The Xenopus Cripto-1 protein is found only in cells comprising the animal hemisphere during embryogenesis because the Cripto-1 mRNA is translationally repressed in the vegetal hemisphere. This paper demonstrates that the RNA binding protein, Bicaudal-C, is responsible for the translational repression. Repression is cap-dependent but not poly(A)-dependent.
Keywords: Bicaudal-C, maternal mRNAs, translation, Xenopus
Abstract
The Xenopus Cripto-1 protein is confined to the cells of the animal hemisphere during early embryogenesis where it regulates the formation of anterior structures. Cripto-1 protein accumulates only in animal cells because cripto-1 mRNA in cells of the vegetal hemisphere is translationally repressed. Here, we show that the RNA binding protein, Bicaudal-C (Bic-C), functioned directly in this vegetal cell-specific repression. While Bic-C protein is normally confined to vegetal cells, ectopic expression of Bic-C in animal cells repressed a cripto-1 mRNA reporter and associated with endogenous cripto-1 mRNA. Repression by Bic-C required its N-terminal domain, comprised of multiple KH motifs, for specific binding to relevant control elements within the cripto-1 mRNA and a functionally separable C-terminal translation repression domain. Bic-C-mediated repression required the 5′ CAP and translation initiation factors, but not a poly(A) tail or the conserved SAM domain within Bic-C. Bic-C-directed immunoprecipitation followed by deep sequencing of associated mRNAs identified multiple Bic-C-regulated mRNA targets, including cripto-1 mRNA, providing new insights and tools for understanding the role of Bic-C in vertebrate development.
INTRODUCTION
Bicaudal-C (Bic-C) RNA binding proteins have vital roles in vertebrate embryogenesis and also in adult tissues, including the kidney, heart, and pancreas (Saffman et al. 1998; Chicoine et al. 2007; Gamberi and Lasko 2012). For example, Bic-C mutant mice develop renal cysts, providing a model for polycystic kidney disease (Maisonneuve et al. 2009; Tran et al. 2010). In Drosophila, where Bic-C was first identified, Bic-C contributes to patterning of the early embryo by repressing specific maternal mRNAs, such as oskar (Saffman et al. 1998; Chicoine et al. 2007). However, in vertebrates the mechanisms of Bic-C action have not been identified nor have mRNA targets been identified systematically.
Translation of the Cripto-1 mRNA (also referred to as xCR1) in Xenopus laevis embryos is confined to the cells of the animal hemisphere (Zhang et al. 2009). This finding explains why xCR1 protein accumulates only within the animal cells even though xCR1 mRNAs are equally distributed in cells of both the animal and vegetal hemispheres (Dorey and Hill 2006). More specifically, we demonstrated that this spatial regulation was achieved by a vegetal cell-specific translational repression mechanism that functioned through specific elements of the xCR1 mRNA 3′ UTR (Zhang et al. 2009). This spatial control of xCR1 translation is consistent with the fact that xCR1 is required for the formation of the anterior nervous system and other structures of the head that develop from cells of the animal hemisphere (Yabe et al. 2003; Tao et al. 2005). Thus, defining the mechanistic basis of spatially controlled xCR1 mRNA translation is important for understanding normal vertebrate embryogenesis.
Here, we show that Xenopus Bic-C RNA binding protein is the key determinant of xCR1 mRNA's spatially regulated translation and functions directly through a regulatory element, called the TCE (translational control element), within xCR1 mRNA's 3′ UTR. Bic-C repression was mediated via separable RNA-binding and effector (repression) domains and, unexpectedly, the Bic-C SAM domain was not essential for repression. Repression was cap- and initiation factor-dependent but poly(A)-independent. Bic-C-directed immunoprecipitation followed by deep sequencing of associated mRNAs revealed many previously unknown Bic-C mRNA targets, several of which, including xCR1 mRNA, encode proteins that have been implicated in developmentally important processes such as the Nodal/TGFβ and Wnt pathways. We conclude that Bic-C functions to influence cell-fate decisions and create embryonic polarities during the maternally controlled stages of vertebrate embryogenesis by direct translational regulation of mRNAs that encode key cell-fate determinants.
RESULTS
Bic-C repressed xCR1 mRNA translation when ectopically expressed in cells of the animal hemisphere
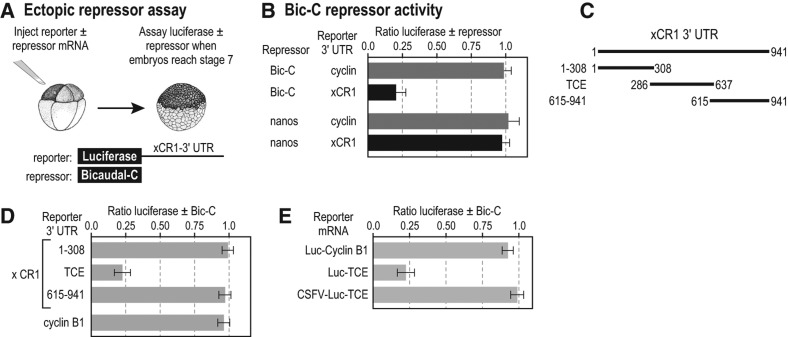
While the Xenopus xCR1 mRNA is distributed throughout all cells of the embryo, only xCR1 mRNA present in animal cells is translated (Dorey and Hill 2006; Zhang et al. 2009). Our previous study demonstrated the existence of a repression mechanism in cells of the vegetal hemisphere that prevents xCR1 mRNA translation (Zhang et al. 2009). To address the mechanism of vegetal-cell repression, we examined the possible role of two known RNA regulatory proteins that are encoded by maternal mRNAs that are themselves localized to vegetal cells—Bicaudal-C (Bic-C) (Wessely and De Robertis 2000) and Nanos1 (previously Xcat2) (Mosquera et al. 1993). We reasoned that if Bic-C and/or Nanos1 were responsible for repression of xCR1 mRNA translation in vegetal cells, then their ectopic expression in animal cells might be able to repress xCR1 mRNA translation in those cells. Therefore, mRNAs encoding Bic-C and Nanos1 were injected into animal cells of eight-cell Xenopus embryos together with a luciferase reporter mRNA containing the 3′ UTR of xCR1 mRNA that is sufficient for vegetal-cell repression (Fig. 1A; Zhang et al. 2009). Repression was quantified as the ratio of luciferase activity observed with the candidate repressor to that obtained without it.
