Abstract
The Drug-Gene Interaction database (DGIdb) mines existing resources that generate hypotheses about how mutated genes might be targeted therapeutically or prioritized for drug development. It provides an interface for searching lists of genes against a compendium of drug-gene interactions and potentially druggable genes. DGIdb can be accessed at dgidb.org.
The druggable genome1, 2 can be defined as the genes or gene products that are known or predicted to interact with drugs, ideally with a therapeutic benefit to patients. We developed the Drug Gene Interaction database (DGIdb) to help researchers interpret the results of genome-wide studies in the context of the druggable genome (Supplementary Figures 1–3). DGIdb organizes genes of the druggable genome into two main classes. The first class includes genes with known drug interactions obtained by literature mining or by parsing publicly available databases. The second class includes genes that may not currently be targeted therapeutically but are ‘potentially’ druggable according to their membership in gene categories associated with druggability (e.g., kinases).
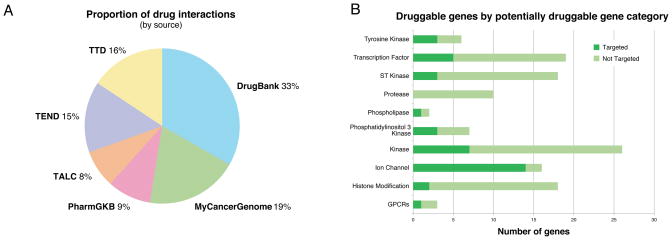
DGIdb integrates data from 13 primary sources (Supplementary Table 1) covering disease-relevant human genes3, 4, drugs5, drug-gene interactions6–10, and potential druggability1, 2, 11, 12. Currently, DGIdb contains over 14,144 drug-gene interactions by 2,611 genes and 6,307 drugs and in addition it includes 6,761 genes belonging to one or more of 39 potentially druggable gene categories (Supplementary Table 2–3). A total of 7,668 unique genes have either known or potential druggability. Each drug-gene or gene-category association is linked to its primary database or literature source. By intersecting the current knowledge of known and potentially druggable genes, DGIdb provides a unique resource for surveying the state of the field of targeted therapies (Supplementary Figure 4). Of the genes in potentially druggable gene categories, only 25.2% (1,704) have a known drug-gene interaction (Supplementary Figure 5) and 5.8% (392) are targeted by an anti-neoplastic agent (Supplementary Table 4). Perhaps unsurprisingly, drug metabolism and drug resistance genes are well represented with 94.1% (32/34) and 57.3% (201/351) of genes respectively having known interactions with drugs. Despite the tremendous interest in kinases as potential drug targets, 561 (68.3%) remain untargeted. Phosphatidylinositol 3-kinases and tyrosine kinases are better represented at 62.5% and 44.6% compared to serine/threonine kinases at 29.5%. Similarly, large fractions (60–70%) of phospholipases, transporters, and metallopeptidases remain untargeted. The most strikingly under-represented druggable gene categories, with as few as 14–27% targeted, include proteases, growth factors, G-protein coupled receptors (GPCR), transcription factors, histone modification genes and protein phosphatases.
To demonstrate the utility of DGIdb we analyzed genes found to be mutated in a cohort of 1,273 breast cancer patients profiled by whole genome and/or exome sequencing13–17 (Supplementary Table 5). For activating mutations, the potential value of targeted therapy is high. However, the most highly recurrently mutated genes in breast cancer, possible drivers of disease and targets for personalized medicine, remain poorly targeted by current drugs. Only 6 of the 31 genes mutated in at least 2.5% of patients (AKT1, CDH1, LRP2, PIK3CA, RYR2, and TP53) have known drug-gene interactions (Supplementary Figure 6A). Expanding the list to the top 1% of recurring mutations increases the number of genes to 315 (Figure 1). 45 (14%) of these genes are targeted by at least one known drug and 132 (42%) belong to one or more potentially druggable gene categories (Figure 1A). All six sources in DGIdb contributed to this list of interactions. However, many interactions (58%) are from sources considered non-curated by DGIdb. Many recurrently mutated genes in key categories are not currently targeted and therefore might be considered high priority for future drug development efforts (Figure 1B). For example, considering genes classified as a kinase according to DGIdb, we can reduce our original list of 315 candidate genes to 26. Only seven of these kinases (AKT1, ERBB2, ERBB3, ERBB4, MTOR, PIK3CA, and PIK3R1) have a known drug interaction (Supplementary Figure 6B). Even among the 45 recurrently mutated genes targeted by known drugs (most of which are not currently used in breast cancer), there may be testable hypotheses that could lead to personalized treatment options for patients with rare activating mutations (Supplementary Figure 6C and Supplementary Table 6). For example, ERBB2 is a well known target of numerous inhibitors when amplified, but only recently was recognized as having recurrent activating mutations in breast cancer18. Numerous candidates for drug development including GATA3, MLL3, CDH1, TLR4, serine/threonine kinases such as MAP3K1, and tyrosine kinases such as ERBB4 stand out as recurrently mutated in breast cancer but poorly targeted by current therapies (Supplementary Figure 7). Ranked according to the type of potentially druggable gene category, the number of supporting sources, patient recurrence rate, and other factors, the researcher can thus use DGIdb to prioritize targets for future drug development efforts.
