Abstract
Connections between inflammation and diseases are suggested important in understanding the genetic mechanisms of diseases. However, studies on the functional cross-links between inflammation and disease genes are still in their early stages. We integrated the protein–protein interaction (PPI), inflammation genes, and gene–disease associations to construct a disease-inflammation network (DIN). We found that nodes, which are both inflammation and disease genes (namely inter-genes), are topologically important in the DIN structure. Via mapping inter-genes to PPI, we classified diseases into two categories, which are significantly different in Intimacy measuring the contribution of inflammation genes to the connections between disease pairs. Furthermore, we constructed a cross-talking subpathways network. As indicated, the cross-subpathway analysis shows great performance in capturing higher-level relationship among inflammation and disease processes. Collectively, The network-based analysis provides us a rather promising insight into the intricate relationship between inflammation and disease genes.
One of the major tasks for contemporary biology and medicine is to decipher the underlying mechanisms of human complex diseases. Inflammation has been proposed as the seventh hallmark of cancer1, which significantly contributes to different development stages of various diseases. Researches on the links between inflammation and disease genes are thus helpful to understand the complex nature of human genetic diseases. However, few systematic studies on the functional links have been reported.
During the past decades, great efforts have been dedicated to identifying disease-related genes, proteins, and metabolites, which directly or indirectly connect to each other through computational or experimentally validated interactions. In an early study on inflammatory diseases, such as rheumatoid arthritis and inflammatory bowel disease, Heller et al.2 identified disease-related genes by using cDNA microarrays. Later, with the advent of sequencing, Jones et al.3 implicated PALB2 as a susceptibility gene of pancreatic cancer with the use of exomic sequencing. An early systemic study of human disease genes completed by Wu et al.4, integrated human protein–protein interactions and known gene-phenotype associations to systematically predict disease genes related to various phenotypes. Turner et al.5 and Furney et al.6 also contributed to the studies of human disease genes. However, few studies have focused on the relationships among genes corresponding to different disease phenotypes. Goh et al.7 constructed a disease phenome network (human disease network, HDN) and a disease genome network (disease gene network, DGN), which indicated a common genetic origin of many diseases through disease-gene association pairs. Recently, inflammation, as an important factor in the initiation and progression of various diseases, has been generally accepted. Donoso et al.8 explored the role of inflammation in age-related macular degeneration. In another study on colorectal cancer, Itzkowitz et al.9 emphasized the important roles of inflammation. Nevertheless, none of the studies have systematically characterized the functional links between inflammation and disease genes at a network level.
In this study, we focused on inflammation, controlled by both environmental and genetic factors and which is one of the most pivotal factors in inducing various diseases, to study the functional cross-links between inflammation and disease genes, as well as the mechanisms of their action. We integrated the human PPI network, inflammation genes, and gene–disease associations to construct a disease-inflammation network (DIN) and then dissected the topological characteristics of the network. In order to describe the relationships between inflammation and disease genes from a systems perspective, we classified diseases into type I diseases, which are significantly associated with inflammation genes, and type II diseases, which are not. Subsequently, we defined Intimacy as the contribution of inflammation genes to the connections between one disease and another, which tends to be higher in type I diseases. Finally, we showed that inflammation genes make great contributions to connections between most genetic diseases, especially in the associations between infection and immunity, as well as between infection and cancer, at a network level and a subpathway network level.
Results
Construction of the DIN
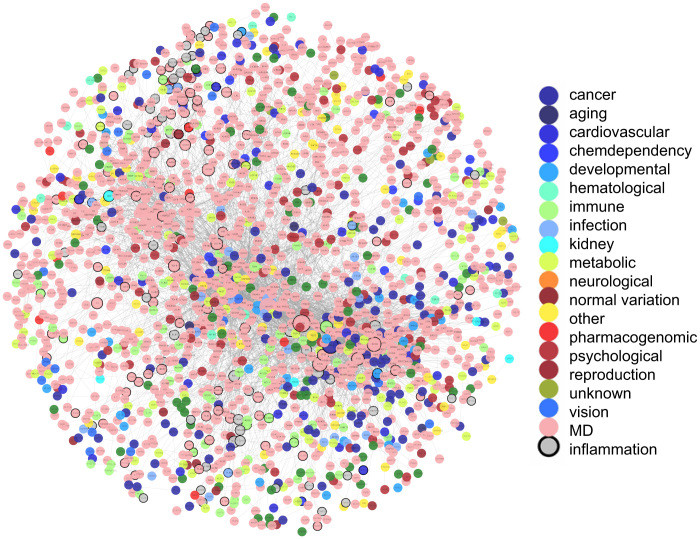
The local inflammatory microenvironment, which is essential for the initiation and progression of various diseases, can be depicted by representative inflammation genes. To interpret the functional links between genetic diseases and inflammatory microenvironment, we used disease and inflammation genes to construct a disease-inflammation network (DIN), integrating PPI information and gene–disease associations. There are 2831 disease-related genes curated by the GAD and 231 inflammation genes generated from the GO database10. We mapped all these genes to the PPI network of the HPRD and then extracted the maximal connected component as the DIN (Figure 1).
Figure 1. The disease and inflammation network (DIN).
The network is constructed by mapping disease genes and inflammation genes to the PPI network and then generating the maximal connected component as the DIN. Inflammation genes are colored in grey and disease genes are colored according to their classes. Those genes that are both inflammation genes and disease genes are drawn with black border. MD denotes genes involved in multiple disease classes.
As shown in Figure 1, the DIN contained 1867 nodes with 6252 interactions. Finally, 1815 disease genes and 160 inflammation genes were included.
