FMRP is an RNA binding protein whose loss causes fragile X syndrome. This study shows that FMRP regulates actin organization and neurite outgrowth by repressing the translation of the mRNA encoding the armadillo protein p0071. A variety of approaches demonstrate that p0071 is a novel and important target of FMRP. Moreover, the studies strongly suggest that impaired actin cytoskeletal functions contribute to fragile X syndrome.
Keywords: FMRP, p0071, actin organization, neuromorphogenesis, dendrite formation
Abstract
Loss of fragile X mental retardation protein (FMRP) causes synaptic dysfunction and intellectual disability. FMRP is an RNA-binding protein that controls the translation or turnover of a subset of mRNAs. Identifying these target transcripts is an important step toward understanding the pathology of the disease. Here, we show that FMRP regulates actin organization and neurite outgrowth via the armadillo protein p0071. In mouse embryonic fibroblasts (MEFs) lacking FMRP (Fmr1−), the actin cytoskeleton was markedly reorganized with reduced stress fibers and F-actin/G-actin ratios compared to fibroblasts re-expressing the protein. FMRP interfered with the translation of the p0071 mRNA in a 3′-UTR-dependent manner. Accordingly, FMRP-depleted cells revealed elevated levels of p0071 protein. The knockdown of p0071 in Fmr1− fibroblasts restored stress fibers and an elongated cell shape, thus rescuing the Fmr1− phenotype, whereas overexpression of p0071 in Fmr1+ cells mimicked the Fmr1− phenotype. Moreover, p0071 and FMRP regulated neurite outgrowth and branching in a diametrically opposed way in agreement with the negative regulation of p0071 by FMRP. These results identify p0071 as an important and novel FMRP target and strongly suggest that impaired actin cytoskeletal functions mediated by an excess of p0071 are key aspects underlying the fragile X syndrome.
INTRODUCTION
Loss of fragile X mental retardation protein (FMRP) causes fragile X syndrome (FXS), a genetic disease characterized by intellectual disability and autism spectrum disorders. In most fragile X syndrome patients, an amplification of a CGG repeat region in the 5′ UTR of the fragile X mental retardation gene (Fmr1) exists which leads to hypermethylation and epigenetic silencing of Fmr1, resulting in the loss of FMRP. FMRP is an mRNA binding protein that functions predominantly as a translational repressor, although stimulation of translation and mRNA stabilization by FMRP have also been described (Santoro et al. 2012). FMRP is enriched at the postsynaptic sites of dendritic spines, where it is proposed to inhibit the translation of a subset of localized mRNAs. After stimulation by appropriate synaptic signals, the translational block imposed by FMRP is abolished, allowing the spatiotemporal control of target mRNA translation and, thus, the fine tuning of protein expression that directs synaptic plasticity (Pfeiffer and Huber 2009).
Dendritic spines are the recipients of excitatory synaptic signals. Alterations in their density, size, or turnover, or defects in their generation result in neuronal dysfunction and lead to cognitive defects (Irwin et al. 2000; Humeau et al. 2009; Cruz-Martín et al. 2010; Kasai et al. 2010; Penzes et al. 2011; Pavlowsky et al. 2012). In FXS as well as in the Fmr1 knockout (KO) mouse, neurons show a high density of long, thin, and immature dendritic spines (Comery et al. 1997; Irwin et al. 2001). The highly dynamic cytoskeleton of dendritic spines is essential for synaptic plasticity (Matus 2000; Dent et al. 2011), and the actin cytoskeleton directly determines the morphology of the spine (Ethell and Pasquale 2005; Cingolani and Goda 2008). In agreement, actin regulators such as the small GTPases Rac1, RhoA, and Cdc42, modulate actin organization and, as a consequence, dendritic spine morphology (Newey et al. 2005; Bosch and Hayashi 2012).
RhoA was implicated in limiting dendrite branching, whereas Rac1 and Cdc42 promote neurite outgrowth. In view of the importance of actin cytoskeleton remodeling in dendrite formation and of FMRP in regulating these structures, it appears essential to characterize the functional link between FMRP and actin regulatory proteins. Based on the severe up-regulation of Rac1 observed in the brain of Fmr1−/− mice, it was suggested that FMRP might act as a negative regulator of Rac1 (Bongmba et al. 2011). However, how FMRP influences Rac1 at the molecular level remains elusive, since the Rac1 mRNA was not identified as a direct target of FMRP in mammals (Schenck et al. 2001; Bongmba et al. 2011), although the Drosophila dRac1 mRNA has been reported to associate with the dFMRP complex (Lee 2003).
To understand the molecular basis of FMRP function, several attempts were made to identify mRNAs that bind to and are directly regulated by FMRP. These approaches include FMRP immunoprecipitation followed by microarray analysis of coprecipitating mRNAs (Brown et al. 2001), high-throughput sequencing of RNAs isolated through cross-linking immunoprecipitation (HITS-CLIP and photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation, PAR-CLIP) (Darnell et al. 2011; Ascano et al. 2012) and yeast three-hybrid analyses (Zou et al. 2008). Several hundreds of mRNAs that potentially associate with FMRP were identified, although to date, only very few of these putative targets have been validated (Miyashiro et al. 2003; Castets et al. 2005; Zalfa et al. 2007; Bechara et al. 2009; Davidovic et al. 2011; Gross et al. 2011; Jung et al. 2012; Kwan et al. 2012; Santoro et al. 2012). The Rho-GTPases were not identified as putative mRNA targets of FMRP in any of these screens, suggesting that FMRP modulates GTPases indirectly by controlling the expression of their regulators. In support of this view, p0071 and its closest relative, δ-catenin/NPRAP, were identified as candidate target mRNAs of FMRP by ribonucleoprotein immunoprecipitation–microarray (RIP-Chip) and/or HITS-CLIP studies (Brown et al. 2001; Darnell et al. 2011). Both proteins, p0071 and δ-catenin/NPRAP, belong to the p120-catenin (p120ctn) family of armadillo-related proteins. Several members of this protein family have been shown to regulate the Rho-family GTPases (Reynolds and Roczniak-Ferguson 2004; Hatzfeld 2005). P120ctn down-regulates RhoA activity (Noren et al. 2000; Anastasiadis and Reynolds 2001; Grosheva et al. 2001) and has been implicated in the regulation of Rac-Rho crosstalk (Wildenberg et al. 2006). In the nervous system, p120ctn gene deletion resulted in reduced spine and synapse densities caused by the de-regulation of Rho-GTPases, with decreased Rac1 and increased RhoA activity (Elia et al. 2006). δ-catenin/NPRAP is almost exclusively expressed in neuronal cells where it regulates spine density and synapse morphogenesis (Arikkath et al. 2009). δ-catenin KO mice revealed abnormalities in synaptic plasticity resulting in impaired cognitive function (Israely et al. 2004), whereas in humans, deletion of δ-catenin causes intellectual disability in Cri-du-Chat syndrome (Medina et al. 2000). P0071 is widely expressed with high expression in neuronal cells. It functions in Rho-signaling by controlling RhoA activity during cytokinesis (Wolf et al. 2006). At the molecular level, p0071 associates with RhoA and its activator, Ect2, thereby stimulating RhoA activity. Its function in neuronal cells has not been addressed so far. Although the role of p120ctn-family proteins in regulating actin dynamics and organization via Rho-GTPases has been characterized in various systems, it remains essentially unknown how this function is controlled by upstream modulators.
Here, we show that FMRP regulates the actin cytoskeleton in fibroblasts and neuronal cells via p0071. FMRP associates with and inhibits the translation of the p0071 mRNA. Rescue of p0071 levels in FMRP-overexpressing or FMRP KO mouse embryonic fibroblasts (MEFs) abolished the FMRP-mediated modulation of actin organization. Similar effects were also observed in neuroblastoma-derived cells and primary hippocampal neurons. These results indicate that p0071 is a direct target of FMRP and plays an essential role in FMRP-mediated regulation of actin organization and neuronal morphogenesis.
RESULTS
FMRP regulates actin organization in mouse embryonic fibroblasts
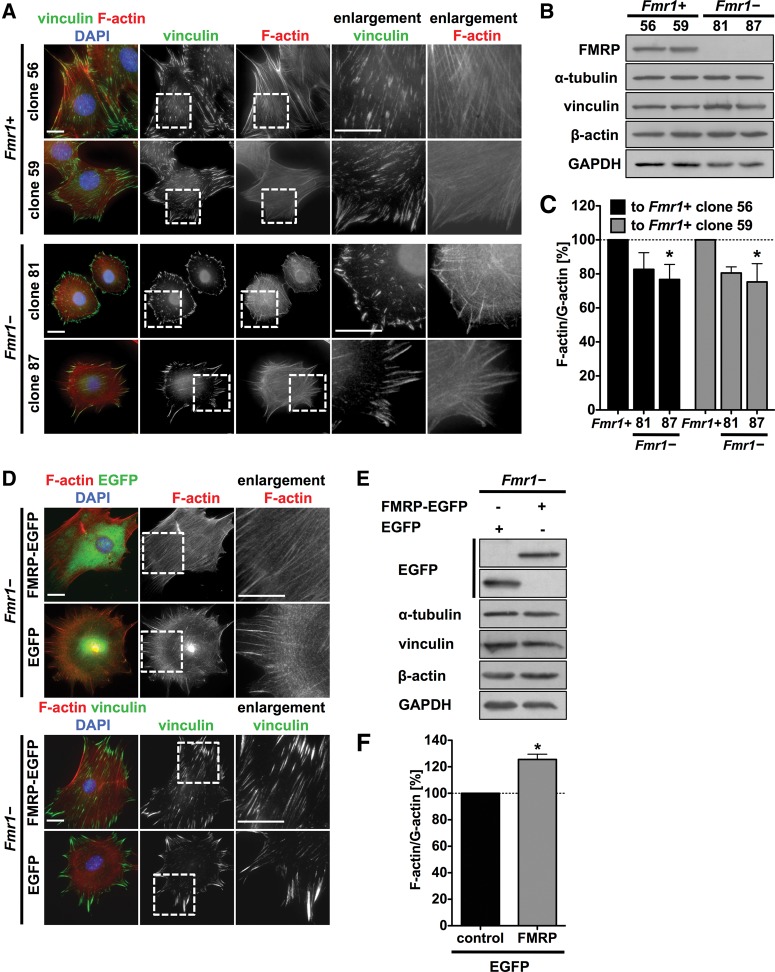
It has been reported that, in patients with FXS as well as in Fmr1−/− mice, dendritic spines are altered (Comery et al. 1997; Irwin et al. 2000), and it was suggested that FMRP modulates actin organization via a negative regulation of Rac1 synthesis (Bongmba et al. 2011). Using immortalized mouse embryonic fibroblasts derived from Fmr1−/− mice (Castets et al. 2005), we observed a considerably altered cell shape and actin organization when compared to Fmr1+ MEFs re-expressing the protein (Fig. 1A,B). FMRP-expressing fibroblasts had an elongated cell shape and contained numerous stress fibers and focal contacts, whereas the Fmr1− fibroblasts were rather round with less stress fibers and focal contacts essentially restricted to the cell periphery. These differences in morphology were observed in two independent Fmr1− clones (clones 81, 87) vs. FMRP re-expressing MEFs (clones 56, 59). To quantify the changes in actin organization, we determined the ratio of filamentous (F-) vs. globular (G-) actin. These experiments revealed that Fmr1− cells showed an ∼25% reduction in F-actin/G-actin ratio compared to cells re-expressing the protein (Fig. 1C).
