Abstract
Proteasome is the major component of the crucial nonlysosomal protein degradation pathway in the cells, but the detailed reaction pathway is unclear. In this study, first-principles quantum mechanical/molecular mechanical free energy calculations have been performed to explore, for the first time, possible reaction pathways for proteasomal proteolysis/hydrolysis of a representative peptide, succinyl-leucyl-leucyl-valyl-tyrosyl-7-amino-4-methylcoumarin (Suc-LLVY-AMC). The computational results reveal that the most favorable reaction pathway consists of six steps. The first is a water-assisted proton transfer within proteasome, activating Thr1-Oγ. The second is a nucleophilic attack on the carbonyl carbon of a Tyr residue of substrate by the negatively charged Thr1-Oγ, followed by the dissociation of the amine AMC (third step). The fourth step is a nucleophilic attack on the carbonyl carbon of the Tyr residue of substrate by a water molecule, accompanied by a proton transfer from the water molecule to Thr1-Nz. Then, Suc-LLVY is dissociated (fifth step), and Thr1 is regenerated via a direct proton transfer from Thr1-Nz to Thr1-Oγ. According to the calculated energetic results, the overall reaction energy barrier of the proteasomal hydrolysis is associated with the transition state (TS3b) for the third step involving a water-assisted proton transfer. The determined most favorable reaction pathway and the rate-determining step have provided a reasonable interpretation of the reported experimental observations concerning the substituent and isotopic effects on the kinetics. The calculated overall free energy barrier of 18.2 kcal/mol is close to the experimentally-derived activation free energy of ~18.3–19.4 kcal/mol, suggesting that the computational results are reasonable.
Keywords: Proteasome, proteolytic mechanism, QM/MM, peptide hydrolysis
Introduction
The 20S proteasome which has been found in all the kingdoms of life, including eukaryotes from yeast to humans, as well as prokaryotes such as archaebacteria and eubacteria,1, 2 is a major cellular constituent and comprises ~1% of the protein in most cells.3 It was discovered independently by several labs, and had been given as many as 22 different names in the literature.3,4 It was eventually denoted as ‘proteasome’ to indicate a particle with the proteolytic function.4 Noteworthy, it is the major component of the nonlysosomal protein degradation pathway in the cells. This pathway plays a primary role in the degradation of the bulk of proteins in mammalian cells, as well as the degradation of abnormal proteins, and thus produces most of the antigenic peptides presented to the immune system by MHC (Major Histocompatibility Complex) class I molecules.1 However, the detailed reaction mechanism for proteasomal proteolysis of these proteins is still mysterious. As well known, protein synthesis and protein degradation are two universal complementary processes, permanently occurring in a living cell, and understanding the principles behind these processes remains to be among the most challenging questions in modern cell biology and biochemistry.1
Proteasome is not simply a ‘garbage disposal unit’, but also has functions in the control of the cell cycle and immune responses.3 In fact, proteasome is involved in the turnover of many critical proteins involved in the control of cell growth, cell differentiation, or metabolic adaptation. Thus, proteasome is a crucial protein which plays a central role in such processes as maintaining cellular homeostasis, controlling the cell cycle, removing misfolded proteins that can be toxic, and regulating the immune system.5 Dysregulation of the proteasome system may result in a variety of human diseases including cancer. Due to its central role in maintaining cellular homeostasis, it is not surprising to note that proteasome has been implicated as an important drug target in the development of many human diseases. In 2003, the Food and Drug Administration (FDA) has approved the use of the proteasome inhibitor bortezomib in the treatment of multiple myeloma.6 In addition, the early proteasome inhibitors are contributing to the development of some new anticancer drugs (CEP-18770, Carfilzomib, and NPI-0052).7 In fact, proteasome has been recognized as a significant target in the search for novel cancer therapeutics,7–10 and there has been increasing interest in studying the proteasome function.11 So far, a variety of proteasome inhibitors have been reported in literature,12–40 including covalent inhibitor Epoxomicin (EPX) which irreversibly inhibits proteasome,41 and the proteasome-EPX reaction mechanism has been examined in our recent computational study.42
Based on the above background, it is crucial for both understanding the intracellular protein degradation and related therapeutic development to uncover the detailed proteolytic mechanism of proteasome. The X-ray crystal structure study reveals that the mammalian constitutive (or regular) 20S proteasome is composed of 28 subunits, including four homoheptameric rings (α7β7β7α7) and each homoheptameric ring consists of seven subunits.43 There are three types of proteasome β-type subunits, i.e. β1, β2, and β5 that have caspase-like (C-L), trypsin like (T-L), and chymotrypsin-like (CT-L) activities, respectively. So, a total of six active sites of proteasome, including two β1 sites, two β2 sites, and two β5 sites, are functionally independent. All of them have an N-terminal threonine residue (Thr1) which can initiate a nucleophilic attack on substrate protein or small peptide. It has been known that the X-ray crystal structure of proteasome shows that each active site exists in the interface of two neighboring subunits.44 Many X-ray crystal structures of proteasome complexes with the various inhibitors have been reported.12, 23, 24, 34, 36, 41, 45–47 However, the X-ray crystal structure analysis cannot determine the details of the reaction process, although it has been carried out to detect the enzyme-ligand binding complexes36 or their reaction products.12, 23, 24, 34, 41, 45–47
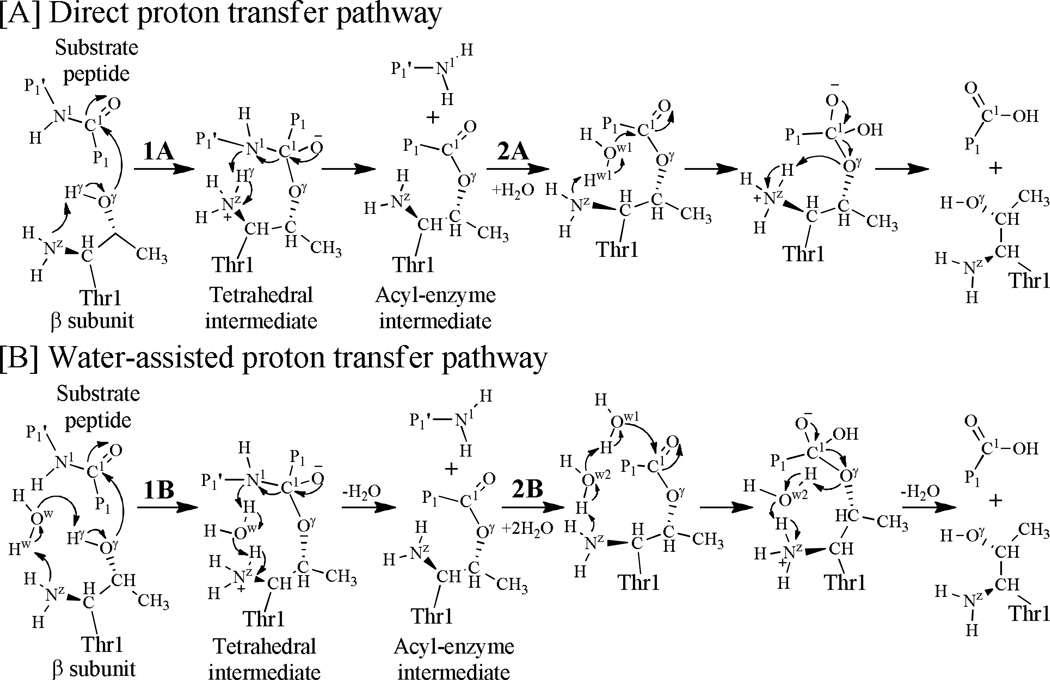
In the past decades, the detailed proteolytic mechanism of proteasome was hotly debated (see Scheme 1 for the reaction mechanisms suggested).7, 24, 47–51 According to the suggested mechanisms, the initial step of the reaction is a proton transfer between Thr1-Oγ and Thr1-Nz within proteasome before the formation of a tetrahedral intermediate. Notably, it has been unclear whether the nucleophile Thr1-Oγ is activated by its N-terminal amino group (Thr1-NzH2) directly without a water molecule (as depicted in Scheme 1A)51 or indirectly via a water molecule nearby (as depicted in Scheme 1B).7, 24, 47–50 The other reaction steps may also involve a proton transfer which may or may not be assisted by an additional water molecule. The key question between these possible reaction pathways is whether an additional water molecule is necessary to mediate the proton transfer processes involved in the proteasomal proteolytic reaction. None of these proposed reaction pathways has been confirmed by experiment or computational modeling so far. Here we report the first computational study on the detailed reaction pathways for the proteasomal proteolysis of a representative substrate.
Scheme 1.
Possible mechanisms leading to substrate peptide bond hydrolysis by the N-terminal Thr1 residue of proteasome. The Pn or Pn′ represents an amino acid residue, and the number n (n=1) refers to the nth residue to the cleavage site: P1 providing the carbonyl and P1′ the amino component.
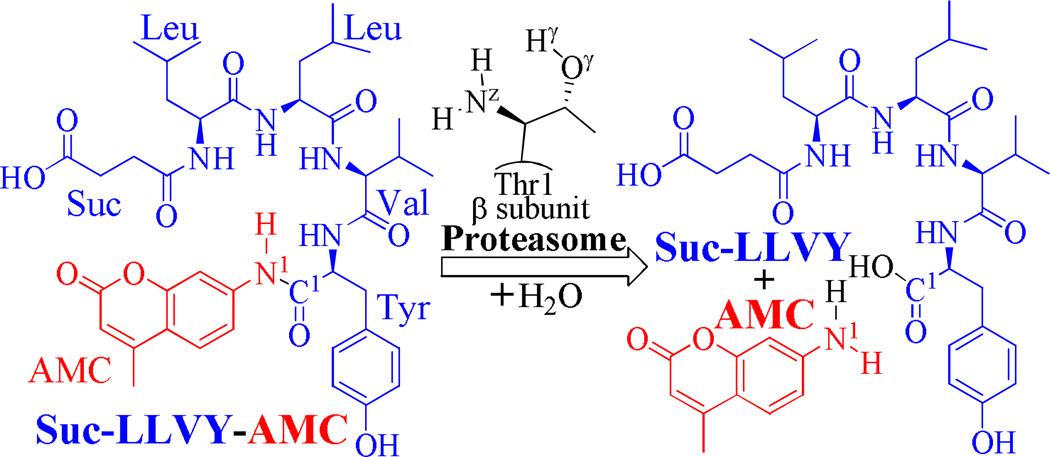
The representative substrate of proteasome examined in this computational study was a well-known fluorogenic peptide succinyl-leucyl-leucyl-valyl-tyrosyl-7-amino-4-methylcoumarin (Suc-LLVY-AMC), because the proteasomal hydrolysis of Suc-LLVY-AMC has been widely studied in experiments and the catalytic rate constant (kcat) for this reaction has been characterized.51, 52 We explored the possible reaction pathways for proteasomal hydrolysis of Suc-LLVY-AMC to generate the carboxylic acid Suc-LLVY and the amine AMC (see Scheme 2 for the reaction)2 by performing molecular dynamics (MD) simulations and first-principles quantum mechanical/molecular mechanical (QM/MM)-free energy (QM/MM-FE) calculations. By using the QM/MM-FE approach, first-principles QM/MM reaction-coordinate calculations were followed by free energy perturbation (FEP) simulations to account for the dynamic effects of the protein environment on the free energy profile for the enzymatic reaction process. Our QM/MM-FE calculations were based on the pseudobond first-principles QM/MM approach53–55 and the revised pseudobond QM/MM-FE implementation.56–61 The computational results clearly reveal the most favorable reaction pathway and the corresponding free energy profile. Based on the calculated free energy profile for the reaction process, the rate-determining step is identified, and the roles of essential residues are discussed on the basis of the QM/MM-optimized geometries.
Scheme 2.
The proteasomal proteolysis/hydrolysis of Suc-LLVY-AMC.