Figure 1. Druggability of genes recurrently mutated in breast cancer.
315 genes recurrently mutated in breast cancer patients were analyzed by DGIdb. A. The number of candidate breast cancer genes that are considered potentially druggable according to six sources. B. The numbers of genes in potentially druggable categories (from dGene) and the numbers of genes in these categories that are targeted by a known drug.
Potential use cases for DGIdb are abundant. A user may enter a single gene to explore the current state of knowledge regarding druggability of that gene. Alternatively they might input a large list of genes to identify the subset with potential druggability. In another use case, researchers may simply want a list of genes belonging to druggable categories of interest. DGIdb provides a bridge between previously inaccessible data on gene druggability and those seeking to understand the significance of genomic variation in human disease.
Online methods
Data sources
Each potential DGIdb data source was evaluated initially for ease of obtaining information and consistency of information stored. Currently, six sources have been identified for known drug-gene interactions (Supplementary Tables 1 and 2). PharmGKB7 collects, encodes, and disseminates knowledge about the impact of human genetic variations on drug response. They curate primary genotype and phenotype data, annotate gene variants and gene-drug-disease relationships via literature review, and summarize important pharmacogenomic genes and drug pathways. PharmGKB has an excellent interface; information is well organized and integrated. Some information is available for download in simple flat files and there is also a Perl API for searching the website. However, neither the flat files nor the API permit easy retrieval of drug-to-gene target relationships. The Therapeutic Target Database (TTD)10 provides information about known and explored therapeutic protein and nucleic acid targets, the targeted disease, pathway information and the corresponding drugs directed at each of these targets. Their complete database is available as flat file downloads providing gene names and synonyms, drug names and synonyms, and drug-gene associations. The DrugBank database6 combines detailed drug (i.e. chemical, pharmacological and pharmaceutical) data with comprehensive drug target (i.e. sequence, structure, and pathway) information. The complete database is available in XML format, allowing automated parsing. Primary records are drug centric but links to targets are provided by ‘partner ids’. The targeted agents in lung cancer (TALC)8 publication reports the results of an annual meeting of lung cancer experts who have summarized, in PDF tables, those targeted agents that are or have been evaluated in lung cancer and have entered clinical trials. In another publication describing trends in the exploitation of novel drug targets (TEND)9 the authors analyzed drugs that were approved by the US Food and Drug Administration during the past three decades and examined the interactions of these drugs with therapeutic targets using the DrugBank database and extensive manual curation. Finally, the list of targeted therapies provided at My Cancer Genome19 represents the combined effort of a team of volunteer editors, each a cancer domain expert, to document targeted therapies.
In addition to the sources of drug-gene interactions documented above, we also investigated sources of potentially druggable genes. Currently, four sources have been included in DGIdb. The concept of the druggable genome was first introduced by Hopkins and Groom (2002)1 who reviewed literature and databases at the time to define a set of 399 known molecular targets which met their ‘rule-of-five’ criteria for oral bioavailability and other constraints. An examination of the sequences of binding domains for those proteins identified 130 protein families as defined by their InterPro20 domains. Extending to all members of these 130 families produced a list of 3,051 proteins that they defined as the “druggable genome”. In 2005, Russ and Lampel2 published an update of this work using a similar approach. They identified 182 PFAM21 protein domain classifications that were approximately equivalent to Hopkins and Groom’s InterPro domains (many of which were in fact based on PFAM). After manual curation and removal of false positives, they reported a list of 2,917 druggable genes. Most recently Kumar and Chang et al (2013) developed the dGene12 list that updates the concept of the druggable genome with a specific focus on cancer. They started by defining a new set of 10 druggable gene classes based on current drug development efforts. Those classes were then populated with 2,257 genes by extensive manual curation of literature, reviews, and existing databases. In addition to these expert-curated sources, the Gene Ontology (GO)11 is possibly the most commonly used resource for characterization of genes into functional categories. Thus, selected terms from GO were also included (Supplementary Table 3).