FIGURE 1.
The loss of FMRP alters the morphology and F-actin/G-actin ratios of MEFs. (A) Immunofluorescence studies of FMRP re-expressing MEF clones (Fmr1+, clones 56, 59, top panels) with parallel stress fibers and Fmr1− cell clones (clones 81, 87, bottom panels) with reduced stress fiber formation. Enlargements of boxed regions are shown in the right panels; bar, 20 µm. (B) Representative Western blotting of Fmr1+ and Fmr1− cell lysates confirming FMRP loss in the Fmr1− cell clones and FMRP re-expression in clones 56 and 59. (C) F-actin/G-actin assay of Fmr1+ and Fmr1− cell clones showing a reduced F-actin/G-actin ratio in Fmr1− cell clones. The F-actin organization of both Fmr1− cell clones (81 and 87) was compared to both WT cell clones re-expressing FMRP (56 and 59). n = 3, (*) P ≤ 0.05. (D) Fluorescence microscopy studies of Fmr1− cells after lentiviral transduction with the indicated constructs showing F-actin- (top panels) and vinculin-staining (bottom panels). Re-expression of FMRP (FMRP-EGFP) reverted the Fmr1− phenotype compared to control transduction (EGFP); bar, 20 µm. (E) Representative Western blotting of Fmr1− cell lysates after lentiviral transduction with FMRP-EGFP or EGFP alone confirming FMRP re-expression in Fmr1− cells. (F) F-actin/G-actin assay of lentivirally transduced Fmr1− MEFs showing an increased F-actin/G-actin ratio in FMRP-EGFP-expressing cells. n = 3, (*) P ≤ 0.05.
To further exclude bias by clonal variability, we analyzed whether FMRP expression in Fmr1− cells after lentiviral transduction without clonal selection rescues the changes in actin organization. Expression of FMRP-EGFP in the Fmr1− cells induced an increase in stress fibers and an elongated cell shape along with a redistribution of focal contacts which was not observed after the expression of EGFP alone (Fig. 1D,E). The expression of FMRP-EGFP increased F-actin/G-actin ratios by ∼25% (Fig. 1F).
These analyses identify FMRP as a potent regulator of actin organization and cellular F-actin/G-actin ratios as well as of cellular morphology of mouse embryonic fibroblasts.
FMRP inhibits p0071 mRNA translation
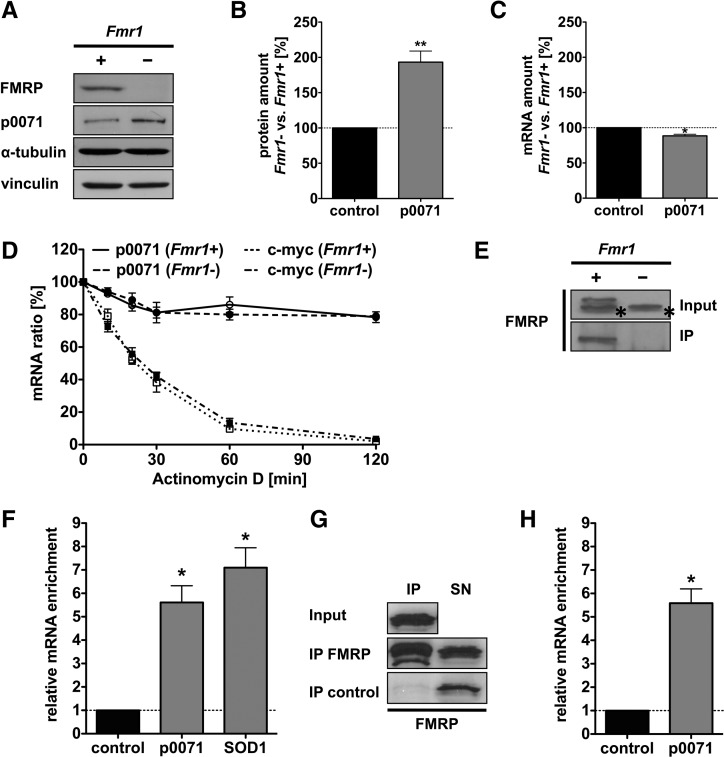
P0071 has been shown to modulate the actin cytoskeleton by regulating small Rho-GTPases and was identified as a FMRP candidate target transcript by three distinct screens (Brown et al. 2001; Darnell et al. 2011; Ascano et al. 2012). Therefore, we aimed at characterizing whether FMRP modulates actin organization by regulating p0071 at the post-transcriptional level. The quantification of p0071 protein and mRNA levels at steady-state revealed that p0071 abundance was markedly increased in the Fmr1− MEFs (Fig. 2A,B). On the contrary, p0071 steady-state mRNA levels as well as p0071 mRNA decay remained essentially unaffected by FMRP depletion (Fig. 2C,D). This suggested that FMRP inhibited p0071 protein synthesis.
FIGURE 2.
FMRP associates with the p0071 mRNA and regulates p0071 expression. (A,B) Representative Western blotting (A) and quantification (B) of p0071 protein levels in Fmr1+ and Fmr1− cell lysates indicating an increased expression of p0071 in Fmr1− cells. n = 4, (**) P ≤ 0.005. (C) Quantification of p0071 mRNA levels in Fmr1+ and Fmr1− cells indicating a slightly reduced p0071 mRNA level in Fmr1− cells. mRNA levels were calculated via the ΔΔCt-method and normalized to the control mRNA encoding RPLP0. n = 4, (*) P ≤ 0.05. (D) For p0071 mRNA decay analyses, cells were treated with 2.5 µM Actinomycin D, and RNA levels were determined by qRT-PCR. p0071 mRNA decayed with the same rate in both, Fmr1+ (black line) and Fmr1− cells (black dashed line). C-myc mRNA stability was not affected by FMRP and served as a control. n = 3. (E) Western blotting of FMRP coimmunoprecipitation experiments. FMRP was specifically precipitated from Fmr1+ cells but not from Fmr1− cells (top panel). The asterisk denotes an unspecific side reaction detected in the input fraction only. (F) qRT-PCR analysis of mRNAs that coprecipitated with FMRP in Fmr1+ vs. Fmr1− cells, showing a p0071- and SOD1 mRNA enrichment. Enrichment was calculated via the ΔΔCt-method and normalized to the control mRNA encoding CyclophilinA. n = 3, (*) P ≤ 0.05. (G) Western blotting of FMRP immunoprecipitated from HEK293 cells with immobilized FMRP antibody. A control fraction was incubated with beads in the absence of FMRP antibody. (IP) immunoprecipitation, (SN) supernatant. (H) qRT-PCR analysis revealed an enrichment of the p0071 mRNA after FMRP precipitation but not in the bead control. Enrichment was calculated via the ΔΔCt-method using CyclophilinA mRNA abundance for normalization. n = 3, (*) P ≤ 0.05.
In order to distinguish between a direct and an indirect effect of FMRP on p0071, we tested whether FMRP associates with the p0071 mRNA as previously suggested (Brown et al. 2001; Darnell et al. 2011). FMRP was immunoprecipitated from FMRP re-expressing Fmr1− MEFs, and copurified RNAs were analyzed by qRT-PCR (Fig. 2E,F). Quantitative assessment of copurified RNAs by qRT-PCR confirmed a significant enrichment of the p0071 mRNA and the reported target transcript SOD1 (Fig. 2F; Bechara et al. 2009). Fmr1− MEFs served as the negative control for the immunoprecipitations. In contrast to p0071 mRNA, qRT-PCRs of unrelated mRNAs, such as CyclophilinA, did not reveal any increase upon FMRP deficiency. Copurification of p0071 mRNA with FMRP was further confirmed for endogenous FMRP isolated from HEK293 cells (Fig. 2G,H). Hence, FMRP associated with the p0071 mRNA, suggesting that the protein inhibits p0071 mRNA translation.
