Argonaute (Ago) proteins are the key effectors of small RNA-mediated gene silencing in eukaryotes. In mammalian cells, Ago2 is the only Ago with dual function in both RNAi and miRNA-mediated pathways. The loading of small RNAs into Ago2 is a regulated process that requires the function of the Hsc70/Hsp90 chaperone complex. The Ago2 apo-enzyme is unstable due to degradation via cellular autophagy. Here the authors identify the Hsp90 co-chaperones, Fkbp4 and Fkbp5, as Ago2-associated proteins. Both loss-of-function and gain-of-function experiments indicate that Fkbp4/5 co-chaperones influence the post-transcriptional stabilization of Ago2 and, consequently, the abundance of cellular miRNAs. These results support a role for Fkbp4/5 in the assembly of the RNA-induced silencing complex (RISC).
Keywords: Ago2, Fkbp4, Fkbp5, Hsp90, microRNA (miRNA), RNA interference (RNAi)
Abstract
Argonaute2 (Ago2) protein and associated microRNAs (miRNAs) or small interfering RNAs (siRNAs) form the RNA-induced silencing complex (RISC) for target messenger RNA cleavage and post-transcriptional gene silencing. Although Ago2 is essential for RISC activity, the mechanism of RISC assembly is not well understood, and factors controlling Ago2 protein expression are largely unknown. A role for the Hsc70/Hsp90 chaperone complex in loading small RNA duplexes into the RISC has been demonstrated in cell extracts, and unloaded Ago2 is unstable and degraded by the lysosome in mammalian cells. Here we identify the co-chaperones Fkbp4 and Fkbp5 as Ago2-associated proteins in mouse embryonic stem cells. Pharmacological inhibition of this interaction using FK506 or siRNA-mediated Fkbp4/5 depletion leads to decreased Ago2 protein levels. We find FK506 treatment inhibits, whereas Fkbp4/5 overexpression promotes, miRNA-mediated stabilization of Ago2 expression. Simultaneous treatment with a lysosome inhibitor revealed the accumulation of unloaded Ago2 complexes in FK506-treated cells. We find that, consistent with unloaded miRNAs being unstable, FK506 treatment also affects miRNA abundance, particularly nascent miRNAs. Our results support a role for Fkbp4/5 in RISC assembly.
INTRODUCTION
Argonaute (Ago) proteins are the key effectors of post-transcriptional gene silencing (PTGS) triggered by microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Hammond et al. 2001; Meister 2013). Both siRNAs and miRNAs are loaded into Ago proteins as short duplexes of ∼22 bp, and subsequently, one strand, the guide, is stably retained while the other (passenger or miRNA*) strand gets degraded (Tomari et al. 2004; Matranga et al. 2005; Rand et al. 2005). Loaded Ago serves then to guide the RNA-induced silencing complex (RISC) to target messenger RNAs (mRNAs), inducing their cleavage in the case of extensive sequence complementary between the guide strand and its target or translation repression and mRNA decay in the case of most miRNA/mRNA pairings (Chekulaeva and Filipowicz 2009). Of the four Ago proteins expressed in mammalian cells, Ago2 is the sole family member capable of endonucleolytic cleavage of a complementary target mRNA and is essential for RNA interference (RNAi) (Liu et al. 2004; Meister et al. 2004).
Loading small RNA duplexes into Ago proteins requires the action of the Hsc70/Hsp90 chaperone machinery in cell-free systems (Iki et al. 2010; Iwasaki et al. 2010; Miyoshi et al. 2010). Chemical inhibition of Hsc70/Hsp90 in mammalian cells leads to decreased levels of Ago2 protein (Tahbaz et al. 2001; Pare et al. 2009; Johnston et al. 2010; Martinez and Gregory 2013). Since unloaded Ago2 is unstable and degraded by the lysosome (and possibly also the proteasome), these findings are consistent with a role for the Hsc70/Hsp90 complex in the homeostatic control of Ago2 expression (Gibbings et al. 2012; Martinez and Gregory 2013; Smibert et al. 2013). Analogous to the regulation of steroid receptors, it has been speculated that the Hsp90 complex uses ATP to mediate a conformational opening in Ago proteins to allow the incorporation of small RNA ligands (Pratt et al. 2008; Iki et al. 2010; Iwasaki et al. 2010; Miyoshi et al. 2010). Here, through the large-scale isolation and characterization of Ago2-containing complexes, we identified the Hsp90 co-chaperone Fkbp5 (also known as Fkbp51) as a novel Ago2-associated protein in mouse embryonic stem cells (ESCs). Fkbp5 is a member of the immunophilin family of proteins, previously shown to interact with Hsp90 to dynamically regulate the ligand-binding activity of steroid receptors (Pratt et al. 2008). Interestingly, other members of the immunophilin family have been recently implicated in the activity and/or biogenesis of small RNAs. In plants, SQUINT (SQN), which encodes the Arabidopsis ortholog of cyclophilin 40 (CyP40), was initially shown to promote miRNA-mediated gene repression in vivo (Smith et al. 2009). sqn mutants resembled weak alleles of ago1 mutants with higher levels of known miRNA-regulated genes and slightly reduced miRNA levels compared with that of wild-type plants. However, AGO1 protein levels were unchanged between sqn mutants and wild-type plants (Smith et al. 2009). Subsequently, Cyp40 was shown to physically interact with AGO1 and facilitate small RNA loading in plant extracts (Iki et al. 2012). Finally, the mouse co-chaperone Fkbp6 and its Drosophila ortholog Shutdown were shown to play a role in the biogenesis of germline-specific small RNAs (PIWI-associated RNAs or piRNAs) (Olivieri et al. 2012; Preall et al. 2012; Xiol et al. 2012).
We found that pharmacological inhibition of the Fkbp–Ago2 interaction by the immunosuppressant FK506 or by Fkbp5 knockdown leads to decreased Ago2 protein expression in mouse and human cells. Conversely ectopic Fkbp5 leads to elevated Ago2 protein levels in a miRNA-dependent fashion. Similarly, we found that loss- and gain-of function examination of the related co-chaperone Fkbp4 (also known as Fkbp52) leads to decrease or elevated Ago2 protein levels, respectively. Further supporting a role of the co-chaperones in small RNA loading, we find that FK506 treatment blocks miRNA-dependent stabilization of Ago2 expression, and isolated Ago2 complexes from treated cells were found to contain substantially reduced miRNA levels. Finally, consistent with the coupling of Ago2-miRNA levels, we find that FK506 treatment leads to a decrease in miRNA expression. Altogether our results support a role for the co-chaperones Fkbp4 and Fkbp5 as new members of the Ago2 small RNA loading complex.
