (1.63, 7.68, 0.63, 1.06) |
| 15 | - | - | - | - | 165.3 | C | - | - | - | - | - | - | 165.3 | C | - | - |
| 16 | - | - | - | - | 61.5 | CH2 | 4.052 | q (7.01) | - | - | - | - | 62.9 | CH2 | 4.040 | q (7.12) |
| 17 | - | - | - | - | 12.6 | CH3 | 0.966 | t (7.01) | - | - | - | - | 14.0 | CH3 | 0.957 | t (7.12) |
1H NMR data were obtained for the synthetic compounds.
13C NMR chemical shifts were obtained from DEPTQ.
The J-coupling values resolved from iterative full 1H spin analysis using PERCH, given with 10 mHz precision.
Ethyl 6-chloro-2-oxo-4-phenyl-2H-chromene-3-carboxylate (2).21
An amorphous white solid: [α]25D 0 (c 0.001, MeOH); UV/Vis (MeOH) λmax (log ε) 257 (3.16), 265 (3.52), 289 (3.95), 294 nm (3.94); IR (neat) ν max 1745, 1724, 1599, 1244, 1039 cm−1; 1H/DEPTQ 13C NMR (900/225 MHz, MeOH-d4), see Table 1; HRMS m/z 328.0515 [M+H]+ (calculated for C18H13ClO4, 328.0502). Synthetic methodology is available in Supporting Information (S24).
7-Chloro-4-phenyl-2H-chromen-2-one (3).20, 22
An amorphous white solid: [α]25D 0 (c 0.001, MeOH); UV/Vis (MeOH) λmax (log ε) 257 (3.17), 265 (3.55), 289 (3.89), 294 nm (3.89); IR (neat) ν max 1743, 1725, 1601, 1279, 1037 cm−1; 1H/DEPTQ 13C NMR (900/225 MHz, MeOH-d4), see Table 1; HRMS m/z 256.0298 [M+H]+ (calculated for C15H9ClO2, 256.0291). Synthetic methodology is available in Supporting Information (S24).
Ethyl 7-chloro-2-oxo-4-phenyl-2H-chromene-3-carboxylate (4).21, 22
An amorphous white solid: [α]25D 0 (c 0.001, MeOH); UV/Vis (MeOH) λmax (log ε) 257 (3.17), 265 (3.55), 289 (3.89), 294 nm (3.89); IR (neat) ν max 1743, 1725, 1601, 1279, 1037 cm−1; 1H/DEPTQ 13C NMR (900/225 MHz, MeOH-d4), see Table 1; HRESIMS m/z 328.0516 [M+H]+ (calculated for C18H13Cl2O4, 328.0502). Synthetic methodology is available in Supporting Information (S24).
Bacterial Strain Preparation for anti-TB Bioassay and Minimum Inhibitory Concentration (MIC)
M. tuberculosis H37Rv ATCC 27294 was purchased from American Type Culture Collection (ATCC) and cultured to late log phase in the 7H12 media, Middlebrook 7H9 broth supplemented with 0.2% (vol/vol) glycerol, 0.05% Tween80, and 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC). The culture was harvested and resuspended in phosphate-buffered saline. Suspensions were then filtered through 8 μm filter membranes and frozen at 80 °C. Prior to use of bacterial stocks for the anti-TB assay, CFUs were determined by plating on 7H11 agar media. The MIC is defined here as the lowest concentration resulting in ≥90% growth inhibition of the bacteria relative to untreated controls. MIC against replicating M. tuberculosis was measured by the Microplate Alamar Blue Assay (MABA).23, 24
Low Oxygen Recovery Assay
The luciferase reporter gene luxAB recombinant M. tuberculosis was prepared as previously reported.25 The bacteria were adapted to low oxygen during culture in a BioStatQ fermenter. The low oxygen-adapted culture was exposed to the test samples in 96-well microplates for 10 days at 37 °C in a hypoxic environment created with an Anoxomat (WS-8080, MART Microbiology). The cultures were then transferred to a normoxic environment at 37 °C for 28 hours. Viability was assessed by the measurement of luciferase-mediated luminescence. The LORA MIC was defined as the lowest concentration effecting a reduction of luminescence of ≥90% relative to untreated cultures.