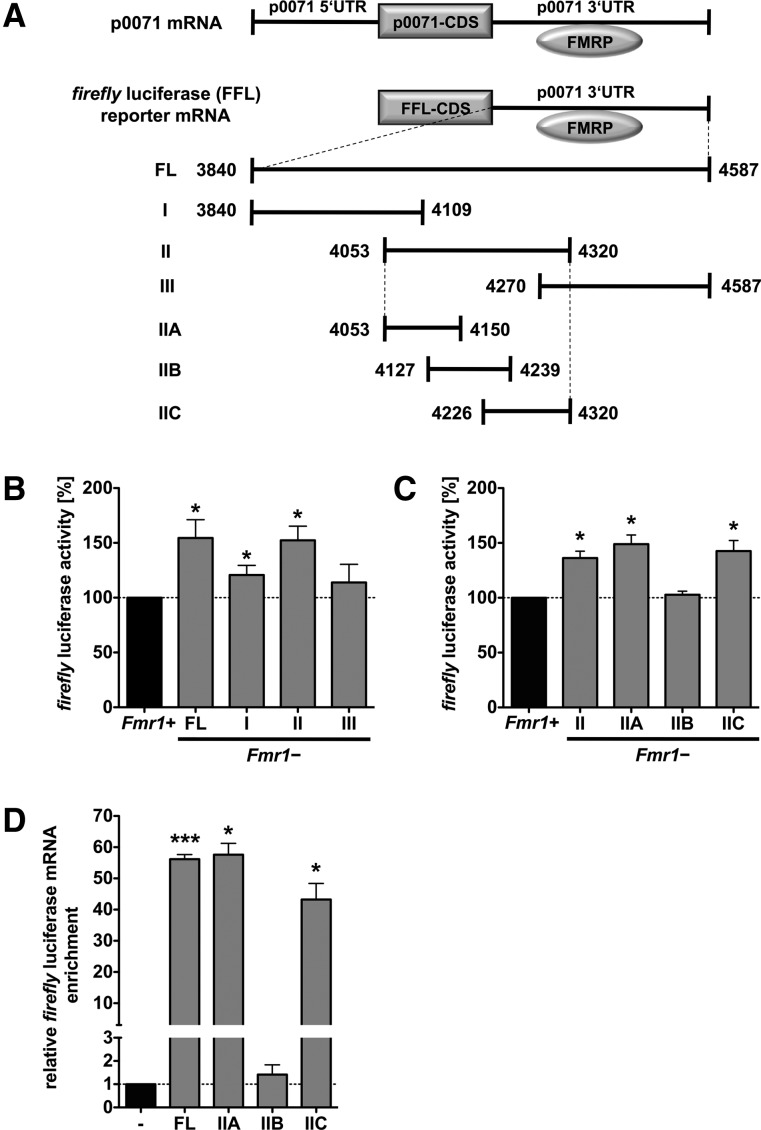
To test this in further detail, we explored how FMRP modulates the translation of luciferase reporter transcripts harboring the p0071 3′ UTR (Fig. 3A). Notably, FMRP was reported to control mRNA translation from the 3′ end, although some recent data suggest that FMRP also targets coding sequences (Darnell et al. 2011; Ascano et al. 2012). The normalized activity of reporters comprising the full-length 3′ UTR (nt 3840–4587) or fragment II (nt 4053–4320) was significantly increased in Fmr1− MEFs (Fig. 3A,B). This was in agreement with up-regulated p0071 protein levels in the Fmr1− MEFs and suggested that FMRP inhibits p0071 mRNA translation via the 3′ UTR.
FIGURE 3.
FMRP represses p0071 mRNA translation in a 3′-UTR-dependent manner. (A) Schematic of luciferase reporters used to determine FMRP-dependent regulation of p0071 mRNA translation. Constructs used in a reporter assay to analyze the effect of FMRP binding to the p0071 3′ UTR. (5′ UTR) 5′ untranslated region, (3′ UTR) 3′ untranslated region, (CDS) coding sequence, (FFL) firefly luciferase, (FL) full length. Nucleotide numbers refer to the p0071 mRNA accession number of NM_003628. (B) Luciferase reporter assays performed in Fmr1+ vs. Fmr1− cells after transfection with the indicated constructs revealed increased FFL activity in Fmr1− cells expressing the full-length p0071 3′ UTR or fragment II. FFL activity was normalized to Renilla luciferase activity. n = 3, (*) P ≤ 0.05. (C) Luciferase reporter assays performed in Fmr1+ vs. Fmr1− cells after transfection with the indicated constructs. Elevated FFL activity was observed with fragments IIA and IIC. FFL activity was normalized to Renilla luciferase activity. n = 3, (*) P ≤ 0.05. (D) qRT-PCR analysis of mRNAs that coprecipitated with FMRP in HEK293 cells transfected with the indicated reporter constructs harboring fragments of the p0071 3′ UTR. Enrichment was calculated via the ΔΔCt-method and normalized to the control mRNA encoding CyclophilinA. n = 3, (***) P ≤ 0.0005, (*) P ≤ 0.05.
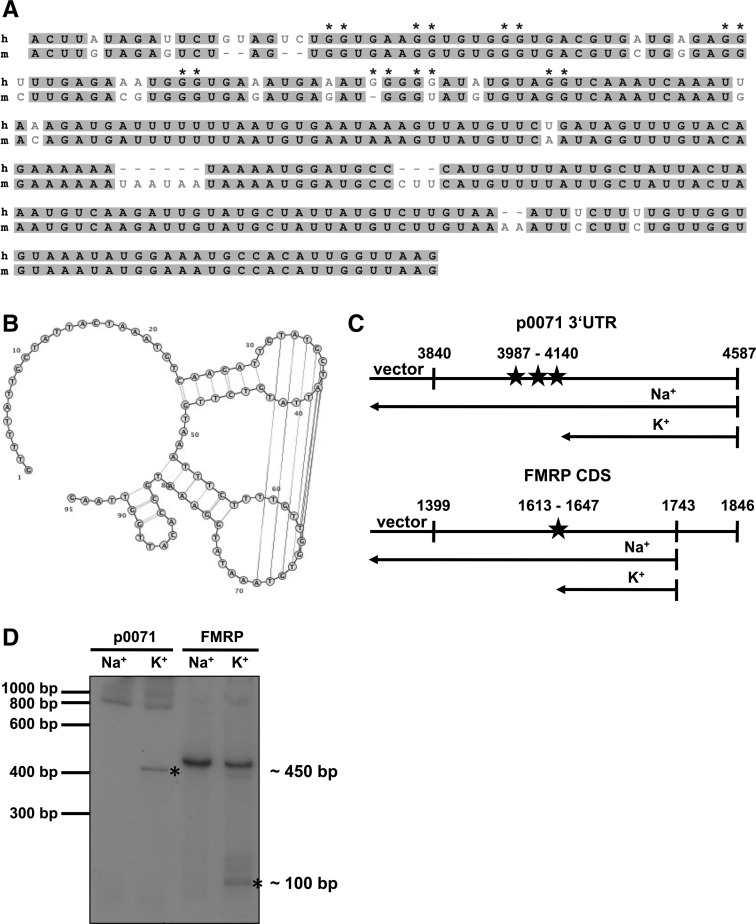
To further characterize the FMRP binding site in the p0071 3′ UTR, we dissected fragment II into three smaller segments, IIA (nt 4053–4150), IIB (nt 4127–4239), and IIC (nt 4226–4320). Fragments IIA and IIC suppressed translation of the luciferase reporter in Fmr1+ cells, whereas fragment IIB had no effect (Fig. 3C). To confirm that this was due to direct binding of FMRP to the p0071 3′-UTR fragments, we immunoprecipitated FMRP and analyzed the precipitates for the presence of luciferase mRNA. In agreement with the reporter activity analyses, fragments IIA and IIC were enriched in the precipitates, whereas fragment IIB did not associate with FMRP (Fig. 3D). We therefore conclude that FMRP directs translational inhibition via at least two regulatory cis-elements in the p0071 3′ UTR. In support of this, the p0071 3′ UTR is highly conserved among mammals (Fig. 4A).
FIGURE 4.
The FMRP p0071 mRNA association depends on two cis-elements in the p0071 3′ UTR. (A) Sequence homology of the human (h) and mouse (m) fragment II nucleotide sequences of the p0071 3′ UTR. Fragments IIA and IIC of the 3′ UTR encompass a G- and U-rich region, respectively, which could function as potential FMRP-binding sites. Asterisks indicate putative G-quadruplex-forming nucleotides as identified by the QGRS-mapper program (Kikin et al. 2006) (http://bioinformatics.ramapo.edu/QGRS/analyze.php). (B) Prediction of RNA secondary structures for the fragment IIC sequence using the in silico prediction program CyloFold (http://cylofold.abcc.ncifcrf.gov; Bindewald et al. 2010) suggests formation of two stem–loops in the p0071 3′ UTR that serve as putative recognition sites for the KH-domains of FMRP, the “kissing complex” motif. (C) Schematic of the p0071 3′ UTR and the FMRP CDS with the position of putative G-quadruplexes (asterisks, p0071) and a known G-quadruplex motif (asterisk, FMRP) and the expected products after reverse transcription in the presence of K+ or Na+ ions. (D) Reverse transcription in the presence of K+ or Na+ shows pausing of the reverse transcriptase in the presence of K+, indicative of a G-quadruplex structure in the p0071 3′ UTR. The FMRP fragment, which is known to contain a G-quadruplex, served as a positive control. Asterisks denote incomplete products originating from reverse transcriptase pausing at G-quadruplex structures in the presence of K+.