RESULTS
Fkbp5 associates with Ago2 in mouse ESCs
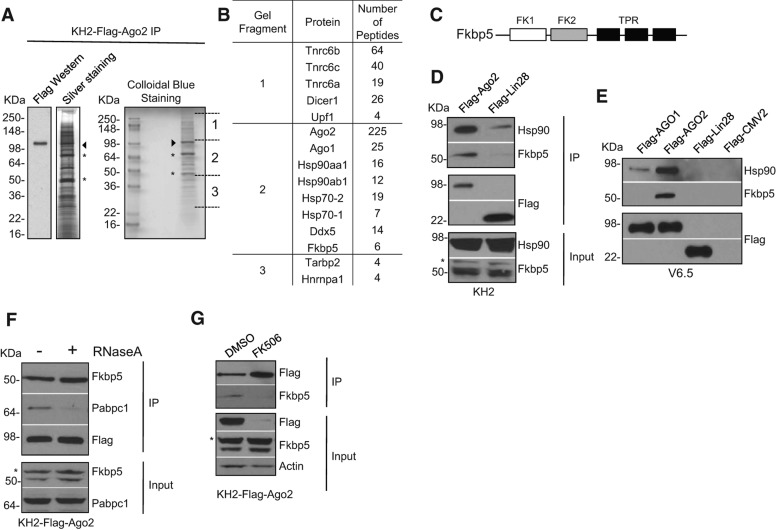
To identify novel proteins that associate with Ago2, we performed large-scale Flag immunoprecipitation (IP) from a stable mouse ESC line (KH2-Flag-Ago2) that expresses Flag-Ago2 under the control of the tetracycline promoter (Chang et al. 2012). The Flag-affinity purified eluate was analyzed by Flag Western, silver staining, and colloidal blue staining and revealed the presence of multiple Ago2-associated proteins (Fig. 1A). Sections 1 through 3 of the colloidal blue-stained gel were subjected to mass spectrometry analysis (Fig. 1B). Several Ago2-interacting proteins that have been previously implicated in miRNA and siRNA pathways were identified, including members of the Hsp70/Hsp90 chaperone machinery known to assist Ago proteins in the loading of small RNA ligands in multiple organisms (Fig. 1B; Hock et al. 2007; Landthaler et al. 2008). In addition, we identified the co-chaperone Fkbp5, a member of the immunophilin family of proteins. Fkbp5 is characterized at the N terminus by two FKBP12-like domains (FK); however, only the first domain confers peptidyl prolyl cis-trans isomerase (PPIase) activity (Sinars et al. 2003; Lu et al. 2007). PPIases are known to catalyze the cis-trans isomerization of prolines to induce conformational changes in client proteins (Wang et al. 2010). This first FK domain binds to and is inhibited by the immunosuppressant FK506 (Sinars et al. 2003; Wu et al. 2004). The C terminus encodes a tetratricopeptide repeat (TPR) motif, which has been shown to interact with the C-terminal end of Hsp90 (Fig. 1C; Scheufler et al. 2000; Pratt and Toft 2003; Zeytuni and Zarivach 2012). To validate the specific association of Fkbp5 with Ago2-containing complex(es), we performed co-IPs using cell lysates prepared from KH2-Flag-Ago2 ESCs or a negative control ESC line that expresses Flag-Lin28 (KH2-Flag-Lin28). Endogenous Fkbp5 protein was detected in Flag-Ago2 but not in Flag-Lin28 immunoprecipitates. In accordance, Hsp90 was also enriched in Flag-Ago2 compared with Flag-Lin28 immunoprecipitates (Fig. 1D). To test whether Fkbp5 associates with other Ago proteins, we next examined the possible interaction between Ago1 and Fkbp5. We transfected V6.5 ESCs with plasmids expressing Flag-Ago1, a positive control Flag-Ago2, a negative control Flag-Lin28, or an empty vector (pFlag-CMV2). Consistent with our previous results, Flag-Ago2 was able to efficiently pull down Hsp90 and Fkbp5. In contrast, however, less Hsp90 was found associated with Ago1, and we were unable to detect Fkbp5 in Flag-Ago1 immunoprecipitates (Fig. 1E), Thus, Fkbp5 appears to selectively associate with Ago2. We next tested whether the Ago2–Fkbp5 interaction was RNA-dependent by performing co-IPs using cell lysates from KH2-Flag-Ago2 cells treated with or without RNase A. We found the Fkbp5–Ago2 association to be resistant to RNase A treatment (Fig. 1F). In contrast, the interaction between Ago2 and Pabpc1 was abolished in the absence of RNA, as previously shown (Fabian et al. 2009). Altogether, these results demonstrate that Fkbp5 is a bona fide Ago2 partner and suggest that this interaction can take place independently of RNA.
FIGURE 1.
The co-chaperone Fkbp5 interacts with Ago2 in mouse ESCs. (A) Cell extracts from KH2-Flag-Ago2 ESCs grown in dox-containing media were subjected to Flag-affinity purification. The affinity eluate was analyzed by SDS-PAGE followed by immunoblotting (IB) using α-Flag antibody, silver staining, and colloidal blue staining followed by mass spectrometry analysis. Arrowhead depicts Ago2, and asterisks depict common contaminants. (B) Partial list of proteins identified by mass spectrometry. (C) Schematic representation of Fkbp5 domains (FK indicates FKBP12-like domain; TPR, tetratricopeptide repeat). (D) Cell extracts from KH2-Flag-Ago2 and control KH2-Flag-Lin28 grown in dox-containing media were subjected to Flag-immunoprecipitation (IP). Associated proteins were detected by Western blotting using α-HSP90 and α-FKBP5 antibodies. Asterisk depicts unspecific protein detected by the α-FKBP5 antibody. (E) Cell extracts from V6.5 ESCs transfected with pFlag-Ago1, pFlag-Ago2, or pFlag-Lin28 expressing constructs or empty vector (pFlag-CMV2), were subjected to Flag IP. Associated proteins were detected by Western blotting using α-HSP90 and α-FKBP5 antibodies. (F) Cell extracts from KH2-Flag-Ago2 cells grown in dox-containing media were subjected to Flag-affinity purification. Where indicated, extracts were treated overnight with RNase A (20 µg/mL). Associated proteins were detected using the indicated antibodies. Asterisk depicts unspecific protein detected by the α-FKBP5 antibody. (G) FK506 blocks the interaction between Ago2 and Fkbp5. KH2-Flag-Ago2 cells were treated with 1 μg/mL dox and (10 µM) FK506 or DMSO for 24 h. Cell extracts were subsequently subjected to Flag-affinity purification. Because the overall levels of Flag-Ago2 are more abundant in extracts from DMSO-treated cells compared with extracts from FK506-treated cells, Flag-IP eluates were diluted by a factor of eight (1:8) for DMSO sample, while Flag-IP eluates from FK506-treated cells were kept undiluted. The resulting Flag IPs were then assessed for associated Fkbp5 by SDS-PAGE and immunoblotting using an α-FKBP5 antibody. An asterisk depicts unspecific protein detected by the α-FKBP5 antibody.
Cyp40, like Fkbp5 and other FKBPs, binds to Hsp90 in steroid receptor complexes (Pratt and Toft 1997, 2003). However, its PPIase domain is structurally distinct from that of FKBPs and does not bind FK506 but instead binds cyclosporine A (CsA). Plant Cyp40, was shown to physically interact with AGO1 in vitro, and this interaction was specifically inhibited by CsA treatment (Iki et al. 2012). Utilizing the same in vitro system, Iki et al. (2012), showed that FK506 slightly inhibited the interaction between FKBP62 and AGO1. Hence, we tested whether the interaction between mouse Ago2 and Fkbp5 could be inhibited by FK506. To this end, we treated KH2-Flag-Ago2 cells with FK506 while simultaneously activating the expression of Flag-Ago2 by addition of doxycycline (dox) to the media. We then performed co-IPs and measured the association of Fkbp5 with Flag-Ago2. Interestingly, we found that FK506 treatment substantially reduced the levels of Flag-Ago2 protein (see input lanes in Fig. 1G), and this prompted us to further examine the consequences of FKBP-inhibition on Ago2 and miRNA expression (see below). However when we loaded eightfold more Flag affinity eluate for the FK506-treated than DMSO-treated samples to normalize the levels of Flag-Ago2, we found that FK506 treatment diminished the association of Fkbp5 with Ago2. These results indicate that FK506 is able to effectively abrogate the interaction between Fkbp5 and Ago2 (Fig. 1G).