Computational Methods
Preparation of the Initial Enzyme-Substrate Structure
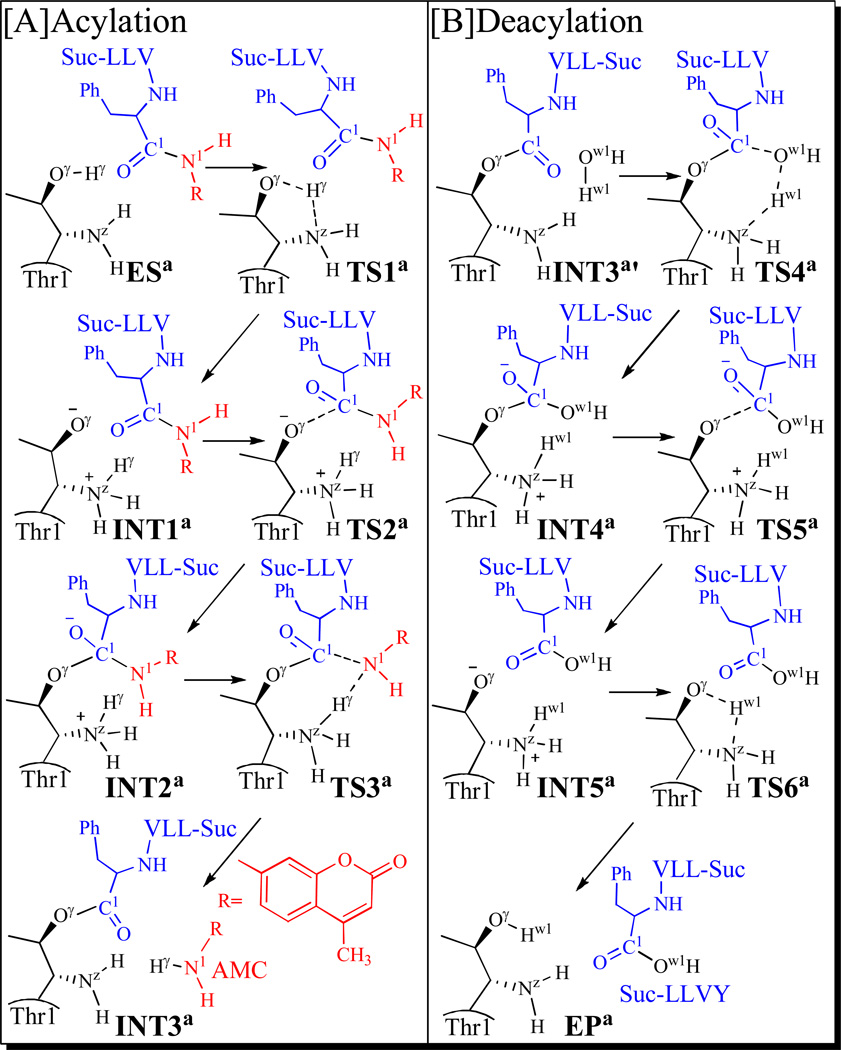
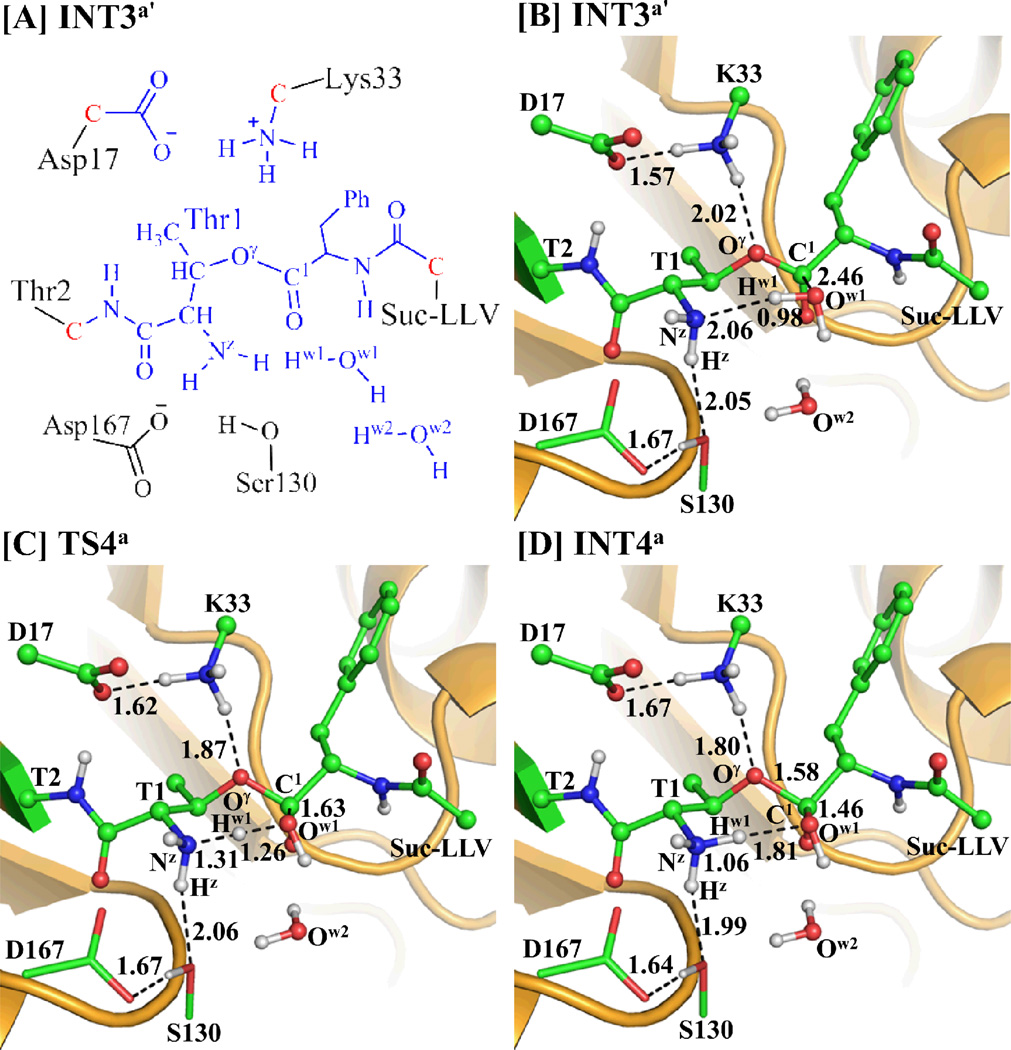
The initial structure of the enzyme-substrate (ES) complex used in the QM/MM reaction-coordinate calculations was constructed from the X-ray crystal structure of proteasome-inhibitor (EPX) complexes (PDB ID: 1G65)41 and our previously performed MD simulation on EPX binding with proteasome in the active site.42, 62 A snapshot from the MD simulation of protein-EPX complex62 was chosen. All the chloride ions and water molecules were removed and, then, EPX was replaced by the complete structure of peptide Suc-LLVY-AMC optimized at the ab initio HF/6-31G* level using the Gaussian03 program.63 The atomic charges of the substrate used in the MD simulation and the subsequent QM/MM calculations were the restrained electrostatic potential (RESP) charges that were determined by first performing the electrostatic potential calculations at the HF/6-31G* level using the Gaussian03 program63 and then fitting with the standard RESP procedure implemented in the Antechamber module of the AMBER11 program.64 As noted above, proteasome consists of 28 subunits (α7β7β7α7) and has six active sites (including two β1 sites, two β2 sites, and two β5 sites) that are functionally independent. All of the active sites are very similar; each active site exists in the interface of two neighboring subunits, has a reactive residue Thr1, and has the same catalytic activity. The β5 active site exists in the interface of the subunits β5 and β6, according to the X-ray crystal structure of proteasome-EPX complex.41, 62 Hence, the initial structure of ES was constructed by retaining only two subunits (β5 and β6) and superimposing the substrate (Suc-LLVY-AMC) with EPX according to the X-ray crystal structure of proteasome-EPX complex. We also considered possible alternative conformations, but noted that none of the other conformations is suitable for the desirable enzymatic reaction. The initial ES binding complex structure (see Figure S1 of Supporting Information) was neutralized by adding four chloride ions and was solvated in an orthorhombic box of TIP3P water molecules65 with a minimum solute-wall distance of 10 Å. The solvated system was refined by performing a ~50 ns MD simulation in the same way as described in our previous computational study on another complex between the enzyme and inhibitor.62 As seen in Scheme 3, there are two stages in the proteasomal hydrolysis of the substrate. The amine AMC leaves the system after the acylation stage (depicted in Scheme 3A). Consequently, we constructed the structure of INT3´ (depicted in Scheme 3B) by removing the amine AMC from the QM/MM-optimized INT3 structure. The constructed INT3´ structure was then relaxed by performing ~50 ns of MD simulation in which the system was also solvated in an orthorhombic box of TIP3P water molecules,65 with a minimum solute-wall distance of 10 Å.
Scheme 3.
Possible reaction pathway for the acylation (A) and deacylation (B) stages of the proteosomal hydrolysis of Suc-LLVY-AMC.
For both the acylation and deacylation stages, the last snapshots of the MD simulations were used to prepare the first-principles QM/MM calculations because the structure of the last snapshot was close to the average structure simulated. Since we are interested in the reaction center, the water molecules beyond 50 Å of the carbonyl carbon (C1) of substrate were removed, leaving the QM/MM system with 3,819 water molecules and a total of 17,960 atoms for the acylation stage, and 2,601 water molecules and a total of 14,317 atoms for the deacylation stage. A pseudobond approach53–55 was used to treat the QM-MM interface; see below for the boundary of the QM-MM system. Prior to the QM/MM geometry optimization, the initial structure of the whole reaction system was energy-minimized by using the MM method and the AMBER11 program,64 and the convergence criterion for the energy gradient of 0.1 kcal·mol−1·Å−1 was achieved.
Minimum-Energy Path of the Enzymatic Reaction
By using a reaction-coordinate driving method and an iterative energy minimization procedure,53 the enzymatic reaction path was determined through the pseudobond QM/MM calculations at the B3LYP/6-31G*:AMBER level. In the QM/MM calculations, the QM calculation was performed at the B3LYP/6-31G* level of theory by using a modified version56 of Gaussian03 program63 and the MM calculation was performed by using a modified version56 of the AMBER8 program.66 Following the geometry optimizations, normal mode analysis was performed to verify/confirm the reactant, intermediates, transition sates, and the final product of the reaction process. Further, the QM/MM-optimized geometries were used to carry out single-point energy calculations at the QM/MM(B3LYP/6-31++G**:AMBER) level. For all of the QM/MM calculations, the improved pseudobond parameters54 were used to treat the boundary carbon atoms, and no cutoff for non-bonded interactions was used. For geometry optimization on the QM subsystem, the convergence criterion followed the original Gaussian03 defaults. For the geometry optimization on the MM subsystem, the convergence criterion was when the root-mean-square deviation (rmsd) of energy gradient was ≤ 0.1 kcal·mol−1·Å−1. The atoms within 20 Å of C1 atom in substrate (Scheme 3) were allowed to move while the other atoms were kept frozen in all the QM/MM calculations. The QM and MM subsystems were energy-minimized iteratively during the QM/MM geometry optimization.
Free Energy Perturbation
Free energy perturbation (FEP) can be used to reasonably evaluate free energy change caused by a small structural change.67–70 This method, in combination with the MD or Monte Carlo (MC) simulation, has been used extensively in computational studies on organic reactions,67, 70–72 protein-ligand interactions,70, 73–78 and protein stability.79, 80 In this computational study, after the minimum-energy path was determined by the QM/MM calculations, the free energy changes associated with the QM-MM interactions were determined by using the FEP implementation (in our revised version of the AMBER program).57 The sampling of the MM subsystem was performed with the QM subsystem frozen at each state along the reaction path. During the FEP calculations, the used point charges on the frozen QM atoms were determined by fitting the electrostatic potential (ESP) in the QM part of the QM/MM single-point calculations. The total free energy change from one state to another was evaluated by using the same procedure as used in our previous computational studies on other enzymatic reaction systems.56–60, 81–83 The MD-based FEP calculations enabled us to more reasonably evaluate the relative free energy changes due to the interactions between the QM and MM subsystems. The final (relative) free energy obtained from the QM/MM-FE calculations was the QM part of the QM/MM energy (excluding the Coulombic interaction energy between the point charges of the MM atoms and the ESP charges of the QM atoms) plus the relative free energy change evaluated by the FEP calculations at 298.15 K. The time step used in the MD-based FEP calculations was 2 fs, and the lengths of all covalent bonds involving a hydrogen atom were constrained. Each one of the MD-based FEP calculations consisted of 50 ps of equilibration and 300 ps of sampling.
All MD simulations and QM/MM-FE calculations were performed on a Dell supercomputer (X-series Cluster with 384 nodes or 4,768 processors) at the University of Kentucky’s Computer Center. The other modeling and computations were performed on SGI Fuel workstations in our own laboratory at the University of Kentucky.
Results and Discussion
Mechanistic insights from MD simulations
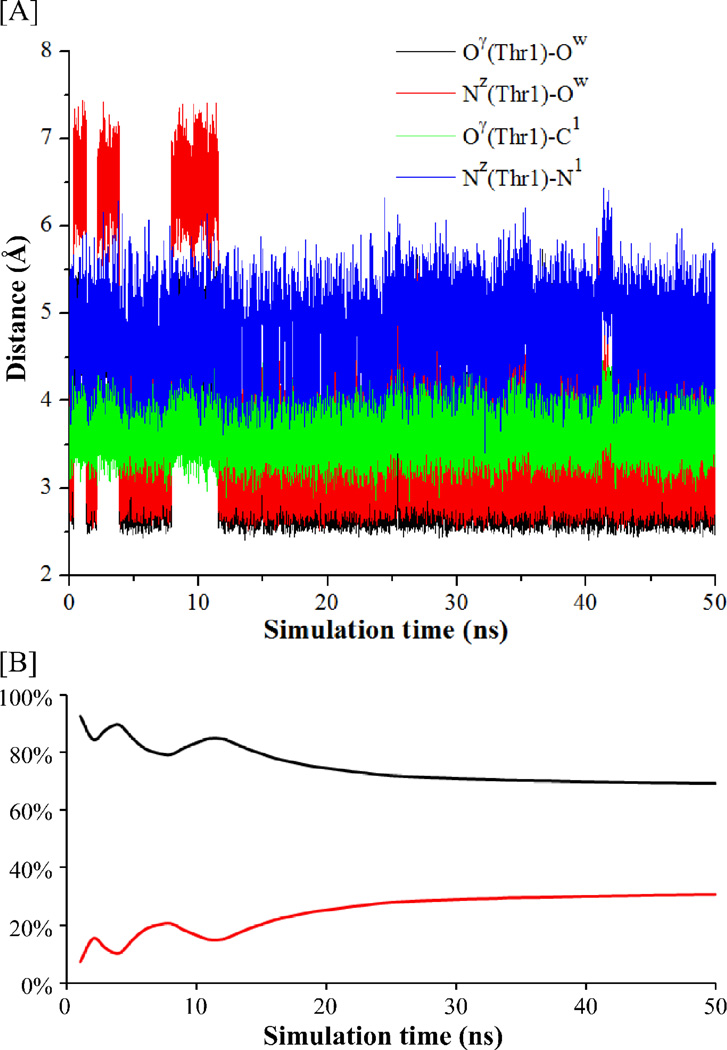
According to the previously proposed enzymatic reaction mechanism,7, 24, 47–50 a solvent water molecule may or may not exist in the reaction between the Thr1-Oγ atom and the Thr1-NzH2 group of the enzyme, and such a water molecule can mediate the first proton transfer process (see Scheme 1). To examine whether a water molecule can exist between the Thr1-Nz and Thr1-Oγ atoms, the ES structure was relaxed by performing the MD simulation for ~50 ns. After the MD simulation, we noted that only one water molecule could stay close to both the Thr1-Nz and Thr1-Oγ atoms at the same time. Depicted in Figure 1 are the tracked changes (time courses) of two key internuclear distances, i.e. the Oγ(Thr1)–Ow distance (between Thr1-Oγ and the oxygen atom of the water molecule) and the Nz(Thr1)–Ow distance (between Thr1-Nz and the oxygen atom of the water molecule). In addition, we also tracked the changes of the Oγ(Thr1)–C1 distance (between the Thr1-Oγ atom and the C1 atom of substrate) and the Nz(Thr1)–N1 distance (between the Thr1-Nz atom and the N1 atom of substrate) in Figure 1, as the new covalent bond C1–Oγ is formed and a proton transfers from Thr1-Nz to N1 of substrate during the hydrolysis reaction.