FMRP contains two KH-type and one RGG-type RNA binding motifs and has been shown to bind to G-quadruplex structures (Darnell et al. 2001), U-rich sequences (Chen et al. 2003), or kissing complex motifs (Darnell et al. 2005). Accordingly, we analyzed the p0071 3′-UTR fragments that associate with FMRP for putative FMRP binding motifs. Fragment IIA is G-rich and might contain G-quadruplex motifs (Fig. 4A). In contrast, fragment IIC might be able to form a kissing complex-like structure (Fig. 4B). To validate a putative G-quadruplex structure in the p0071 3′ UTR, we analyzed cation-dependent termination of reverse transcription. Since G-quadruplexes are stabilized by K+ but not by Na+, reverse transcriptase pauses at G-quadruplexes only in the presence of K+ (Castets et al. 2005). FMRP with a known G-quadruplex structure in its coding sequence served as a positive control. This assay strongly supports the existence of the predicted G-quadruplex in the p0071 3′ UTR (Fig. 4C,D).
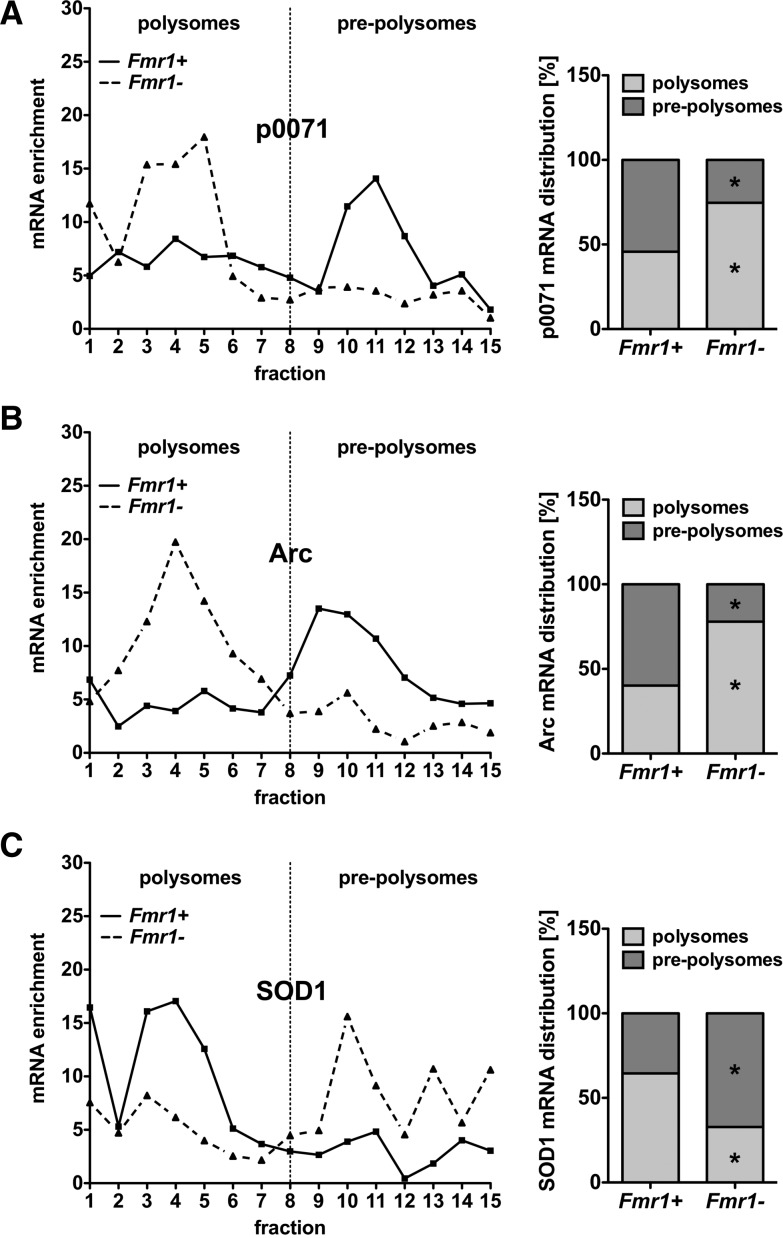
The role of FMRP in inhibiting p0071 mRNA translation was further analyzed by sucrose gradient centrifugations to determine the polysomal abundance of the p0071 mRNA. The latter, as well as the Arc mRNA which was reported to be inhibited by FMRP at the translational level (Park et al. 2008), were enriched in the polysomal fractions in Fmr1− MEFs (Fig. 5A,B). In contrast, FMRP was reported to promote the translation of the SOD1 mRNA (Bechara et al. 2009). In agreement, the abundance of this mRNA was reduced in the polysomal fraction of Fmr1− MEFs (Fig. 5C). These experiments strongly support our notion that FMRP suppresses p0071 mRNA translation.
FIGURE 5.
p0071 mRNA distribution is shifted to polysomal fractions in Fmr1− cells. (A–C) Cytoplasmic Fmr1+ and Fmr1− cell extracts were fractionated by centrifugation on a linear 10%–45% sucrose gradient. Diagrams on the right show mean values of three independent experiments with pre-polysomal vs. polysomal distribution of the investigated mRNAs. n = 3, (*) P ≤ 0.05. (A) The p0071 mRNA distribution revealed a shift to polysomal fractions in Fmr1− cells (black dashed line) in comparison to Fmr1+ cells (black line), indicating a translational repression of the p0071 mRNA by FMRP. (B) The Arc mRNA is known to be translationally repressed by FMRP and reveals a similar shift to polysomal fractions in Fmr1− cells (black dashed line). (C) The SOD1 mRNA translation is positively regulated by FMRP and served as a control for translational repression as indicated by an increased pre-polysomal mRNA abundance in Fmr1− cells (black dashed line).
The FMRP S499A mutant fails to regulate p0071 mRNA translation
Phosphorylation of FMRP at S500 (S499 in mice) by Casein kinase II (Siomi et al. 2002) has been shown to modulate FMRP function. Studies on the FMRP-directed regulation of Arc protein synthesis suggested that phosphorylated FMRP suppresses the constitutive translation of the Arc mRNA. In view of the similarities in the regulation of Arc- and p0071 mRNAs, we investigated whether phosphorylation of FMRP likewise modulates p0071 mRNA translation. Luciferase reporter assays revealed that the FMRP S499A mutant which mimics the nonphosphorylated form of FMRP was not able to repress translation via the p0071 3′ UTR (Fig. 6A). This suggested that phosphorylation at S499 enhanced FMRP-directed inhibition of p0071 mRNA translation. Moreover, the FMRP S499A mutant failed to rescue cell morphology and actin filament organization when expressed in Fmr1− MEFs (Fig. 6B). Taken together, this provides strong evidence that the FMRP-dependent regulation of actin organization essentially relies on the FMRP-dependent regulation of p0071 protein synthesis.
FIGURE 6.
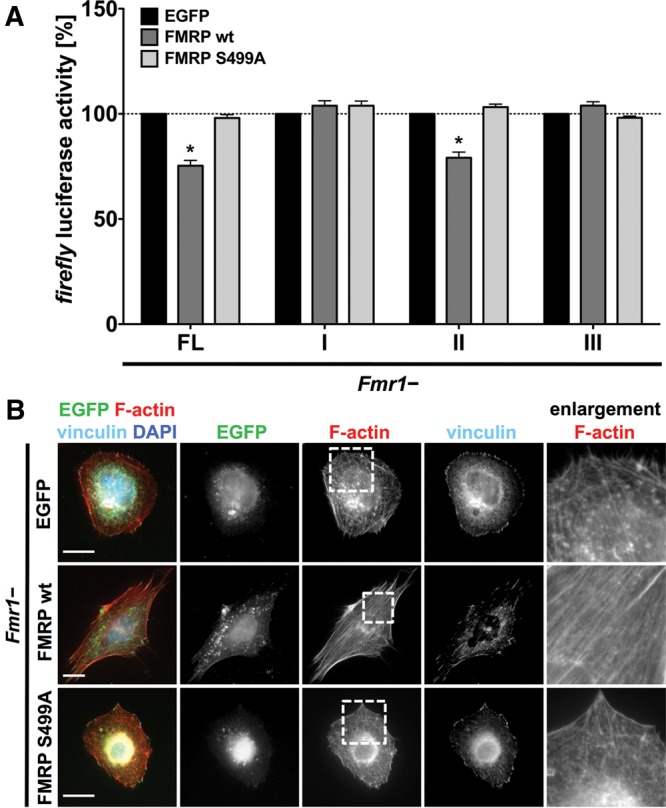
FMRP controls p0071 protein synthesis and actin organization in a S499-dependent manner. (A) Luciferase reporter assay with the indicated constructs (as described in Fig. 3A) showing that translational repression of the p0071 3′ UTR depends on S499 of FMRP. Transfection of the FMRP S499A mutant in Fmr1− MEFs had no influence on luciferase activity. n = 3, (*) P ≤ 0.05. (B) Immunofluorescence studies of Fmr1− MEFs overexpressing EGFP, EGFP-FMRP WT, or EGFP-FMRP S499A. Whereas the expression of FMRP WT promotes parallel organization of stress fibers and elongated cell morphology, expression of FMRP S499A fails to modulate actin organization or cell shape. Bars, 20 µm.