Data curation and import
Interactions in DGIdb are defined as a relationship between a gene and a drug with an associated interaction type (e.g., inhibitor) from a specified source. Because data sources behind DGIdb have different definitions of genes and drugs, and the same gene or drug may be represented by a variety of names, DGIdb unifies reports of the same gene or drug by different names into a single entity for search purposes, locatable by any of its aliases. Genes can have multiple alternate names such as gene description, gene synonyms, gene symbol, and gene identifiers (Ensembl, Uniprot, Entrez, etc.). Genes may also have additional meta-data such as gene biotypes. Drugs, similarly, can also have multiple alternate names such as trade names, drug synonyms, etc. and meta-data such as drug class. Druggable gene categories in DGIdb are defined as a relationship between a gene concept and a category deemed to be of interest for druggability from a specified source. Associations between genes and druggable categories are imported from a variety of sources. Wherever possible, categories from different sources were merged into a single consistent category for use in DGIdb. The final druggable gene category lists (e.g., tyrosine kinases) consist of the union of all genes listed in that category from each source (Supplementary Table 3). All interaction and category relationships are linked to their source details providing a full citation and links to the primary data. The complete database schema is provided as Supplementary Figure 1. In all cases data were obtained from primary sources or publications, manually curated (in the case of PDF documents) or parsed with Perl or Ruby to an intermediate tab-delimited file and then imported into a PostgreSQL database with a Ruby importer. Gene targets were mapped from unofficial or source-specific identifiers to Entrez Gene identifiers and symbols by manual curation and synonyms entered as alternate names. In many cases this involved one-to-many mappings from complexes or pathways (e.g., ‘Proteasome’) to individual gene members. Similarly, primary drug names were assigned using generic name, trade name, or development names in that order of preference and all others entered into the database as alternate names. Importers were developed to automatically or semi-automatically import data from raw sources to facilitate regular update where possible. All importer code is available at https://github.com/genome/dgi-db along with instructions for creating custom importers. A brief description of import strategies for each specific data source follows.
Entrez Gene
Gene records were imported from Entrez Gene4 using the ‘gene_info’ and ‘gene2accession’ files obtained from the NCBI ftp site [webref1]. Specifically, Entrez id, symbol, synonyms, Ensembl gene id (from xrefs), description, and protein accessions were imported for each gene in the ‘gene_info’ file. These gene records formed the basis for all gene concepts in DGIdb to which all other gene instances were mapped using one or more of the underlying identifiers. In addition, gene-gene interactions were imported from Entrez Gene using the ‘interactions’ file obtained from the NCBI ftp site [webref2]. For each gene concept in DGIdb, all human interacting gene partners were associated as gene-gene interactions. At the time of import, NCBI human gene interactions were based on data provided by the BIND, BioGRID, and HPRD databases.
Ensembl
Gene records were imported from Ensembl3 using the transcript GTF file available through Ensembl’s ftp site [webref3]. Currently version 68_37 has been imported. Ensembl gene id was imported as a primary gene id and gene name as an alternate name. Gene biotype was imported as meta-data.
PubChem
Drug alternate names were imported from PubChem5 using the ‘CID-Synonym-filtered’ file obtained from the NCBI ftp site [webref4]. Due to the tremendous size of this data source, the only drugs that were imported were drugs that had one or more names matching an existing drug alias from another data source. For each of the drugs meeting these criteria, the drug’s PubChem id, primary name, and a list of aliases was obtained.
dGene
The complete list of potentially druggable genes from dGene12 was obtained from the authors. Druggable gene categories were used as provided by dGene class. Entrez gene id and symbol were also used as provided.
Russ and Lampel
Russ and Lampel’s (2005) list of druggable genes was obtained directly from the authors (personal communication). The file is not to our knowledge available otherwise2. Individual gene categories or protein families were not provided. Therefore all genes from this list were added to a single generic category called “Druggable Genome”. Ensembl gene id was imported as provided as the primary gene id. Display id and description were also imported as alternate names.
Hopkins and Groom
The “original” definition of the druggable genome was adopted and updated from the Hopkins and Groom (2002) publication1. First, their list of druggable protein families was obtained from the journal’s supplementary information page [webref5]. These InterPro20 identifiers were manually updated to account for cases where ids have been deprecated, replaced or split into multiple new ids using the InterPro website [webref6]. Each InterPro id was then queried against the ‘InterPro/UniProt’ database using the BioMART perl API to obtain UniProtKB protein accessions for each protein family. All non-human records, proteins not in the ‘complete human proteome’ and those without Swiss-Prot (Reviewed) status and without protein-level or transcript-level evidence were removed. Interpro families were then manually assigned to new or existing druggable gene categories in DGIdb where possible and also all assigned to the generic “druggable genome” category. For small families (less than 25 proteins), which could not be mapped to an existing category, a new category was not created. Those proteins appear only in the generic “druggable genome” category. The resulting 2,810 proteins were imported into DGIdb with their corresponding druggable gene category. Uniprot identifiers were imported as primary gene id and also mapped to Entrez and Ensembl gene identifiers. InterPro type, name, accession, short name, UniProt status and evidence were also imported as gene meta-data.