Figure 1.
[A] Key internuclear distances vs the simulation time (t) in the MD-simulated ES structure. [B] The calculated percentage of the snapshots of the MD-simulated ES structure that favor the direct proton-transfer pathway (black line) or the water-assisted proton-transfer pathway (red line) in the acylation stage. The percentage given at each time point (t) refers to that based on all of the snapshots collected between 0 and t, showing the convergence of the MD simulation in terms of the distribution of the two types of snapshots.
As seen in Figure 1A, the simulated average Oγ(Thr1)–Ow, Nz(Thr1)–Ow, Oγ(Thr1)–C1, and Nz(Thr1)–N1 distances are ~3.17, ~3.61, ~3.70, and ~4.64 Å, respectively. Overall, the average Nz(Thr1)–Ow and Oγ(Thr1)–Ow distances are too long to mediate the proton transfer between Thr1-Nz and Thr1-Oγ, suggesting that the water molecule was not in a favorable position to assist the proton transfer. Checking the distances in all of the collected 50,000 snapshots (one per ps) depicted in Figure 1, only 15,421 snapshots (~30.8%, depicted in Figure 1B) might be suitable for the water-assisted proton-transfer when both the Nz(Thr1)–Ow and Oγ(Thr1)–Ow distances were shorter than 3.2 Å. In the remaining 34,579 snapshots (~69.2%, depicted in Figure 1B), the Nz(Thr1)–Ow and Oγ(Thr1)–Ow distances were too long for the water-assisted proton-transfer. So, ~69.2% snapshots favor the pathway of activating Thr1-Oγ directly by Thr1-Nz. This reaction pathway is associated with a direct proton transfer from Thr1-Oγ to Thr1-Nz. According to the insights obtained from the MD simulation and the previously proposed mechanisms (as mentioned above), both the possible direct proton-transfer and water-assisted proton-transfer pathways were examined. For convenience of discussion below, superscript “a” is always used to represent the stationary states associated with the direct proton-transfer reaction pathway and superscript “b” always refers to the stationary states corresponding to the water-assisted proton-transfer reaction pathway.
Reaction pathway for the direct proton transfer
In light of the results from the MD simulation, one may first reasonably assume that the hydroxyl oxygen of Thr1 side chain (Thr1-Oγ) might be activated directly by its N-terminal amino group (Thr1-NzH2). Further, we had a working hypothesis of the possible hydrolysis pathway of the substrate as depicted in Scheme 3. The possible reaction pathway shown in Scheme 3 has been confirmed by our QM/MM reaction-coordinate calculations, as discussed below.
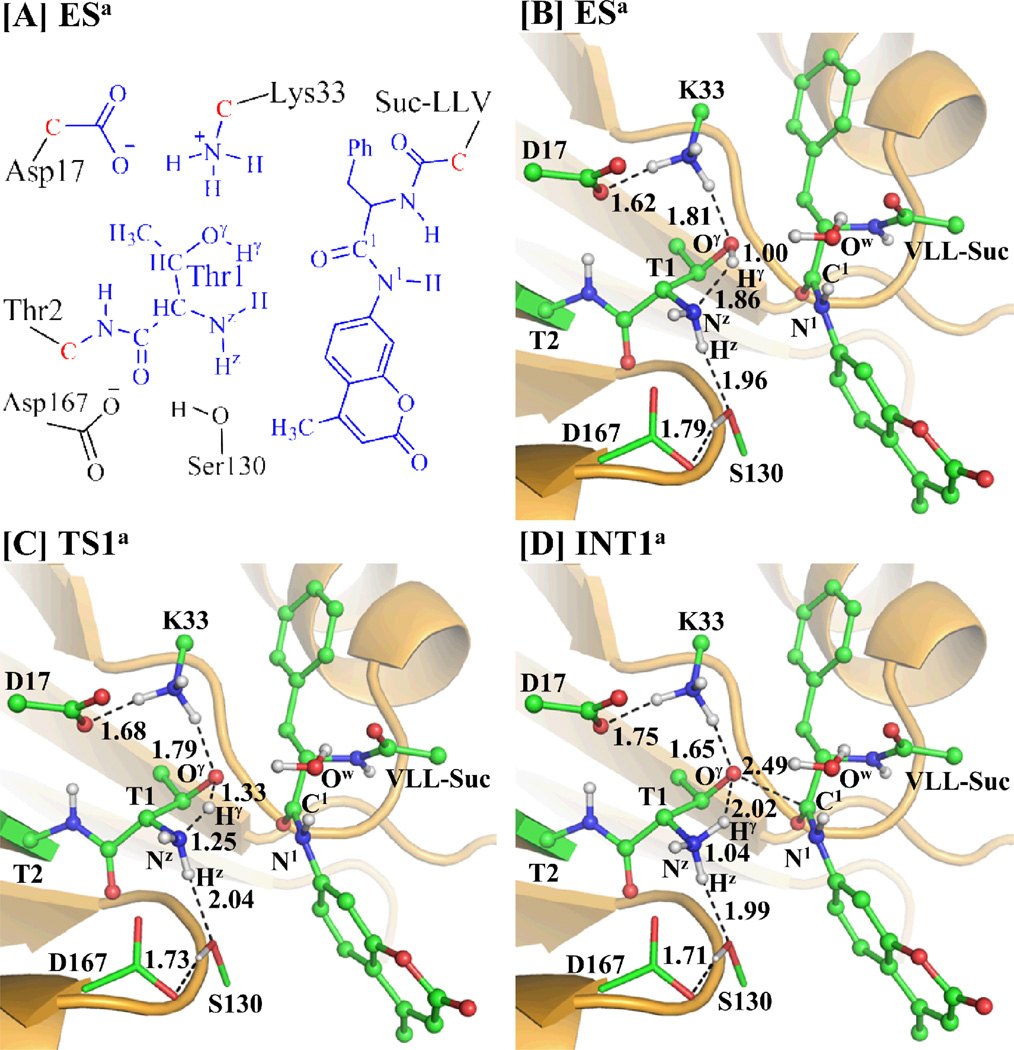
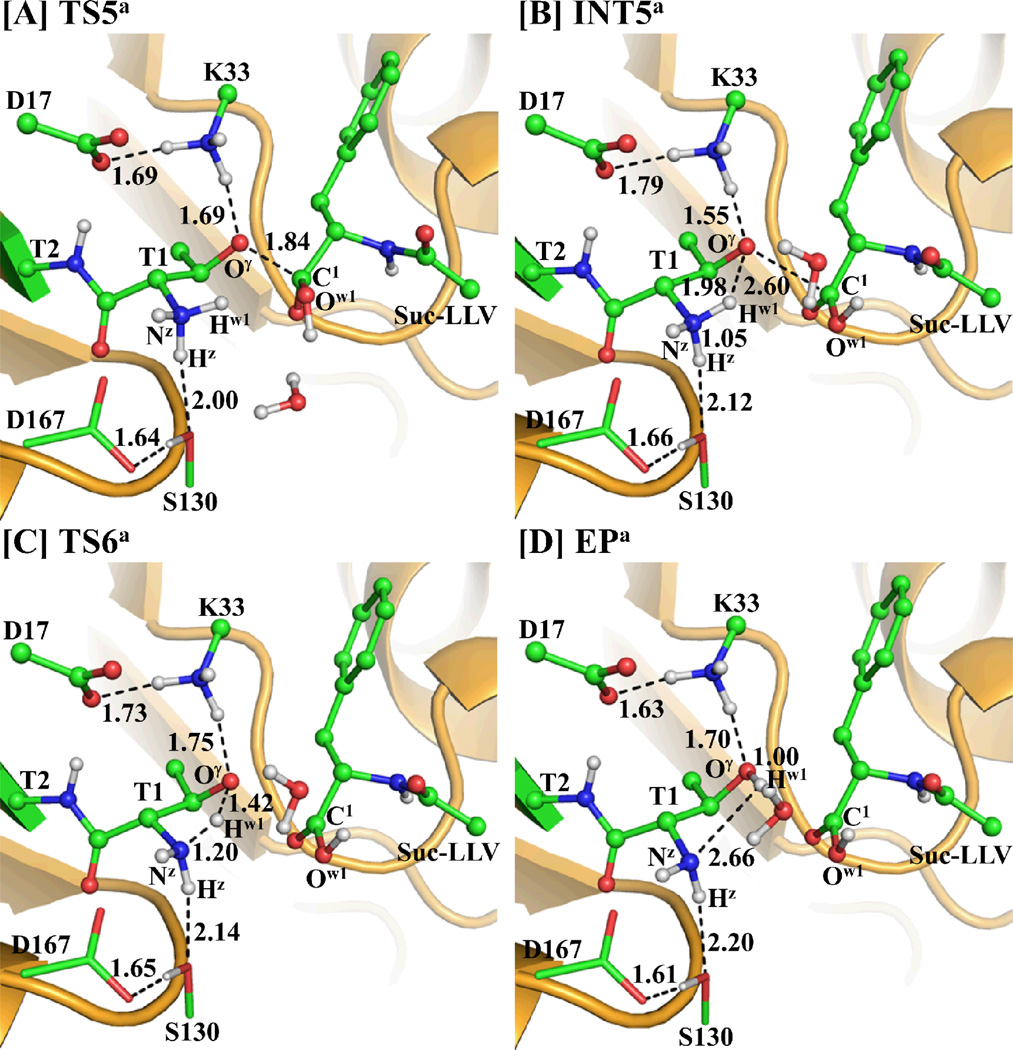
Starting from our QM/MM-optimized ESa structure in the acylation stage and INT3a´ structure in the deacylation stage, the QM/MM reaction-coordinate calculations were performed at the B3LYP/6-31G*:AMBER level. The results obtained from the QM/MM calculations revealed that the hydrolysis of substrate indeed should consist of six reaction steps, including the acylation stage (steps 1 to 3) depicted in Scheme 3A and the deacylation stage (steps 4 to 6) as depicted in Scheme 3B. The first reaction step is a direct proton (Hγ) transfer from the Thr1-Oγ atom to the Thr1-Nz atom within the enzyme, forming a zwitterionic intermediate INT1a via transition state TS1a. The second reaction step is a nucleophilic attack on the C1 atom of substrate by the activated Thr1-Oγ via transition state TS2a. The third reaction step is a proton (Hγ) transfer from Thr1-Nz to the nitrogen atom (N1) of substrate, coupled with the breaking of the C1–N1 bond via transition state TS3a. The fourth reaction step is a concerted process, i.e. nucleophilic attack on the C1 atom of substrate by the oxygen (Ow1) atom of a water molecule, accompanied with a proton transfer from WAT-Ow1 to Thr1-Nz, resulting in the formation of another zwitterionic intermediate INT4a via transition state TS4a. The fifth reaction step is breaking of the C1–Oγ bond, which proceeds from intermediate INT4a to the zwitterionic intermediate INT5a via transition state TS5a. The sixth reaction step is the regeneration of Thr1 by a proton transfer from Thr1-Nz to Thr1-Oγ via transition state TS6a. Depicted in Figures 2 to 5 are the QM/MM-optimized geometries of the reactant, intermediates, transition states, and product of the enzymatic reaction process.
Figure 2.
(A) Division of the QM/MM system. Atoms in blue color were treated as QM part. The boundary carbon atoms colored in red were treated with the improved pseudobond parameters. All of the other atoms were considered as the MM subsystem. (B to D) Optimized geometries for the key states during the acylation stage of the proteosomal hydrolysis of substrate in the β5 active site. The geometries were optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The key distances in the figures are in Å. Carbon, oxygen, nitrogen, and hydrogen atoms are colored in green, red, blue, and white, respectively. The backbone of the protein is rendered as ribbon and colored orange. The QM atoms are represented as balls and sticks and the surrounding residues are rendered as sticks or lines. For clarity, nonpolar hydrogen atoms in the QM part have been hidden in the structures.
Figure 5.
Optimized geometries for the key states during the deacylation stage of the proteosomal hydrolysis of substrate. The QM part is the same with Figure 4A and the geometries were also optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
During the QM/MM calculations for the acylation stage (consisting of steps 1 to 3), as shown in Figure 2A, the atoms colored in blue were treated by the QM method, the boundary atoms colored red in Figure 2A were treated with the improved pseudobond parameters,54 and the other atoms of the reaction system were considered as the MM subsystem. In addition, a water molecule nearby the active site was also included in the QM part, which may or may not mediate the proton transfer in the previously proposed mechanism (depicted in Scheme 1).