Dissection of the DIN
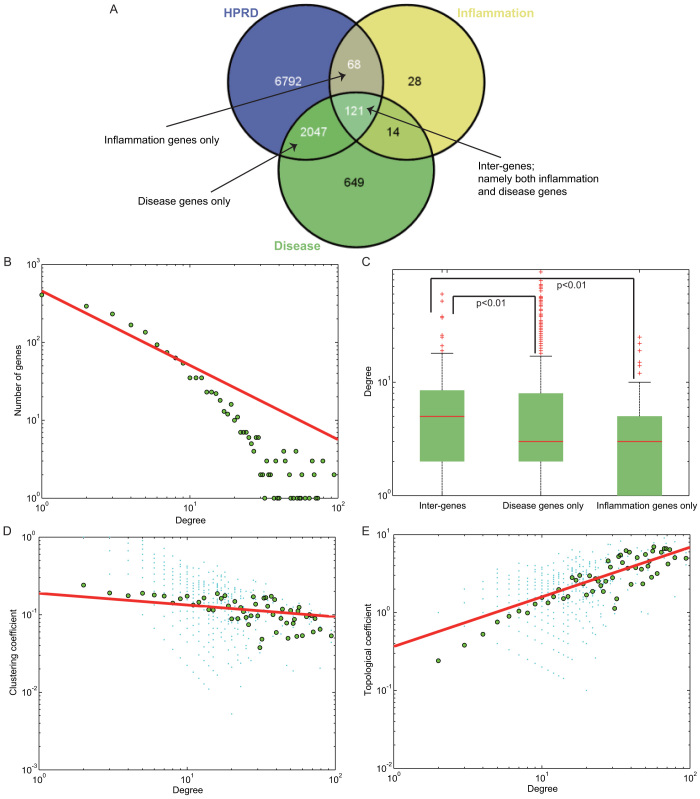
To delicately depict the functional cross-links between disease and inflammation genes in the network, we first examined the overlap among disease genes, inflammation genes, and PPI nodes (Figure 2A). As shown, we could classify the nodes in the DIN as disease genes only, inflammation genes only, or both disease and inflammation genes (henceforth, inter-genes). Subsequently, we determined the topological characteristics of the DIN, such as the degree, clustering, and topological coefficient. The degree distribution followed p(k) ∝ k−0.9536 using all the nodes in the DIN (Figure 2B), which showed that the DIN is a scale-free biological network. In a scale-free network11, most of the nodes have only a few interactions, whereas a few nodes with a large number of interactions are tended to be hubs. We further examined the degree distributions of three kinds of nodes in the DIN. As shown, the general degree distribution of nodes that are inter-genes was significantly greater than that of nodes that are disease genes only, with a p-value of 0.005 (Wilcoxon's Rank-Sum Test, Figure 2C). Similarly, it was also significantly higher, when comparing the degree distribution of inter-genes with inflammation genes only (p = 0.0012, Wilcoxon's Rank-Sum Test, Figure 2C). While those nodes in the DIN that are inflammation genes only had a median degree of 3, which is definitely equal to that of nodes that are disease genes only (p = 0.0628, Wilcoxon's Rank-Sum Test, Figure 2C). Besides, in order to properly control the analysis of inflammation genes and disease genes, we again compared them with non-disease genes in the original PPI network. As indicated, the general degree distribution is significantly higher when comparing inflammation genes with non-disease genes (p = 6.8293e-07, Wilcoxon's Rank-Sum Test, Supplementary Figure 1), as well as comparing disease genes with non-disease genes (p = 2.4687e-75, Wilcoxon's Rank-Sum Test, Supplementary Figure 1). Moreover, in the DIN, the shortest paths show a similar tendency just as the degree distributions for the three kinds of nodes, with nodes that are inter-genes having significantly shorter length of paths.
Figure 2. Topological analysis of the DIN.
The number of overlapping genes among the nodes of the PPI network, disease genes, and inflammation genes are shown in (A). Black arrow indicates the three categories of inter-genes, inflammation genes only and disease genes only in the DIN. (B) Degree distribution for all the nodes in the DIN is plotted on the x-axis, and the numbers of genes are plotted on the y-axis. (C) Degree distribution for three categories of nodes in the DIN. Significance tests were based on the Wilcoxon's Rank-Sum Test and p values for comparisons between inter-genes and disease genes only, and between inter-genes and inflammation genes only are both less than 0.01. Clustering coefficients (D) and topological coefficients (E) for all the nodes in the DIN are plotted on the y-axis, and the corresponding degrees are plotted on the x-axis.
The clustering coefficient of each node in the network is a measure of the tendency of nodes in a network to form clusters or groups12. In Figure 2D, we found that, with an increase in the node degree, the clustering coefficient decreases. In addition to clustering coefficient, the topological coefficient is also computed13, which is used to measure the extent to which a node shares links with the others in a network. As shown in Figure 2E, with an increase in the node degree, the topological coefficient increases, which was consistent with previous researches on biological network structures14,15,16. Additionally, in consideration of the accuracy of topological analysis, we also compared mean values of degree (Supplementary Figure 2), clustering coefficient (Supplementary Figure 3), and topological coefficient (Supplementary Figure 4) of all nodes in the DIN with random distribution, separately. 1000 random DINs were extracted from 1000 random PPI networks, using edge permutation with degree of each node in the original PPI unchanged. As shown, all these topological parameters were significantly higher as comparing with the random distributions.