FK506 treatment leads to decreased Ago2 protein levels
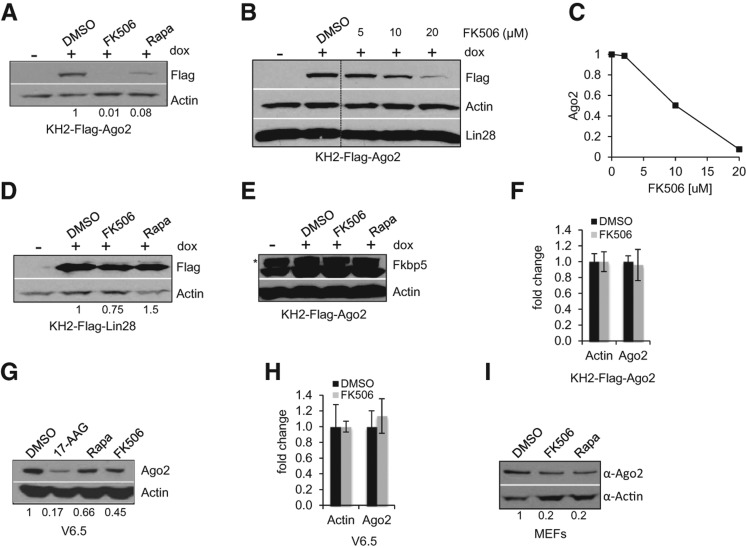
It has been shown that mammalian cells treated with geldanamycin (or its analog 17-Allylamino-17-demethoxygeldanamycin [17-AAG]) or 2-phenylethynesulfonamide (PES) to specifically inhibit the function of Hsp90 and Hsc70, respectively, exhibited reduced levels of Ago2 (Tahbaz et al. 2001; Johnston et al. 2010; Martinez and Gregory 2013). Considering also recent findings that unloaded Ago2 is unstable and gets degraded, likely, via the autophagy pathway (Gibbings et al. 2012; Martinez and Gregory 2013), we examined the effect of inhibiting the interaction between Ago2 and Fkbp5 using immunosuppressant FK506 on the expression of newly synthesized Ago2 protein. We also utilized the related immunosuppressant Rapamycin as a positive control, which has been shown to decrease Ago2 levels in HeLa cells via inhibition of the mTOR pathway and subsequent activation of autophagy (Gibbings et al. 2012). We treated KH2-Flag-Ago2 ESCs with the immunosuppressants at the same time we induced the expression of the Flag-Ago2 transgene by the addition of dox to the media. Treatment with either FK506 or Rapamycin resulted in a dramatic and dose-dependent reduction of Flag-Ago2 levels detected by Western blot (Fig. 2A–C). Importantly, addition of FK506 or Rapamycin to the control KH2-Flag-Lin28 cell line did not significantly affect levels of Flag-Lin28, thereby excluding possible nonspecific effects of these chemicals on general protein stability (Fig. 2D). It is worth noting that neither FK506 nor Rapamycin caused reduced Fkbp5 protein expression in treated ESCs (Fig. 2E). qRT-PCR analysis revealed that FK506 treatment did not influence Ago2 mRNA levels, further supporting a post-transcriptional control of Ago2 protein expression (Fig. 2F). We next examined the effect of FK506 treatment on the steady-state levels of endogenous Ago2 expression. We observed decreased expression of endogenous Ago2 in FK506-treated ESCs as well as with the positive control inhibitors (Fig. 2G), whereas levels of Ago2 mRNA were unaffected (Fig. 2H). To investigate whether the observed effects on Ago2 levels is a phenomenon specific to ESCs, we treated mouse embryonic fibroblasts (MEFs) with FK506 and Rapamycin. Like in ESCs, both inhibitors reduced Ago2 protein levels (Fig. 2I).
FIGURE 2.
FK506 treatment leads to decreased Ago2 protein levels. (A) KH2-Flag-Ago2 cells were treated with 1 μg/mL dox and the indicated inhibitor (20 µM) 17-AAG, (5 µg/mL) Rapamycin (Rapa), (20 µM) FK506, or control DMSO. Whole-cell extracts were assessed for Flag-Ago2 protein levels by Western blotting. The numbers on the bottom indicate the relative Flag-Ago2 abundance. (B) KH2-Flag-Ago2 cells were treated for 24 h with 1 μg/mL dox and the indicated concentration of FK506. Whole-cell extracts were assessed for Flag-Ago2 or endogenous Lin-28 protein levels by Western blotting. Actin serves as loading control. (C) Relative Flag-Ago2 abundance quantified from B. (D) Control KH2-Flag-Lin28 cells were treated with 1 μg/mL dox and the indicated inhibitor as in A. Whole-cell extracts were assessed for Flag-Lin28 protein levels by Western blotting. Actin serves as loading control. The numbers on the bottom indicate the relative Flag-Lin28 abundance. (E) KH2-Flag-Ago2 cells were treated with 1 μg/mL dox and the indicated inhibitor as in A. Whole-cell extracts were assessed for endogenous Fkbp5 protein levels. Asterisk depicts unspecific protein detected by the α-FKBP5 antibody. (F) KH2-Flag-Ago2 cells were treated with control DMSO or (10 µM) FK506 for 24 h. Total RNA was collected for analysis of Ago2 mRNA by qRT-PCR. Actin was used for normalization control. Error bars, mean ± SD with n = 3. (G) V6.5 ESCs were treated for 24 h with (20 µM) 17-AAG, (5 µg/mL) Rapa, (10 µM) FK506, or control DMSO. Whole-cell extracts were assessed for endogenous Ago2 protein levels by Western blotting. Actin serves as loading control. The numbers on the bottom indicate the relative Ago2 abundance. (H) KH2-Flag-Ago2 cells were treated with control DMSO or (10 µM) FK506 for 24 h. Total RNA was collected for analysis of Ago2 mRNA by qRT-PCR. Actin was used for normalization control. Error bars, mean ± SD with n = 3. (I) MEFs were treated for 24 h with either control DMSO, (10 µM) FK506, or (5 µg/mL) Rapa. Whole-cell extracts were assessed for endogenous Ago2 protein levels by Western blotting. Actin serves as loading control. The numbers on the bottom indicate the relative Ago2 abundance.