FIGURE 1.
xCR1 mRNA translation can be repressed by Bicaudal-C. (A) Diagram of repression assay. Luciferase reporter mRNAs Luc/xCR1 (contains the 3′ UTR of the xCR1 mRNA) or Luc/Cyclin-BI (contains the 3′ UTR of the cyclinB1 mRNA) were injected into Xenopus embryos with or without the mRNA encoding candidate repressors (HA-Bic-C or MYC-Nanos1). Injected embryos were assayed for luciferase (Sheets et al. 1994; Fritz and Sheets 2001; Zhang et al. 2009), and the ratio of luciferase with and without a putative repressor was calculated as a measure of repression. (B) Bic-C specifically repressed the Luc/xCR1 reporter mRNA. (C,D) Regions of the xCR1 3′ UTR used in luciferase reporter mRNAs. The TCE (translational control element) was previously referred to as the Mut2 region (Zhang et al. 2009) and shown to be sufficient for repression in vegetal cells. Reporter mRNAs were analyzed for repression by Bic-C as described in A and B. (E) Repression by Bicaudal-C required a 5′ cap. The CSFV-Luc/xCR1-TCE reporter mRNAs contain the IRES from the CSFV 5′ of the luciferase coding region and the TCE of the xCR1 3′ UTR. The reporter mRNAs were analyzed for repression by Bic-C as described in A and B.
Ectopic Bic-C repressed translation of the Luc-xCR1 reporter mRNA, while Nanos1 did not (Fig. 1B). Both Bic-C and Nanos1 proteins were expressed, as measured by immunoblotting (Supplemental Fig. 1A). Repression required elements within the xCR1 3′ UTR, as a cyclin B1 3′-UTR reporter was unaffected (Fig. 1B; Sheets et al. 1994). Levels of both reporter mRNAs were unaffected by Bic-C; thus, Bic-C did not cause mRNA decay (Supplemental Fig. 1B,C).
To identify RNA sequences required to direct repression by Bic-C in animal cells, we analyzed reporters bearing different segments of the xCR1 3′ UTR in the ectopic repression assay. A central region of the xCR1 mRNA 3′ UTR, termed the TCE (nt 286–637) (Zhang et al. 2009), was essential for repression in animal cells: reporters carrying the TCE were repressed by Bic-C, while reporters bearing the 3′-UTR regions to its 5′ (nt 1–308) or 3′ (nt 615–941) sides were not (Fig. 1C). Additional control reporters, each with a different 3′ UTR, were insensitive to Bic-C (Supplemental Fig. 1D). These data identify a repressive element that responds to Bic-C in animal cells. That same element also mediates repression by endogenous factors in vegetal cells (Zhang et al. 2009).
Bic-C-mediated translation repression operated through the translation-initiation complex
In our previous study of xCR1 mRNA translation, we showed that the xCR1 3′ UTR directed vegetal-cell repression through a mechanism that required the 5′ cap and initiation factors eIF4F and eIF3. To begin to discern Bic-C's mechanism of action, we analyzed reporter RNAs bearing an ApppG cap and the CSFV IRES in animal cells of Xenopus embryos expressing Bic-C (Fig. 1E). The ApppG cap prevents cap-dependent translation, while the CSFV IRES bypasses the requirement for the initiation factors eIF4F and eIF3 (Otero et al. 2001; Kieft 2008). The CSFV IRES abolished Bic-C-mediated repression (Fig. 1E). Thus, Bic-C-dependent repression, as measured in the animal cell repression assay, recapitulated the vegetal-cell repression mechanism mediated through the xCR1 3′ UTR in terms of its dependence on the normal 5′ mRNA translation initiation complex.
The C-terminal region of vertebrate Bic-C possessed the repressive activity that functioned independent of a 3′ poly(A) tail
The protein domains essential for the translational repressor functions of Bic-C are unknown in any organism. To identify repression domains in Bic-C, we tethered segments of Bic-C to reporter mRNAs via MS2 coat protein (Coller et al. 1998; Coller and Wickens 2007). SinceMS2 protein provides the RNA binding activity, repression can be assayed independent of Bic-C's own RNA binding activity (Fig. 2A–C). We analyzed different segments of Bic-C as MS2-fusion proteins: the N-terminal half (N-term) that is comprised of multiple hnRNP-K-homolog (KH) domains, which are recognized RNA binding modules, the C-terminal half (C-term) that includes the sterile alpha motif (SAM) domain, the C-terminal half lacking the SAM domain (C-term ΔSAM), and finally the isolated SAM domain (Fig. 2A). Only the C-terminal portion of Bic-C repressed the reporter as efficiently as full-length Bic-C (Fig. 2C). The C-terminal region lacking the SAM domain (C-term ΔSAM) also repressed translation, albeit less efficiently. The N-terminal half and the isolated SAM domain exhibited no repression activity. We conclude that the C-terminal region of Bic-C outside of the SAM domain possesses repression activity, while the SAM domain contributes to but is not sufficient for repression. This region of Bic-C, which lacks similarity to known protein domains, exhibits >50% amino acid identity among vertebrate Bic-C proteins (Gamberi and Lasko 2012).