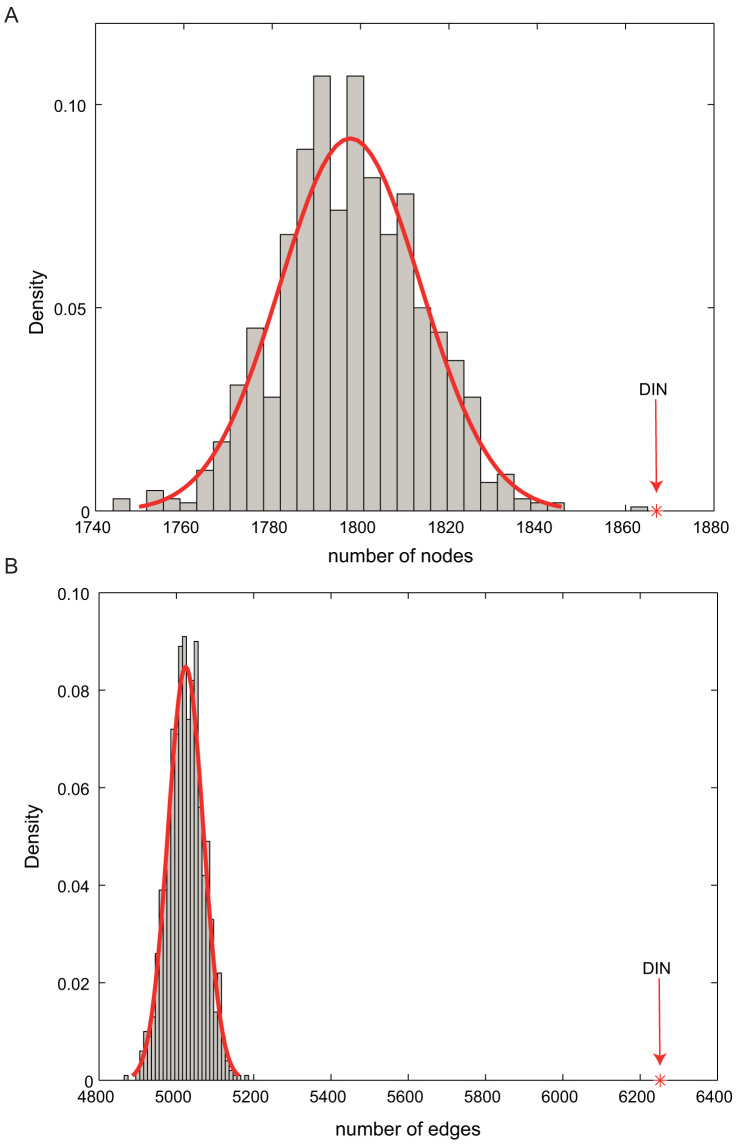
To further confirm the reality of the DIN, the number of nodes and edges in the real DIN were also compared with the random distribution. As shown in Figure 3, the average number of nodes and edges across 1000 random DINs were significantly smaller than that of the real DIN.
Figure 3. The further analysis of the DIN.
Density plot of the random number of nodes (A) and edges (B) in 1000 random DINs, with the real values of the number of nodes and edges in the DIN indicated by a red downward arrow.
Characterization of the functional cross-links between disease and inflammation genes at a network level
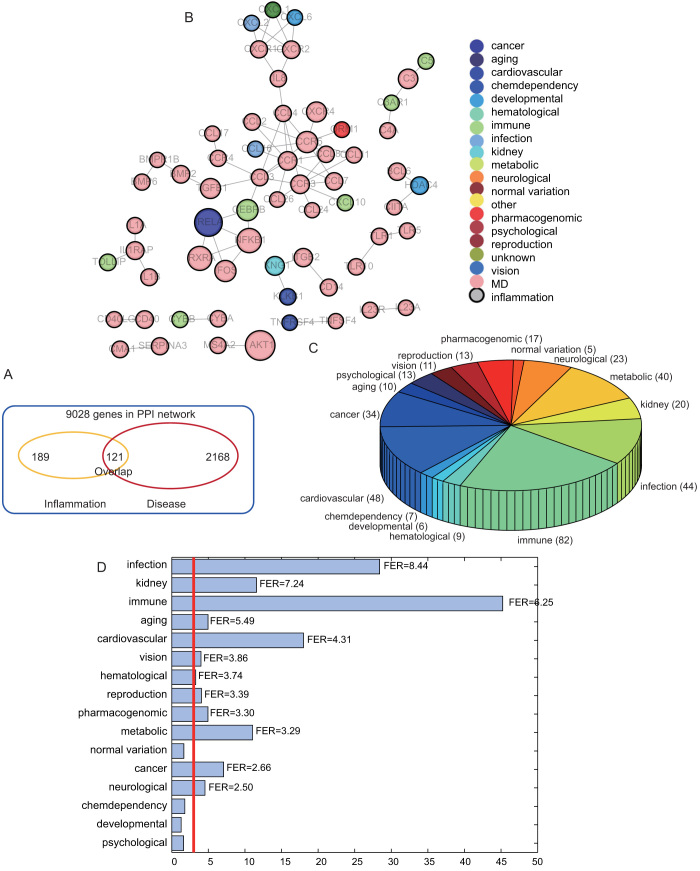
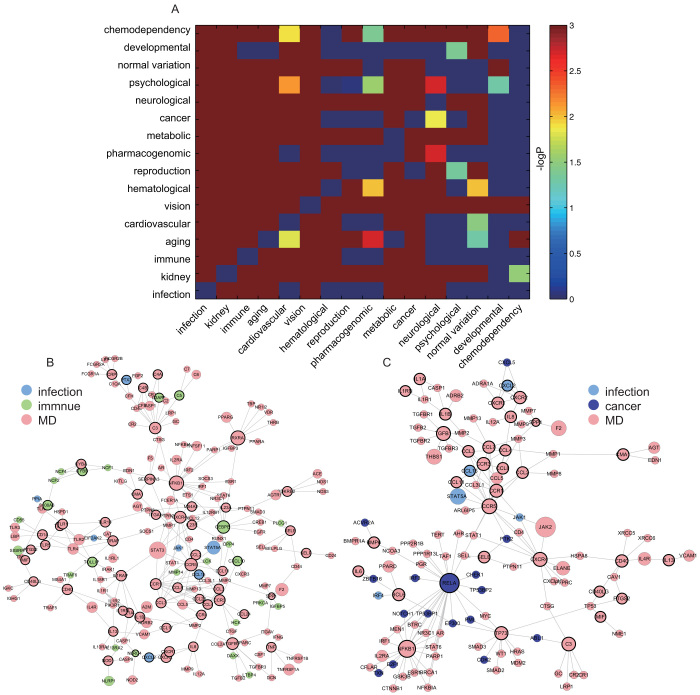
To explore the functional cross-links between disease and inflammation genes, we further mapped 121 inter-genes to the human PPI network and then calculated the significance of the overlap between inter-genes and the PPI network through hypergeometric distribution. With 121 inter-genes mapped to the human PPI network, a significant overlap was observed with a p-value of 3.6713e-32 (Figure 4A, hypergeometric distribution). Subsequently, we focused on the 121 overlapping genes, which formed a new network with the maximal connected component containing 32 genes and 49 relationships (Figure 4B). As shown by the result, inflammation genes tend to be associated with multiple disease genes, which indicates an important role of inflammation genes in contributing to genetic diseases in the network. We also examined the distribution of 121 genes into 18 disease classes according to the GAD classification system by excluding the classes “unknown” and “other” (Figure 4C). We showed that the class “immune” overlaps with most inflammation genes, which suggests that alterations in the focal inflammatory microenvironment are more associated with immunity. Furthermore, some inter-genes are shared by different disease classes, termed as multiple diseases (MD), as illustrated in Figure 4B, which shows the multiple functions of inflammation in various genetic diseases (Figure 4C).