FMRP modulates actin organization by controlling p0071 expression
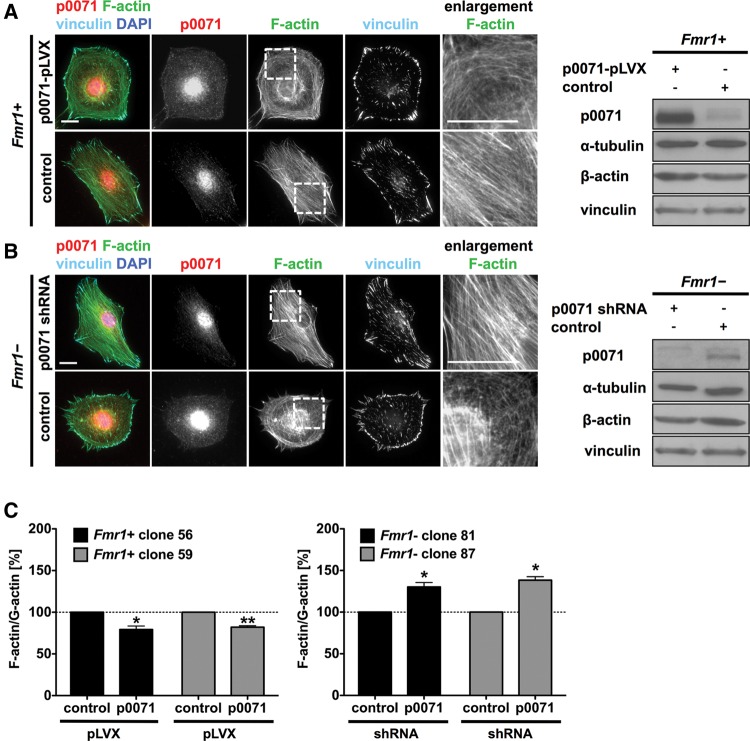
Having shown that FMRP controls p0071 mRNA translation, we aimed at analyzing whether this regulatory role is essential for the FMRP-directed control of actin organization. Cell morphology as well as actin organization observed upon the overexpression of p0071 in FMRP re-expressing cells was reminiscent of the phenotype observed for Fmr1− MEFs (cf. Fig. 7A and Fig. 1A). P0071 overexpression in the Fmr1+ MEFs induced a rounded cell morphology, a peripheral accumulation, and smaller size of focal contacts, as well as a severe reduction of stress fibers. The latter was confirmed by determining cellular F-actin/G-actin ratios which were reduced by p0071 overexpression (Fig. 7C). Thus, p0071 overexpression in FMRP re-expressing MEFs mimicked the effect of FMRP depletion, suggesting p0071 as a major player in the regulation of actin organization downstream from FMRP. In support of this, the depletion of p0071 by shRNA-mediated knockdown in Fmr1− MEFs rescued the wild-type phenotype. P0071 knockdown induced an elongated cell shape with increased numbers of parallel actin stress fibers (Fig. 7B), elongated and larger focal contacts, as well as elevated F-actin/G-actin ratios (Fig. 7C). Taken together, these findings indicate that the inhibition of p0071 mRNA translation by FMRP is essentially involved in modulating cell morphology and actin organization in MEFs.
FIGURE 7.
p0071 overexpression and knockdown mimic the Fmr1− and Fmr1+ phenotype. (A,B) Immunofluorescence studies and Western blotting of Fmr1+ cells and Fmr1− cells after lentiviral transduction with the indicated vectors. Overexpression of EGFP-p0071 in Fmr1+ cells mimicked the Fmr1− phenotype, whereas the expression of EGFP alone had no effect (A). The shRNA-mediated knockdown of p0071 in Fmr1− cells mimicked the Fmr1+ phenotype (B). Bars, 20 µm. (C) F-actin/G-actin assay after lentiviral p0071 overexpression in Fmr1+ cells (left panel) and after p0071 knockdown in Fmr1− cells (right panel). Overexpression of p0071 reduced the F-actin/G-actin ratio in both Fmr1+ MEF clones (56 and 59), whereas the ratio was increased upon p0071 depletion in both Fmr1− MEF clones (81 and 87). n = 3, (**) P ≤ 0.005, (*) P ≤ 0.05.
P0071 and FMRP modulate neurite outgrowth and dendritic arbor formation
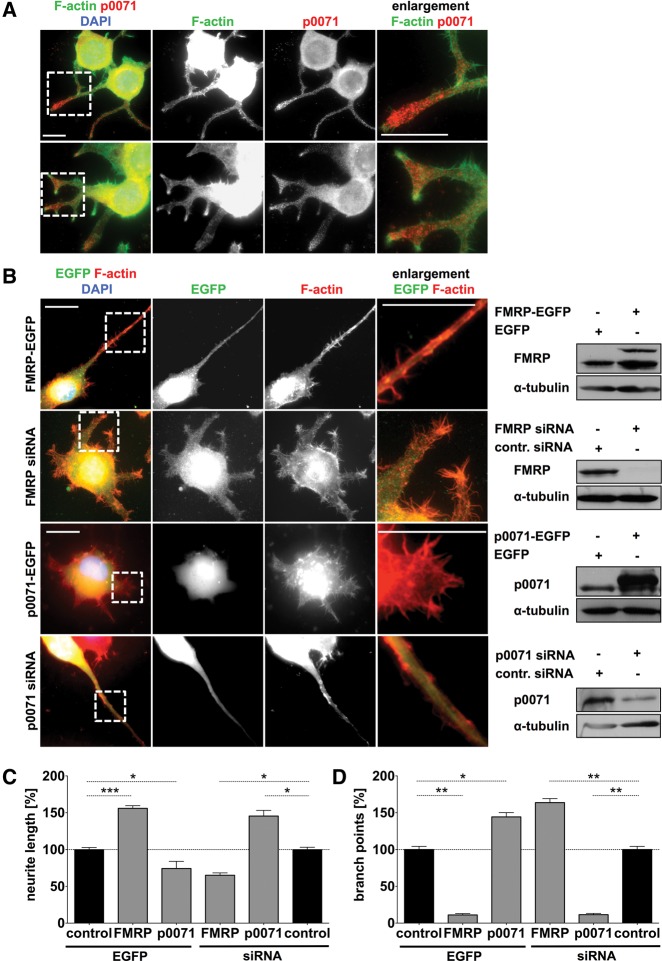
In view of the role of FMRP in controlling actin organization and cell morphology in MEFs via p0071 and the high expression levels of FMRP and p0071 in neurons, we speculated that FMRP and p0071 may control neurite outgrowth in an antagonistic manner. This was first analyzed in Neuro-2a cells upon the induction of neurite outgrowth by retinoic acid. In these neuroblastoma cells, p0071 was observed in the cell body as well as in neurites, suggesting that p0071 may play a role in neurite outgrowth (Fig. 8A). To test this in further detail, we analyzed how the depletion or overexpression of FMRP vs. p0071 affects neurite morphology. FMRP overexpression induced the formation of long and unbranched neurites, whereas the opposite was observed upon the knockdown of FMRP (Fig. 8B–D). On the contrary, neurite length was decreased by p0071 overexpression, whereas branching was increased. Again, the opposite was observed upon p0071 depletion. Moreover, the knockdown of FMRP as well as p0071 overexpression led to severely altered morphology of the actin cytoskeleton, with the appearance of F-actin-rich microspike-decorated protrusions (Fig. 8B).
FIGURE 8.
Role of p0071 in Neuro-2a neuroblastoma derived cells. (A) Immunofluorescence studies of differentiated Neuro-2a cells showing p0071 localization. F-actin was labeled with Alexa Fluor 594-conjugated Phalloidin. Bar, 20 µm. (B) Immunofluorescence studies and Western blotting of differentiated Neuro-2a cells after transfection with the indicated constructs. Bars, 20 µm. (C,D) Quantitation of neurite length (C) and branch point number (D) of 100 transfected cells as shown in B, compared to pEGFP-C2 or control siRNA transfection (control). n = 3, (***) P ≤ 0.0005, (**) P ≤ 0.005, (*) P ≤ 0.05.
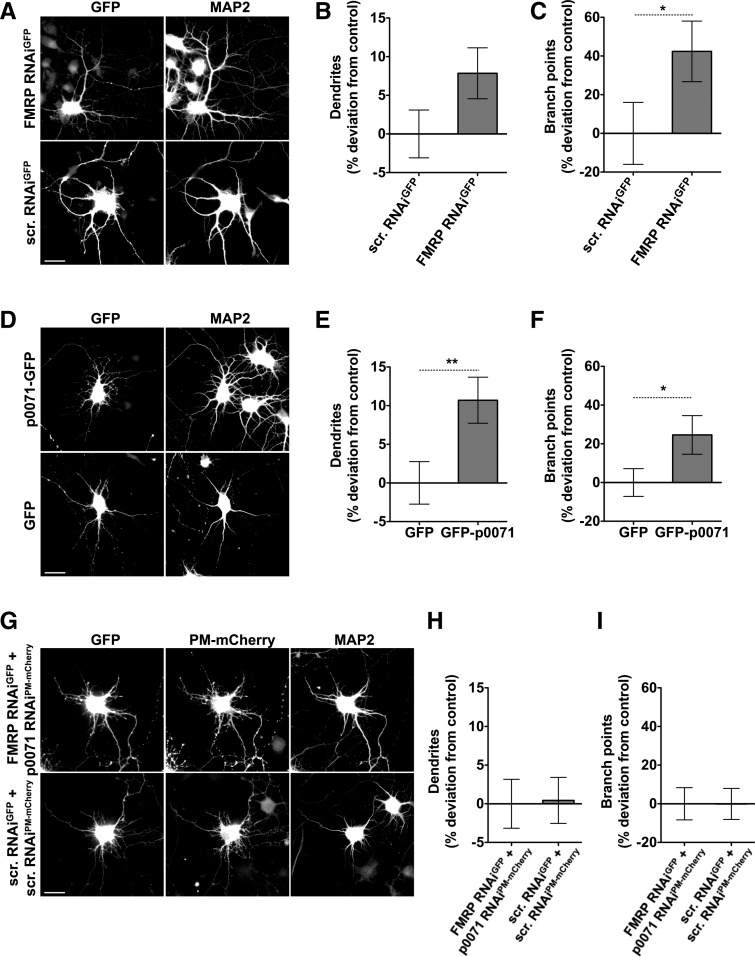
To further analyze whether increased p0071 expression could be responsible for morphological defects observed in FMRP-deficient neurons, we compared the effects of FMRP and p0071 depletion on rat primary hippocampal neurons. FMRP depletion by RNAi promoted dendritogenesis. Whereas the average number of MAP2-positive neurites, i.e., dendrites, per cell showed an 8% increase that did not reach statistical significance, the number of dendritic branches was significantly increased by ∼40% (Fig. 9A–C).