GO
Manually selected categories (terms) and their corresponding protein products were imported from the Gene Ontology (GO)11. See Supplementary Table 3 for the GO terms selected for import. XML files were downloaded for each term by automated query of the AMIGO web interface [webref7]. GO gene names were imported as primary gene name and UniProtKB ids as alternate names. GO ids, secondary GO ids, reactome id, and supporting PMIDs were imported as gene meta-data.
My Cancer Genome
The complete list of targeted therapeutics from the My Cancer Genome resource19 was obtained by parsing the web content at mycancergenome.org [webref8] with a custom Ruby module. Each target was mapped to Entrez Gene identifiers and symbols by manual curation. In many cases this involved one-to-many mappings from complexes or pathways (e.g., Proteasome) to individual gene members. A primary drug name was assigned using the first generic name, trade name, or development name in that order and all others entered into the database as alternate names. Drug classes were also imported and used to infer interaction type. Problematic characters were also removed manually.
TALC
The ‘targeted agents in lung cancer’ (TALC) publication8 was obtained from primary literature. Drugs, genes and interactions were manually curated from PDF tables to a tab-delimited file for import into the database. Gene target names were mapped to Entrez Gene identifiers and symbols by manual curation. A primary drug name was assigned using the first generic name, trade name, or development name in that order and all others entered into the database as alternate names. Drug class and drug type were assigned based on details in each record. DrugBank identifier and CAS ids were also assigned where possible.
TEND
The ‘trends in the exploitation of novel drug targets’ (TEND) publication9 was obtained from the journal online. Drugs, genes and interactions were manually curated from PDF tables to a tab-delimited file for import into the database. Gene target ids provided as UniProt accessions were mapped to Entrez Gene and Ensembl gene identifiers and symbols by manual curation. Target main class, target subclasses and transmembrane helix count were imported as gene meta-data. Year of approval and indications (called drug class in DGIdb) were imported as drug meta-data.
PharmGKB
The complete current dataset of PharmGKB7 (as of July 12, 2012) was obtained with permission from PharmGKB. Gene and drug data files were downloaded directly from www.pharmgkb.org [webref9]. Relationship (i.e., interactions) data files were obtained by request. Only relationships which linked drug entities to gene entities and were indicated as “associated” as opposed to “ambiguous” or “not associated” were imported. PharmGKB accession ids were used for primary gene id but Ensembl gene id, Entrez gene id, gene symbol, gene name, alternate names and alternate symbols were also imported. Variant annotation and VIP status were also imported as gene meta-data. For drug details, PharmGKB drug name was imported as a primary drug id but additional name(s), generic name(s), trade name(s), brand name(s), drug cross-references, and the SMILES string were imported as alternate names. Drug type and external vocabulary were imported as drug meta-data.
TTD
The complete current dataset of the Therapeutic Targets Database (TTD)10 was downloaded from bidd.nus.edu.sg[webref10]. Interactions were obtained from the ‘TTD_download.txt’ file for any entries that had ‘Drug(s)’ listed. Also determined from this file were target name, UniProt ID, synonyms and interaction time. The latter was determined by comparison of TTD attributes against a manually predefined list of inhibitor type values. UniProt ids were further mapped to Entrez and Ensembl identifiers using the HUMAN_9606_idmapping_selected.tab file available at ftp.uniprot.org [webref11]. For each drug, the corresponding drug name, CAS number, PubChem CID, and PubChem SID were obtained from the ‘TTD_crossmatching.txt’ file and drug synonyms obtained from the ‘Synonyms.txt’ file.
DrugBank
The complete current dataset of DrugBank6 including all drugs and targets was obtained from the drugbank.xml file available at drugbank.ca [webref12]. Drugs, genes, and interactions were parsed from this XML file. Gene data obtained included DrugBanks’s partner identifier, UniProtKB identifier and gene symbol which were further mapped to Entrez and Ensembl identifiers as described for TTD. Drug data obtained included DrugBank’s drug identifier, drug name, drug synonyms, CAS number, drug brands, drug type, drug groups, drug categories, target partner ids, and target count. Drug-gene interactions were determined by linking drug target partner ids to gene partner ids and also included target actions (interaction type). Problematic characters (e.g., tabs) were stripped from affected data. Missing values were specified as “N/A”.