Figure 4. Analysis for the importance of inter-genes at the level of network.
(A) The number of the overlapping genes between inflammation and disease in human PPI network. (B) A subnetwork was generated by mapping inter-genes to the human PPI network. (C) Distribution of inter-genes into different disease classes. (D) Fold enrichment ratios (FERs) were computed according to the overlapping between inflammation genes and genes from different disease classes.
To show the statistical significance of the overlap between inflammation and disease genes, the p-value and fold-enrichment ratios (FER) were calculated (Figure 4D). Only four classes, namely chemo-dependency, developmental, normal variation, and psychological disease, were not significant (p > 0.001). We termed these diseases as non-inflammation-related diseases (NIRD). On the contrary, those diseases with a significant overlap with inflammation were termed as inflammation-related diseases (IRD). In addition to the most highly associated class “immune”, the disease class cardiovascular, infection, and cancer were also in relation to inflammation, which has been supported by literature17,18. In order to measure the contribution of inflammation genes to the connections between one disease to another, we further examined the Intimacy of each pair of diseases in the two disease categories of IRD and NIRD (details in Methods). For each pair of diseases, we constructed 1000 random pseudo-inflammation gene sets, and then computed the random Intimacy values to construct a random distribution. By comparing the real Intimacy with the random distribution of Intimacy, we could define the significance of the Intimacy for the pair of diseases (Figure 5A).
Figure 5. Intimacy heatmap of disease pairs and examples indicating the functional cross-links between inflammation genes and disease genes.
(A) Examination of the Intimacy bridged by inflammation genes using all disease pairs from different disease classes. Diseases are ordered by their FER values (fold enrichment ratios), and minus 10-based logarithm p-value is showed as the right-side color bar. (B ~ C) Examples show the functional roles of inflammation genes in bridging the connections between disease pairs. Gene modules were generated from disease pair of infection and immunity, and disease pair of infection and cancer, separately.
As indicated by the results, Intimacy, among IRD tended to be higher than that in NIRD, which suggests the potential functions of inflammation genes in bridging the connection between each pair of diseases. Biologically, the computation of Intimacy is designed to take the direction of disease conversion between each pair of diseases into consideration. Therefore, in order to measure the general level of Intimacy between each disease pair, we ranked all disease pairs based on the sum of Intimacy for each disease pair with direction information (i.e. sum of Intimacy from disease A to B and that from disease B to A; Supplementary Table 1). The most associated disease classes bridged by inflammation genes are “immune” and “infection”, which has already been supported by literature19,20,21. Therefore, Intimacy defined by inflammation genes, which are also part of immune response and infection, contributing to the connections between immune and infection is unsurprisingly the most relevant one. Others ranking in the top 5 includes disease pair of cardiovascular and metabolic (Supplementary Figure 5)22,23, cardiovascular and immune (Supplementary Figure 6)24,25, and aging and cancer (Supplementary Figure 7)26,27, whose connections have already been shown in relation with the bridgeness of inflammation genes. Disease pair of normal variation and metabolic was suggested by us to be newly involved. As supported by the literature28,29, metabolic system is one of the most fundamental requirements for survival, whose proper function is mutually dependent on immune response. Inflammation could cause disequilibrium of the mutual dependence of metabolic and immune systems and then lead to chronic disorders of homeostasis. Beta-glucuronidase belonging to the disease class of normal mutation, which is generally known to be associated with inflammation in the exudates from gingival30. We therefore inferred the disease pair of normal variation and metabolic could be bridged by inflammation genes in proper conditions.
To further explain in detail the Intimacy bridged by inflammation genes, we focused on two specific disease pairs. The maximal connected component of the genes of each given pair of diseases was generated after mapping these genes to the human PPI network and then defined as the gene module of this pair of disease classes. We took the gene modules of two pairs of diseases as examples: the pair of infection and immunity (Figure 5B) and the pair of infection and cancer (Figure 5C). Generally, genes with higher degree, such as NFKB1 (Figure 5B), RXRA (Figure 5B), RELA (Figure 5C) and CCR5 (Figure 5C), tend to be hubs in the gene modules, and are believed to have much more impact on the global structure of the module networks. The transcription factor NFKB1 (Figure 5B) is the most abundant form of NF-kappa-B, which is complexed with the product of the gene RELA (Figure 5C). NF-kappa-B31,32 is a transcription factor that is activated by various intra- and extracellular stimuli, such as cytokines, oxidant free radicals, ultraviolet irradiation, and bacterial or viral products. The expression of genes regulated by Rel/NFKB members is involved in immunity and apoptotic and oncogenic processes; thus, NFKB133, as an inflammation gene, is important in linking infection and immunity. Because NF-KB plays a well-known function in the regulation of inflammation, we thus reasoned that inflammation bridges infection and immunity. Epidemiologic studies34,35 have shown that chronic inflammation predisposes individuals to various types of cancer. Furthermore, it is estimated that the underlying infections that could cause chronic inflammation are linked to approximately 15% of all deaths from cancer worldwide36. RELA, as an important inflammatory factor, is involved in linking infection and cancer through the mediator of inflammation37,38,39. We thus conclude that the inflammation genes NFKB1 and RELA are important in the link between infection and immunity, as well as between infection and cancer. Additionally, disease gene RXRA (Figure 5B) and CCR5 (Figure 5C) have been partially validated to be linked to the process of immune40,41 and infection, respectively42,43. Nevertheless, further researches on these two and some other genes are needed to understand the cellular and molecular mechanisms mediated by them underlying those complex diseases.