FIGURE 2.
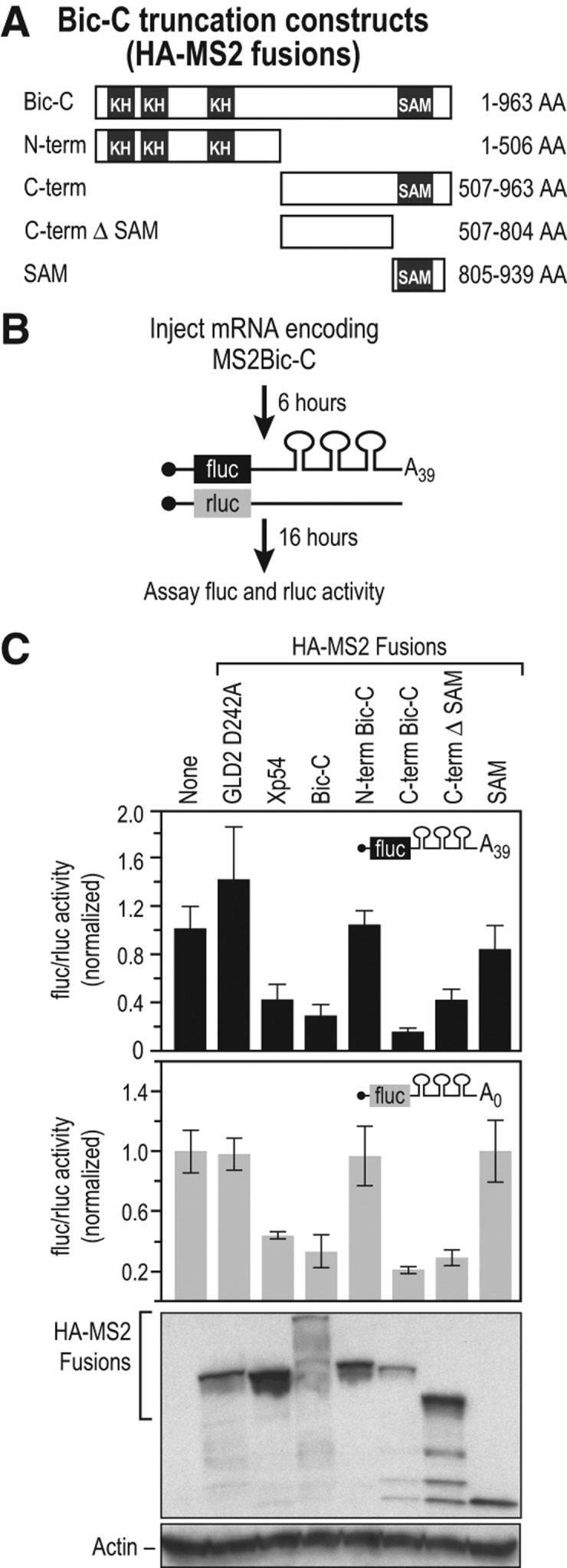
The C-terminal region of Bic-C contains translational repression activity. (A) The regions of Bic-C fused to MS2. (B) Diagram of the tethered translation assay (Coller et al. 1998; Coller and Wickens 2002, 2007). mRNAs encoding different Bic-C MS2 fusions were injected into oocytes, followed by injection of two reporter mRNAs; the 3′ UTR of the firefly luciferase reporter mRNA contained three MS2-binding sites and a 39-nt poly(A) tail; the Renilla luciferase reporter mRNA lacked MS2 sites. (C) The C-terminal half of Bic-C repressed translation independent of poly(A). Top panel: relative repression of firefly luciferase poly A39 mRNA translation by each protein. Middle panel: translational repression of firefly luciferase mRNA lacking a poly(A) tail by each protein. Bottom panel: analysis of fusion protein expression by immunoblotting with α–HA and α–actin antibodies.
Translational repression by Drosophila Bic-C requires that the target mRNA possess a 3′ poly(A) tail (Chicoine et al. 2007). However, in our previous study, vegetal-cell repression of xCR1 mRNA was poly(A) tail-independent (Zhang et al. 2009). These data suggest that vertebrate Bic-C-mediated repression is different from that in Drosophila in that it does not require the presence of a poly(A) tail on the target mRNA. Therefore, we compared Bic-C reporter mRNA repression targets that differed only in terms of containing or lacking a poly(A) tail in the tethered translation assay (Fig. 2C). Bic-C-dependent repression occurred efficiently regardless of whether the target mRNA contained a poly(A) tail. In addition, a human MS2-C-term-Bic-C fusion protein behaved similarly to the Xenopus MS2-C-term-Bic-C fusion protein in these experiments (Supplemental Fig. 2). We conclude that Bic-C-mediated translational repression in vertebrates does not require that the target mRNA contain a poly(A) tail.