Cytotoxicity
Cytotoxicity26, 27 was assessed using Vero (ATCC CRL-1586) cells. Vero cells were cultured in 10% fetal bovine serum (FBS) in Eagle minimum essential medium. The culture was incubated at 37 °C under 5% CO2 in air and then diluted with phosphate-buffered saline to 106 cells/mL. In a transparent 96-well plate (Falcon Microtest 96), 2-fold serial dilutions of testing samples with a final volume of 200 μL cell culture suspension was prepared. After 72 h incubation at 37 °C, the medium was removed and monolayers were washed twice with 100 μL of warm Hanks balanced salt solution (HBSS). One hundred μL of medium and 20 μL of MTS-PMS (Promega) were added to each well. Plates were then incubated for 3 hours, and cytotoxicity was determined by the measurement of absorbance at 490 nm.
MIC against Drug-resistant M. tuberculosis Isolates
M. tuberculosis strains individually resistant to rifampin (RMP, ATCC 35838), isoniazid (INH, ATCC 35822), streptomycin (SM, ATCC 35820), Cycloserine (CS, ATCC 35826) and kanamycin (KAN, ATCC 35827) were obtained from the American Type Culture Collection (ATCC). Cultures were prepared and MICs against drug-resistant M. tuberculosis isolates were determined by the MABA as described above for M. tuberculosis H37Rv.
MIC against Non-tuberculous Mycobacteria
M. abscessus (ATCC19977), M. chelonae (ATCC35752), M. avium (ATCC15769), M. marinum (ATCC927), M. kansasii (ATCC12478), and M. bovis BCG (ATCC35734) were purchased from ATCC. Cultures were prepared and MICs against these Mycobacteria were determined by the MABA as described above for M. tuberculosis H37Rv. M. abscessus was incubated with 7H12 medium at 37 °C for 3 days and for additional 4 hours after adding 12 μL of 20% Tween80 and 20 μL of Alamar blue dye. M. bovis was incubated with 7H12 medium at 37 °C for 7 days, and an additional 1 day of incubation after adding 12 μL of 20% Tween80 and 20 μL of Alamar blue dye. M. chelonae was incubated with 7H9 medium at 30°C for 3 days, plus additional 6 days of incubation after adding 12 μL of 20% Tween80 and 20 μL of Alamar blue dye. M. marinum was incubated with 7H9 medium at 30 °C for 5 days, and an additional 1 day of incubation after adding 12 μL of 20% Tween80 and 20 μL of Alamar blue dye. M. avium and M. kansasii were incubated with 7H9 media at 37 °C for 6 days, plus an additional day of incubation after adding 12 μL of 20% Tween80 and 20 μL of Alamar blue dye. Viability was assessed by measuring fluorescence at 530 nm excitation/590 nm emission with a Victor3 multilabel reader (PerkinElmer).
Spectrum of Activity
Staphylococcus aureus (ATCC29213), Candida albicans (ATCC90028), Escherichia coli (ATCC25922), M. smegmatis MC2155, Streptococcus pneumoniae (ATCC49619), Enterococcus faecalis (ATCC29212), and Pseudomonas aeruginosa (ATCC27853) were purchased from ATCC. The relative activity against S. aureus, C. albicans, S. pneumoniae (with 2% HBL), E. faecalis, P. aeruginosa, and E. coli was determined by broth micro dilution with a spectrophotometric readout at 570 nm (S. pneumonia at 490 nm) as described in the National Committee on Clinical Laboratory Standards.28, 29 The activity against M. smegmatis was determined by MABA, incubating 3 days at 37 °C plus additional 4 hours of incubation after adding 12 μL of 20% Tween80 and 20 μL of Alamar blue dye. Viability was assessed by the measurement of fluorescence with the Victor 3.
Supplementary Material
Acknowledgments
We are grateful to Dr. Regan Nally from Fungi Perfecti LLC., Olympia for the preparation of the extracts. The construction of the UIC CSB and the 900 MHz (21.1 T) NMR spectrometer were funded by NIGMS grant P41 GM068944.