FIGURE 9.
Promoted dendritogenesis observed upon FMRP RNAi is phenocopied by p0071 overexpression and suppressed by p0071 RNAi. (A,D,G) Dissociated primary hippocampal neurons were transfected at DIV4 and processed for immunofluorescence microscopy after 48 h. MAP2 staining was used for morphological evaluation of transfected neurons. Bars, 20 µm. (B,C,E,F,H,I) Quantitative analyses evaluating number of dendrites (B,E,H) and dendritic branch points (C,F,I), respectively. Data represent mean ± SEM. FMRP RNAiGFP, n = 76; scrambled (scr.) RNAiGFP, n = 73; GFP-p0071, n = 80; GFP, n = 80; FMRP RNAiGFP + p0071 RNAiPM-mCherry, n = 70; scrambled (scr.) RNAiGFP + scrambled (scr.) RNAiPM-mCherry, n = 66. (*) P < 0.05, (**) P < 0.01. FMRP-deficient neurons showed a trend toward an increased number of dendrites and a significant increase in dendritic branch points compared to control cells (A–C). Neurons transfected with GFP-p0071 mimicked this morphological phenotype. The amount of dendritic branch points as well as of dendrites was significantly higher in comparison to GFP-transfected cells (D–F). If both FMRP and p0071 are knocked down, the phenotype is rescued, showing dendrite number and dendritic branch points at control level (G–I).
In line with our finding that FMRP represses the translation of p0071, GFP-p0071 overexpression led to a promotion of dendritogenesis, with both the number of dendrites (+11%; P < 0.01) and the number of branches (+25%; P < 0.05) being significantly increased when compared to control transfected neurons (Fig. 9D–F). An excess of p0071, thus, phenocopied FMRP loss of function in neuronal cells.
If FMRP mediated repression of p0071 function is, indeed, critical for proper dendrite formation during neuronal network development, a knockdown of p0071 should suppress the FMRP loss-of-function phenotype. In order to address this hypothesis, we used two sets of RNAi tools against FMRP and p0071 driven by distinct promoters (H1.1 and U6.1). The use of distinct promoters excludes that reduced RNAi effects caused by exhaustion of endogenous factors associating with two instead of one plasmid feign a putative phenotype suppression. The additional use of two different fluorescent reporters (GFP and PM-mCherry, respectively) ensured that double-transfection of neurons can be validated (Haag et al. 2012).
Cotransfection of hippocampal neurons with FMRP RNAi and p0071 RNAi effectively suppressed the promotion of dendritogenesis observed upon knocking down FMRP alone. Both the dendrite number and the branch points of the dendritic arbor of double-transfected neurons were indistinguishable from those coexpressing the two scrambled RNAi constructs (Fig. 9G–I).
Taken together, these findings are consistent with our initial studies in MEFs and support the view that FMRP-regulated expression of p0071 is an important molecular mechanism for proper neuromorphogenesis derailed upon FMRP deficiency.
DISCUSSION
In this study, we show that FMRP regulates actin organization via the p120ctn-family member p0071. Fmr1− MEFs revealed not only altered cell morphology and actin organization with reduced stress fibers but also a reduced F-actin/G-actin ratio. This phenotype was fully mimicked by the overexpression of p0071 in FMRP re-expressing MEFs. Moreover, the knockdown of p0071 in Fmr1− MEFs rescued the FMRP loss-of-function phenotypes with respect to cell shape, actin organization, and F-actin/G-actin ratios. Also, in primary neurons, p0071 RNAi fully suppressed defects in neuronal morphogenesis caused by loss of FMRP. Taken together, this indicates that p0071 is an important downstream mediator of FMRP-dependent remodeling of the actin cytoskeleton in MEFs as well as in neuronal cells.
The regulation of neuronal morphology and synaptic modifications depends on the remodeling of the actin cytoskeleton. A link between FMRP and actin remodeling via the small GTPase Rac1 was suggested by the observation that activity of the GTPase was stimulated in Fmr1−/− mice (Bongmba et al. 2011). It was proposed that FMRP modulates Rac1 signaling indirectly via the translational repression of the PP2A phosphoserine/threonine phosphatase. PP2A, in turn, could dephosphorylate cofilin, a downstream target of Rac1 which enhances actin depolymerization and causes actin filament branching. Thus, decreased phosphorylation of cofilin via increased PP2A would account for altered actin organization in Fmr1− cells (Castets et al. 2005). However, the activation of Rac1 itself and its effector p21-activated kinase (PAK) appears impaired in Fmr1−/− mice (Hayashi et al. 2007; Chen et al. 2010). How FMRP modulates Rho-signaling upstream of the GTPases and their targets remains poorly understood. Here, we propose p0071 as a direct target of FMRP that functions upstream of Rho-GTPases and mediates actin remodeling in fibroblasts as well as in neuronal cells.
We show that FMRP inhibits p0071 mRNA translation resulting in up-regulated p0071 protein synthesis in Fmr1− MEFs. In agreement with RIP as well as HITS-CLIP/PAR-CLIP studies (Brown et al. 2001; Darnell et al. 2011; Ascano et al. 2012), we observed an association of FMRP with the p0071 mRNA. Luciferase reporter studies suggest that FMRP facilitates inhibition of p0071 mRNA translation via two regulatory cis-determinants in the p0071 3′ UTR. The most 5′ element is located between nt 4053 and 4150 of the 3′ UTR and was found to be G-rich. Although the potential for G-quadruplex formation in the 3′ UTR of p0071 by a genome-wide in silico screen was proposed to be only moderate (Eddy and Maizels 2006), the QGRS-Mapper, an in silico prediction algorithm for G-quadruplexes, identified three putative G-quadruplex motifs in the p0071 3′ UTR (Kikin et al. 2006), two of which are positioned in the G-rich element of fragment IIA (Fig. 4A,C) essential for mediating FMRP-dependent inhibition of p0071 mRNA translation (Fig. 3C,D). The presence of a G-quadruplex motif in fragment IIA was experimentally verified (Fig. 4D). The second more 3′-located regulatory cis-determinant is U-rich (∼44%) and might form a kissing complex-type secondary structure (Fig. 4B). Recently, Ascano et al. (2012) reported the discovery of two distinct RNA-recognition elements, the ACUK and WGGA motifs (in which K = G or U and W = A or U) in ∼50% of FMRP mRNA binding sites. We find an ACUK element in fragment IIA and a WGGA motif in fragment IIC. However, fragment IIB also contains a WGGA motif, indicating that this short-sequence motif alone is not sufficient to mediate FMRP-dependent regulation of target mRNAs. FMRP contains one RGG-box and two KH-domains which have been implicated in RNA-binding of the protein. While the RGG-box was shown to bind G-quadruplex RNAs, the KH2-domain was suggested to mediate association with the kissing complex-like elements. Association with the latter is abrogated by the I304N mutation in the KH2-domain observed in one severely affected patient, pointing to the importance of this domain (Darnell et al. 2005). Moreover, the KH-domains appear to be essential for polyribosome association, whereas deletion of the RGG-box did not affect the polysomal association of FMRP (Mazroui et al. 2003; Darnell et al. 2005). Based on these findings, it was suggested that FMRP could interact with both motifs at the same time, either in two distinct RNA molecules or in a single transcript containing both G-quadruplex and kissing complex RNA motifs. Whereas the interaction with the KH-domains is essential for translational control and is shared with the FMRP-related proteins FXR1 and FXR2, the G-quadruplex could be important for directing substrate-specificity, since the ability to recognize G-quadruplex structures is unique to FMRP (Darnell et al. 2009). Thus, the identification of these two types of RNA motifs in the p0071 mRNA suggests p0071 as a highly specific target of FMRP, a conclusion supported by the identification of this target mRNA by RIP as well as CLIP (Brown et al. 2001; Darnell et al. 2011).
FMRP plays a role in mRNA transport where it represses translation of target mRNAs (Dictenberg et al. 2008). Phosphorylation of FMRP by Casein kinase II (Siomi et al. 2002) and S6K1 (Narayanan et al. 2008) controls the activity of FMRP as a translational repressor modulating neuronal function, and phosphorylated FMRP may associate with stalled ribosomes (Ceman et al. 2003). Phosphorylation of S499 (S500 in humans) appears to be important for translational repression since a phospho-mimetic mutant of FMRP suppressed steady-state dendritic Arc levels, whereas a nonphosphorylatable mutant (S500A) did not (Niere et al. 2012). According to this model, phosphorylated FMRP functions to suppress steady-state translation of target mRNAs. Upon mGluR stimulation, PP2A becomes activated and rapidly dephosphorylates FMRP (Narayanan et al. 2007), inducing the rapid synthesis of target mRNAs leading to local protein synthesis in dendritic spines (Niere et al. 2012). A recent study has shown that FMRP phosphorylation also modulates miRNA-dependent regulation of target RNAs. Dissociation of the RNA-induced silencing complex (RISC) from FMRP-bound mRNA which activates mRNA translation depended on FMRP de-phosphorylation (Muddashetty et al. 2011). We show here that the p0071 mRNA is translationally repressed by FMRP but not by the FMRP S499A mutant, suggesting that de-phosphorylation of FMRP leads to the release of p0071 translational repression. This model implies that FMRP could mediate spatial p0071 mRNA delivery for local protein synthesis in response to synaptic stimulation. P0071 mRNA translation would be repressed during transport and released at the dendrites upon stimulation. In support of this, the p0071 mRNA was found to be localized at the leading edge of migrating fibroblasts, was highly enriched in pseudopodia in response to migratory stimuli (Mili et al. 2008), and was also localized to cellular protrusions of astrocytes (Thomsen and Lade Nielsen 2011).