The N-terminal region of Bic-C bound the same region of 3′-UTR mRNA that was required for translational repression of xCR1 mRNA
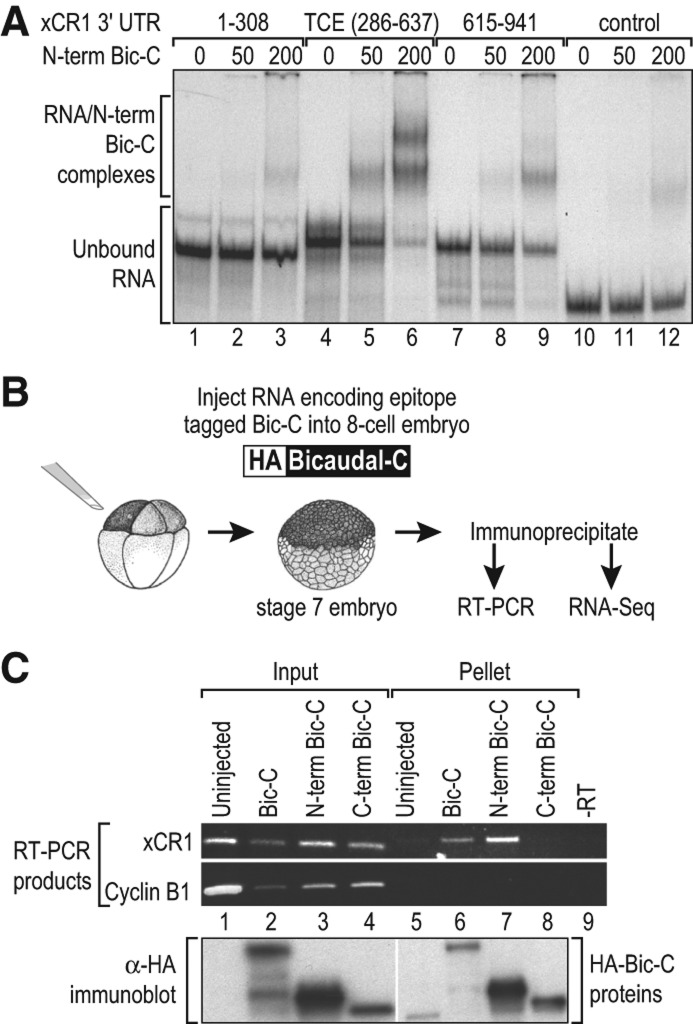
The N-terminal region of Xenopus Bic-C is comprised of multiple KH domains, which often mediate protein-RNA interactions (Wessely and De Robertis 2000; Valverde et al. 2008; Gamberi and Lasko 2012; Muller-McNicoll and Neugebauer 2013). To determine whether Bic-C bound RNA elements within the xCR1 3′ UTR directly, we measured the interaction between recombinant Bic-C protein and various RNA substrates in vitro. We purified bacterially expressed protein that comprised the N-terminus of Bic-C, including the KH domains (aa 1–506), as a GST fusion protein (N-Bic-C). In electrophoretic mobility shift assays, the N-Bic-C protein bound a radiolabeled RNA consisting of the TCE, the RNA element mediating repression by Bic-C in animal cells (Fig. 1B), and the same RNA region required to mediate vegetal-cell repression of xCR1 mRNA reporters in vivo (Fig. 3A; Zhang et al. 2009). In contrast, RNAs from the 5′ or 3′ side of the TCE (Fig. 3A, 1–308 RNA, lanes 1–3 and 615–941 RNA, lanes 7–9) or a control RNA lacking xCR1 mRNA 3′-UTR sequences (Fig. 3A, lanes 10–12) were bound only weakly by N-Bic-C. We conclude that the N-terminal region of Bic-C contains the protein's RNA binding function and is capable of binding specifically to a region of the xCR1 3′ UTR sufficient for translational repression.
FIGURE 3.
The N-terminal region of Bic-C specifically binds the TCE region of the xCR1 3′ UTR that directs translational repression. (A) Radiolabeled RNAs were mixed with 0, 50, and 200 nM GST-N-term Bic-C protein, and binding was analyzed by native gel electrophoresis. The 1–308, TCE (286–637), and 615–941 radiolabeled RNAs were derived from the xCR1 3′ UTR (see Fig. 2). The negative control RNA was derived from the pSTBlue plasmid. (B) Xenopus embryos injected with mRNAs encoding HA-tagged versions of Bic-C, the N-terminal half of Bic-C, or the C-terminal half of Bic-C. The Bic-C proteins were immunoprecipitated from blastula stage injected embryos with αHA antibodies, and the mRNAs present were analyzed using RT-PCR and RNA-seq. (C) The presence of the xCR1 and cyclinB1 mRNAs in the input and pellet fractions as assayed with RT-PCR. The input samples were unfractionated extract, and the pellet samples were the HA immunoprecipitates. Proteins from the input and pellet samples were analyzed by immunoblotting and the HA antibody (lower panel). The low molecular weight species detected in the uninjected sample (lane 5) is a cross-reacting species with the HA antibody.
Bicaudal-C binds xCR1 mRNA in vivo
If Bic-C mediates repression in vivo, then ectopic Bic-C should bind the endogenous xCR1 mRNA in animal cells. To test this prediction, we expressed HA-tagged full-length Bic-C (HA-Bic-C) and the N-terminal (N-term) and C-terminal (C-term) halves of Bic-C in embryos by injecting mRNAs encoding the various Bic-C proteins into the animal cells of an eight-cell embryo, The tagged Bic-C proteins were immunoprecipitated from Stage 7 embryos (∼ 256 cells) with an anti-HA antibody (Fig. 3B). The presence of specific mRNAs in the immunoprecipitates (pellet fraction) was assayed using RT-PCR (Fig. 3C; Cooke et al. 2010). Endogenous xCR1 mRNA was coimmunoprecipitated by HA-Bic-C, while the highly abundant cyclin B1 mRNA was not (Fig. 3C). Moreover, the N-terminal half of Bic-C efficiently bound the xCR1 mRNA, while the C-terminal half did not. Our results are consistent with recent studies in mammalian kidney cells that found that the N-terminal domain of human Bic-C was also sufficient to bind the adenylate cyclase-6 mRNA target (Piazzon et al. 2012). Thus, Bic-C associated specifically with the endogenous xCR1 mRNA in embryos, consistent with its role as a direct translational repressor. The N-terminal half of the protein was sufficient for binding to a specific RNA substrate in vivo as well as in vitro.