Fkbp4 and Fkbp5 control Ago2 expression
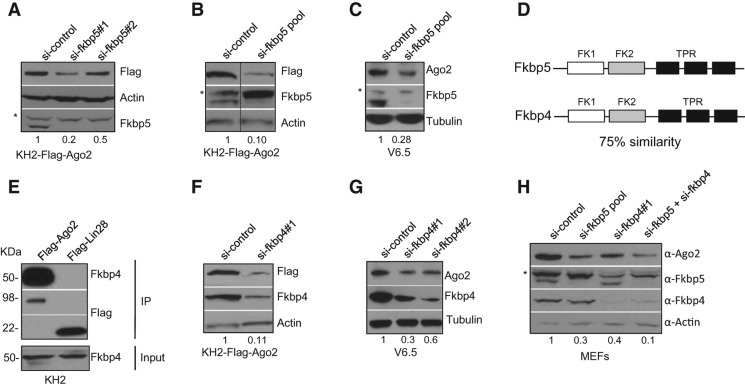
To directly investigate the role of Fkbp5 in Ago2 regulation, we next performed siRNA-mediated knockdown experiments to deplete Fkbp5 expression in ESCs. KH2-Flag-Ago2 ESCs were transfected with either control or Fkbp5 siRNAs for 48 h and then treated with dox for an additional 24 h to induce Flag-Ago2 expression. Western blot revealed Flag-Ago2 levels were reduced in si-Fkbp5 knockdown samples compared with controls (Fig. 3A,B). We also found that endogenous Ago2 levels are similarly reduced upon Fkbp5 knockdown in V6.5 ESCs (Fig. 3C). Altogether, these results indicate that Fkbp5 plays a role in the regulation of Ago2 protein levels. Other TPR-containing immunophilins such as Fkbp4, Cyp40, and PP5 can also associate with steroid–receptor complexes. Thus, different co-chaperones may interact with Ago2 and exert distinct effects on Ago2 protein levels. Although we did not find any other co-chaperone in our mass spectrometry analysis, we decided to investigate the potential role of Fkbp4 in Ago2 regulation. Fkbp4 (also called Fkpb52) is the closest homolog of Fkbp5, with 75% of sequence similarity, and shares a similar domain structure (Fig. 3D). First, to determine whether Fkbp4 physically associates with Ago2 complexes, we performed co-IPs using cell lysates prepared from inducible ESCs expressing Flag-Ago2 or negative control Flag-Lin28. Similar to our Fkbp5 analysis, endogenous Fkbp4 protein was detected in Flag-Ago2 but not Flag-Lin28 immunoprecipitates (Fig. 3E). In support of a role in Ago2 regulation, Fkbp4 knockdown led to reduced levels of Flag-Ago2 transgene in the dox-inducible cell line as well as endogenous Ago2 protein in wild-type ESCs (Fig. 3F,G). These results indicate that analogous to Fkbp5, Fkbp4 also influences Ago2 protein levels. Furthermore, siRNA-mediated knockdown of Fkbp5 and Fkbp4 led to significant reduction of Ago2 levels in MEFs (Fig. 3H). These results indicate that the co-chaperones likely associate with and regulate Ago2 levels in undifferentiated and differentiated cell types. Interestingly, MEFs treated with siRNAs against both Fkbp4 and Fkpb5 displayed Ago2 protein levels that are further reduced compared with either siRNA alone, indicating that Fkbp4 and Fkbp5 function, at least partially, redundantly in the regulation of Ago2 (Fig. 3H).
FIGURE 3.
Fkbp4 and Fkbp5 control Ago2 expression. (A,B) KH2-Flag-Ago2 ESCs were pretreated with 50 nM of either control siRNA, si-Fkbp5 (A), or si-Fkbp5 pool (B) for 48 h. Subsequently, 1 μg/mL dox was added for 24 h. Whole-cell extract was analyzed for Flag-Ago2 and Fkbp5 levels by Western blotting. Actin was used as loading control. The numbers on the bottom indicate the relative Flag-Ago2 abundance. (C) V6.5 ESCs were treated with 50 nM of either si-Fkbp5 or control siRNA for 60 h. Whole-cell extract was analyzed for endogenous Ago2 and Fkbp5 levels by Western blotting. Actin was used as loading control. The numbers on the bottom indicate the relative Ago2 abundance. (A–C) Asterisk depicts unspecific protein detected by the α-FKBP5 antibody. (D) Schematic representation of Fkbp5 and Fkbp4 domains (FK indicates FKBP12-like domain; TPR, tetratricopeptide repeat). (E) Cell extracts from KH2-Flag-Ago2 and control KH2-Flag-Lin28 grown in dox-containing media were subjected to Flag IP. Associated proteins were detected by Western blotting using α-FKBP4 antibody. Note that Flag-IP panel is the same as Figure 1D. (F) KH2-Flag-Ago2 ESCs were pretreated with 50 nM of either control siRNA or si-Fkbp4 for 48 h. Subsequently, 1 μg/mL dox was added for 24 h. Whole-cell extract was analyzed for Flag-Ago2 and Fkbp4 levels by Western blotting. Actin was used as loading control. The numbers on the bottom indicate the relative Flag-Ago2 abundance. (G) V6.5 ESCs were treated with 50 nM of either si-Fkbp4 or control siRNA for 60 h. Whole-cell extract was analyzed for endogenous Ago2 and Fkbp4 levels by Western blotting. Actin was used as loading control. The numbers on the bottom indicate the relative Ago2 abundance. (H) MEFs were treated with 50 nM of control siRNA, si-Fkbp5, or si-Fkbp4 for 60 h. Whole-cell extract was analyzed for endogenous Ago2, Fkbp5, and Fkbp4 levels by Western blotting. Actin was used as loading control. The numbers on the bottom indicate the relative Ago2 abundance. An asterisk depicts unspecific protein detected by the α-FKBP5 antibody.