In conclusion, we have shown here that FMRP directly regulates p0071 protein synthesis by binding to its 3′ UTR and that p0071 is an essential downstream target of FMRP that mediates FMRP-dependent cytoskeletal organization and thereby has a profound effect on neuronal morphogenesis. Antagonistic functions of p0071 and FMRP in primary hippocampal neurons and the fact that a p0071 knockdown rescues the FMRP knockdown induced phenotype in neuronal cells strongly support the relevance of these findings in the context of the neurological disorder caused by lack of FMRP. Future work needs to assess whether p0071 mRNA and protein levels are up-regulated in FMRP knockout mice and human patients and if p0071 might qualify as a pharmacological target in the treatment of fragile X syndrome.
MATERIALS AND METHODS
Cell culture and transfection
Embryonic fibroblasts from Fmr1−/− mice (Castets et al. 2005) were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 150 µg/mL hygromycin B (PAA). HEK293 cells were grown in DMEM with 10% FCS and 1% sodium pyruvate (PAA). Neuro-2a cells were grown in DMEM with 5% FCS and 1% nonessential amino acids (PAA). For differentiation, Neuro-2a cells were maintained in DMEM with 2% FCS, 1% nonessential amino acids, 20 µM retinoic acid (Sigma Aldrich), and 3 µM aphidicolin (Sigma Aldrich). Cells were transfected with Lipofectamine 2000 or Lipofectamine LTX (Invitrogen) according to the manufacturer's protocol.
Plasmids, shRNAs, and lentiviruses
The p0071 3′ UTR and 3′-UTR fragments were amplified from HeLa cDNA and inserted at the 3′ end of a firefly luciferase coding sequence in pcDNA3.1 (Invitrogen). Lentiviruses for cDNA transduction were generated using the Lenti-X Lentiviral Expression System (Clontech). The p0071 wild-type sequence was subcloned from p0071-EGFP (Wolf et al. 2006) into pLVX-puro (Clontech). The shRNA plasmids were constructed with Clontech's shRNA Sequence Designer. For virus production, HEK293T cells were cotransfected with pMD2.G (Addgene, no. 12259), psPAX2 (Addgene, no. 12260), and pLVX-puro plasmids containing EGFP, EGFP-FMRP, p0071 WT, or p0071 shRNAs. Lentiviruses were purified 48 h post-transfection by using the Lenti-X Concentrator (Clontech).
To generate RNAi constructs directed against FMRP and p0071, the following oligonucleotides were annealed and cloned into pRNAT-H1.1/Hygro-GFP (GenScript) and pRNAT-U6.1/Neo-GFP (GenScript), respectively:
FMRP RNAi:
5-gatccGCACCAAGTTGTCTCTTATACTTCAAGAGAGTATAAGAGACAACTTGGTGCTTTTTTACGCGTa-3 and
5-agcttACGCGTAAAAAAGCACCAAGTTGTCTCTTATACTCTCTTGAAGTATAAGAGACAACTTGGTGCg-3;
p0071 RNAi:
5-gatccAGGAGCTTCAGTTTCAACGTTCAAGAGACGTTGAAACTGAAGCTCCTTTTTTTACGCGTa-3 and
5-agcttACGCGTAAAAAAAGGAGCTTCAGTTTCAACGTCTCTTGAACGTTGAAACTGAAGCTCCTg-3.
The pRNAT-U6.1-driven p0071 RNAi construct coexpressing plasma membrane mCherry (PM-mCherry) (p0071 RNAiPM-mCherry) was generated by replacing GFP with PM-mCherry subcloned from a vector containing 20 aa comprising farnesylation signal from c-Ha-Ras fused to the C terminus of mCherry (PM-mCherry; provided by M. Korte).
A nonsilencing RNAi control (scrambled RNAi2 in pRNAT-H1.1GFP; scr. RNAiGFP) was generated with the following oligonucleotides:
5-gatccCACAGCGTCTGAACGTAGTTTCAAGAGAACTACGTTCAGACGCTGTGTTTTTa-3 and
5-agcttAAAAACACAGCGTCTGAACGTAGTTCTCTTGAAACTACGTTCAGACGCTGTGg-3,
and integrated into pRNAT-H1.1/Hygro-GFP (GenScript). A scrambled RNAi control driven by the U6.1 promoter (scrambled RNAi1 in pRNAT-U6.1PM-mCherry; scr. RNAiPM-mCherry) was generated by replacing GFP with PM-mCherry in scrambled RNAiGFP (U6.1) (Haag et al. 2012).
Luciferase reporter assay
Sixteen hours after cotransfection of Fmr1+ and Fmr1− cells with firefly luciferase reporter and Renilla luciferase plasmids, luciferase activity was measured using Dual-Glo (Promega). Firefly luciferase activities were normalized to Renilla luciferase activities.
qRT-PCR and Western blotting
Total RNA was extracted using TRIzol (Invitrogen) and transcribed with random or oligo-dT primers and SuperScript II-RT into cDNA (Invitrogen). The qRT-PCR was performed based on the SYBR Green technology using 2×PCR Master Mix (Promega) supplemented with SYBR Green (iQ5; Bio-Rad cycler). Real-time PCR quantification was performed according to the ΔΔCt method (Livak and Schmittgen 2001).
For Western blotting, total protein extracts were analyzed with the following antibodies: anti-FMRP rabbit (Abcam), anti-p0071-tail#10 guinea pig (Peptide Specialty Laboratories), anti-tubulin clone DM1A (Sigma Aldrich), anti-vinculin mouse (Sigma Aldrich), anti-β-actin clone AC-74 (Sigma Aldrich), anti-GAPDH mouse (Sigma Aldrich), and anti-GFP rabbit (Invitrogen).
Coimmunoprecipitation
For the analysis of protein-RNA complexes, Fmr1+ and Fmr1− cells or HEK293 cells were lysed in TapII-buffer (10 mM HEPES pH 7.6, 10 mM KCl, 3 mM MgCl2, 0.5% NP-40, 5% glycerol supplemented with protease inhibitors (Sigma Aldrich), RNaseOUT (Invitrogen), and yeast t-RNA (10 mg/mL; Invitrogen). The cleared cell lysates were supplemented with KCl to a final concentration of 150 mM before incubation with pre-immobilized anti-FMRP (Abcam) on protein A beads (Pierce). RNA was eluted by the addition of 0.2% SDS and β-mercaptoethanol (Sigma Aldrich) before the addition of proteinase K (Fermentas). After phenol-extraction, RNA was precipitated and reverse transcribed. Precipitation of FMRP was analyzed by Western blotting from aliquots recovered before proteinase-K treatment.
Immunofluorescence
Cells grown on cover slips were rinsed in PBS and fixed with 3.7% formaldehyde in PBS for 10 min at 4°C, and permeabilized at 4°C in 0.5% Triton X-100 in PBS for 15 min. Alternatively, cells were fixed and permeabilized in 2% formaldehyde and 0.5% Triton X-100 in PBS for 10 min at 4°C. Cells were washed in PBS and blocked for 30 min in PBS containing 1% (w/v) dry milk before antibody application and mounted in Mowiol.
All image acquisition was performed using standardized settings on a microscope (E600; Nikon) with a 60× objective (Apochromat TIRF, Nikon) connected to a CCD camera (CCD-1300QLN; VDS Vosskühler) using NIS Elements AR 3.10 software (Nikon).
Cell culture, immunofluorescence microscopy, and morphometric analyses of primary hippocampal neurons
Dissociated primary hippocampal neurons were prepared and grown on poly-D-lysine-coated glass cover slips as described previously (Kessels et al. 2001). Neurons were transfected at 4 d in vitro (DIV4) with the calcium phosphate method and fixed 48 h later with 4% formaldehyde for 10 min. After incubation with 25 mM glycine in PBS for 30 min, cells were blocked and permeabilized for 1 h in 2% BSA/10% horse serum in PBS (block solution) containing 0.25% Triton X-100. Incubation with polyclonal rabbit anti-MAP2 antibody in block solution was performed for 1 h. The cover slips were then washed three times for 10 min with block solution and incubated with secondary Alexa Fluor 647 goat anti-rabbit antibody in block solution for 1 h. Finally, samples were washed twice with block solution and two times with PBS (10 min each) and mounted onto glass slides using Mowiol.
Images were recorded by AxioVision using a Zeiss AxioObserver.Z1 microscope equipped with Zeiss Plan-Apochromat 40×/1.3 objective and AxioCam MRm CCD camera (Zeiss) and processed by ImageJ and Adobe Photoshop.
Morphometric measurements of transiently transfected hippocampal neurons were performed with ImageJ according to Schwintzer et al. (2011). Transfected neurons identified by anti-MAP2 staining were imaged in systematic sweeps across the cover slips of repeated transfections with independent neuronal preparations. The number of dendrites and the number of dendritic branching points were determined from 66 to 80 DIV6 neurons for each condition. A minimal length of 10 µm was used as a threshold for dendrite and dendritic branch consideration. All data were normalized to the corresponding controls run in parallel in each individual experimental set. Statistical analysis was performed using the two-tailed Student's t-test.
F-actin/G-actin ratio
Fmr1+ or Fmr1− cells (5 × 103 each) were allowed to adhere overnight in 96-well plates, and then fixed and processed for F-actin labeling by Alexa Fluor 594-conjugated Phalloidin (Invitrogen) or for G-actin labeling by Alexa Fluor 594-conjugated DNaseI (Invitrogen). F-actin/G-actin ratios were determined by fluorescence spectrometry using DAPI staining for normalization. Triplicates of each sample were analyzed.