Identification of Bic-C mRNA targets
To identify additional mRNA targets of Bic-C without bias, we immunoprecipitated Bic-C and identified associated mRNAs by deep sequencing (Fig. 3B). The RNAs from an immunoprecipitation of embryos not expressing HA-Bic-C was used as a negative control. Sequence reads were mapped to reference transcripts from the Xenopus laevis UniGene contigs and transcript abundances were estimated for each RNA-seq sample using RSEM (Li and Dewey 2011). Statistical analysis of the RNA sequences from the Bic-C immunoprecipitates compared to the control sample identified 62 mRNAs enriched in the Bic-C sample using a false discovery rate cutoff of 0.05 (Supplemental Table 1). The enrichment of specific mRNAs in the Bic-C immunoprecipitate was confirmed by assaying for individual mRNAs using quantitative RT-PCR (Supplemental Fig. 3).
In addition to the expected xCR1 mRNA, many of the mRNAs associated with Bic-C encode proteins that function in developmentally relevant pathways. For example, the Dpy30 mRNA encodes a histone methyltransferase important for cell fate decisions in ES cells (Jiang et al. 2011), while the BCCIP mRNA encodes a protein that guides progenitor cells in neural development (Huang et al. 2012). Furthermore, several of the Bic-C mRNA targets encode proteins implicated in Nodal/TGFβ signaling; Smad4b (Chang et al. 2006) is a pathway transcription factor, Oct25 is a transcription factor antagonist (Cao et al. 2008), and Coco is a secreted signaling antagonist (Bell et al. 2003; Vonica and Brivanlou 2007; Supplemental Table 2). These results suggest that Bic-C regulates multiple maternal mRNAs that encode critical proteins important for early vertebrate development.
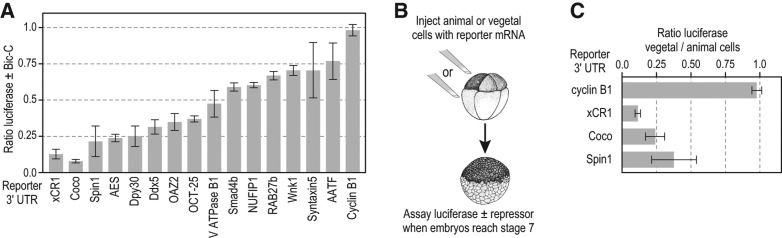
If the mRNAs we had identified were bona fide targets in vivo, then they were likely to be repressed by Bic-C in the ectopic repression assay (Fig. 1A). Therefore, the 3′ UTRs from 14 different Bic-C-HA-associated mRNAs were used to generate luciferase reporter mRNAs. Since the 3′ UTRs of most Xenopus mRNAs are poorly characterized, we chose 14 potential Bic-C mRNA targets based primarily on the availability of 3′-UTR sequence information for cloning. Bic-C repression of these reporter mRNAs was assessed using the ectopic Bic-C expression animal cap assay described above (Fig. 1A). Each of the 14 reporters was repressed by Bic-C, though the extent of repression varied from 25% to 90% (Fig. 4A). These data indicate that Bic-C association with these mRNAs was biologically relevant to their translational regulation.
FIGURE 4.
Identification of Bicaudal-C-regulated mRNAs. (A) Reporter mRNAs created with the 3′ UTRs of 14 different Bic-C-associated mRNAs identified by RIP-seq were assayed as described in Figure 1A. Reporter mRNAs containing the xCR1 TCE and the cyclin B1 3′ UTR serve as positive and negative controls, respectively. (B,C) Diagram of vegetal cell-specific repression assay (Zhang et al. 2009). Reporter mRNAs are injected into animal cells or vegetal cells of separate eight-cell embryos. When embryos reach stage 7, luciferase activity is assayed and the ratio in vegetal cells vs. animal cells calculated.
The normal mechanism of xCR1 translational repression is vegetal cell-specific, consistent with Bic-C protein's localization to these cells. While the additional Bic-C mRNA targets identified above were isolated in animal cell experiments, another prediction was that their 3′ UTRs would direct vegetal cell-specific repression. We tested this prediction by analyzing the repression guided by the 3′ UTRs of the Coco and Spin reporter mRNAs in vegetal cells (Fig. 4B). These 3′ UTRs were chosen for analysis because they were efficiently repressed by ectopic Bic-C in animal cells (Fig. 4A). Reporter mRNAs containing 3′ UTRs from the Coco and Spin mRNAs were repressed in vegetal cells as predicted for bona fide regulatory targets of Bic-C translational repression (Fig. 4C).
DISCUSSION
Bic-C protein is responsible for spatial regulation of xCR1 mRNA in the Xenopus embryo. Its ectopic presence in the animal hemisphere is sufficient to repress an mRNA bearing the 3′-UTR elements of xCR1 mRNA. Further, Bic-C associates with a battery of other mRNAs and mediates regulation of reporters through their 3′ UTRs. Many of the target mRNAs have vital roles in development, including the Nodal pathway in which xCR1 protein participates. We suggest that maternal Bic-C helps generate embryonic polarities and influence cell-fate decisions during early vertebrate embryogenesis by direct translational regulation of mRNAs that encode cell-fate determinants.