FK506 impairs the loading of miRNAs into Ago2
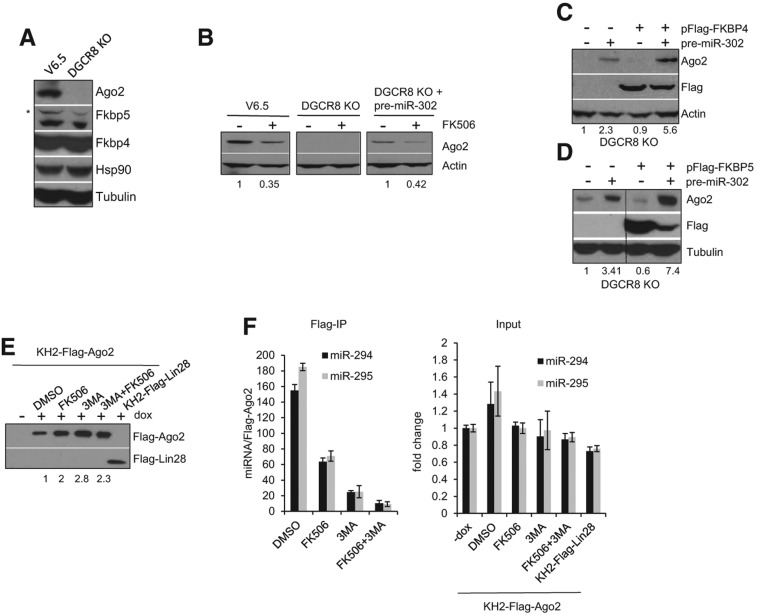
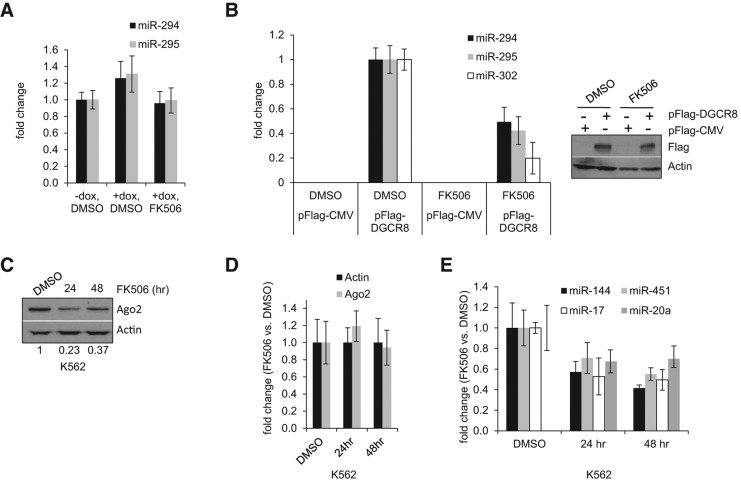
We have previously shown that miRNA ligands post-transcriptionally regulate the levels of Ago2 and that unloaded Ago2 protein is unstable (Martinez and Gregory 2010). Similar results were also recently reported in Drosophila (Smibert et al. 2013). DGCR8-deficient mouse ESCs, which lack mature miRNAs, display dramatically reduced Ago2 levels compared with their wild-type counterparts (Fig. 4A). Levels of the co-chaperone machinery, including Hsp90, Fkbp5, and Fkbp4, do however not depend on cellular miRNA levels since DGCR8 Knock Out (KO) and V6.5 wild-type ESCs have comparable levels of these proteins (Fig. 4A). Ago2 protein level in DGCR8 KO ESCs can be rescued by restoring miRNA levels, for instance, by transfection of small RNA duplexes or pre-miRNAs (Martinez and Gregory 2013). In agreement with our previous findings, DGCR8 KO ESCs displayed low levels of Ago2 compared with wild-type ESCs (Fig. 4B), and Ago2 levels were restored by transfection of pre-miR-302a (Fig. 4B). Interestingly, and consistent with a role for Fkbp4/5 in Ago2 loading, this miRNA-dependent stabilization of Ago2 protein expression was blocked by FK506 treatment (Fig. 4B). We additionally performed gain-of-function experiments in DGCR8 KO ESCs and found that overexpression of Fkbp4 (Fig. 4C) or Fkbp5 (Fig. 4D) resulted in an increase in Ago2 levels only when miRNA expression was rescued by overexpression of pre-miR-302a. We have previously shown that overexpression of HSP90 in DGCR8 KO cells increased Ago2 levels only when miRNA levels had been restored (Martinez and Gregory 2013). Altogether, these results implicate Fkbp4/5 and the Hsc70/Hsp90 complex in the loading of Ago2 with miRNA duplexes and the consequent stabilization of Ago2 expression.
FIGURE 4.
FK506 impairs the miRNA loading and stabilization of Ago2. (A) Whole-cell extracts from V6.5 and miRNA-deficient DGCR8 KO ESCs were assessed for endogenous Ago2, Fkbp5, Fkbp4, and Hsp90 protein levels by Western blotting. An asterisk depicts unspecific protein detected by the α-FKBP5 antibody. Tubulin serves as loading control. (B) V6.5, DGCR8 KO, and DGCR8 KO ESCs previously transfected with pre-miR-302a (30 nM final) for 24 h were treated with (10 µM) FK506. Ago2 levels in total cell extracts were analyzed by Western blot. Tubulin serves as loading control. The numbers on the bottom indicate the relative Ago2 abundance. (C) DGCR8 KO ESCs were co-transfected with 20 nM of pre-miR-302a and 0.5 ng/μL of either empty pFlag-CMV2 or pFlag-FKBP4 expressing constructs as indicated. Ago2 and Flag-FKBP4 levels in total cell extracts were analyzed by Western blot. Actin serves as loading control. (D) As in C except Fkbp5 was expressed and tubulin serves as loading control. The numbers on the bottom indicate the relative Ago2 abundance. (E) FK506 impairs miRNA loading into Ago2. KH2-Flag-Ago2 cells were treated for 24 h with 1 μg/mL dox and either DMSO, (5 µM) FK506, (5 mM) 3-MA, or combination of FK506 and 3-MA as indicated. The control KH2-Flag-Lin28 cell line was treated with 1 μg/mL dox for 24 h. Cell extracts were subsequently subjected to Flag-affinity purification. Flag-IP eluates were diluted as in Figure 1G to obtain comparable amounts of Flag tagged protein for each condition. Numbers on the bottom indicate the relative levels of immunoprecipitated Flag-Ago2 for each condition compared with DMSO control. (F, left) Associated RNA from Flag IP eluates in E was analyzed by TaqMan qRT-PCR for the indicated miRNAs. For each condition, the miRNA/protein ratio was calculated by dividing the fold enrichment of the indicated miRNA (compared to −dox treatment) by the Flag-Ago2 protein level (relative to DMSO). Error bars, mean ± SD with n = 3. (Right) Total RNA from the cell lysate (input) assessed for levels of indicated miRNAs by TaqMan qRT-PCR. U6 was used for normalization control. Error bars, mean ± SD with n = 3.
To directly investigate whether FK506 inhibits loading of miRNAs into Ago2 complexes, we isolated Flag-Ago2 containing ribonucleoprotein complexes from KH2-Flag-Ago2 treated with control (DMSO) or FK506. Since unloaded Ago2 is unstable and gets degraded in cells via the autophagy pathway, we also treated cells with the autophagy inhibitor 3-methylamphetamine (3-MA) to overcome the degradation of unloaded Ago2. We isolated associated RNAs from normalized levels of Flag-Ago2 (or Flag-Lin28 negative control) affinity-purified complexes and quantified miRNA levels by qRT-PCR (Fig. 4E,F). Consistent with the reported role of autophagy pathway in degrading unloaded Ago2, we found that Ago2 complexes isolated from 3-MA–treated cells contained reduced miRNA levels. This result is expected due to the stabilization of unloaded Flag-Ago2 protein. FK506 treatment alone modestly (∼25%) reduced the level of associated miRNAs compared with DMSO treatment (Fig. 4F). However, if we simultaneously inhibited Ago2 loading and stabilized unloaded Ago2 by treating cells with both FK506 and 3-MA, we found greatly diminished levels of Ago2-associated miRNAs (Fig. 4F). Importantly, this dramatic reduction in loaded Ago2 is not simply due to a reduced availability of miRNAs in the FK506- and 3-MA–treated cells since the overall abundance of miRNAs in these lysates was unaltered by these treatments (Fig. 4F, right panel). These results provide direct evidence that (1) unloaded Ago2 can be stabilized by inhibition of the lysosome and (2) FK506 treatment blocks Ago2 loading in mouse ESCs.