Additional sources considered for import
Other sources considered for future import as interactions include the Cancer Commons [cancercommons.org], the Clearity Foundation [clearityfoundation.org], STITCH22, SuperTarget23, ChEMBL24, Promiscuous25 and CTD26. Commercial sources considered include MetaDrug from Thomson Reuters [thomsonreuters.com] and Pharmaco Atlas from NextBio Research [www.nextbio.com]. Other sources considered for future import as potentially druggable genes include the Potential Drug Target Database (PDTD)27, KinBase28, Integrated Druggable Genome Database (IDGD) from Sophic Systems Alliance [www.sophicalliance.com], and Novartis’ druggable genome list29. Future plans to expand DGIdb also include empirical drug-gene association mapping based on compound screening datasets such as ConnectivityMap30, BindingDB31, the Sanger Institute’s Genomics of Drug Sensitivity in Cancer32, and Broad Institute’s Cancer Cell Line Encyclopedia33. Other areas for improvement include capturing information regarding genes that mediate adverse responses and pharmacogenetic relationships. Such relationships might be obtained from T3DB34, SuperCYP35, SIDER36, and the adverse drug reaction study by Lounkine et al (2012)37.
Gene grouping
A major challenge in identifying drug-gene interactions is the unification of gene and drug identifiers. This was accomplished through a grouping approach in which primary gene and drug concepts were first defined according to Entrez gene4 and PubChem5, respectively. Our current approach to gene grouping occurs in three steps: preprocessing, group creation, and group population. The preprocessing stage aims to create two sets of mappings to aide in grouping. These two mappings encompass every known gene alias in the system except for a few one or two character aliases, which are ignored. The first maps Entrez gene name strings to the system entities that represent them. The second, referred to as the “default mapping,” links unknown gene aliases to their system entity representations. These mappings are used for lookups later in the grouping process. The group creation stage has the goal of creating a system level cluster (gene object) for each Entrez gene name. For each of these Entrez gene names from the preprocessing stage, we create a gene object to represent the concept of a biological gene represented in different ways across various data sources. We then add the system entries that we mapped to each Entrez gene name to their gene object. The end result of this process is a set of gene objects where there exists one gene object for each official Entrez gene name. This currently constitutes the complete set of gene objects in the system. The group population stage has the goal of attempting to unambiguously map all of the genes in each data source to one of our new gene objects. We begin this stage by taking the system entity for each gene listed in a data source (gene claim object), and enumerating them one by one. We first skip any gene claim objects that already belong to a gene object. This is usually the result of having already been added to a group in the group creation stage. We then look for gene objects with the same name as the gene claim object or any of its aliases. We refer to these as direct gene objects, and keep a record of each gene object we found and how many times we found it. We then take all of the aliases for the gene claim objects and check them against the default mapping we created in the preprocessing stage. We take these alias objects from the default mapping and attempt to map them to their corresponding gene claim objects. If any of these gene claim objects are part of a gene object, we refer to the gene objects as indirect gene objects and keep track of each one and the number of times we found it. At this point, we finally attempt to add the gene claim to a gene based on several rules. If we found exactly one direct gene object, we add the gene claim to the direct gene and move on to the next gene claim. If we found no direct gene objects and exactly one indirect group, we add the gene claim to the indirect gene and continue on to the next gene claim. For all other cases we are either unable to find any gene objects to add the gene claim to or there are several gene objects we could add the gene claim to. In this case, we skip the gene claim and move on to the next one.
Drug grouping
Our current approach to drug grouping occurs in three steps: preprocessing, group creation, and group population. The preprocessing stage aims to create two sets of mappings to aide in grouping. These two mappings encompass every known drug alias in the system except for a few one or two character aliases, which are ignored. The first maps PubChem primary drug name strings to the system entities that represent them. The second, referred to as the “default mapping,” links unknown drug aliases to their system entity representations. These mappings are used for lookups later in the grouping process. The group creation stage has the goal of creating a system level cluster (drug object) for each PubChem drug name. For each of these PubChem drug names from the preprocessing stage, we create a drug object to represent the concept of a drug represented in different ways across various data sources. We then add the system entries that we mapped to each PubChem drug name to their drug object. The end result of this process is a set of drug objects where there exists one drug object for each official PubChem drug name. This currently constitutes the complete set of drug objects in the system. The group population stage has the goal of attempting to unambiguously map all of the drugs in each data source to one of our new drug objects. We begin this stage by taking the system entity for each drug listed in a data source (drug claim object), and enumerating them one by one. We first skip any drug claim objects that already belong to a drug object. This is usually the result of having already been added to a group in the group creation stage. We then look for drug objects with the same name as the drug claim object or any of its aliases. We refer to these as direct drug objects, and keep a record of each drug object we found and how many times we found it. We then take all of the aliases for the drug claim objects and check them against the default mapping we created in the preprocessing stage. We take these alias objects from the default mapping and attempt to map them to their corresponding drug claim objects. If any of these drug claim objects are part of a drug object, we refer to the drug objects as indirect drug objects and keep track of each one and the number of times we found it. At this point, we finally attempt to add the drug claim to a drug based on several rules. If we found exactly one direct drug object, we add the drug claim to the direct drug and move on to the next drug claim. If we found no direct drug objects and exactly one indirect group, we add the drug claim to the indirect drug and continue on to the next drug claim. For all other cases we are either unable to find any drug objects to add the drug claim to or there are several drug objects we could add the drug claim to. In this case, we skip the drug claim and move on to the next one.