Step 1: Proton Hγ Transfers from Thr1-Oγ to Thr1-Nz
As shown in Figure 2B for the QM/MM-optimized ESa structure, both the internuclear distance between the Thr1-Oγ and the closest hydrogen atom of the Lys33-NH3+ group and the internuclear distance between the Thr1-Hz and Ser130-O atoms are shorter than 2.00 Å. The internuclear distance between the Lys33-NH3+ hydrogen and Asp17-CO2− oxygen atoms is 1.62 Å, while the internuclear distance between the Ser130-H (hydroxyl hydrogen) and the Asp167-CO2− oxygen is 1.79 Å, revealing a hydrogen-bond network in the reaction center.
As shown in Scheme 3A, the reaction step 1 involves breaking of the Hγ–Oγ bond and formation of the Hγ–Nz bond. Therefore, RHγ-Oγ–RHγ-Nz was used as the reaction coordinate for the QM/MM reaction-coordinate calculations on reaction step 1. As shown in the QM/MM-optimized geometries (Figure 2B to 2D), the RHγ-Oγ elongates from 1.00 Å in ESa (Figure 2B) to 1.33 Å in TS1a (Figure 2C), while the RHγ-Nz shortens from 1.86 Å in ESa to 1.25 Å in TS1a and then to 1.04 Å in INT1a. Noteworthy, during this reaction step, the hydrogen bond between Thr1-Oγ and Lys33-NH3+ group is strengthened, which helps to stabilize the zwitterionic intermediate INT1a.
Step 2: Nucleophilic Attack on C1 of Substrate by Thr1-Oγ
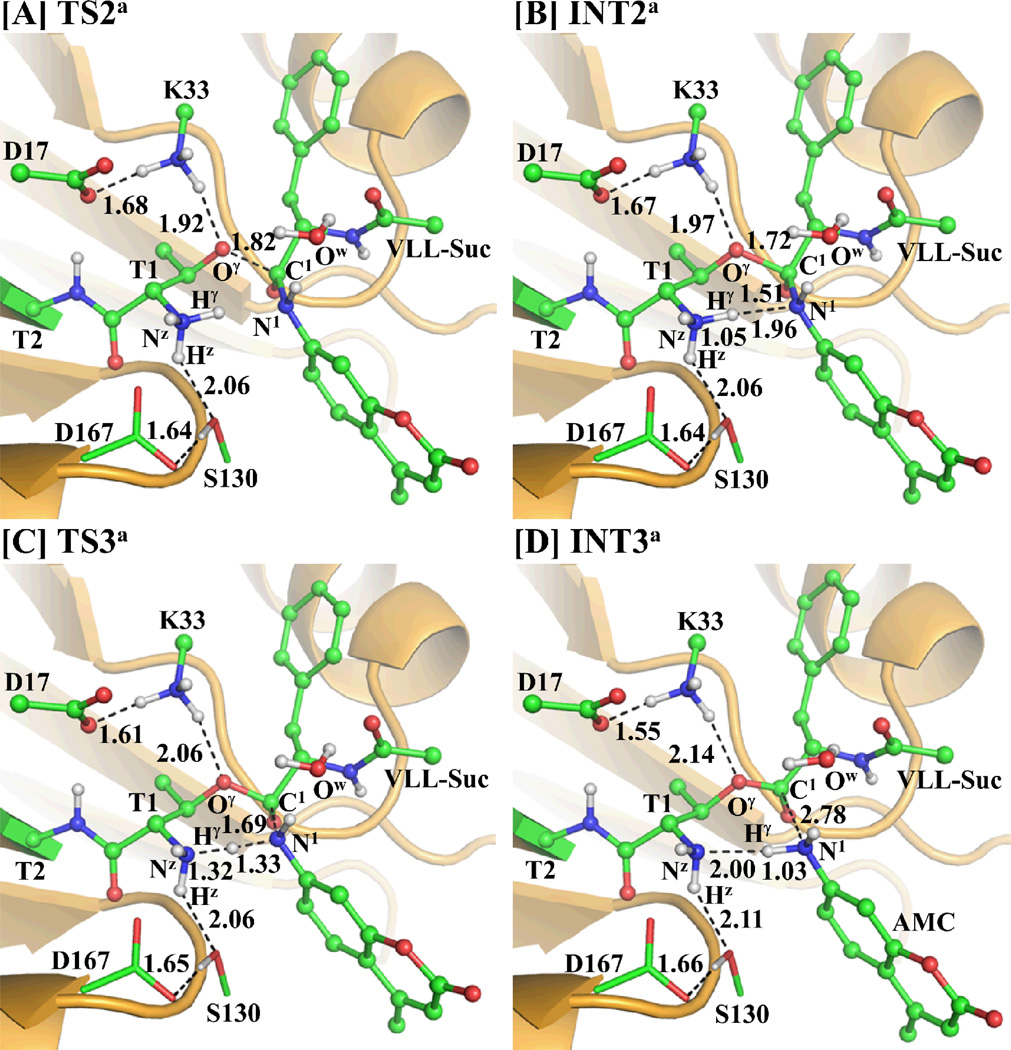
In this reaction step, the nucleophilic attack on C1 atom of substrate is initiated by the negatively charged Thr1-Oγ atom and, thus, the Oγ–C1 bond is formed. This reaction step is featured by the formation of the Oγ–C1 bond and, therefore, the reaction coordinate for this reaction step was chosen as ROγ-C1. The distance ROγ-C1 is shortened from 2.49 Å in INT1a (Figure 2D) to 1.82 Å in TS2a (Figure 3A) and then to 1.72 Å in a charged tetrahedral intermediate INT2a (Figure 3B).
Figure 3.
Optimized geometries for the key states during the acylation stage of the proteosomal hydrolysis of substrate. The QM part is the same with Figure 2A and the geometries were also optimized at QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
Step 3: Dissociation of the Amine AMC
In this reaction step, a proton (Hγ) transfers from Thr1-Nz to N1 of substrate, coupled with the breaking of the C1–N1 bond. This reaction step involves the breaking of the Nz–Hγ and C1–N1 bonds, and the formation of the N1–Hγ bond. The nature of the reaction process can be represented by the changes of the Nz–Hγ distance (RNz-Hγ), C1–N1 distance (RC1–N1), and N1–Hγ distance (RN1-Hγ). Thus, the reaction coordinate for this step was chosen as RNz-Hγ+ RC1–N1–RN1-Hγ. During this reaction step, the distance RN1-Hγ is shortened from 1.96 Å in INT2a (Figure 3B) to 1.33 Å in TS3a (Figure 3C) and then to 1.03 Å in intermediate INT3a (Figure 3D). Meanwhile, RNz-Hγ changes from 1.05 Å in INT2a to 1.32 Å in TS3a and then to 2.00 Å in INT3a, while RC1–N1 changes from 1.51 Å in INT2a to 1.69 Å in TS3a and then to 2.78 Å in INT3a.
During the QM/MM calculations for the deacylation stage (steps 4 to 6), as shown in Figure 4A, the atoms colored in blue were treated by the QM method, the boundary atoms colored red in Figure 4A were treated with the improved pseudobond parameters,54 and the other atoms of the reaction system were considered as the MM subsystem. The amine AMC was removed from the above discussed QM/MM-optimized geometry of INT3a to construct the structure of INT3a´, which was then relaxed by performing the ~50 ns MD simulation. Based on the relevant internuclear distances in the collected 50,000 snapshots (one per ps), 48,459 snapshots (~96.9%) might be suitable for the deacylation reaction when at least one water molecule was close to the active site (where the distance between Nz(Thr1) and a water oxygen was shorter than 4.0 Å). Within the 48,459 snapshots suitable for the deacylation reaction, only 2,088 snapshots (~4.2%) might be suitable for the water-assisted proton-transfer because at least two water molecules were close to the active site (where the distances between Nz(Thr1) and two water oxygen atoms were all shorter than 6.0 Å). Moreover, two water molecules (depicted in Figure 4) close to the carbonyl carbon (C1) of substrate were included in the QM part. One of the two water molecules would initiate nucleophilic attack on the carbonyl carbon C1 of the substrate, and the other one may mediate the proton transfer process according to one of the above-mentioned possible mechanisms (depicted in Scheme 1).
Figure 4.
(A) Division of the QM/MM system. Atoms in blue color were treated as QM part. The boundary carbon atoms colored in red were treated with the improved pseudobond parameters. All of the other atoms were considered as the MM subsystem. (B to D) Optimized geometries for the key states during the deacylation stage of the proteosomal hydrolysis of substrate in the β5 active site. The geometries were optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
Step 4: Nucleophilic Attack on C1 of Substrate by Ow1 of a Water Molecule and Proton Transfer from Ow1 to Thr1-Nz
Along with the nucleophilic attack on the C1 atom of substrate by the Ow1 atom of a water molecule, the proton Hw1 transfers from the Ow1 atom to the Nz atom at the same time to generate a zwitterionic intermediate INT4a. Such concerted process involves the breaking of the Ow1–Hw1 bond and the formation of the C1–Ow1 and Nz–Hw1 bonds (Scheme 3B). Thus, the distances ROw1–Hw1, RC1–Ow1, and RNz–Hw1 were chosen to represent the reaction coordinate as ROw1–Hw1–RC1–Ow1–RNz–Hw1 for the current reaction step. The distance ROw1–Hw1 elongates from 0.98 Å in INT3a´ (Figure 4B) to 1.26 Å in TS4a (Figure 4C) and then to 1.81 Å in INT4a (Figure 4D), while the distances RC1–Ow1 and RNz–Hw1 shorten respectively from 2.46 and 2.06 Å in INT3a´ to 1.63 and 1.31 Å in TS4a and then to 1.46 and 1.06 Å in INT4a.
Step 5: Breaking of the C1–Oγ bond
As seen from Scheme 3B, the C1–Oγ bond is broken in this step. The change in distance RC1–Oγ reflects the nature of the current reaction step. Thus, the reaction coordinate for the current reaction step was expressed as RC1–Oγ. The distance RC1–Oγ elongates from 1.58 Å in INT4a to 1.84 Å in TS5a (Figure 5A), and then to 2.60 Å in INT5a (Figure 5B). Moreover, the hydrogen-bond interaction between Thr1-Oγ and residue Lys33 becomes stronger from 1.80 Å in INT4a to 1.55 Å in INT5a via 1.69 Å in TS5a, which is helpful to stabilize the transition state TS5a and the zwitterionic intermediate INT5a.
Step 6: Regeneration of the Residue Thr1
As seen from Scheme 3B, starting from intermediate INT5a, the negatively charged Oγ atom abstracts a proton (Hw1) from the positively charged Nz atom. Accompanied by breaking of the Nz–Hw1 bond, the Hw1–Oγ bond is formed. The changes in distances RNz-Hw1 and RHw1-Oγ reflect the nature of the current reaction step. Thus, the reaction coordinate for the current reaction step was expressed as RNz-Hw1–RHw1-Oγ. While the distance RNz-Hw1 changes from 1.05 Å in INT5a to 1.20 Å in TS6a (Figure 5C) and then to 2.66 Å in EPa (Figure 5D), the distance RHw1-Oγ shortens from 1.98 Å in INT5a to 1.42 Å in TS6a and then to 1.00 Å in EPa.
Reaction pathway for the water-assisted proton transfer
As indicated in Scheme 1, the proton transfer during the hydrolysis reaction between the peptide and proteasome may also be mediated by an additional water molecule. For this reason, we also accounted for the possibility of the water-assisted proton-transfer pathway for each of the relevant reaction steps corresponding to the first, third, fourth, and sixth steps of the fundamental reaction pathway (corresponding to the direct proton transfer) shown in Scheme 3. Depicted in Scheme 4 are the possible water-assisted proton-transfer pathways associated with these steps.
Scheme 4.
The possible water-assisted proton transfer pathways for the first (A), third (B), fourth (C), and sixth (D) reaction steps depicted in Scheme 3.
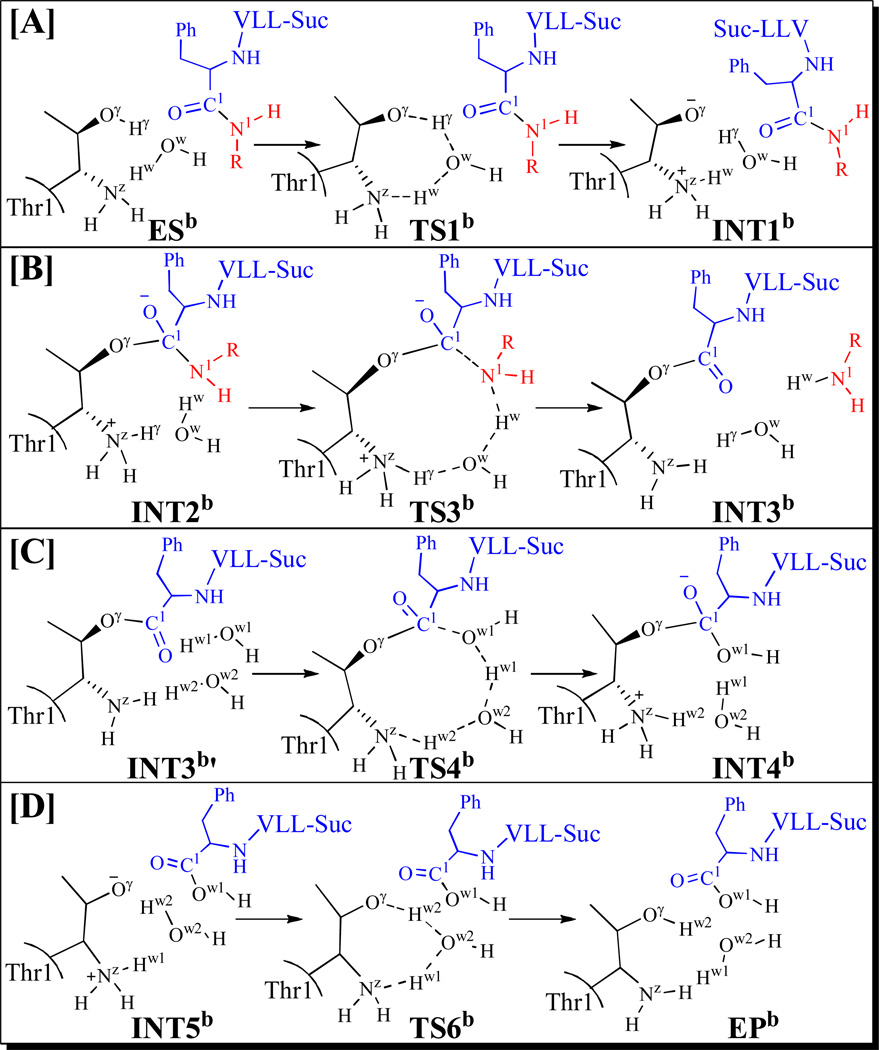
The possible water-assisted proton transfer depicted in Scheme 4A involves breaking of the Oγ–Hγ and Ow–Hw bonds and formation of the Ow–Hγ and Nz–Hw bonds. Hence, the internuclear distances ROγ-Hγ, ROw-Hw, ROw-Hγ, and RNz-Hw were used to represent the reaction coordinate as ROγ-Hγ+ROw-Hw–ROw-Hγ–RNz-Hw for the QM/MM reaction-coordinate calculations. Such a water-assisted proton-transfer pathway was confirmed by the QM/MM reaction-coordinate calculations. According to the QM/MM reaction-coordinate calculations, the distances ROw-Hγ and RNz-Hw are shortened respectively from 1.62 and 1.67 Å in ESb (Figure 6A) to 1.28 and 1.16 Å in TS1b (Figure 6B), and then to 1.04 and 1.06 Å in INT1b (Figure 6C). Meanwhile, the corresponding distances ROγ-Hγ and ROw-Hw are elongated respectively from 1.00 and 1.02 Å in ESb to 1.15 and 1.38 Å in TS1b, and then to 1.46 and 1.62 Å in INT1b.