FK506 treatment leads to decreased miRNA levels
Levels of Ago proteins within the cell have been shown to influence global miRNA levels. For instance, it has been reported that overexpression of Ago proteins increases mature miRNA levels, while Ago2-deficient MEFs and hematopoietic cells display reduced levels of mature miRNAs (Diederichs and Haber 2007; O'Carroll et al. 2007). In addition, it has been reported that in sqn mutant plants, the levels of certain miRNAs are reduced compared with wild-type plants (Smith et al. 2009). We therefore next examined whether treatment of ESCs with FK506 would also lead to changes in miRNA levels. We treated KH2-Flag-Ago2 ESCs with dox in the presence or absence of FK506. Addition of dox to induce overexpression of Flag-Ago2 modestly increased the levels of miR-294 and miR-295, two miRNAs expressed specifically in ESCs (Fig. 5A). Simultaneous addition of dox and FK506 decreased miRNA levels to the basal level (−dox) (Fig. 5A). Although miRNA levels increased upon Flag-Ago2 overexpression and decreased upon addition of FK506, these changes were very modest (note similar results in Fig. 4F, right panel). This could be due to either the relatively low level of Flag-Ago2 overexpression compared with endogenous Ago2 levels in this system and/or the possibility that the level of miRNAs required to stabilize ectopic Ago2 is limiting in these cells. Moreover, the relatively long half-life of preloaded Ago2-miRNA complexes might mask effects of inhibiting Ago2 loading over the relatively short time-course of these experiments. We therefore utilized a more sensitive experimental system where we could specifically monitor the effect of FK506 treatment on the expression of nascent miRNAs. DGCR8 KO ESCs were transfected with either a control pFlag-CMV plasmid or a rescuing pFlag-DGCR8 plasmid together with the indicated control DMSO or FK506. As expected, FK506 abrogated the accumulation of mature miRNAs in DGCR8-rescued cells, presumably by blocking miRNA association with Ago2 in the RISC (Fig. 5B).
FIGURE 5.
FK506 treatment leads to decreased miRNA levels. (A) Total RNA from KH2-Flag-Ago2 ESCs treated for 24 h with 1 μg/mL dox and either (10 µM) FK506 or control DMSO was assessed for miR-294 and miR-295 levels by TaqMan qRT-PCR. snoRNA142 was used for normalization control. Error bars, mean ± SD with n = 3. (B) Dgcr8 KO ESCs were transfected with either empty vector (pFlag-CMV2) or a pFlag-DGCR8 expressing construct for 10 h and subsequently treated with either (10 µM) FK506 or control DMSO for 24 h. (Left) Total RNA was assessed for miR-294, miR-295, and miR-302a levels by TaqMan qRT-PCR. snoRNA142 was used for normalization control. Error bars, mean ± SD with n = 3. (Right) Whole-cell extracts were assessed for Flag-DGCR8 levels by Western blotting. (C) K562 cells treated with DMSO or (10 µM) FK506 for 24 h or 48 h. Whole-cell extracts were analyzed for endogenous Ago2 by Western blotting. Actin was used as loading control. The numbers on the bottom indicate the relative Ago2 abundance. (D) Total RNA from samples in C was assessed for Ago2 transcript level by qRT-PCR. Actin was used for normalization control. Error bars, mean ± SD with n = 3. (E) Total RNA from samples in C was assessed for levels of the indicated miRNAs by TaqMan qRT-PCR. snoRNA142 was used for normalization control. Error bars, mean ± SD with n = 3.
Of the four Ago proteins (Ago1–4) in mammals, Ago2 is the only one that retains endonuclease activity encoded by its PIWI domain (Rivas et al. 2005). This endonucleolytic activity is required for the biogenesis of miR-451, a miRNA important for erythropoiesis (Cheloufi et al. 2010). Human K562 cells are of the erythroleukemia type and express high levels of miR-451. We therefore treated K562 cells with FK506 to inhibit co-chaperone function. As in mouse cells, FK506 treatment led to decreased Ago2 protein expression (Fig. 5C) with no effect on Ago2 mRNA levels (Fig. 5D). In these cells, however, a more substantial effect on the steady-state miRNA levels was also observed (Fig. 5E). FK506 treatment led not only to reduced miR-451 levels but also to reduced levels of miR-144, a miRNA expressed from the same locus as miR-451 but whose maturation does not require Ago2 (Cheloufi et al. 2010). Likewise, miR-17 and miR-20a were down-regulated in FK506-treated samples. Hence, the reduction in miRNA levels we observed after co-chaperone inhibition and concomitant decrease in Ago2 protein levels most likely reflects a destabilization of mature miRNAs and does not depend on Ago2 catalytic activity. This is in agreement with a recent report indicating that Ago proteins stabilize mature miRNAs independently of active endonucleolytic activity (Winter and Diederichs 2011). The more substantial effect of FK506 treatment on miRNA expression in these cells compared with mouse ESCs might reflect a more rapid turnover of miRNAs in K562 cells and/or a greater dependence on Ago2 compared with other Ago family members in erythroid lineages (O'Carroll et al. 2007).
DISCUSSION
In this study, we identify Fkbp4 and Fkbp5 as novel components of the Ago2 RISC loading complex. This assertion is based on the RNA-independent physical association of Fkbp5 with Ago2-containing complexes isolated from mouse ESCs. Since unloaded Ago2 is unstable in cells and degraded by the lysosome, inhibition of this interaction with FK506 treatment or Fkbp4/5 knockdown leads to decreased Ago2 levels, and FK506 treatment blocks the miRNA-mediated stabilization of Ago2. Moreover, direct examination of the miRNA content of Ago2 complexes isolated from cells simultaneously treated with a lysosome inhibitor and FK506 revealed the accumulation of unloaded Ago2 complexes. Finally we provide evidence that FK506 treatment impacts miRNA expression. Thus our data provide new insight into the factors required for RISC loading and the control of Ago2 expression with implications for RNAi.
The role of Hsp90 has been well studied in the context of steroid receptors. It has been shown that Hsp90 functions to open the receptor's ligand-binding cleft. To do so, Hsp90 dynamically binds, in addition to the receptor, a set of cofactors and co-chaperones that are part of this assembly machinery. First, Hsp90 binds to the client proteins with ordered assistance of Hsp40, Hsp70, and the TPR domain-containing protein Hop. Then, Hop is replaced by other TPR domain-containing co-chaperones, also called immunophilins, such as Fkbp5, Fkbp4, Cyp40, and PP5 (Pratt and Toft 1997, 2003; Grad and Picard 2007). These co-chaperones enter into steroid receptor complexes through a direct and competitive binding at the C terminus of Hsp90 via their essential TPR domains (Cheung-Flynn et al. 2003). Hsp90 is a major component of Ago-containing complexes and functions to assist Ago2 in the loading of small RNA ligands (Iki et al. 2010; Iwasaki et al. 2010; Miyoshi et al. 2010). If unloaded, Ago2 is unstable and gets degraded via a mechanism that likely involves the autophagy pathway (Gibbings et al. 2012; Martinez and Gregory 2013). Our findings point to the Hsp90 co-chaperones Fkbp4/5 as bona fide Ago2 interacting partners in mouse ESCs, which is in agreement with a recently published large-scale proteomic study (Frohn et al. 2012). In that report, the investigators employed a large-scale mass spectrometry analysis to identify Ago2-binding proteins in the presence or absence of miRNAs in mouse ESCs. However, the significance of this interaction remained unexplored. While our manuscript was in preparation, another study found Fkbp4 as an Ago2-associated factor in HeLa cells and that Fkbp4 knockdown compromises RNAi efficiency (Pare et al. 2013). In contrast to our findings, however, Fkbp4 knockdown did not seem to influence Ago2 levels (Pare et al. 2013). We find that siRNA mediated knockdown of Fkbp4/5 results in reduced Ago2 protein levels. Moreover, we find that FK506 abrogates the interaction between Ago2 and Fkbp4/5 and renders Ago2 unstable. It is then tempting to speculate that Fkbp4/5 functions to assist Hsp90 in Ago2 loading. Consistent with this idea, dual treatment of KH2-Flag-Ago2 ESCs with FK506 and the autophagy inhibitor 3-MA, severely impairs the association of Ago2 with miRNAs. Our model proposes that Fkbp4/5 associate with Ago2–Hsp90 complexes to help open the small RNA binding cleft to facilitate small RNA loading and then dynamically dissociate after siRNA/miRNAs are incorporated. Alternatively, Fkbp4/5 could associate with Hsp90 to regulate other aspects of Ago2 function, such as protein folding. Indeed, the Hsp90 machinery has been reported to control client proteins at many levels, including folding, function, and turnover. Future studies are needed to undercover the precise mechanism of Fkbp4/5 action.