Anti-neoplastic filtering
Due to strong interest in cancer-specific gene targeted therapies we created an anti-neoplastic filter. The intent of this filter is to remove drug-gene interactions from results that do not explicitly involve an anti-cancer agent. Interactions from sources such as DrugBank cover a comprehensive range of diseases and conditions and many genes identified in a cancer study may have interactions with drugs not deemed suitable or interesting in a cancer context. All drugs from TALC and My Cancer Genome were considered anti-neoplastic since documenting such drugs is the stated purpose of those resources. For all other sources, drugs were only considered antineoplastic if they were annotated as such with any meta-data terms that were identified by manual review as likely to indicate relevance to cancer treatment (Supplementary Table 4).
Source trust level
Data sources imported in DGIdb were divided into two basic trust level classes. ‘Expert-curated’ are those such as dGene, the publications from Russ and Lampel or Hopkins and Groom, MyCancerGenome, TALC and TEND, which are primarily the result of expert curation of the literature or expert knowledge. ‘Non-curated’ sources such as GO, PharmGKB, TTD and DrugBank, were deemed to be more comprehensive and inclusive of putative interactions and do not meet the same standard of trust as those classed as expert-curated. In addition to this categorization, sources were further ranked by trust level within the trust classes. It should be noted that DGIdb’s definitions of expert-curated versus non-curated are only within the specific context of therapy-relevant drug-gene interactions or druggability with a bias towards cancer therapies. In fact, a great deal of expert curation has gone into GO, PharmGKB, TTD and DrugBank and this distinction is not meant to lessen the value of those excellent resources. The distinction is only meant as a useful sorting tool for our specific purposes. Researchers interested in more accepted therapeutic options for a gene might sometimes wish to limit results to the expert-curated category whereas those interested in more experimental options for hypothesis generation might include all sources.
DGIdb analysis of a breast cancer meta-dataset
To demonstrate the utility of DGIdb for druggable gene analysis in the context of a large-scale cancer genome sequencing initiative, mutation annotation format (MAF) files were obtained for mutations observed in several large-scale breast cancer sequencing projects13–17. The MAF files were merged into a single list containing 65,880 mutations observed in one or more of 1,273 patients. This large resource combines data from multiple sources using different sequencing and variant calling protocols. It is not the complete picture of mutations in breast cancer but it does represent a good sample case. A list of candidate genes was extracted by first removing silent mutations and then determining those genes mutated in at least 1% or 2.5% of patients. The resulting candidate gene lists of 315 and 31 genes respectively were used as input to the DGIdb ‘search interactions’ tool. Interactions were selected with the following options: without filtering, expert curated interactions only, anti-neoplastic drugs only, and only those with a defined mechanism of action. Each of these results was exported as a TSV file and imported into R for generation of visualizations. We summarized the genes mutated in each tumor by mutation type (missense, in-frame insertion, etc.) and drugs available (Figure 2E and Supplementary Figures 6 A–C, and Supplementary Figure 7). The list of 315 candidate genes mutated in breast cancer was also used as input to the DGIdb ‘search categories’ tool. The subset of candidate genes predicted to be potentially druggable were exported as a TSV file and imported into R for visualization. A druggability ‘score’ was calculated for each gene by taking the maximum of drug count or mutation recurrence rate. Heat maps were generated using the ‘plotrix’ R library and all other plots were created using the ‘ggplot2’ R library.
Implementation
DGIdb is built in Ruby on Rails with PostgreSQL as the primary data store. Memcached is utilized heavily for caching, as the data is largely static between new source imports. The site is served with Apache and Phusion Passenger on a server running Ubuntu 12.04 LTS (Precise Pangolin). The code itself is divided into two primary components – the web application itself and the libraries that handle the importing and normalization of new sources.
The web application is organized in a Model-View-Controller (MVC) architecture with a couple of notable exceptions. In an effort to keep application logic out of the view templates, presenter objects are utilized to decorate domain models with view logic while still allowing access to the underlying models through delegation. Additionally, most domain logic is pulled out into command and helper classes. This allows for a separation of concerns between the persistence layer (data model) and business logic of the application. This architecture also makes the API implementation simpler. The same back-end code runs to produce the result for both the API and the web site. At render time, the result is simply wrapped in a different presenter object and sent to a JSON template instead of an HTML template.
Two of the web application’s primary pieces of functionality are its gene name matching algorithm and its implementation of filtering. The gene name matching process attempts to account for potential ambiguity in user search terms. It first attempts to make an exact match on Entrez gene symbols. If it finds such a match, it assumes it to be what the user meant. If it is unable to find an exact Entrez match for a search term, it reverts to searching through all reported aliases for gene clusters in the system. If the system finds more than one gene cluster that matches the search term, it will classify the result as ambiguous and return all potential gene group matches. The ambiguity is expressed in both the user interface and API responses in order to help the user decide which gene they meant.