Sucrose gradient centrifugation
Fmr1+ and Fmr1− cells were incubated with cycloheximide (100 µg/mL) for 15 min before preparing cytoplasmic lysates. After determining protein concentration, 2 mg protein were loaded on a linear 10%–45% (w/v) sucrose gradient and centrifuged in a Beckmann SW40Ti rotor for 2 h at 40,000 rpm. Gradients were fractionated using a collection system, and RNA was isolated using phenol/chloroform extraction. RNA levels in each fraction were determined by qRT-PCR.
Primer extension
Primer extensions were performed as previously described by Schaeffer et al. (2001). In brief, RNA (3 pmol) and 105 cpm of [γ-32P]ATP-labeled primer were heated to 95°C for 1 min and annealed at 20°C for 5 min in 6 µL of 50 mM HEPES-Na pH 7.0, 100 mM NaCl or KCl as indicated, and 50 mM EDTA. Extensions were carried out for 30 min at 37°C by adding 9 µL of a mixture containing 50 mM Tris-HCl pH 8.3 at 37°C, 6 mM MgCl2, 40 mM NaCl or KCl, 1 mM of dNTP mix, and 6 Units of AMV reverse transcriptase (Promega). Reaction products were analyzed on 8% polyacrylamide-8 M urea gels followed by autoradiography.
ACKNOWLEDGMENTS
We thank Barbara Bardoni (IPMC-CNRS, Valbonne) for providing Fmr1− and FMRP re-expressing MEFs, M. Korte (TU Braunschweig, Germany) for providing a plasmid encoding for plasma membrane-targeted (farnesylated) mCherry, and A. Kreusch and B. Schade for technical help. This work was supported by the DFG GRK 1591 grants to S.H. and M.H., by the DFG SFB 610 to M.H., by grants from the Schram Foundation to B.Q., and from the DFG to M.M.K. and to B.Q.
Author contributions: A.N. performed most experiments and prepared the corresponding figures, J.S. performed experiments on primary neuronal cells and prepared the corresponding figure, R.K. analyzed G-quadruplex structures, and M.L. performed the lentiviral transductions. M.H. and S.H. planned and supervised the project, M.M.K. and B.Q. planned and supervised the experiments on primary hippocampal neurons, and M.H. wrote the manuscript.
REFERENCES
- Anastasiadis PZ, Reynolds AB 2001. Regulation of Rho GTPases by p120-catenin. Curr Opin Cell Biol 13: 604–610 [DOI] [PubMed] [Google Scholar]
- Arikkath J, Peng I-F, Ng YG, Israely I, Liu X, Ullian EM, Reichardt LF 2009. δ-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J Neurosci 29: 5435–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492: 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara EG, Didiot M-C, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, et al. 2009. A novel function for Fragile X Mental Retardation Protein in translational activation. PLoS Biol 7: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E, Kluth T, Shapiro BA 2010. CyloFold: Secondary structure prediction including pseudoknots. Nucleic Acids Res 38: W368–W372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongmba OYN, Martinez LA, Elhardt ME, Butler K, Tejada-Simon MV 2011. Modulation of dendritic spines and synaptic function by Rac1: A possible link to Fragile X syndrome pathology. Brain Res 1399: 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y 2012. Structural plasticity of dendritic spines. Curr Opin Neurobiol 22: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487 [DOI] [PubMed] [Google Scholar]
- Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel J-L, Bardoni B 2005. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet 14: 835–844 [DOI] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST 2003. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet 12: 3295–3305 [DOI] [PubMed] [Google Scholar]
- Chen L, Yun SW, Seto J, Liu W, Toth M 2003. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience 120: 1005–1017 [DOI] [PubMed] [Google Scholar]
- Chen LY, Rex CS, Babayan AH, Kramár EA, Lynch G, Gall CM, Lauterborn JC 2010. Physiological activation of synaptic Rac>PAK (p-21 activated kinase) signaling is defective in a mouse model of fragile X syndrome. J Neurosci 30: 10977–10984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y 2008. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 9: 344–356 [DOI] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT 1997. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc Natl Acad Sci 94: 5401–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Martín A, Crespo M, Portera-Cailliau C 2010. Delayed stabilization of dendritic spines in fragile X mice. J Neurosci 30: 7793–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107: 489–499 [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB 2005. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev 19: 903–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Darnell RB 2009. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet 18: 3164–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovic L, Navratil V, Bonaccorso CM, Catania MV, Bardoni B, Dumas M-E 2011. A metabolomic and systems biology perspective on the brain of the fragile X syndrome mouse model. Genome Res 21: 2190–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Merriam EB, Hu X 2011. The dynamic cytoskeleton: Backbone of dendritic spine plasticity. Curr Opin Neurobiol 21: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ 2008. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell 14: 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy J, Maizels N 2006. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res 34: 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia LP, Yamamoto M, Zang K, Reichardt LF 2006. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron 51: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Pasquale EB 2005. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol 75: 161–205 [DOI] [PubMed] [Google Scholar]
- Grosheva I, Shtutman M, Elbaum M, Bershadsky AD 2001. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: A link between cell-cell contact formation and regulation of cell locomotion. J Cell Sci 114: 695–707 [DOI] [PubMed] [Google Scholar]
- Gross C, Yao X, Pong DL, Jeromin A, Bassell GJ 2011. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J Neurosci 31: 5693–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag N, Schwintzer L, Ahuja R, Koch N, Grimm J, Heuer H, Qualmann B, Kessels MM 2012. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J Neurosci 32: 17842–17856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M 2005. The p120 family of cell adhesion molecules. Eur J Cell Biol 84: 205–214 [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Rao BSS, Seo J-S, Choi H-S, Dolan BM, Choi S-Y, Chattarji S, Tonegawa S 2007. Inhibition of p21-activated kinase rescues symptoms of fragile X syndrome in mice. Proc Natl Acad Sci 104: 11489–11494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau Y, Gambino F, Chelly J, Vitale N 2009. X-linked mental retardation: Focus on synaptic function and plasticity. J Neurochem 109: 1–14 [DOI] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT 2000. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex 10: 1038–1044 [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. 2001. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: A quantitative examination. Am J Med Genet 98: 161–167 [DOI] [PubMed] [Google Scholar]
- Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X 2004. Deletion of the neuron-specific protein δ-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol 14: 1657–1663 [DOI] [PubMed] [Google Scholar]
- Jung K-M, Sepers M, Henstridge CM, Lassalle O, Neuhofer D, Martin H, Ginger M, Frick A, DiPatrizio NV, Mackie K, et al. 2012. Uncoupling of the endocannabinoid signalling complex in a mouse model of fragile X syndrome. Nat Commun 3: 1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J 2010. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 33: 121–129 [DOI] [PubMed] [Google Scholar]
- Kessels MM, Engqvist-Goldstein AE, Drubin DG, Qualmann B 2001. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J Cell Biol 153: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O, D'Antonio L, Bagga PS 2006. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 34: W676–W682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Johnson MB, Dube U, Shim S, Rašin M-R, Sousa AMM, Fertuzinhos S, Chen J-G, Arellano JI, et al. 2012. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A 2003. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130: 5543–5552 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matus A 2000. Actin-based plasticity in dendritic spines. Science 290: 754–758 [DOI] [PubMed] [Google Scholar]
- Mazroui R, Huot M-E, Tremblay S, Boilard N, Labelle Y, Khandjian EW 2003. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum Mol Genet 12: 3087–3096 [DOI] [PubMed] [Google Scholar]
- Medina M, Marinescu RC, Overhauser J, Kosik KS 2000. Hemizygosity of δ-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics 63: 157–164 [DOI] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG 2008. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J 2003. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37: 417–431 [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ 2011. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell 42: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST 2007. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by Group I mGluR and mediated by PP2A. J Neurosci 27: 14349–14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST 2008. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J Biol Chem 283: 18478–18482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, Govek E-E, Van Aelst L 2005. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol 64: 58–74 [DOI] [PubMed] [Google Scholar]
- Niere F, Wilkerson JR, Huber KM 2012. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci 32: 5924–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Liu BP, Burridge K, Kreft B 2000. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol 150: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park JM, Kim S, Kim J-A, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. 2008. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 59: 70–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlowsky A, Chelly J, Billuart P 2012. Emerging major synaptic signaling pathways involved in intellectual disability. Mol Psychiatry 17: 682–693 [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen J-E, Woolfrey KM 2011. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 14: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM 2009. The state of synapses in fragile X syndrome. Neuroscientist 15: 549–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Roczniak-Ferguson A 2004. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 23: 7947–7956 [DOI] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST 2012. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu Rev Pathol 7: 219–245 [DOI] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J 20: 4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL 2001. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci 98: 8844–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B 2011. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J 30: 3147–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Higashijima K, Ishizuka A, Siomi H 2002. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Mol Cell Biol 22: 8438–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R, Lade Nielsen A 2011. A Boyden chamber-based method for characterization of astrocyte protrusion localized RNA and protein. Glia 59: 1782–1792 [DOI] [PubMed] [Google Scholar]
- Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB 2006. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell 127: 1027–1039 [DOI] [PubMed] [Google Scholar]
- Wolf A, Keil R, Götzl O, Mun A, Schwarze K, Lederer M, Hüttelmaier S, Hatzfeld M 2006. The armadillo protein p0071 regulates Rho signalling during cytokinesis. Nat Cell Biol 8: 1432–1440 [DOI] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SGN, et al. 2007. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci 10: 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K, Liu J, Zhu N, Lin J, Liang Q, Brown WT, Shen Y, Zhong N 2008. Identification of FMRP-associated mRNAs using yeast three-hybrid system. Am J Med Genet B Neuropsychiatr Genet 147B: 769–777 [DOI] [PubMed] [Google Scholar]