We found that Fkbp5 preferentially associates with Ago2- but not Ago1-containing complexes. A report by Johnston et al. (2010) showed that Hsp90 inhibition by geldanamycin treatment results in significant reduction of Ago1 protein levels in HeLa cells, indicating that, similar to Ago2, Ago1 loading is assisted by Hsp90. Similarly, our unpublished data suggest that Ago1 protein is also destabilized by 17-AAG treatment in V6.5 cells; however, in contrast to Ago2, Ago1 is not destabilized in the absence of miRNAs, since DGCR8 KO ESCs display even higher levels of Ago1 protein compared with wild-type ESCs. Altogether, these findings would indicate that different Ago proteins could potentially associate with distinct Hsp90/co-chaperones complexes or have different affinities for the same Hsp90/co-chaperone complex. A comprehensive analysis of Ago1- and Ago2-associated co-chaperones would be necessary to address this question.
Is the PPIase activity of Fkbp4/5 important for Ago2 regulation? FK506 binds to active PPIase domains, and since FK506 treatment reduces Ago2 levels, it could be argued that Fkbp5 isomerase activity is important for Ago2 regulation. However, mutagenesis analysis of plant Cyp40 indicates that both the PPIase and TPR domains are essential for facilitation of AGO1 loading, but the PPIase activity itself may not be as important in this regard (Iki et al. 2012). Similarly, Fkbp6 reportedly plays a role in the biogenesis of piRNAs even though it possesses an inactive PPIase domain (Olivieri et al. 2012; Preall et al. 2012; Xiol et al. 2012). Future studies are needed to conclusively determine a role for Fkbp4/5 PPIase activity in Ago2 regulation, but an interesting starting point might be the two prolines (P63 and P622) that are in the cis-configuration in the RNA-bound Ago2 structure (Elkayam et al. 2012; Schirle and MacRae 2012).
Although our mass spectrometry analysis only found Fkbp5 binding to Ago2 in ESCs, it is feasible that Ago2 binds to other TPR-containing co-chaperones. Indeed, plant AGO1 was shown to associate with multiple TPR-containing proteins, albeit in vitro (Iki et al. 2012). We have extended our analysis to Fkbp4, the closest Fkbp5 homolog. Similar to Fkbp5, Fkbp4 associates to and regulates Ago2 protein levels. Moreover, double knockdown of Fkbp5 and Fkbp4 in MEFs indicates the existence of some level of redundancy between both co-chaperones in the regulation of Ago2. fkbp5 KO and fkpb4 KO mice have been previously generated to study the in vivo role of co-chaperones in the regulation of the androgen receptor. Although fkpb5 KO mice appear normal, fkbp4 KO mice have defects in male reproductive organs, likely due to the role of Fkbp4 in androgen signaling. However, double fkbp5/fkbp4 KO are embryonic lethal (dying before embryonic day 7.5), indicating that Fkbp5 and Fkbp4 are functionally redundant with respect to an early cellular function that is critical to embryonic development (Yong et al. 2007). In the future, it will be interesting to address the individual as well as combinatorial contribution of all different TPR-containing co-chaperone to mammalian Ago2 stability and function. Finally, we have shown that human K562 cells also down-regulate Ago2 and miRNA levels when exposed to FK506, indicating that Ago2 regulation by Hsp90-associated co-chaperones is likely a highly conserved mechanism.
MATERIALS AND METHODS
Cell culture, cell lines, and inhibitor treatment
ESCs (V6.5, DGCR8 KO, KH2-Flag-Ago2, and KH2-Flag-Lin28) were maintained in DMEM with ESGRO (1000 units/mL; Millipore no. ESG1107), supplemented with antibiotics (GIBCO no. 15140) and with 15% (v/v) Stasis Stem Cell fetal bovine serum (GEMINI no. 100-125). MEFs were maintained in DMEM supplemented with antibiotics and 10% (v/v) fetal bovine serum (GIBCO no. 26140). K562s were maintained in RPMI media. Dox-inducible KH2-Flag-Lin28 and KH2-Flag-Ago2 cells have been previously described (Hagan et al. 2009; Chang et al. 2012). To induce expression of Flag-tagged transgenes, dox was added to the media at 1 μg/mL for the indicated period of time. Rapamycin (Sigma no. R0392) and 17-AAG (17-demethoxy-17-allylaminogeldanamycin; LC LABS no. A-6880) were dissolved in DMSO and added to cells for 24 h to a final concentration of 5 µg/mL and 20 µM, respectively. FK506 (Sigma no. F4679) was also dissolved in DMSO and added to cells for 24 h to a final concentration of 10 μM, unless otherwise noted.
DNA constructs and RNA
The cDNA of human FKBP5 and FKBP4 was PCR amplified from the Human ORFeome Collaboration Clones (Open Biosystems item nos. OHS5894-99860672 and OHS5894-99170149) using the primers hFKBP4 Fw, 5′-CATAAGATATCGACAGCCGAGGAGATGAAGGCGACC-3′, and Rv, 5′-GCGCTCTAGATTATGCTTCTGTCTCCACCTGAGACT-3′; hFKBP5 Fw, 5′-CATAAGATATCGACTACTGATGAAGGTGCCAAGAAC-3′, and Rv, 5′-GCGCTCTAGATTATACGTGGCCCTCAGGTTTCTCTTC-3′ and cloned into the EcoRV and XbaI sites of pFlag-CMV2. The constructs used in Figure 1E were obtained as follows: pIRESneo-Flag/HA-AGO2 and pIRESneo-Flag/HA-AGO1 were obtained from Addgene (plasmid nos. 10822 and 10820, respectively), and Flag-DGCR8 and pFlag-Lin28 have been previously described (Gregory et al. 2004; Viswanathan et al. 2008). Precursor miRNA for mmu-miR-302a was purchased from Ambion. Control (D-001810-01), mmu-Fkbp4 (si-GENOME mFkbp4 set of 4 no. D-062559-01-04), and mmu-Fkbp5 (si-GENOME SMARTpool mFkbp5 no. M-040262-00-0005 and si-GENOME mFkpb5 set of 4 no. MQ-040262-00-0002) siRNAs were purchased from Dharmacon.