Footnotes
The 1H, 13C, COSY, HSQC, and HMBC NMR spectra of isolated 1 and 2. The 1H, 13C NMR spectra, full 1H NMR spin analysis, synthesis of 1–4. These materials are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dye C. Lancet. 2006;367:938–940. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- 2.Dmitriev VA. Antibiot Khimioter. 2008;53:3–6. [PubMed] [Google Scholar]
- 3.Ginsberg AM, Spigelman M. Nat Med. 2007;13:290–294. doi: 10.1038/nm0307-290. [DOI] [PubMed] [Google Scholar]
- 4.Abdool Karim SS, Churchyard GJ, Abdool Karim Q, Lawn SD. Lancet. 2009;374:921–933. doi: 10.1016/S0140-6736(09)60916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abubakar I, Moore J, Drobniewski F, Kruijshaar M, Brown T, Yates M, Anderson C, Smith EG, Magee J, Lipman M, McMenamin J, Ruddy M, Watson JM. Thorax. 2009;64:512–515. doi: 10.1136/thx.2008.108712. [DOI] [PubMed] [Google Scholar]
- 6.Donald PR, van Helden PD. N Engl J Med. 2009;360:2393–2395. doi: 10.1056/NEJMp0903806. [DOI] [PubMed] [Google Scholar]
- 7.Farnsworth NR, Akerele O, Bingel AS, Soejarto DD, Guo Z. Bull World Health Organ. 1985;63:965–981. [PMC free article] [PubMed] [Google Scholar]
- 8.Stamets P. Int J Med Mushr. 2005;7:495– 506. [Google Scholar]
- 9.Stamets P. Int J Med Mushr. 2002;5:179– 192. [Google Scholar]
- 10.Grazywnowicz K. Int J Med Mushr. 2001;3:2–3. [Google Scholar]
- 11.Wu X, Yang JS, Yan M. Chem Pharm Bull (Tokyo) 2009;57:195–197. doi: 10.1248/cpb.57.195. [DOI] [PubMed] [Google Scholar]
- 12.Anderson C. Phytochem. 1971;10:2713–2717. [Google Scholar]
- 13.Anderson C. Phytochem. 1972;11:2847–2852. [Google Scholar]
- 14.Epstein W. J Am Chem Soc. 1979;101:2748–2750. [Google Scholar]
- 15.Wu X, Yang JS, Zhou L, Dong YS. Chem Pharm Bull. 2004;52:1375–1377. doi: 10.1248/cpb.52.1375. [DOI] [PubMed] [Google Scholar]
- 16.Napolitano JG, Godecke T, Rodriguez-Brasco MF, Jaki BU, Chen SN, Lankin DC, Pauli GF. J Nat Prod. 2012;75:238–248. doi: 10.1021/np200949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina-Salinas GM, Rivas-Galindo VM, Said-Fernandez S, Lankin DC, Munoz MA, Joseph-Nathan P, Pauli GF, Waksman N. J Nat Prod. 2011;74:1842–1850. doi: 10.1021/np2000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher JM, Schinkovitz A, Zapp J, Wang Y, Franzblau SG, Becker H, Lankin DC, Pauli GF. J Nat Prod. 2010;73:656–663. doi: 10.1021/np900806j. [DOI] [PubMed] [Google Scholar]
- 19.Inui T, Wang Y, Nikolic D, Smith DC, Franzblau SG, Pauli GF. J Nat Prod. 2010;73:563–567. doi: 10.1021/np900674d. [DOI] [PubMed] [Google Scholar]
- 20.Cresente-Campo J. Eur J Org Chem. 2010;2010:4130– 4135. [Google Scholar]
- 21.Tawada H. Chem Pharm Bull. 1995;26:616–625. doi: 10.1248/cpb.43.616. [DOI] [PubMed] [Google Scholar]
- 22.Gabbutt CD, Heron BM, Instone AC. Tetrahedron. 2006;62:737–745. [Google Scholar]
- 23.Collins L, Franzblau SG. Antimicrob Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. J Clin Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SH, Warit S, Wan B, Hwang CH, Pauli GF, Franzblau SG. Antimicrob Agents Chemother. 2007;51:1380–1385. doi: 10.1128/AAC.00055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantrell CL, Lu T, Fronczek FR, Fischer NH, Adams LB, Franzblau SG. J Nat Prod. 1996;59:1131–1136. doi: 10.1021/np960551w. [DOI] [PubMed] [Google Scholar]
- 27.Falzari K, Zhu Z, Pan D, Liu H, Hongmanee P, Franzblau SG. Antimicrob Agents Chemother. 2005;49:1447–1454. doi: 10.1128/AAC.49.4.1447-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards (NCCLS); Villanova, PA. 1990. Approved standard M7-A2. [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards (NCCLS); Villanova, PA. 1991. Informational Supplement M100-S3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.