Figure 6.
Optimized geometries of intermediates ESb, INT1b, and the transition state TS1b. The QM part is the same with Figure 2A and the geometries were optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
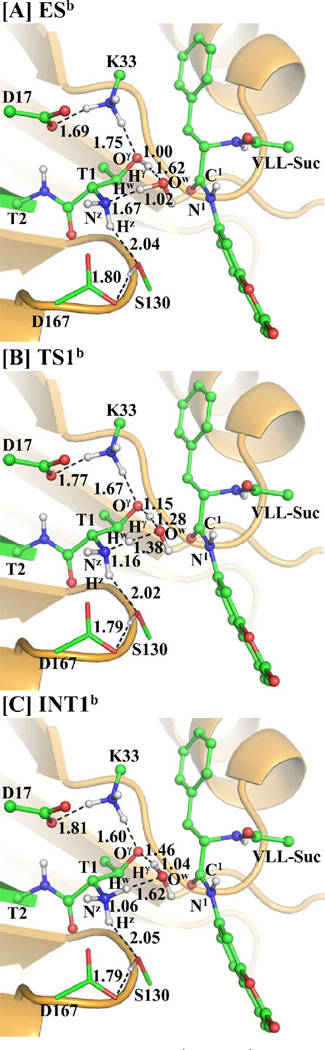
A water molecule may also assist the proton transfer during the structural change from INT2b to INT3b via transition state TS3b, as depicted in Scheme 4B. This possible water-assisted proton-transfer pathway involves the breaking of the Nz–Hγ, Ow–Hw, and C1–N1 bonds and the formation of the Ow–Hγ and N1–Hw bonds. Thus, the distances RNz-Hγ, ROw-Hw, RC1-N1, ROw-Hγ, and RN1-Hw were chosen to represent the reaction coordinate as RNz-Hγ+ROw-Hw+RC1-N1–ROw-Hγ–RN1-Hw in the QM/MM reaction-coordinate calculations. According to the reaction-coordinate calculations, the distances ROw-Hγ and RN1-Hw are shortened respectively from 1.52 and 1.69 Å in INT2b (Figure 7A) to 1.39 and 1.44 Å in TS3b (Figure 7B), and then to 1.01 and 1.03 Å in INT3b (Figure 7C), while the corresponding distances RNz-Hγ, ROw-Hw, and RC1-N1 are elongated respectively from 1.07, 1.00, and 1.52 Å in INT2b to 1.13, 1.09, and 2.14 Å in TS3b and then to 1.66, 1.74, and 3.07 Å in INT3b.
Figure 7.
Optimized geometries of intermediates INT2b, INT3b, and transition state TS3b. The QM part is the same with Figure 2A and the geometries were also optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
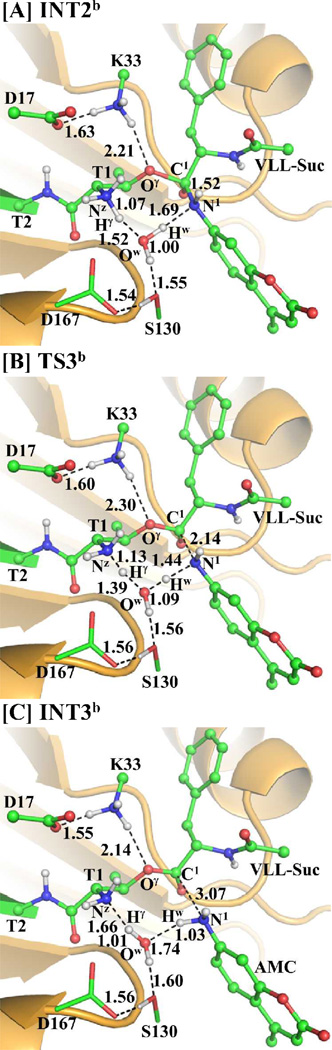
Besides the water molecule for nucleophilic attack on the carbonyl carbon C1 of substrate in the step 4, an additional water molecule may also mediate the proton transfer during the transformation from INT3b´ to INT4b via transition state TS4b, as depicted in Scheme 4C. This possible water-mediated proton-transfer pathway involves the breaking of the Ow1–Hw1 and Ow2–Hw2 bonds and the formation of the Nz–Hw2, Ow2–Hw1, and C1–Ow1 bonds. Thus, the distances ROw1-Hw1, ROw2-Hw2, RNz-Hw2, ROw2-Hw1, and RC1-Ow1 were chosen to represent the reaction coordinate as ROw1-Hw1+ ROw2-Hw2–RNz-Hw2–ROw2-Hw1–RC1-Ow1 in the QM/MM reaction-coordinate calculations. According to the reaction-coordinate calculations, the distances ROw1-Hw1 and ROw2-Hw2 are elongated respectively from 0.99 and 0.99 Å in INT3b´ (Figure 8A) to 1.29 and 1.18 Å in TS4b (Figure 8B), and then to 1.85 and 1.75 Å in INT4b (Figure 8C), while the corresponding distances RNz-Hw2, ROw2-Hw1, and RC1-Ow1 are shortened respectively from 1.90, 1.77, and 2.56 Å in INT3b´ to 1.36, 1.15, and 1.72 Å in TS4b and then to 1.06, 0.98, and 1.47 Å in INT4b.
Figure 8.
Optimized geometries of intermediates INT3b´, INT4b, and transition state TS4b. The QM part is the same with Figure 4A and the geometries were also optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
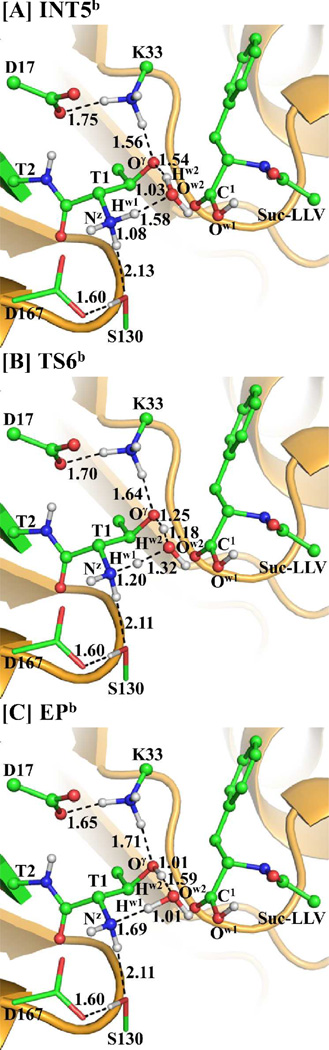
Similarly, an additional water molecule may also mediate the proton transfer during the transformation from INT5b to EPb via transition state TS6b, as depicted in Scheme 4D. This possible water-mediated proton-transfer pathway involves the breaking of the Nz–Hw1 and Ow2–Hw2 bonds and the formation of the Ow2–Hw1 and Oγ–Hw2 bonds. Thus, the distances RNz-Hw1, ROw2-Hw2, ROw2-Hw1, and ROγ–Hw2 were chosen to represent the reaction coordinate as RNz-Hw1+ ROw2-Hw2–ROw2-Hw1–ROγ–Hw2 in the QM/MM reaction-coordinate calculations. According to the reaction-coordinate calculations, the distances RNz-Hw1 and ROw2-Hw2 are elongated respectively from 1.08 and 1.03 Å in INT5b (Figure 9A) to 1.20 and 1.18 Å in TS6b (Figure 9B), and then to 1.69 and 1.59 Å in EPb (Figure 9C), while the corresponding distances ROw2-Hw1 and ROγ–Hw2 are shortened respectively from 1.58 and 1.54 Å in INT5b to 1.32 and 1.25 Å in TS6b and then to 1.01 and 1.01 Å in EPb.
Figure 9.
Optimized geometries of intermediates INT5b, EPb, and transition state TS6b. The QM part is the same with Figure 4A and the geometries were also optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. The color scheme is the same as that of Figure 2.
It should be noted that we have discussed the possible alternative (water-assisted proton transfer) pathways for the first, third, fourth, and sixth steps of the reaction. This is because the second and fifth reaction steps do not involve a proton transfer.
Free energy profiles
As discussed above, the QM/MM reaction-coordinate calculations at the B3LYP/6-31G*:AMBER level have revealed six reaction steps (steps 1 to 3 in the acylation stage and steps 4 to 6 in the deacylation stage) for the fundamental proteosomal hydrolysis pathway of substrate. Further, the QM/MM single-point energy calculations were performed at the B3LYP/6-31++G**:AMBER level for each QM/MM optimized geometry along the minimum-energy path to determine the free energy profile of the reaction process. For each structure along the reaction path, the ESP charges calculated in the QM part of the QM/MM calculation were used in the subsequent FEP simulations. Depicted in Figures 10 and 11 are the free energy profiles for the competing reaction pathways associated with the direct and water-mediated proton-transfer processes. The free energy profiles were determined by the QM/MM-FE calculations first without the zero-point and thermal corrections for the QM subsystem, and then with the zero-point and thermal corrections for the QM subsystem (values given in parentheses).
Figure 10.
Free energy profile of the acylation determined by the QM/MM-FE calculations for the reaction pathways associated with the direct proton transfer (A) and the water-assisted proton transfer (B). There were 84 points (structures) in Figure 10A and 96 points (structures) in Figure 10B along the reaction coordinate used in the FEP calculations. The relative free energies were determined first without zero-point and thermal corrections, and then corrected with the zero-point and thermal corrections for the QM subsystem (values in parentheses). The QM/MM-FE calculations were performed at the B3LYP/6-31++G**:AMBER level for each QM/MM-optimized geometry along the reaction path.
Figure 11.
Free energy profile of the deacylation determined by the QM/MM-FE calculations for the reaction pathways associated with the direct proton transfer (A) and the water-assisted proton transfer (B). There were 89 points (structures) in Figure 11A and 72 points (structures) in Figure 11B along the reaction coordinate used in the FEP calculations. The relative free energies were determined first without the zero-point and thermal corrections, and then corrected with the zero-point and thermal corrections for the QM subsystem (values in parentheses). The QM/MM-FE calculations were performed at the B3LYP/6-31++G**:AMBER level for each QM/MM-optimized geometry along the reaction path.