Further dissection of the functional cross-links based on disease- and inflammation-related subpathways
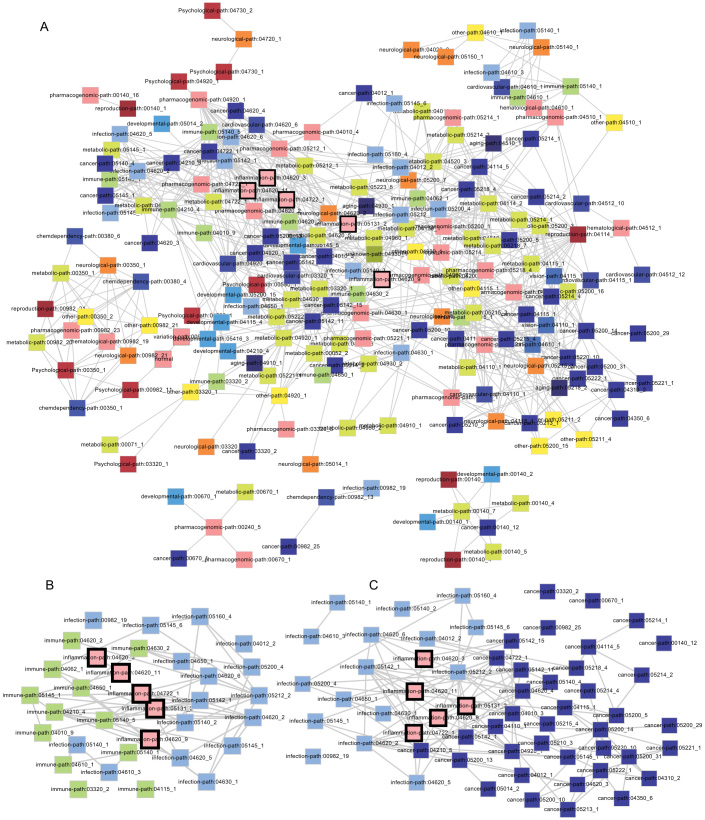
To further assess the functional cross-links between disease and inflammation genes, we constructed a subpathway–subpathway network based on disease-related subpathways, inflammation-related subpathways, and pathway structure data. Using the iSubpathwayMiner software package, disease class-related and inflammation-related subpathways were generated according to 15149 unique gene-disease associations involving 18 disease classes and 2831 disease genes. Any two subpathways that are significantly enriched for common genes were connected by an edge in the final subpathway–subpathway network if the gene overlapping between them was significant (p < 0.01, hypergeometric distribution). We thus constructed a subpathway–subpathway cross-talk network (Figure 6A) with 202 subpathway nodes and 716 edges.
Figure 6. The cross-talking subpathway network based on disease- and inflammation-related subpathways.
The whole subpathway–subpathway cross-talking network contained 202 subpathway nodes and 716 edges. The rectangles in the network correspond to disease- and inflammation-related subpathways. The nodes are colored according to their categories, which contains 18 disease classes obtained from the GAD database (A). (B ~ C) Examples of subpathway-supathway network showing functional connections bridged by inflammation genes. The network in (B) was generated by extracting inflammation-, immune-, and infection-related subpathways, and the network in (C) was generated through extracting inflammation-, cancer-, and infection-related subpathways.
In parallel with the two gene modules (Figure 5B and 5C) generated from the PPI network, two disease- and inflammation-related subpathway-based subnetworks extracted from the subpathway network were much more representative of the bridgeness of disease and inflammation genes. For example, five inflammation-related subpathways (path:04620_3, path:04620_11, path:04722_1, path:05131_2, and path:04620_9) mediated the connections between infection and immunity (Figure 6B). In agreement with Figure 5B, NFKB1 and RELA also emerged as the essential genes in linking infection and immunity when searching for the common genes shared by both inflammation- and infection-related subpathways, as well as inflammation- and immune-related subpathways. Some new genes were also included, such as IRAK4, IRAK1, and MYD88. Shared by multiple immune-, inflammation-, and infection-related subpathways, such as path:05145_1, path:04722_1, and path:04620_5, the inflammation gene IRAK144 is a critical mediator of innate immunity, which is also important in the immune response corresponding to viral infection45,46. Similarly, another IRAK family member, IRAK4, is also in control of the immune response to intracellular infection, such as Chlamydia pneumonia47. Furthermore, the sharing of path:05145_1, path:04620_3, and path:04620_11 by the inflammation gene MYD88 is also critical in the connection between infection and immunity mediated by inflammation. Takeuchi et al.48 confirmed that mice with MyD88 deficiency are highly susceptible to Staphylococcus aureus infection and that immune cells can be activated through the TLR7 MyD88-dependent signaling pathway49. With regard to the connection between infection and cancer mediated by inflammation (Figure 6C), the IL1B and TNF contained in path: 04620_9 are also implicated in the link between inflammation and cancer, in addition to the above-discussed inflammation genes NFKB1, RELA, MyD88, IRAK4, and IRAK1. Gene polymorphisms in IL1B and TNF could induce cancer risk through an inflammatory microenvironment in several different populations50,51. In addition, the NFKB1 and RELA that reside in the NF-kB node of path:04620_9 and the NF-kB signaling pathway that resides upstream of path:04620_9 are important in the tumor-promoting processes activated by inflammation and infection52.