Cell transfections
Lipofectamine 2000 (Invitrogen no. 11668-019) was used according to the manufacturer's instructions to transfect both DNA and RNA into ESCs. Lipofectamine RNAiMAX (Invitrogen, no. 56531) was used to transfect MEFs. Otherwise noted, cells were collected 48 h after transfection.
Cell extracts, Flag IP, and antibodies
Cells were harvested in lysis buffer (20 mM Tris-HCl at pH 8.0, 137 mM NaCl, 1 mM EDTA, 1% [v/v] Triton X-100, 10% [v/v] glycerol, 1.5 mM MgCl2, 1 mM DTT, 0.2 mM PMSF). For Flag IP, Flag-tagged proteins were affinity-purified using α-Flag M2 Agarose beads (Sigma) and washed six times with BC-100 buffer (20 mM Tris at pH 7.6, 100 mM KCl, 10 mM β-mercaptoethanol, 10% [v/v] glycerol, 0.2% [v/v] Nonidet P-40, 0.2 mM PMSF) before elution with 0.5 mg/mL Flag peptide. For the co-IP experiment with RNase A treatment, 20 μg/mL of RNase A was added to whole-cell extracts and incubated overnight at 4°C prior to IP. Antibodies used in this study are listed as follows: α-Tubulin, (Abcam no. ab6046), α-Ago2 (Cell Signaling no. C34C6), α-Flag (Sigma no. A8592), α-HSP90 (Cell Signaling no. 4874), α-FKBP4 (ProteinTech Group no. 10655-1-AP), α-FKBP5 (ProteinTech Group no. 14155-1-AP), α-AGO1 (Sigma no. SAB4200084), a-Lin28A (Cell Signaling no. A177), and α-Actin (Sigma no. A2066). α-Pabpc1 was published previously (Kimura et al. 2009). All Western blots were developed by enhanced chemiluminescence and quantified using the ImageJ software (National Institutes of Health).
RNA quantification
Total RNA was isolated using TRIzol reagent (Invitrogen no. 15596-018). For miRNA analysis, 5 ng of total RNA was reverse transcribed using gene-specific stem–loop RT primers and Multiscribe reverse transcriptase (Applied Biosystems no. 4311235). The resulting cDNAs were mixed with TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems no. 4324018). Mouse snoRNA-142 was used as normalization control. For mRNA measurement, 1 μg of total RNA was reverse transcribed using random hexamers and SuperScript III reverse transcriptase (Invitrogen no. 18080). PCR products were detected using iQ SYBR Green Supermix (Bio-Rad no. 170-8880) using the following primers: mAgo2 Fw, 5′-TGCCATGGTACGAGAGTTGCTCAT-3′, and mAgo2 Rv, 5′-ACGATGAACGTGATTCCTGGCTGA-3′; mActin Fw, 5′-CAGAAGGAGATTACTGCTCTGGCT-3′, and mActin Rv, 5′-TACTCCTGCTTGCTGATCCACATC-3′; hAgo2 Fw, 5′-GGTCCGCGAGCTCCTCATCCAG-3′, and hAgo2 Rv, 5′-CAGAAGAGCCGGGTGTGGTGCC-3′; and hActin Fw, 5′-TGAAGTGTGACGTGGACATC-3′, and hActin Rv, 5′-GGAGGAGCAATGATCTTGAT-3′. Quantitative PCR for both miRNAs and mRNAs was performed using iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad).
ACKNOWLEDGMENTS
We thank Robert Blelloch (UCSF) for DGCR8 KO ESCs and Tadashi Baba (UT, Japan) for α-Pabpc1 antibody. We thank Robinson Triboulet for helpful comments on the manuscript and Piotr Sliz for analysis of published Ago2 structures. We thank Boston Children's Hospital/IDDRC Proteomics Core for mass spectrometry. R.I.G. was supported by a grant from the US National Institute of General Medical Sciences (NIGMS; R01GM086386). N.J.M. is supported by NIGMS post-doctoral fellowship (1F32GM095102).
Author contributions: N.J.M. designed and performed all of the experiments in Figures 1 through 5 with assistance from J.deR.B. H.-M.C. generated the KH2-Flag-Ago2 cells and performed the large-scale isolation of Ago2-containing complexes from mouse ESCs. R.I.G. and N.J.M. wrote the paper.
REFERENCES
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI 2012. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 3: 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W 2009. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 21: 452–460 [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung-Flynn J, Roberts PJ, Riggs DL, Smith DF 2003. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J Biol Chem 278: 17388–17394 [DOI] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L 2012. The structure of human argonaute-2 in complex with miR-20a. Cell 150: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohn A, Eberl HC, Stohr J, Glasmacher E, Rudel S, Heissmeyer V, Mann M, Meister G 2012. Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol Cell Proteomics 11: 1442–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O 2012. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol 14: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad I, Picard D 2007. The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol 275: 2–12 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokohama E, Mitsuhara IL, Meshi T, Ishikawa M 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Meshi T, Ishikawa M 2012. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J 31: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G 2010. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ishida K, Kashiwabara S, Baba T 2009. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod 80: 545–554 [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Lu KP, Finn G, Lee TH, Nicholson LK 2007. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 3: 619–629 [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Gregory RI 2010. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell 7: 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Gregory RI 2013. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA 19: 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G 2013. Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet 14: 447–459 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026 [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A 2007. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 47: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC 2009. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell 20: 3273–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Lapointe P, Hobman TC 2013. Hsp90 cochaperones p23 and FKBP4 physically interact with hAgo2 and activate RNA interference–mediated silencing in mammalian cells. Mol Biol Cell 24: 2303–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev 18: 306–360 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Toft DO 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 228: 111–133 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y 2008. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem 283: 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18: 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I 2000. Structure of TPR domain–peptide complexes: Critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 101: 199–210 [DOI] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ 2012. The crystal structure of human Argonaute2. Science 336: 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinars CR, Cheung-Flynn J, Rimerman RA, Scammell JG, Smith DF, Clardy J 2003. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc Natl Acad Sci 100: 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Yang JS, Azzam G, Liu JL, Lai EC 2013. Homeostatic control of Argonaute stability by microRNA availability. Nat Struct Mol Biol 20: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Willmann MR, Wu G, Berardini TZ, Moller B, Weijers D, Poethig RS 2009. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci 106: 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N, Carmichael JB, Hobman TC 2001. GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem 276: 43294–43299 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD 2004. A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ 2010. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 141: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Diederichs S 2011. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 8: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Wu B, Li P, Liu Y, Lou Z, Ding Y, Shu C, Ye S, Bartlam M, Shen B, Rao Z 2004. 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc Natl Acad Sci 101: 8348–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47: 970–979 [DOI] [PubMed] [Google Scholar]
- Yong W, Yang Z, Periyasamy S, Chen H, Yucel S, Li W, Lin LY, Wolf IM, Cohn MJ, Baskin LS, et al. 2007. Essential role for co-chaperone Fkbp52 but not Fkbp51 in androgen receptor-mediated signaling and physiology. J Biol Chem 282: 5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20: 397–405 [DOI] [PubMed] [Google Scholar]