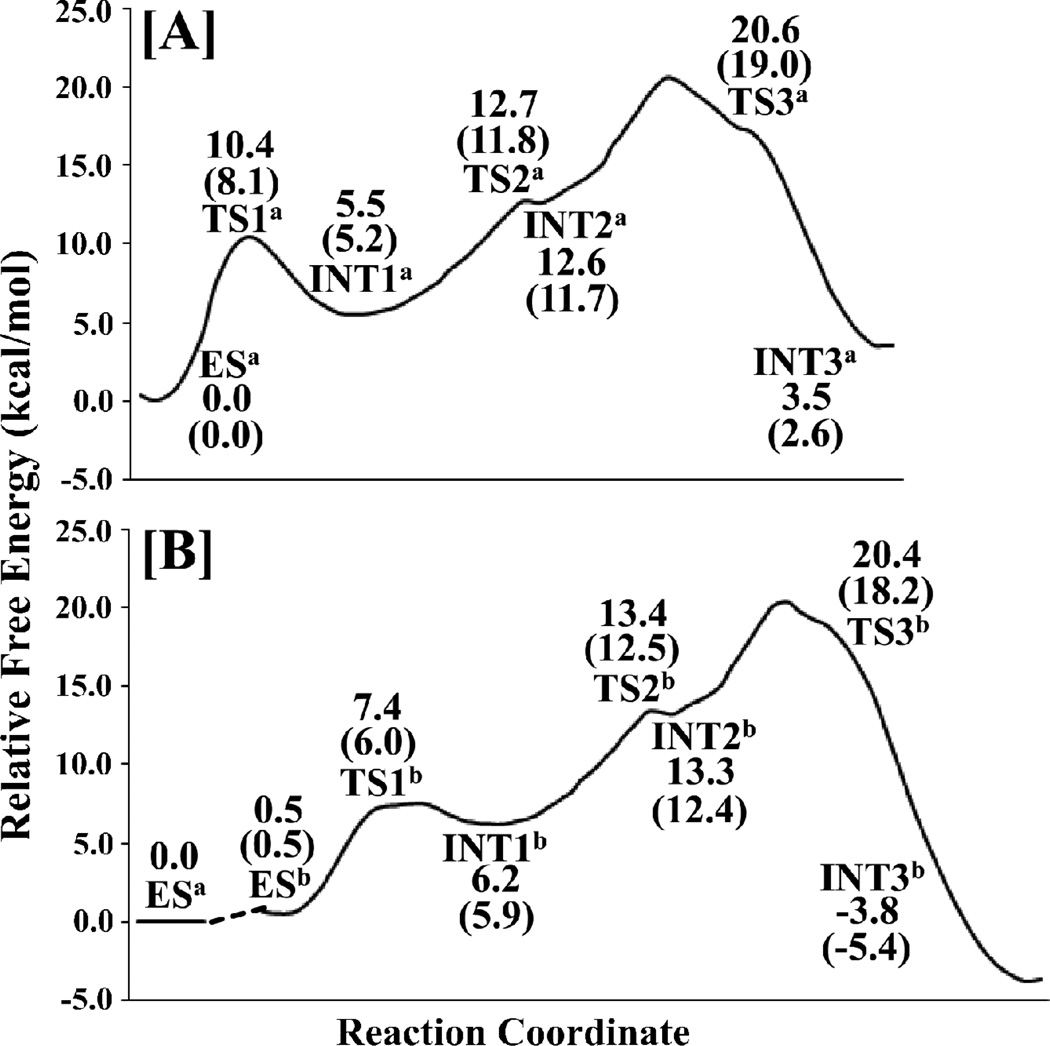
The free energy profile of the acylation stage for the reaction pathway associated with the direct proton transfer is shown in Figure 10A. Without the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the first to third reaction steps are 10.4, 7.2, and 8.0 kcal/mol, respectively. With the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the first to third reaction steps become 8.1, 6.6, and 7.3 kcal/mol, respectively. It should be noted that the energy barrier of the entire acylation stage should be the energy difference between the lowest state ESa and the highest state TS3a and the difference is 19.0 kcal/mol (with the zero-point and thermal corrections for the QM subsystem).
The free energy profile of the acylation stage for the reaction pathway associated with the water-assisted proton transfer is depicted in Figure 10B. Without the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the first to third reaction steps are 6.9, 7.2, and 7.1 kcal/mol, respectively. With the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the first to third reaction steps become 5.5, 6.6, and 5.8 kcal/mol, respectively. The overall free energy change from ESb to TS3b is 17.7 kcal/mol. Noteworthy, the aforementioned MD simulations have demonstrated that ~69.2% snapshots may be considered as ESa and ~30.8% snapshots may be considered as ESb, suggesting that the Gibbs free energy of ESb is about ~0.5 kcal/mol higher than that of ESa, i.e. ΔΔG = ΔG(ESb) – ΔG(ESa) = ~0.5 kcal/mol, according to the well-known Boltzmann distribution. Accounting for the free energy difference between ESa and ESb, the overall free energy barrier for the acylation stage should be the free energy change from ESa to TS3b, i.e. 0.5 + 17.7 = 18.2 kcal/mol. The calculated overall free energy barrier of 18.2 kcal/mol for the water-assisted proton transfer pathway is lower than that for the direct proton transfer pathway by only 0.8 kcal/mol. The difference of 0.8 kcal/mol might be insignificant in consideration of the possible computational errors. Nevertheless, we may comfortably conclude that both the direct and water-assisted proton transfer pathways could significantly contribute to the acylation, although the computational data indicate that the water-assisted proton transfer pathway is slightly more favorable than the direct proton transfer pathway.
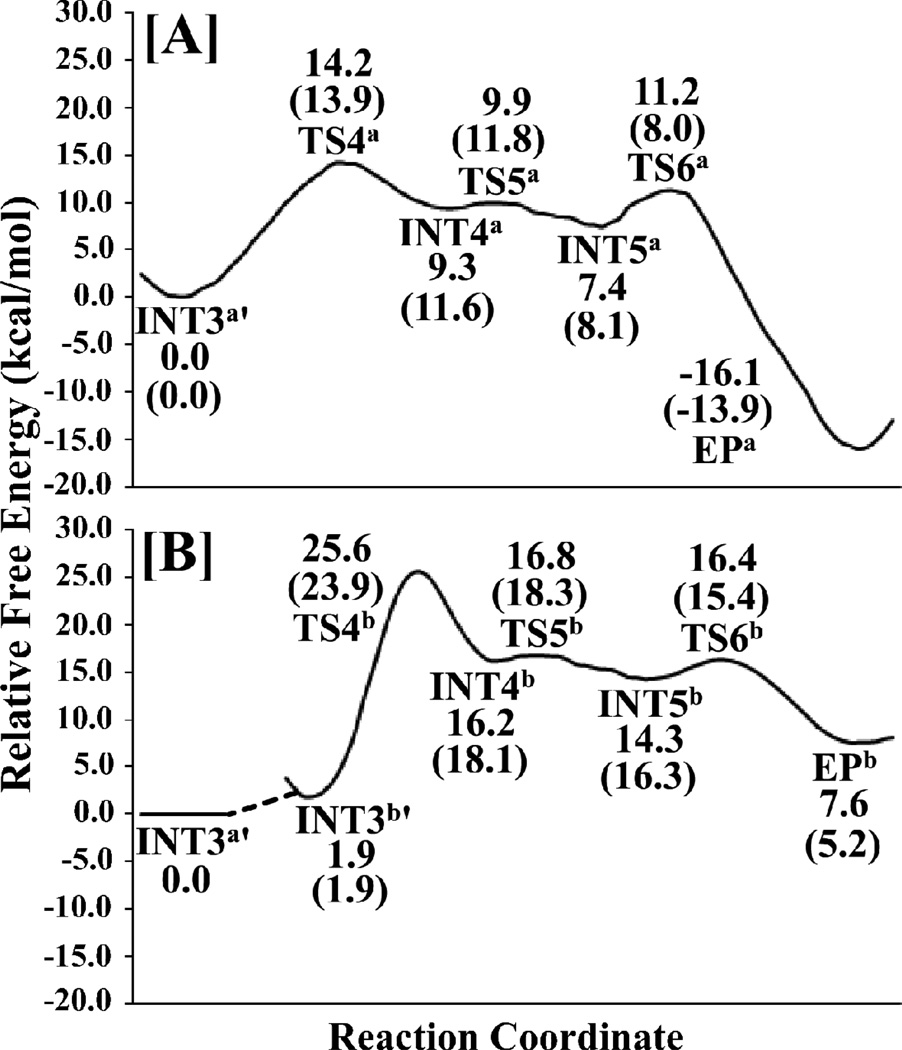
The free energy profile of the deacylation stage for the reaction pathway associated with the direct proton transfer is depicted in Figure 11A. Without the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the fourth to sixth reaction steps are 14.2, 0.6, and 3.8 kcal/mol, respectively. With the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the fourth and fifth reaction steps become 13.9 and 0.2 kcal/mol, respectively. The free energy change from INT5a to TS6a is a negative value (−0.1 kcal/mol) after the zero-point and thermal corrections, which indicates that the sixth reaction step is actually barrierless and the zwitterionic intermediate INT5a is very unstable and cannot really exist. The highest free energy barrier of the deacylation stage (associated with TS4a) is 13.9 kcal/mol, as seen in Figure 11A.
Depicted in Figure 11B is the free energy profile of the deacylation stage for the reaction pathway associated with the water-assisted proton transfer. As seen in Figure 11B, without the zero-point and thermal corrections, the free energy barriers calculated for the fourth to sixth steps of the water-assisted proton-transfer pathway are 23.7, 0.6, and 2.1 kcal/mol, respectively. With the zero-point and thermal corrections for the QM subsystem, the free energy barriers calculated for the fourth and fifth reaction steps become 22.0 and 0.2 kcal/mol, respectively. The free energy change from INT5b to TS6b is a negative value (−0.9 kcal/mol) after the zero-point and thermal corrections, which suggests that the sixth reaction step is actually barrierless and the zwitterionic intermediate INT5b is very unstable and cannot really exist. Noteworthy, the aforementioned MD simulations suggested that, within all of the snapshots that might be suitable for the deacylation reaction, ~4.2% snapshots may be considered as INT3b´, whereas the remaining ~96.8% snapshots may be considered as INT3a´, suggesting that the Gibbs free energy of INT3b´ is about ~1.9 kcal/mol higher than that of INT3a´, i.e. ΔΔG = ΔG(INT3b´) – ΔG(INT3a´) = ~1.9 kcal/mol, according to the well-known Boltzmann distribution. Accounting for the free energy difference between INT3a´ and INT3b´, the overall free energy barrier for the deacylation stage should be the free energy change from INT3a´ to TS4b, i.e. 1.9 + 22.0 = 23.9 kcal/mol. The highest free energy barrier calculated for the deacylation stage associated with the water-assisted proton transfer pathway is 23.9 kcal/mol, about 10.0 kcal/mol higher than that associated with the direct proton transfer pathway. Based on the calculated energy barriers, the direct proton transfer is energetically more favorable in the deacylation stage, although the water-assisted proton transfer might be slightly more favorable in the acylation stage. Further, within the most favorable reaction pathway, the overall (highest) energy barrier of 13.9 kcal/mol (associated with TS4a) calculated for the deacylation stage is lower than that (18.2 kcal/mol associated with TS3b) calculated for the acylation stage by about 4.3 kcal/mol, indicating that the rate-determining step is the third step (associated with TS3b) in the acylation stage.
We note that our calculated results are consistent with the available experimental observations. First, the proteasomal hydrolysis reactions of different types of substrates (with the same P1´, but slightly different P1 for the peptide substrate depicted in Scheme 1) that produce the same acyl-enzyme intermediate (associated with the intermediate INT3) are hydrolyzed at different rates, suggesting that the deacylation stage cannot be rate-determining.51 In addition, the proteasomal hydrolysis of peptide Suc-LLVY-AMC was decreased by more than 2-fold in heavy water, indicating that the rate-determining step should involve a proton from solvent water.51 This reported kinetic isotopic effect is consistent with our computational finding that the rate-determining step associated with TS3b is a water-assisted proton transfer process.
It is interesting to note that the currently studied proteasomal hydrolysis reaction of the substrate and the previously examined proteasome inhibition reaction with EPX42 have a common first reaction step (i.e. the activation of Thr1-Oγ) when we ignore the identity of the ligand (substrate or inhibitor) in the active site, although the subsequent steps of the two reactions are totally different. In comparison of the data from the studies on the two types of reactions, one can note that the most favorable pathway for the first reaction step of the proteasomal hydrolysis reaction is different from that of the proteasome inhibition reaction. The lowest free energy barrier is associated with the water-assisted proton transfer for the proteasomal hydrolysis reaction, whereas the lowest free energy barrier is associated with the direct proton transfer for the proteasome inhibition reaction.42
Based on the free energy profiles depicted in Figures 10B and 11A for the most favorable reaction pathway, the overall free energy barrier for the entire reaction process is determined by the free energy change (18.2 kcal/mol) from ESa to TS3b. We wanted to know whether the calculated overall free energy barrier of 18.2 kcal/mol is reasonably consistent with the available experimental reaction rate constant (kcat) or not. According to the reported experimental data,51, 52 kcat = 0.21 – 0.03 s−1, which is associated with an activation free energy of ~18.3 – 19.4 kcal/mol at room temperature (25°C) according to the conventional transition state theory.84 Our calculated free energy barrier of 18.2 kcal/mol is close to the experimentally derived activation free energy of ~18.3 – 19.4 kcal/mol, suggesting that the computational results are reasonable.
Finally, we would like to address a potential question concerning whether the QM/MM scheme used in the above-discussed QM/MM calculations is reasonable or not. As shown in Figure 2A and 4A, the QM/MM boundary atoms in the above QM/MM scheme (denoted as QM/MM scheme I) are directly bonded to the charged functional groups of residues Asp17 and Lys33. For comparison, we also tested another QM/MM scheme (denoted as QM/MM scheme II) with an extended QM subsystem which includes an additional CH2 group from each of the two charged residues (Asp17 and Lys33). By using QM/MM scheme II, we re-optimized the geometries of ESa, ESb, TS3a, and TS3b at the B3LYP/6-31G*:AMBER level, followed by the single-point energy calculations at the QM/MM(B3LYP/6-31++G**:AMBER) level. The calculated energetic data are summarized in Table 1 for comparison. As seen in Table 1, changing QM/MM scheme I to QM/MM scheme II only slightly and systematically decreases the calculated energy barrier, i.e. 0.1 kcal/mol for E(TS3b) – E(ESb) and 0.2 kcal/mol for E(TS3a) – E(ESa), suggesting that QM/MM scheme I is acceptable for the QM/MM calculations on the enzymatic reaction system concerned in the present study.
Table 1.