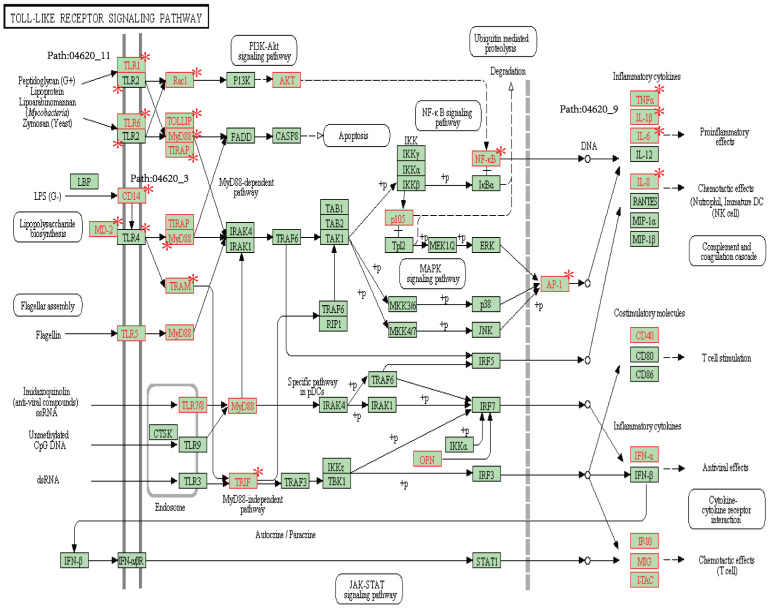
Interestingly, three subpathways (path:04620_3, path:04620_9, and path:04620_11) reside in the same KEGG pathway, path:04620 (Toll-like receptor signaling pathway; Figure 7), which is associated with infection, inflammation, and cancer53,54. Furthermore, two inflammation-related subpathways, path:04620_3 and path:04620_11, which are associated with immunity and infection mediated by MYD88, are located upstream of path:04620. Moreover, the subpathway path:04620_9 is located downstream of path:04620. Collectively, path:04620_9 is related to the promotion of tumor, which is tightly associated with an inflammatory microenvironment. This might thus suggest that path:04620 (Toll-like receptor signaling pathway) could be a good exhibitor of cancer progression activated by inflammation and infection.
Figure 7. Detailed information of subpathways (i.e. path:04620_3, path:04620_11, and path:04620_9) in the KEGG (i.e. path:04620).
Nodes marked by red asterisk are genes significantly overlapping with the corresponding subpathways.
Discussion
The identification of human disease-associated genes has long been of central importance in the study of human genetics. With the establishment of the human disease–disease network, a shift has been seen from the study of disease genes to the study of associations between various disease genes. Because the close relationship between inflammation and various disease phenotypes has been widely accepted55, studies of the roles of inflammation in carcinogenesis have emerged. However, systematic studies on the functional links remain in their early stages.
Inflammation contributes to the diverse progression of human complex diseases, such as immunity and cancer. To study the functional cross-links and the underlying mechanisms of their action, we integrated the PPI network, gene–disease information, and inflammation genes to construct a DIN network. By further dissecting topological parameters, such as the shortest paths of the DIN, we found that nodes that are inter-genes are important in the maintenance of the network structure, which is consistent with a previous study7. In the present study, we confirmed the topological importance of disease genes in the DIN. In addition, we showed that inter-genes, as both inflammation and cancer genes, are topologically important. After mapping inter-genes to the PPI network, we computed the FER values between inter-genes and different disease gene sets and then classified diseases as either IRD or NIRD. Based on the two classifications, we further examined the Intimacy of each pair of diseases mediated by inflammation genes. As shown by the results, the Intimacy of IRD was found to be higher than that of NIRD, which is a good indicator of the functional cross-links between inflammation and various disease phenotypes.
We comprehensively examined all the disease pairs via ranking them based on the summed Intimacy for each pair (i.e. sum the Intimacy from disease A to B and disease B to A; disease A and disease B belong to one disease pair; Supplementary Table 1). In total, we found that there were only one pair of inflammatory diseases that ranked in the top 5 of the table, together with other three pairs of inflammation-related diseases; their connections bridged by inflammation were all confirmed by other researches. In addition to those literature-supported pairs, the remaining pair ranking in the top 5 was newly found by us that might be involved in the bridgeness by inflammation genes in proper conditions (i.e. disease pair of normal variation and metabolic). As shown by the examples of gene modules extracted from the PPI network, inflammation was indeed found to be important in mediating between infection and immunity, as well as between infection and cancer. Collectively, as suggested by the results, our integrated approach could not only recur the connections bridged by inflammation genes between those well-known inflammatory disease pairs, but also predict new connections, which could therefore help us to study the functional roles of inflammation genes between disease pairs, via their structural importance at the level of network.
Additionally, we also constructed a subpathway–subpathway network based on disease- and inflammation-related subpathways to characterize the cross-links, as illustrated by the two examples of disease-related subpathways mediated by inflammation-related subpathways. Furthermore, this network-based analysis adds a new layer of complexity to the study of human diseases in that it considers inflammation as an important factor in the initiation and progression of diseases.
Methods
Data
A set of inflammation genes was obtained from the Gene Ontology categories “inflammatory response” (GO:0006954) and “regulation of inflammatory response” (GO:0050727), namely the human inflammation gene set containing 231 genes. The human disease gene set was generated on May 2012 from the Genetic Association Database (GAD, http://geneticassociationdb.nih.gov/), which includes 2831 disease genes corresponding to 18 disease classes. The database56 is an comprehensive archive of associated genes of human complex diseases and disorders, which also includes summary data extracted from published papers on candidate genes and GWAS studies. The human PPI network (http://www.hprd.org)57, which involves 9028 proteins with 35865 high-confidence interactions, is then used to construct DIN. The database seems the most integrated for human proteins in the public domain and widely used in scientific researches58,59.
Fold-enrichment ratio and Intimacy between each disease pair
The fold-enrichment ratio (FER) is defined as the ratio between the observed value and expected value (O/E ratio). An estimator is used to measure whether the observed overlap when mapping inflammation genes to the gene set of each disease class is large enough to be significant.
For the given pair of diseases dk and dj with corresponding disease gene sets gk1, …, gkm and gj1, …, gjn, the Intimacy is defined to describe the contribution of inflammation genes in bridging the connections between disease dk to dj. Considering the disease information passed from dk to dj based on the human PPI network connection, we can define how much dj is influenced by dk by treating genes related to dj as abnormal genes (upregulated or downregulated). Let I(dk → dj) denote the intensity with which dj is influenced by dk, as follows:
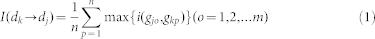 |
where n is the total number of disease genes of dj and 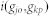 is the Intimacy between the disease pair. By using the network-based method, the shortest path method, we can define
is the Intimacy between the disease pair. By using the network-based method, the shortest path method, we can define 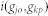 from the following transformation:
from the following transformation:
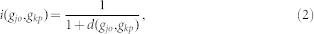 |
where 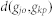 is the shortest path length between
is the shortest path length between 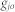 and
and 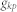 .