Rather than being implemented as SQL WHERE clauses, result filtering is implemented using sets. For interaction filtering, the set of all interactions meeting each possible filter criterion is pre-calculated into a set of ids. Each of these sets can be individually cached for fast retrieval later. Set operations are then utilized to combine filters quickly. For instance, if a user wanted to see only inhibitor interactions that involved kinase genes and are from DrugBank, the following steps would take place. The set of all inhibitor interactions would be intersected with the set of all interactions involving kinases, which would then be intersected with the set of all interactions reported by DrugBank. Each intermediate step as well as the final filter will be cached. Over time, the most common permutations are calculated and cached, making filtering almost instantaneous. Once the final set is calculated, each returned interaction’s id can be checked for presence in the set in constant (O(1)) time.
DGIdb is integrated with The Genome Institute’s Genome Modeling System (manuscript in preparation) and forms an integral part of this pipeline for automated analysis of cancer genomes in a clinical context. Genes identified with potentially relevant cancer-driving events (single nucleotide variants, transcript fusions, etc.) are automatically queried against DGIdb using the API.
Access
The DGIdb web interface allows exploration of the druggable genome through three simple tools (Supplementary Figure 2). The ‘Search Interactions’ web interface allows entry of multiple genes for query against the database of known interactions. Interactions can be filtered by source, source trust level, gene category, interaction type and limited to only anti-cancer drugs. A set of default genes can be entered for illustrative purposes. Once submitted, the results page indicates all known drug-gene interactions for the input gene list. Search terms with ambiguous gene name mapping are shown but indicated as such. Results can be further filtered in real time using the filter results box. Additional display tabs provide a general summary of the search results, and detailed summaries broken down by search term, gene, and source. The ‘Search Categories’ interface performs similarly but instead of returning specific known drug-gene interactions, it returns genes with membership in any of the pre-selected druggable gene categories. Results can again be pre-filtered for specific sources, source trust levels or gene categories. Alternatively, the lists of potentially druggable genes can be browsed directly by going to the ‘Browse Categories’ tab. By default, output is directed to an HTML web view. However, all results pages can be downloaded as a tab-delimited (TSV) text file for exploration in Excel or other software. In addition to the web interface, all data from DGIdb are available as tab-delimited data downloads and also through a web services API. HTTP Get or Post requests can be submitted by URL crafting or with scripting languages (Perl LWP, etc.). Results of submitted gene list queries (after application of any included filter options) are returned in JSON which can also be readily parsed by most programming languages (e.g., Perl, Ruby, Java, Python, etc.). This functionality is meant to allow automation of queries in analysis pipelines. A tutorial, answers to frequently asked questions, source details, downloads, API documentation, and contact details are available under the ‘Help’ menu. The DGIdb API can be used to query for drug-gene interactions in your own applications through a simple JSON based interface. Extensive documentation of the API including functioning code example is maintained at: http://dgidb.genome.wustl.edu/api.
Supplementary Material
Acknowledgments
We would like to thank the creators of each source we used as input and all users of DGIdb who have contributed feedback. We are grateful to Russ Altman, Stanford University, and PharmGKB for granting us a license to mine their data and William Pao of My Cancer Genome for agreeing to collaborate with us. We would also like to thank the patients who participated in research that led to the breast cancer data used to demonstrate the utility of DGIdb. This work was funded by an NHGRI center grant (U54 HG003079) to RKW.
Footnotes
Author contributions
OLG and MG wrote the manuscript. MG and OLG were responsible for the selection and manual curation of data sources, creation of importers, initial design of the web interface, testing, analysis and figure creation for the manuscript. ID and JFM helped with figure creation. ACC and JVW conceived and lead the Ruby implementation of the web interface, database, importers and API. JK, MG, OLG, SMS, ID, and JFM made contributions to the code. JFM was the lead user experience web developer. SMS, JME, JRW and DJD supervised software development. MBC facilitated the public deployment of DGIdb and all related systems requirements. RB, RDK, CAM and DEL provided beta testing feedback. JS and RG provided and curated the TALC dataset. RB and RDK contributed the dGene dataset and the breast cancer data used for analysis demonstration purposes. NCS contributed manual curation of data sources and documentation. SMS, JME, DJD, LD, TJL, ERM, and RKW contributed ideas and use case requirements. ACC, JVW, CAM, RDK, RB, DEL and ERM contributed text and revised the manuscript. ERM and RWK are mentors of MG and OLG.
Contributor Information
Malachi Griffith, Email: mgriffit@genome.wustl.edu.