The conclusion that Bic-C is the difference between animal and vegetal cells responsible for the differential translation of xCR1 mRNA is supported by four lines of evidence. First, Bic-C protein is confined to vegetal cells through localization of its mRNA (Wessely and De Robertis 2000). Second, ectopic expression of Bic-C in animal cells was sufficient to repress a reporter mRNA bearing the regulatory region in the 3′ UTR that is responsible for vegetal repression. Third, Bic-C-mediated repression was dependent on the 5′ cap and initiation factors, as is vegetal cell-specific repression. Fourth, the N-terminal region of Bic-C, containing multiple KH domains, associated with endogenous xCR1 mRNA in vivo and bound with specificity to the relevant control element (TCE) within the xCR1 mRNA 3′ UTR in vitro. Since the mere presence of Bic-C in animal cells is sufficient to drive repression, we conclude that Bic-C protein is the only component missing from animal cap cells necessary for vegetal cell-specific repression.
Repression requires two distinct domains in Bic-C. The N-terminal domain is sufficient for specific binding to xCR1 mRNA but insufficient for repression; conversely, the C-terminal domain is sufficient for repression but fails to bind the RNA. The N-terminal region contains multiple KH domains, which likely mediate interactions with the regulatory element. The C-terminal region contains a SAM domain, a protein–protein interaction module required for Bic-C's association with RNA granules (P-bodies) (Maisonneuve et al. 2009; Tran et al. 2010). However, the SAM domain was neither sufficient nor essential for Bic-C-mediated repression, though it did enhance that activity. Identification of the key region responsible for repression is now critical.
Recent studies in mammalian kidney cells suggest that Bic-C repression involves miRNAs through an unknown mechanism (Tran et al. 2010; Piazzon et al. 2012). However, miRNA-mediated repression is deficient in Xenopus embryos until after the activation of zygotic transcription, when Argonaute protein first appears (Lund et al. 2011). Thus, Bic-C activity in the early stages of development is likely miRNA-independent.
Through immunoprecipitation and deep sequencing, we identified multiple putative mRNAs controlled by Bic-C. Several targets were sensitive to Bic-C-mediated repression in the animal cell assay, and at least two, Coco and Spin1, were robustly repressed in vegetal cells. We propose that Bic-C acts in a post-transcriptional regulatory network that establishes the proper balance of proteins in the embryo essential for normal development. While its precise role(s) remains to be determined, our findings suggest important roles in Nodal signaling.
Cripto proteins were originally discovered as Nodal/TGFβ pathway components (Ding et al. 1998; Gritsman et al. 1999; Schier 2009). In addition to xCR1, several other putative Bic-C mRNA targets identified in this study are strongly implicated in Nodal signaling (Supplemental Table 2). However, the group of mRNAs encodes both activators (e.g., Smad4b) and antagonists (e.g., Coco) of Nodal signaling, indicating that Bic-C's influence on signaling by this pathway may be complex. Bic-C also may influence development through control of other pathways. For example, while Cripto proteins such as xCR1 have been traditionally viewed as exclusive components of Nodal/TGFβ signaling (Schier 2009), other evidence indicates that they also function in other signaling processes, such as the Wnt pathway (Tao et al. 2005; Nagaoka et al. 2012, 2013). Our results connecting xCR1 and Bic-C in Xenopus embryos raise the possibility that analogous regulation occurs in mammalian somatic cells. Indeed, Bic-C's inhibition of Wnt signaling may require repression of mammalian Cripto mRNA (Maisonneuve et al. 2009).
Our study focused on the role of Bic-C present in the maternal embryo, prior to the onset of zygotic transcription. In later mammalian development and in adults, Bic-C continues to be critical, influencing specific organs, particularly the kidney (Maisonneuve et al. 2009; Tran et al. 2010). Some targets regulated in the embryo, identified here, may be controlled by Bic-C in later development as well. For example, Coco mRNA (called Dand5 in mouse and Cerl2 in humans) controls positioning of the visceral organs within the body cavity. Loss-of-function alleles of Coco and Bic-C cause similar defects in organ positioning, raising the possibility that Bic-C may control Coco in somatic cells. Similarly, multiple other targets of Bic-C identified here (e.g., the mRNAs encoding the Wnk1 and V-ATPase B1 proteins) are critical for normal kidney development and function (Karet et al. 1999a,b; Arroyo and Gamba 2012; Naguro et al. 2012). Thus, our findings and the recent demonstration that adenylate cyclase6 mRNA is a Bic-C target in kidney cells (Piazzon et al. 2012) support the idea that translational control by Bic-C plays a key role in renal development and point to relevant mRNA targets worth further inquiry. In other systems, single regulatory proteins often mediate control of hundreds of mRNAs with related functions (Gerber et al. 2004; Ule and Darnell 2006; Richter 2007; Kershner and Kimble 2010). Our results reveal that Bic-C may form an analogous hub during vertebrate development.
MATERIALS AND METHODS
Luciferase reporter mRNA plasmids and mRNA synthesis
Firefly luciferase reporter mRNAs were generated that contained different segments of the xCR1 mRNA 3′ UTR and other 3′ UTRs (Sheets et al. 1994; Fritz and Sheets 2001; Zhang et al. 2009). See Supplemental Methods.
mRNA injections and luciferase assays
Reporter mRNAs were injected into either the animal cells or vegetal cells of eight-cell embryos. When injected embryos reached stage 7, extracts were prepared and analyzed for luciferase activity (Sheets et al. 1994; Fritz and Sheets 2001; Zhang et al. 2009).