.
Generation of random networks
The PPI network was randomized 1000 times using edge permutation, with the degree corresponding to each node in the original PPI network kept unchanged. The edge permutation approach has been widely applied on various kinds of networks to generate randomized networks60. All 1000 random PPI networks were used to construct 1000 random DINs via mapping inflammation and disease genes to those random PPI networks and then extracting the maximal connected component, separately.
Random test and statistical analysis
In order to compute the significance of Intimacy bridged by inflammation genes, we constructed 1000 random pseudo-inflammation gene sets. Each pseudo-inflammation gene set contained the same number of genes as the real inflammation gene set, and each pseudo-inflammation gene had the same degree as the real one. Given a pair of diseases, we computed the real Intimacy bridged by the real inflammation gene set, and then computed a random distribution of Intimacy values using the 1000 pseudo-inflammation gene sets. Subsequently, we could define the significance of the Intimacy for the pair of diseases, via comparing with the random distribution of Intimacy.
The significance of the overlap between inflammation genes and disease genes against nodes of the human PPI network, and the overlap between gene sets from different subpathways, were computed by hypergeometric distribution as follows:
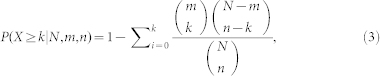 |
considering that a set of N elements has two subsets with m and n elements, respectively. We calculated the probability of containing at least k overlapping elements using the formula.
Construction of subpathway–subpathway network
We used the method that has been incorporated into the CRAN package iSubpathwayMiner (http://cran.r-project.org/web/packages/iSubpathwayMiner/) to identify the disease-related subpathways. In this method, the subpathway regions were located by lenient distance similarity of signature nodes within the pathway structure. Subsequently, we used hypergeometric test to identify disease-related subpathways.
Author Contributions
Y.P.Z. and H.H.F. conceived and designed the research, performed the molecular genetics manipulations and the experiments, analyzed data and wrote the paper. X.L. and Y.X.L. designed the research, analyzed the data and wrote the paper. Y.X. participated in the design of the study and contributed to the mathematical analysis of the data. J.X. contributed to the design of the study and helped to write the paper. Y.J.X. designed the research, analyzed data and wrote the paper. All authors read and approved the final anuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported in part by the Creative Research Groups of The National Natural Science Foundation of China (Grant No. 81121003, 91129710, 91029717, 61170154 and 31200997), the Specialized Research Fund for the Doctoral Program of Higher Education of China (Grant Nos. 20102307110022), the Science Foundation of Heilongjiang Province (Grant Nos. D200834 and C201207) and the Science Foundation of Heilongjiang Province Education Department (Grant Nos. YJSCX2012-230HLJ, Grant Nos.YJSCX2012-252HLJ).
References
- Colotta F., Allavena P., Sica A., Garlanda C. & Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- Heller R. A. et al. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci U S A 94, 2150–2155 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324, 217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Jiang R., Zhang M. Q. & Li S. Network-based global inference of human disease genes. Mol Syst Biol 4, 189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner F. S., Clutterbuck D. R. & Semple C. A. POCUS: mining genomic sequence annotation to predict disease genes. Genome Biol 4, R75 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney S. J., Alba M. M. & Lopez-Bigas N. Differences in the evolutionary history of disease genes affected by dominant or recessive mutations. BMC Genomics 7, 165 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K. I. et al. The human disease network. Proc Natl Acad Sci U S A 104, 8685–8690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso L. A., Kim D., Frost A., Callahan A. & Hageman G. The role of inflammation in the pathogenesis of age-related macular degeneration. Surv Ophthalmol 51, 137–152 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkowitz S. H. & Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol 287, G7–17 (2004). [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Tombor B., Albert R., Oltvai Z. N. & Barabasi A. L. The large-scale organization of metabolic networks. Nature 407, 651–654 (2000). [DOI] [PubMed] [Google Scholar]
- Soffer S. N. & Vazquez A. Network clustering coefficient without degree-correlation biases. Phys Rev E Stat Nonlin Soft Matter Phys 71, 057101 (2005). [DOI] [PubMed] [Google Scholar]
- Xu J. & Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics 22, 2800–2805 (2006). [DOI] [PubMed] [Google Scholar]
- Xu J. & Li Y. Discovering disease-genes by topological features in human protein–protein interaction network. Bioinformatics 22, 2800–2805 (2006). [DOI] [PubMed] [Google Scholar]
- Stelzl U. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 (2005). [DOI] [PubMed] [Google Scholar]
- Ravasz E., Somera A. L., Mongru D. A., Oltvai Z. N. & Barabási A.-L. Hierarchical organization of modularity in metabolic networks. Science 297, 1551–1555 (2002). [DOI] [PubMed] [Google Scholar]
- Infection, inflammation, and mental illness. The body's immune response to infectious agents may imperil the brain. Harv Ment Health Lett 26, 1–3 (2009). [PubMed] [Google Scholar]
- Grivennikov S. I., Greten F. R. & Karin M. Immunity, inflammation, and cancer. Cell 140, 883–899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Lawrence T. & Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell 124, 823–835 (2006). [DOI] [PubMed] [Google Scholar]
- Fox J. G. et al. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces helicobacter-induced gastric atrophy. Nature medicine 6, 536–542 (2000). [DOI] [PubMed] [Google Scholar]
- Lindholt J. S. & Shi G.-P. Chronic inflammation, immune response, and infection in abdominal aortic aneurysms. European journal of vascular and endovascular surgery 31, 453–463 (2006). [DOI] [PubMed] [Google Scholar]