Obi L. Griffith, Email: ogriffit@genome.wustl.edu.
Adam C. Coffman, Email: acoffman@genome.wustl.edu.
James V. Weible, Email: jweible@genome.wustl.edu.
Josh F. McMichael, Email: jmcmicha@genome.wustl.edu.
Nicholas C. Spies, Email: nspies@wustl.edu.
James Koval, Email: james.ross.koval@gmail.com.
Indraniel Das, Email: idas@genome.wustl.edu.
Matthew B. Callaway, Email: mcallawa@genome.wustl.edu.
James M. Eldred, Email: jeldred@genome.wustl.edu.
Christopher A. Miller, Email: cmiller@genome.wustl.edu.
Janakiraman Subramanian, Email: jsubrama@dom.wustl.edu.
Ramaswamy Govindan, Email: rgovinda@dom.wustl.edu.
Runjun D. Kumar, Email: kumarr@wusm.wustl.edu.
Ron Bose, Email: rbose@dom.wustl.edu.
Li Ding, Email: lding@genome.wustl.edu.
David E. Larson, Email: dlarson@genome.wustl.edu.
David J. Dooling, Email: ddgenome@genome.com.
Scott M. Smith, Email: ssmith@genome.wustl.edu.
Timothy J. Ley, Email: tley@dom.wustl.edu.
Elaine R. Mardis, Email: emardis@wustl.edu.
Richard K. Wilson, Email: rwilson@wustl.edu.
References
- 1.Hopkins AL, Groom CR. Nature reviews Drug discovery. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Russ AP, Lampel S. Drug discovery today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 3.Flicek P, et al. Nucleic acids research. 2011;39:D800–806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maglott D, Ostell J, Pruitt KD, Tatusova T. Nucleic acids research. 2011;39:D52–57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, et al. Nucleic acids research. 2012;40:D400–412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knox C, et al. Nucleic acids research. 2011;39:D1035–1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. Biomarkers in medicine. 2011;5:795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Somaiah N, Simon GR. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:S1758–1785. doi: 10.1097/01.JTO.0000407557.30793.a6. [DOI] [PubMed] [Google Scholar]
- 9.Rask-Andersen M, Almen MS, Schioth HB. Nature reviews Drug discovery. 2011;10:579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 10.Zhu F, et al. Nucleic acids research. 2010;38:D787–791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M, et al. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar RD, Chang LW, Ellis MJ, Bose R. PloS one. 2013;8:e67980. doi: 10.1371/journal.pone.0067980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerji S, et al. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan Z, et al. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 16.Shah SP, et al. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens PJ, et al. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose R, et al. Cancer discovery. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh P, et al. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013. [Google Scholar]
- 20.Hunter S, et al. Nucleic acids research. 2012;40:D306–312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punta M, et al. Nucleic acids research. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn M, et al. Nucleic acids research. 2012;40:D876–880. doi: 10.1093/nar/gkr1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hecker N, et al. Nucleic acids research. 2012;40:D1113–1117. doi: 10.1093/nar/gkr912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaulton A, et al. Nucleic acids research. 2012;40:D1100–1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Eichborn J, et al. Nucleic acids research. 2011;39:D1060–1066. doi: 10.1093/nar/gkq1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis AP, et al. Nucleic acids research. 2013;41:D1104–1114. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Z, et al. BMC bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 29.Orth AP, Batalov S, Perrone M, Chanda SK. Expert opinion on therapeutic targets. 2004;8:587–596. doi: 10.1517/14728222.8.6.587. [DOI] [PubMed] [Google Scholar]
- 30.Lamb J, et al. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 31.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. Nucleic acids research. 2007;35:D198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, et al. Nucleic acids research. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barretina J, et al. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim E, et al. Nucleic acids research. 2010;38:D781–786. doi: 10.1093/nar/gkp934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preissner S, et al. Nucleic acids research. 2010;38:D237–243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. Molecular systems biology. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lounkine E, et al. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web references
- 1.ftp://ftp.ncbi.nih.gov/gene/DATA/
- 2.ftp://ftp.ncbi.nlm.nih.gov/gene/GeneRIF/interactions.gz
- 3.http://useast.ensembl.org/info/data/ftp/index.html
- 4.ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound/Extras/
- 5.http://dx.doi.org/10.1038/nrd892
- 6.http://www.ebi.ac.uk/interpro/
- 7.http://amigo.geneontology.org/cgi-bin/amigo/go.cgi
- 8.http://www.mycancergenome.org/content/other/molecular-medicine/targeted-therapeutics
- 9.http://www.pharmgkb.org/downloads.jsp
- 10.http://bidd.nus.edu.sg/group/cjttd/TTD_Download.asp
- 11.ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/by_organism/
- 12.http://www.drugbank.ca/downloads
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.