RNA blot hybridization
Total RNA from embryos injected with reporter mRNAs was analyzed by RNA blot hybridization (Sheets et al. 1994; Zhang et al. 2009) using a radiolabeled probe to detect the firefly luciferase mRNA.
Electrophoretic mobility assays
Recombinant GST-N-term Bic-C fusion protein (residues 1–506) was expressed and purified as described (Hou et al. 2005, 2009). RNA substrates were generated by in vitro transcription with 32P-UTP. The RNAs encoding the xCR1 3′-UTR fragments 1–308, TCE (previously referred to as Mut2), and 615–941 were derived from the xCR1 3′ UTR (Zhang et al. 2009). The NEG control RNA (261 nt) was generated from the pSTBlue-1 plasmid. Binding reactions (20 μL) contained GST-N-term Bic-C protein (either 0, 50, or 200 nM), 10 mM Hepes pH 8.0, 1 mM EDTA, 50 mM KCl, 0.02% Tween-20, 0.2 mg/mL yeast tRNA, 100 μg/mL BSA, 2 mM DTT, and 0.5 nM RNA. Reaction products were analyzed on 4% (1×TBE) native polyacrylamide gels.
Immunoblotting
Immunoblotting was done as described (Kwak et al. 2008) using mouse monoclonal anti-HA-tag antibody and anti-Actin antibody.
Tethered function assays
Tethered function assays were performed in Xenopus oocytes as described (Coller et al. 1998; Coller and Wickens 2002; Cooke et al. 2010). Information on MS2 fusions is found in Supplemental Methods.
Q-PCR
Quantitative PCR to analyze mRNAs associated with Bic-C was performed as described (Park et al. 2011). Information on primers is found in Supplemental Methods.
Immunoprecipitations
Xenopus embryos were injected with mRNA encoding HA-Bic-C (full length Bic-C), HA-N-term Bic-C (the N-terminal half of Bic-C, aa 1–506), or HA-C-term Bic-C (the C-terminal half of Bic-C, aa 507–963). Blastula stage (st.7) injected embryos were lysed in 100 μL of TNMEN-150 buffer (Cooke et al. 2010). The lysate was centrifuged (4°C, 10 min at 5000 rpm) and the supernatant incubated with α-HA antibody coupled to protein-G agarose (2 h, 4°C). The beads were collected (1 min, 3000 rpm) and washed 4× in 1 mL TNMEN 150 buffer. For each wash, the beads were incubated in buffer at 4°C for 5 min, spun at 3000 rpm for 1 min, and supernatant removed. RNA and protein were isolated from the washed beads for analysis.
Construction of RNA-seq libraries and RNA-seq
Immunoprecipitated RNA and a total RNA control were submitted to the University of Wisconsin-Madison Biotechnology Center for RNA-seq library preparation and sequencing. See Supplemental Methods for specifics of library preparation and sequencing.
RNA-seq data analysis
Transcript abundances were estimated for each RNA-seq sample using RSEM (v1.0.2.5) (Hou et al. 2009; Li and Dewey 2011). RSEM was provided with the X. laevis UniGene contigs (Wheeler et al. 2003) to use as reference sequences. The read counts estimated for each transcript by RSEM were rounded and then given as input to the R package DESeq (v1.0.6) for differential expression analysis. BIC-C targets were identified as the transcripts that had positive log-fold change and FDR less than 0.05 in a comparison of the HA-pellet to UN-pellet samples.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Catherine Fox for her insightful and useful comments. We also thank Laura Vanderploeg for preparing figures and Susanne Blaser Imboden for technical assistance. E. De Robertis and Mary Lou King kindly supplied reagents. This work was supported by NSF grant 1050395 (M.D.S.), NIH grants HG005232 (C.N.D.), GM31892, and GM50942 (M.W.), NIH training grant T32 GM07215 (A.C.), and an Advanced Opportunity Fellowship from the University of Wisconsin-Madison (A.C.).