The QM/MM energies calculated on four key states (ESa, ESb, TS3a, and TS3b) at the QM/MM(B3LYP/6-31++G**:AMBER)//QM/MM(B3LYP/6-31G*:AMBER) level using two different QM/MM schemes (I and II).
| QM/MM scheme I | QM/MM scheme II | |
|---|---|---|
| E(ESb) | −2050.837028 au | −2129.531891 au |
| E(TS3b) | −2050.806649 au | −2129.501625 au |
| E(TS3b) – E(ESb) | 19.1 kcal/mol | 19.0 kcal/mol |
| E(ESa) | −2050.884783 au | −2129.498713 au |
| E(TS3a) | −2050.854884 au | −2129.469072 au |
| E(TS3a) – E(ESa) | 18.8 kcal/mol | 18.6 kcal/mol |
Conclusion
The first-principles QM/MM-FE calculations performed in this study have demonstrated the detailed mechanism for the proteasomal hydrolysis of a representative peptide, i.e. Suc-LLVY-AMC. According to the results obtained from the QM/MM-FE calculations, the most favorable reaction pathway is associated with the water-assisted proton transfer in the acylation stage, and associated with the direct proton transfer in the deacylation stage, and the entire reaction process consists of six reaction steps. The reaction is initiated by a water-assisted proton (Hγ) transfer from the Thr1-Oγ atom to the Thr1-Nz atom within the enzyme to activate the Thr1-Oγ. Then, the negatively charged Thr1-Oγ atom initiates the nucleophilic attack on the carbonyl carbon C1 atom of substrate. Subsequently, a water-assisted proton (Hγ) transfers from Thr1-Nz to the N1 atom of substrate, coupled with the dissociation of the amine AMC by breaking the C1–N1 bond. The fourth step is a nucleophilic attack on the carbonyl carbon C1 atom of substrate by a water molecule, accompanied with a proton transfer from the water to Thr1-Nz. The fifth step is the dissociation of the carboxylic acid Suc-LLVY by breaking the C1–Oγ bond. The final step is the regeneration of Thr1 by a direct proton transfer from Thr1-Nz to Thr1-Oγ.
The free energy profile calculated for the most favorable reaction pathway indicates that the free energy barriers for the first to fifth reaction steps are 6.0, 6.6, 5.8, 13.9, and 0.2 kcal/mol, respectively. The sixth step of the reaction associated with the direct proton transfer is barrierless. The overall free energy barrier of the entire proteasomal hydrolysis process should be the free energy change (18.2 kcal/mol) from ESa to TS3b, and the rate-determining step should be the third reaction step (a water-assisted proton transfer associated with TS3b) of the acylation stage. The determined most favorable reaction pathway and the rate-determining step are consistent with available experimental observations concerning the substituent and isotopic effects on the kinetics and, thus, provide a reasonable interpretation of the reported experimental observations. The calculated overall free energy barrier of 18.2 kcal/mol is close to the experimentally derived activation free energy of ~18.3–19.4 kcal/mol, suggesting that the obtained mechanistic insights are reasonable.
Due to the unusual importance of proteasome in the proteolysis/degradation of proteins/peptides in life sciences, the fundamental mechanistic insights obtained from this computational study provide an important base for further studies on molecular mechanisms of a variety of biological processes related to proteasome and/or proteolysis/degradation of proteins/peptides in living systems. Further, as potent proteasome inhibitors could be therapeutically valuable and as the currently used proteasome inhibitors in clinical practice/studies are all peptides, the obtained new mechanistic insights could also be valuable in future mechanism-based design of novel peptide inhibitors of proteasome. In addition, the obtained novel, general insights into the detailed catalytic mechanisms concerning the water-assisted proton transfer versus direct proton transfer could also be valuable for future computational studies on possible reaction pathways of other enzyme reactions.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH (grants R01 DA035552, R01 DA032910, R01 DA013930, and R01 DA025100 to Zhan), the NSF (grant CHE-1111761 to Zhan), the China Postdoctoral Science Foundation (grant No.2013M530340 to Wei), and the National Natural Science Foundation of China (No.21303167 to Wei). Wei worked in Zhan’s laboratory for this project at the University of Kentucky as an exchange graduate student (from the Zhengzhou University) supported by the China Scholarship Council. The authors also acknowledge the Computer Center at the University of Kentucky for supercomputing time on a Dell X-series Cluster with 384 nodes or 4,768 processors.
Footnotes
Supporting Information Available. The complete author lists of the references with more than 10 authors. The initial structure of the ES (Figure S1) constructed by retaining only two subunits β5 and β6 and the substrate (Suc-LLVY-AMC). The absolute QM/MM energies (in Hartree, Table S1) calculated at the QM/MM (B3LYP/6-31++G**:AMBER) level using the geometries optimized at the QM/MM(B3LYP/6-31G*:AMBER) level, the thermal corrections for the QM subsystem calculated at the QM/MM(B3LYP/6-31G*:AMBER) level, coordinates of QM part of the geometries optimized at the QM/MM(B3LYP/6-31G*:AMBER) level. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Borissenko L, Groll M. 20S Proteasome and Its Inhibitors: Crystallographic Knowledge for Drug Development. Chem. Rev. 2007;107(3):687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 2.Groettrup M, Soza A, Kuckelkorn U, Kloetzel PM. Peptide Antigen Production by the Proteasome: Complexity Provides Efficiency. Immun. Today. 1996;17(9):429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg AL. New Insights into Proteasome Function - from Archaebacteria to Drug Development. Chem. Biol. 1995;2(8):503–508. doi: 10.1016/1074-5521(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg AL. The Mechanism and Functions of Atp-Dependent Proteases in Bacterial and Animal-Cells. Eur. J. Biochem. 1992;203(1–2):9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg AL. Protein Degradation and Protection against Misfolded or Damaged Proteins. Nature. 2003;426(6968):895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 6.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, et al. Bortezomib or High-dose Dexamethasone for Relapsed Multiple Myeloma. New Engl. J. Med. 2005;352(24):2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 7.Genin E, Reboud-Ravaux M, Vidal J. Proteasome Inhibitors: Recent Advances and New Perspectives in Medicinal Chemistry. Curr. Top. Med. Chem. 2010;10(3):232–256. doi: 10.2174/156802610790725515. [DOI] [PubMed] [Google Scholar]
- 8.Adams J. The Proteasome: A Suitable Antineoplastic Target. Nat. Rev. Cancer. 2004;4(5):349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg AL, Rock K. Not Just Research Tools--Proteasome Inhibitors Offer Therapeutic Promise. Nature Med. 2002;8(4):338–340. doi: 10.1038/nm0402-338. [DOI] [PubMed] [Google Scholar]
- 10.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: Proteasome Inhibition as an Effective Anticancer Therapy. Annu. Rev. Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 11.Sprangers R, Li X, Mao X, Rubinstein JL, Schimmer AD, Kay LE. TROSY-based NMR Evidence for A Novel Class of 20S Proteasome Inhibitors. Biochemistry. 2008;47(26):6727–6734. doi: 10.1021/bi8005913. [DOI] [PubMed] [Google Scholar]
- 12.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal Structure of the 20S Proteasome from the Archaeon T. Acidophilum at 3.4 A Resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 13.Vinitsky A, Michaud C, Powers JC, Orlowski M. Inhibition of the Chymotrypsin-like Activity of the Pituitary Multicatalytic Proteinase Complex. Biochemistry. 1992;31(39):9421–9428. doi: 10.1021/bi00154a014. [DOI] [PubMed] [Google Scholar]
- 14.Vivier M, Rapp M, Papon J, Labarre P, Galmier MJ, Sauziere J, Madelmont JC. Synthesis, Radiosynthesis, and Biological Evaluation of New Proteasome Inhibitors in a Tumor Targeting Approach. J. Med. Chem. 2008;51(4):1043–1047. doi: 10.1021/jm701419g. [DOI] [PubMed] [Google Scholar]
- 15.Marastoni M, McDonald J, Baldisserotto A, Canella A, De Risi C, Pollini GP, Tomatis R. Proteasome Inhibitors: Synthesis and Activity of Arecoline Oxide Tripeptide Derivatives. Bioorg. Med. Chem. Lett. 2004;14(8):1965–1968. doi: 10.1016/j.bmcl.2004.01.081. [DOI] [PubMed] [Google Scholar]
- 16.Marastoni M, Baldisserotto A, Canella A, Gavioli R, De Risi C, Pollini GP, Tomatis R. Arecoline Tripeptide Inhibitors of Proteasome. J. Med. Chem. 2004;47(6):1587–1590. doi: 10.1021/jm0309102. [DOI] [PubMed] [Google Scholar]
- 17.Aubin S, Martin B, Delcros JG, Arlot-Bonnemains Y, Baudy-Floc'h M. Retro Hydrazino-Azapeptoids as Peptidomimetics of Proteasome Inhibitors. J. Med. Chem. 2005;48(1):330–334. doi: 10.1021/jm049455f. [DOI] [PubMed] [Google Scholar]
- 18.Gaczynska M, Osmulski PA, Gao Y, Post MJ, Simons M. Proline- and Arginine-rich Peptides Constitute A Novel Class of Allosteric Inhibitors of Proteasome Activity. Biochemistry. 2003;42(29):8663–8670. doi: 10.1021/bi034784f. [DOI] [PubMed] [Google Scholar]
- 19.Zhu YQ, Zhao X, Zhu XR, Wu G, Li YJ, Ma YH, Yuan YX, Yang J, Hu Y, Ai L, et al. Design, Synthesis, Biological Evaluation, and Structure-Activity Relationship (SAR) Discussion of Dipeptidyl Boronate Proteasome Inhibitors, Part I: Comprehensive Understanding of the SAR of alpha-Amino Acid Boronates. J. Med. Chem. 2009;52(14):4192–4199. doi: 10.1021/jm9005093. [DOI] [PubMed] [Google Scholar]
- 20.Zhu YQ, Wu G, Zhu XR, Ma YH, Zhao X, Li YJ, Yuan YX, Yang J, Yu S, Shao F, et al. Synthesis, in Vitro and in Vivo Biological Evaluation, and Comprehensive Understanding of Structure-Activity Relationships of Dipeptidyl Boronic Acid Proteasome Inhibitors Constructed from beta-Amino Acids. J. Med. Chem. 2010;53(24):8619–8626. doi: 10.1021/jm1009742. [DOI] [PubMed] [Google Scholar]
- 21.Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, Ploegh HL, Ovaa H, Galardy PJ. Activity Probe for in Vivo Profiling of the Specificity of Proteasome Inhibitor Bortezomib. Nat. Methods. 2005;2(5):357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 22.Milo LJ, Lai JH, Wu WG, Liu YX, Maw H, Li YH, Jin ZP, Shu Y, Poplawski SE, Wu Y, et al. Chemical and Biological Evaluation of Dipeptidyl Boronic Acid Proteasome Inhibitors for Use in Prodrugs and Pro-Soft Drugs Targeting Solid Tumors. J. Med. Chem. 2011;54(13):4365–4377. doi: 10.1021/jm200460q. [DOI] [PubMed] [Google Scholar]
- 23.Groll M, Balskus EP, Jacobsen EN. Structural Analysis of Spiro beta-Lactone Proteasome Inhibitors. J. Am. Chem. Soc. 2008;130(45):14981–14983. doi: 10.1021/ja806059t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groll M, Huber R, Potts BC. Crystal Structures of Salinosporamide A (NPI-0052) and B (NPI-0047) in Complex with the 20S Proteasome Reveal Important Consequences of beta-Lactone Ring Opening and A Mechanism for Irreversible Binding. J. Am. Chem. Soc. 2006;128(15):5136–5141. doi: 10.1021/ja058320b. [DOI] [PubMed] [Google Scholar]
- 25.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of Proteasome Activities and Subunit-specific Amino-terminal Threonine Modification by Lactacystin. Science. 1995;268(5211):726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 26.Nett M, Gulder TA, Kale AJ, Hughes CC, Moore BS. Function-oriented Biosynthesis of beta-Lactone Proteasome Inhibitors in Salinispora Tropica. J. Med. Chem. 2009;52(19):6163–6167. doi: 10.1021/jm901098m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng LH, Mohan R, Kwok BHB, Elofsson M, Sin N, Crews CM. Epoxomicin, a Potent and Selective Proteasome Inhibitor, Exhibits in Vivo Antiinflammatory Activity. Proc. Natl. Acad. Sci. U.S.A. 1999;96(18):10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng L, Kwok BH, Sin N, Crews CM. Eponemycin Exerts Its Antitumor Effect through the Inhibition of Proteasome Function. Canc. Res. 1999;59(12):2798–2801. [PubMed] [Google Scholar]
- 29.Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, Jiang J, Laidig GJ, Lewis ER, Parlati F, et al. Antitumor Activity of PR-171, a Novel Irreversible Inhibitor of the Proteasome. Canc. Res. 2007;67(13):6383–6391. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, Van Leeuwen FW, Chanan-Khan AA, et al. Potent Activity of Carfilzomib, a Novel, Irreversible Inhibitor of the Ubiquitin-Proteasome Pathway, against Preclinical Models of Multiple Myeloma. Blood. 2007;110(9):3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogyo M, McMaster JS, Gaczynska M, Tortorella D, Goldberg AL, Ploegh H. Covalent Modification of the Active Site Threonine of Proteasomal beta Subunits and the Escherichia Coli Homolog HslV by A New Class of Inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1997;94(13):6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nazif T, Bogyo M. Global Analysis of Proteasomal Substrate Specificity using Positional-scanning Libraries of Covalent Inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2001;98(6):2967–2972. doi: 10.1073/pnas.061028898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rydzewski RM, Burrill L, Mendonca R, Palmer JT, Rice M, Tahilramani R, Bass KE, Leung L, Gjerstad E, Janc JW, et al. Optimization of Subsite Binding to the beta 5 Subunit of the Human 20S Proteasome using Vinyl Sulfones and 2-Keto-1,3,4-Oxadiazoles: Syntheses and Cellular Properties of Potent, Selective Proteasome Inhibitors. J. Med. Chem. 2006;49(10):2953–2968. doi: 10.1021/jm058289o. [DOI] [PubMed] [Google Scholar]