- Manabe I. Chronic inflammation links cardiovascular, metabolic and renal diseases. Circ J 75, 2739–2748 (2011). [DOI] [PubMed] [Google Scholar]
- Tracy R. P. Inflammation, the metabolic syndrome and cardiovascular risk. Int J Clin Pract Suppl 10–17 (2003). [PubMed] [Google Scholar]
- Cengel A. Epicardial adipose tissue, metabolic syndrome, inflammation, and cardiovascular risk. Turk Kardiyol Dern Ars 40, 696–698 (2012). [DOI] [PubMed] [Google Scholar]
- Lin G. M., Chu K. M. & Han C. L. The influence of inflammation outweighing the metabolic syndrome on cardiovascular risk and mortality. Int J Cardiol 146, 431 (2011). [DOI] [PubMed] [Google Scholar]
- Caruso C., Lio D., Cavallone L. & Franceschi C. Aging, longevity, inflammation, and cancer. Ann N Y Acad Sci 1028, 1–13 (2004). [DOI] [PubMed] [Google Scholar]
- Vasto S. et al. Inflammation, ageing and cancer. Mech Ageing Dev 130, 40–45 (2009). [DOI] [PubMed] [Google Scholar]
- Hotamisligil G. S. Inflammation and metabolic disorders. Nature 444, 860–867 (2006). [DOI] [PubMed] [Google Scholar]
- Navab M., Gharavi N. & Watson A. D. Inflammation and metabolic disorders. Curr Opin Clin Nutr Metab Care 11, 459–464 (2008). [DOI] [PubMed] [Google Scholar]
- Bang J., Cimasoni G. & Held A. J. Beta-glucuronidase correlated with inflammation in the exudate from human gingiva. Arch Oral Biol 15, 445–451 (1970). [DOI] [PubMed] [Google Scholar]
- Jiang Z., Wu W. & Qian M. L. Cellular damage and apoptosis along with changes in NF-kappa B expression were induced with contrast agent enhanced ultrasound in gastric cancer cells and hepatoma cells. Cancer Cell Int 12, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. E., Boesch-Saadatmandi C., Dose J., Schultheiss G. & Rimbach G. Anti-inflammatory potential of allyl-isothiocyanate--role of Nrf2, NF-(kappa) B and microRNA-155. J Cell Mol Med 16, 836–843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Ross E. J., Wang Y. & Horwitz B. H. Nfkb1 inhibits LPS-induced IFN-beta and IL-12 p40 production in macrophages by distinct mechanisms. PLoS One 7, e32811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- Nathan C. & Ding A. Nonresolving inflammation. Cell 140, 871–882 (2010). [DOI] [PubMed] [Google Scholar]
- Balkwill F. & Mantovani A. Inflammation and cancer: back to Virchow? Lancet 357, 539–545 (2001). [DOI] [PubMed] [Google Scholar]
- Yazdani S. et al. Nuclear factor kappaB1/RelA mediates the inflammation and/or survival of human airway exposed to sulfur mustard. J Recept Signal Transduct Res 31, 367–373 (2011). [DOI] [PubMed] [Google Scholar]
- Steinbrecher K. A., Harmel-Laws E., Sitcheran R. & Baldwin A. S. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J Immunol 180, 2588–2599 (2008). [DOI] [PubMed] [Google Scholar]
- Algul H. et al. Pancreas-specific RelA/p65 truncation increases susceptibility of acini to inflammation-associated cell death following cerulein pancreatitis. J Clin Invest 117, 1490–1501 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang J., Wu Y. & Li J. The regulation of thymic stromal lymphopoietin in gut immune homeostasis. Dig Dis Sci 56, 2215–2220 (2011). [DOI] [PubMed] [Google Scholar]
- Clement S., Pascarella S. & Negro F. Hepatitis C virus infection: molecular pathways to steatosis, insulin resistance and oxidative stress. Viruses 1, 126–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. G. et al. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces helicobacter-induced gastric atrophy. Nat Med 6, 536–542 (2000). [DOI] [PubMed] [Google Scholar]
- Choe H. et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85, 1135–1148 (1996). [DOI] [PubMed] [Google Scholar]
- Gottipati S., Rao N. L. & Fung-Leung W. P. IRAK1: a critical signaling mediator of innate immunity. Cell Signal 20, 269–276 (2008). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S. Innate immune recognition of viral infection. Nat Immunol 7, 131–137 (2006). [DOI] [PubMed] [Google Scholar]
- Saito T. & Gale M. Jr Regulation of innate immunity against hepatitis C virus infection. Hepatol Res 38, 115–122 (2008). [DOI] [PubMed] [Google Scholar]
- Trumstedt C. et al. Role of IRAK4 and IRF3 in the control of intracellular infection with Chlamydia pneumoniae. J Leukoc Biol 81, 1591–1598 (2007). [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Hoshino K. & Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol 165, 5392–5396 (2000). [DOI] [PubMed] [Google Scholar]
- Hemmi H. et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 3, 196–200 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang W. H., Wang X. L., Zhou J., An L. Z. & Xie X. D. Association of interleukin-1B (IL-1B) gene polymorphisms with risk of gastric cancer in Chinese population. Cytokine 30, 378–381 (2005). [DOI] [PubMed] [Google Scholar]
- Garza-Gonzalez E. et al. Role of the polymorphic IL-1B, IL-1RN and TNF-A genes in distal gastric cancer in Mexico. Int J Cancer 114, 237–241 (2005). [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 441, 431–436 (2006). [DOI] [PubMed] [Google Scholar]
- Chen K. et al. Toll-like receptors in inflammation, infection and cancer. Int Immunopharmacol 7, 1271–1285 (2007). [DOI] [PubMed] [Google Scholar]
- Killeen S. D., Wang J. H., Andrews E. J. & Redmond H. P. Exploitation of the Toll-like receptor system in cancer: a doubled-edged sword? Br J Cancer 95, 247–252 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens L. M. & Werb Z. Inflammation and cancer. Nature 420, 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. G., Barnes K. C., Bright T. J. & Wang S. A. The genetic association database. Nat Genet 36, 431–432 (2004). [DOI] [PubMed] [Google Scholar]
- Li Z. et al. Characterization the regulation of herpesvirus miRNAs from the view of human protein interaction network. BMC Syst Biol 5, 93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbe R. K., Koyuturk M. & Chance M. R. An integrative -omics approach to identify functional sub-networks in human colorectal cancer. PLoS Comput Biol 6, e1000639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K. et al. Identification of the proliferation/differentiation switch in the cellular network of multicellular organisms. PLoS Comput Biol 2, e145 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R. et al. Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information