REFERENCES
- Arroyo JP, Gamba G 2012. Advances in WNK signaling of salt and potassium metabolism: Clinical implications. Am J Nephrol 35: 379–386 [DOI] [PubMed] [Google Scholar]
- Bell E, Munoz-Sanjuan I, Altmann CR, Vonica A, Brivanlou AH 2003. Cell fate specification and competence by Coco, a maternal BMP, TGFβ, and Wnt inhibitor. Development 130: 1381–1389 [DOI] [PubMed] [Google Scholar]
- Cao Y, Siegel D, Oswald F, Knochel W 2008. Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J Biol Chem 283: 34168–34177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Brivanlou AH, Harland RM 2006. Function of the two Xenopus smad4s in early frog development. J Biol Chem 281: 30794–30803 [DOI] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P 2007. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704 [DOI] [PubMed] [Google Scholar]
- Coller J, Wickens M 2002. Tethered function assays using 3′ untranslated regions. Methods 26: 142–150 [DOI] [PubMed] [Google Scholar]
- Coller J, Wickens M 2007. Tethered function assays: An adaptable approach to study RNA regulatory proteins. Methods Enzymol 429: 299–321 [DOI] [PubMed] [Google Scholar]
- Coller JM, Gray NK, Wickens MP 1998. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev 12: 3226–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M 2010. Translational repression by deadenylases. J Biol Chem 285: 28506–28513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM 1998. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395: 702–707 [DOI] [PubMed] [Google Scholar]
- Dorey K, Hill CS 2006. A novel Cripto-related protein reveals an essential role for EGF-CFCs in Nodal signalling in Xenopus embryos. Dev Biol 292: 303–316 [DOI] [PubMed] [Google Scholar]
- Fritz BR, Sheets MD 2001. Regulation of the mRNAs encoding proteins of the BMP signaling pathway during the maternal stages of Xenopus development. Dev Biol 236: 230–243 [DOI] [PubMed] [Google Scholar]
- Gamberi C, Lasko P 2012. The bic-C family of developmental translational regulators. Comp Funct Genomics 2012: 141386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF 1999. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97: 121–132 [DOI] [PubMed] [Google Scholar]
- Hou Z, Bernstein DA, Fox CA, Keck JL 2005. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc Natl Acad Sci 102: 8489–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Danzer JR, Mendoza L, Bose ME, Muller U, Williams B, Fox CA 2009. Phylogenetic conservation and homology modeling help reveal a novel domain within the budding yeast heterochromatin protein Sir1. Mol Cell Biol 29: 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Lu H, Liu S, Droz-Rosario R, Shen Z 2012. Requirement of mouse BCCIP for neural development and progenitor proliferation. PloS One 7: e30638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG 2011. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karet FE, Finberg KE, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Medina JF, Lifton RP 1999a. Localization of a gene for autosomal recessive distal renal tubular acidosis with normal hearing (rdRTA2) to 7q33-34. Am J Hum Genet 65: 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez-Soriano J, Santos F, Cremers CW, Di Pietro A, et al. 1999b. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet 21: 84–90 [DOI] [PubMed] [Google Scholar]
- Kershner AM, Kimble J 2010. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci 107: 3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33: 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M 2008. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci 105: 14644–14649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN 2011. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Sheets MD, Imboden SB, Dahlberg JE 2011. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev 25: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, Constam DB 2009. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136: 3019–3030 [DOI] [PubMed] [Google Scholar]
- Mosquera L, Forristall C, Zhou Y, King ML 1993. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 117: 377–386 [DOI] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM 2013. How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet 14: 275–287 [DOI] [PubMed] [Google Scholar]
- Nagaoka T, Karasawa H, Castro NP, Rangel MC, Salomon DS, Bianco C 2012. An evolving web of signaling networks regulated by Cripto-1. Growth Factors 30: 13–21 [DOI] [PubMed] [Google Scholar]
- Nagaoka T, Karasawa H, Turbyville T, Rangel MC, Castro NP, Gonzales M, Baker A, Seno M, Lockett S, Greer YE, et al. 2013. Cripto-1 enhances the canonical Wnt/β-catenin signaling pathway by binding to LRP5 and LRP6 co-receptors. Cell Signal 25: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguro I, Umeda T, Kobayashi Y, Maruyama J, Hattori K, Shimizu Y, Kataoka K, Kim-Mitsuyama S, Uchida S, Vandewalle A, et al. 2012. ASK3 responds to osmotic stress and regulates blood pressure by suppressing WNK1-SPAK/OSR1 signaling in the kidney. Nat Commun 3: 1285. [DOI] [PubMed] [Google Scholar]
- Otero LJ, Devaux A, Standart N 2001. A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA 7: 1753–1767 [PMC free article] [PubMed] [Google Scholar]
- Park S, Patterson EE, Cobb J, Audhya A, Gartenberg MR, Fox CA 2011. Palmitoylation controls the dynamics of budding-yeast heterochromatin via the telomere-binding protein Rif1. Proc Natl Acad Sci 108: 14572–14577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazzon N, Maisonneuve C, Guilleret I, Rotman S, Constam DB 2012. Bicc1 links the regulation of cAMP signaling in polycystic kidneys to microRNA-induced gene silencing. J Mol Cell Biol 4: 398–408 [DOI] [PubMed] [Google Scholar]
- Richter JD 2007. CPEB: A life in translation. Trends Biochem Sci 32: 279–285 [DOI] [PubMed] [Google Scholar]
- Saffman EE, Styhler S, Rother K, Li W, Richard S, Lasko P 1998. Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol 18: 4855–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF 2009. Nodal morphogens. Cold Spring Harb Perspect Biol 1: a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev 8: 926–938 [DOI] [PubMed] [Google Scholar]
- Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, Yan D, Asashima M, Wylie CC, Lin X, Heasman J 2005. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell 120: 857–871 [DOI] [PubMed] [Google Scholar]
- Tran U, Zakin L, Schweickert A, Agrawal R, Doger R, Blum M, De Robertis EM, Wessely O 2010. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development 137: 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Darnell RB 2006. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol 16: 102–110 [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L 2008. Structure and function of KH domains. FEBS J 275: 2712–2726 [DOI] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH 2007. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Dev Biol 303: 281–294 [DOI] [PubMed] [Google Scholar]
- Wessely O, De Robertis EM 2000. The Xenopus homologue of Bicaudal-C is a localized maternal mRNA that can induce endoderm formation. Development 127: 2053–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, et al. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res 31: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe S, Tanegashima K, Haramoto Y, Takahashi S, Fujii T, Kozuma S, Taketani Y, Asashima M 2003. FRL-1, a member of the EGF-CFC family, is essential for neural differentiation in Xenopus early development. Development 130: 2071–2081 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Forinash KD, McGivern J, Fritz B, Dorey K, Sheets MD 2009. Spatially restricted translation of the xCR1 mRNA in Xenopus embryos. Mol Cell Biol 29: 3791–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]