- 34.Groll M, Schellenberg B, Bachmann AS, Archer CR, Huber R, Powell TK, Lindow S, Kaiser M, Dudler R. A Plant Pathogen Virulence Factor Inhibits the Eukaryotic Proteasome by a Novel Mechanism. Nature. 2008;452(7188):755–758. doi: 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
- 35.Baldisserotto A, Ferretti V, Destro F, Franceschini C, Marastoni M, Gavioli R, Tomatis R. alpha,beta-Unsaturated N-Acylpyrrole Peptidyl Derivatives: New Proteasome Inhibitors. J. Med. Chem. 2010;53(17):6511–6515. doi: 10.1021/jm100122e. [DOI] [PubMed] [Google Scholar]
- 36.Groll M, Koguchi Y, Huber R, Kohno J. Crystal Structure of the 20 S Proteasome:TMC-95A Complex: a Non-Covalent Proteasome Inhibitor. J. Mol. Biol. 2001;311(3):543–548. doi: 10.1006/jmbi.2001.4869. [DOI] [PubMed] [Google Scholar]
- 37.Vivier M, Jarrousse AS, Bouchon B, Galmier MJ, Auzeloux P, Sauzieres J, Madelmont JC. Preliminary Studies of New Proteasome Inhibitors in the Tumor Targeting Approach: Synthesis and in Vitro Cytotoxicity. J. Med. Chem. 2005;48(21):6731–6740. doi: 10.1021/jm050181l. [DOI] [PubMed] [Google Scholar]
- 38.Basse N, Montes M, Marechal X, Qin L, Bouvier-Durand M, Genin E, Vidal J, Villoutreix BO, Reboud-Ravaux M. Novel Organic Proteasome Inhibitors Identified by Virtual and in Vitro Screening. J. Med. Chem. 2010;53(1):509–513. doi: 10.1021/jm9011092. [DOI] [PubMed] [Google Scholar]
- 39.Furet P, Imbach P, Noorani M, Koeppler J, Laumen K, Lang M, Guagnano V, Fuerst P, Roesel J, Zimmermann J, et al. Entry into A New Class of Potent Proteasome Inhibitors Having High Antiproliferative Activity by Structure-based Design. J. Med. Chem. 2004;47(20):4810–4813. doi: 10.1021/jm049660v. [DOI] [PubMed] [Google Scholar]
- 40.Adsule S, Barve V, Chen D, Ahmed F, Dou QP, Padhye S, Sarkar FH. Novel Schiff Base Copper Complexes of Quinoline-2 Carboxaldehyde as Proteasome Inhibitors in Human Prostate Cancer Cells. J. Med. Chem. 2006;49(24):7242–7246. doi: 10.1021/jm060712l. [DOI] [PubMed] [Google Scholar]
- 41.Groll M, Kim KB, Kairies N, Huber R, Crews CM. Crystal Structure of Epoxomicin : 20S Proteasome Reveals a Molecular Basis for Selectivity of alpha ',beta '-Epoxyketone Proteasome Inhibitors. J. Am. Chem. Soc. 2000;122(6):1237–1238. [Google Scholar]
- 42.Wei DH, Lei BL, Tang MS, Zhan CG. Fundamental Reaction Pathway and Free Energy Profile for Inhibition of Proteasome by Epoxomicin. J. Am. Chem. Soc. 2012;134(25):10436–10450. doi: 10.1021/ja3006463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The Structure of the Mammalian 20S Proteasome at 2.75 Angstrom Resolution. Structure. 2002;10(5):609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 44.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S Proteasome from Yeast at 2.4 Angstrom Resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 45.Groll M, Berkers CR, Ploegh HL, Ovaa H. Crystal Structure of the Boronic Acid-based Proteasome Inhibitor Bortezomib in Complex with the Yeast 20S Proteasome. Structure. 2006;14(3):451–456. doi: 10.1016/j.str.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Hines J, Groll M, Fahnestock M, Crews CM. Proteasome Inhibition by Fellutamide B Induces Nerve Growth Factor Synthesis. Chem. Biol. 2008;15(5):501–512. doi: 10.1016/j.chembiol.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groll M, Nazif T, Huber R, Bogyo M. Probing Structural Determinants Distal to the Site of Hydrolysis That Control Substrate Specificity of the 20S Proteasome. Chem. Biol. 2002;9(5):655–662. doi: 10.1016/s1074-5521(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 48.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic Mechanism and Assembly of the Proteasome. Chem. Rev. 2009;109(4):1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 49.Duggleby HJ, Tolley SP, Hill CP, Dodson EJ, Dodson G, Moody PC. Penicillin Acylase Has A Single-amino-acid Catalytic Centre. Nature. 1995;373(6511):264–268. doi: 10.1038/373264a0. [DOI] [PubMed] [Google Scholar]
- 50.Myung J, Kim KB, Crews CM. The Ubiquitin-Proteasome Pathway and Proteasome Inhibitors. Med. Res. Rev. 2001;21(4):245–273. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kisselev AF, Songyang Z, Goldberg AL. Why Does Threonine, and Not Serine, Function as the Active Site Nucleophile in Proteasomes? J. Biol. Chem. 2000;275(20):14831–14837. doi: 10.1074/jbc.275.20.14831. [DOI] [PubMed] [Google Scholar]
- 52.Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma-Acidophilum - a Threonine Protease. Science. 1995;268(5210):579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 53.Zhang YK, Liu HY, Yang WT. Free Energy Calculation on Enzyme Reactions with an Efficient Iterative Procedure to Determine Minimum Energy Paths on a Combined ab initio QM/MM Potential Energy Surface. J. Chem. Phys. 2000;112(8):3483–3492. [Google Scholar]
- 54.Zhang YK. Improved Pseudobonds for Combined ab initio Quantum Mechanical/Molecular Mechanical Methods. J. Chem. Phys. 2005;122(2):024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 55.Zhang YK. Pseudobond ab initio QM/MM Approach and Its Applications to Enzyme Reactions. Theor. Chem. Acc. 2006;116(1–3):43–50. [Google Scholar]
- 56.Zheng F, Yang WC, Ko MC, Liu JJ, Cho H, Gao DQ, Tong M, Tai HH, Woods JH, Zhan CG. Most Efficient Cocaine Hydrolase Designed by Virtual Screening of Transition States. J. Am. Chem. Soc. 2008;130(36):12148–12155. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu JJ, Hamza A, Zhan CG. Fundamental Reaction Mechanism and Free Energy Profile for (−)-Cocaine Hydrolysis Catalyzed by Cocaine Esterase. J. Am. Chem. Soc. 2009;131(33):11964–11975. doi: 10.1021/ja903990p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu JJ, Zhang YK, Zhan CG. Reaction Pathway and Free-Energy Barrier for Reactivation of Dimethylphosphoryl-Inhibited Human Acetylcholinesterase. J. Phys. Chem. B. 2009;113(50):16226–16236. doi: 10.1021/jp9055335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu JJ, Zhao XY, Yang WC, Zhan CG. Reaction Mechanism for Cocaine Esterase-Catalyzed Hydrolyses of (+)- and (−)-Cocaine: Unexpected Common Rate-Determining Step. J. Phys. Chem. B. 2011;115(17):5017–5025. doi: 10.1021/jp200975v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Zhan CG. Reaction Pathway and Free Energy Profile for Cocaine Hydrolase-catalyzed Hydrolysis of (−)-Cocaine. J. Chem. Theory Comput. 2012;8:1426–1435. doi: 10.1021/ct200810d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li DM, Huang XQ, Han KL, Zhan CG. Catalytic Mechanism of Cytochrome P450 for 5 '-Hydroxylation of Nicotine: Fundamental Reaction Pathways and Stereoselectivity. J. Am. Chem. Soc. 2011;133(19):7416–7427. doi: 10.1021/ja111657j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lei B, Abdul Hameed MD, Hamza A, Wehenkel M, Muzyka JL, Yao XJ, Kim KB, Zhan CG. Molecular Basis of the Selectivity of the Immunoproteasome Catalytic Subunit LMP2-Specific Inhibitor Revealed by Molecular Modeling and Dynamics Simulations. J. Phys. Chem. B. 2010;114(38):12333–12339. doi: 10.1021/jp1058098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, udin KN, Burant JC, et al. Gaussian 03, Version C.02. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 64.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. AMBER11. San Francisco: University of California; 2010. [Google Scholar]
- 65.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 66.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, et al. AMBER8. San Francisco: University of California; 2004. [Google Scholar]
- 67.Chandrasekhar J, Smith SF, Jorgensen WL. Sn2 Reaction Profiles in the Gas-Phase and Aqueous-Solution. J. Am. Chem. Soc. 1984;106(10):3049–3050. [Google Scholar]
- 68.Kollman P. Free-Energy Calculations - Applications to Chemical and Biochemical Phenomena. Chem. Rev. 1993;93(7):2395–2417. [Google Scholar]
- 69.Beveridge DL, DiCapua FM. Free Energy via Molecular Simulation: Applications to Chemical and Biomolecular Systems. Annu. Rev. Biophys. Biophys. Chem. 1989;18:431–492. doi: 10.1146/annurev.bb.18.060189.002243. [DOI] [PubMed] [Google Scholar]
- 70.Acevedo O, Jorgensen WL. Advances in Quantum and Molecular Mechanical (QM/MM) Simulations for Organic and Enzymatic Reactions. Acc. Chem. Res. 2010;43(1):142–151. doi: 10.1021/ar900171c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Acevedo O, Jorgensen WL. Cope Elimination: Elucidation of Solvent Effects from QM/MM Simulations. J. Am. Chem. Soc. 2006;128(18):6141–6146. doi: 10.1021/ja057523x. [DOI] [PubMed] [Google Scholar]
- 72.Acevedo O, Jorgensen WL. Influence of Inter- and Intramolecular Hydrogen Bonding on Kemp Decarboxylations from QM/MM Simulations. J. Am. Chem. Soc. 2005;127(24):8829–8834. doi: 10.1021/ja051793y. [DOI] [PubMed] [Google Scholar]
- 73.Jarmula A, Cieplak P, Les A, Rode W. Relative Free Energies of Binding to Thymidylate Synthase of 2- and/or 4-Thio and/or 5-Fluoro Analogues of dUMP. J. Comput. Aided Mol. Des. 2003;17(10):699–710. doi: 10.1023/b:jcam.0000017377.07094.2e. [DOI] [PubMed] [Google Scholar]
- 74.Udier-Blagovic M, Tirado-Rives J, Jorgensen WL. Structural and Energetic Analyses of the Effects of the K103N Mutation of HIV-1 Reverse Transcriptase on Efavirenz Analogues. J. Med. Chem. 2004;47(9):2389–2392. doi: 10.1021/jm0303507. [DOI] [PubMed] [Google Scholar]
- 75.Zhang W, Hou TJ, Qiao XB, Huai S, Xu XJ. Binding Affinity of Hydroxamate Inhibitors of Matrix Metalloproteinase-2. J. Mol. Mod. 2004;10(2):112–120. doi: 10.1007/s00894-004-0181-1. [DOI] [PubMed] [Google Scholar]
- 76.Alexandrova AN, Rothlisberger D, Baker D, Jorgensen WL. Catalytic Mechanism and Performance of Computationally Designed Enzymes for Kemp Elimination. J. Am. Chem. Soc. 2008;130(47):15907–15915. doi: 10.1021/ja804040s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guimaraes CR, Udier-Blagovic M, Jorgensen WL. Macrophomate Synthase: QM/MM Simulations Address the Diels-Alder Versus Michael-Aldol Reaction Mechanism. J. Am. Chem. Soc. 2005;127(10):3577–3588. doi: 10.1021/ja043905b. [DOI] [PubMed] [Google Scholar]
- 78.Guimaraes CR, Boger DL, Jorgensen WL. Elucidation of Fatty Acid Amide Hydrolase Inhibition by Potent alpha-Ketoheterocycle Derivatives from Monte Carlo Simulations. J. Am. Chem. Soc. 2005;127(49):17377–17384. doi: 10.1021/ja055438j. [DOI] [PubMed] [Google Scholar]
- 79.Danciulescu C, Nick B, Wortmann FJ. Structural Stability of Wild Type and Mutated alpha-Keratin Fragments: Molecular Dynamics and Free Energy Calculations. Biomacromolecules. 2004;5(6):2165–2175. doi: 10.1021/bm049788u. [DOI] [PubMed] [Google Scholar]
- 80.Funahashi J, Sugita Y, Kitao A, Yutani K. How Can Free Energy Component Analysis Explain the Difference in Protein Stability Caused by Amino Acid Substitutions? Effect of Three Hydrophobic Mutations at the 56th Residue on the Stability of Human Lysozyme. Protein Eng. 2003;16(9):665–671. doi: 10.1093/protein/gzg083. [DOI] [PubMed] [Google Scholar]
- 81.Chen X, Fang L, Liu J, Zhan CG. Reaction Pathway and Free Energy Profile for Butyrylcholinesterase-catalyzed Hydrolysis of Acetylcholine. J. Phys. Chem. B. 2011;115(5):1315–1322. doi: 10.1021/jp110709a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen X, Zhao X, Xiong Y, Liu J, Zhan CG. Fundamental Reaction Pathway and Free Energy Profile for Hydrolysis of Intracellular Second Messenger Adenosine 3',5'-Cyclic Monophosphate (cAMP) Catalyzed by Phosphodiesterase-4. J. Phys. Chem. B. 2011;115(42):12208–12219. doi: 10.1021/jp205509w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen X, Fang L, Liu J, Zhan CG. Reaction Pathway and Free Energy Profiles for Butyrylcholinesterase-catalyzed Hydrolysis of Acetylthiocholine. Biochemistry. 2012;51(6):1297–1305. doi: 10.1021/bi201786s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alvarez-Idaboy JR, Galano A, Bravo-Perez G, Ruiz ME. Rate Constant Dependence on the Size of Aldehydes in the NO(3) + Aldehydes Reaction. An Explanation via Quantum Chemical Calculations and CTST. J. Am. Chem. Soc. 2001;123(34):8387–8395. doi: 10.1021/ja010693z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.