Abstract
The non-invasive high-resolution spatial mapping of cell metabolism within tissues could provide substantial advancements in assessing the efficacy of stem cell therapy and understanding tissue development. Here, using two-photon excited fluorescence microscopy, we elucidate the relationships among endogenous cell fluorescence, cell redox state, and the differentiation of human mesenchymal stem cells into adipogenic and osteoblastic lineages. Using liquid chromatography/mass spectrometry and quantitative PCR, we evaluate the sensitivity of an optical redox ratio of FAD/(NADH + FAD) to metabolic changes associated with stem cell differentiation. Furthermore, we probe the underlying physiological mechanisms, which relate a decrease in the redox ratio to the onset of differentiation. Because traditional assessments of stem cells and engineered tissues are destructive, time consuming, and logistically intensive, the development and validation of a non-invasive, label-free approach to defining the spatiotemporal patterns of cell differentiation can offer a powerful tool for rapid, high-content characterization of cell and tissue cultures.
The ability of stem cells to self-renew and differentiate into specialized cell types presents a number of opportunities for treating or regenerating tissue compromised by disease or trauma1. However, the complex relationships among different signaling pathways, the extracellular microenvironment, and metabolic requirements of the cell, present a significant challenge in efficiently controlling stem cell differentiation in stem cell therapy and tissue engineering1,2. To assess cell differentiation and function at specific time points within tissues, techniques such as Western blots, quantitative polymerase chain reaction (qPCR) and immunohistochemistry (IHC), are most commonly utilized3,4. Although such approaches are highly sensitive to specific cell characteristics, their destructive nature does not allow for dynamic or real-time assessments of cell function within tissues. Non-invasive characterizations of cell function are possible through metabolomic and proteomic analyses of cell secretions5, but these techniques cannot provide a spatial map of functional development within inherently heterogenous populations. As a result, there is a need for a repeatable, non-destructive imaging approach capable of quantitatively mapping cell differentiation and function within intact three-dimensional (3D) tissues.
Two-photon excited fluorescence (TPEF) microscopy offers a number of advantages over traditional microscopy for imaging 3D biological samples. In TPEF microscopy, fluorophores that emit in the visible range are excited by the simultaneous absorption of two near-infrared (NIR) photons, which enables deeper light penetration of up to 0.5–1 mm, intrinsic depth sectioning, efficient light collection, and reduced out-of-focus photodamage compared to confocal microscopy6,7. Using NIR excitation, the endogenous fluorescence of nicotinamide and flavin adenine dinucleotides (NADH and FAD, respectively) can be detected, primarily emanating from mitochondria7,8,9. These enzymatic cofactors participate ubiquitously in cellular metabolism, coordinating the regulation of virtually every major metabolic pathway. The redox cofactors serve a particularly important role in mitochondria as electron carriers linking the tricarboxylic acid (TCA) cycle to the electron transport chain. While they exist in either oxidized (NAD+, FAD) or reduced (NADH, FADH2) forms, only NADH and FAD yield significant autofluorescence7,10. Despite an emerging understanding of the different metabolic profiles of stem cells and their differentiated progeny, the use of NADH and FAD fluorescence to analyze the metabolic basis of differentiation is just beginning.
Quantifying NADH or FAD autofluorescence and interpreting its relationship to cell metabolism can be challenging for a number of reasons. Additional cellular fluorophores can contribute to the measured signal intensity; NADPH, in particular, cannot be distinguished from NADH due to its similar fluorescence lifetime and emission spectrum7,11. Furthermore, when cofactors are bound to metabolic enzymes, NADH fluorescence quantum yield increases, while FAD quantum yield decreases, which causes variation in the measured fluorescence intensities12,13,14,15. Furthermore, NADH and FAD intensity can vary as a function of depth within 3D tissues due to the absorption or scattering properties of the tissue. In an important step to account for potential absorption or scattering effects, Chance et al. proposed the use of a ratio of FAD/NADH fluorescence as a measure of cell redox state13. A normalized variation of this ratio, FAD/(NADH + FAD), has also been utilized because it provides an upper and lower bound for ratio values, which is attractive for traditional statistical comparisons that assume a normal distribution7. These different variants of an optical redox ratio have been employed as a measure of metabolic activity in a range of studies over the last half century7,8,11,12,16,17, and optical redox ratios involving FAD and NADH fluorescence intensities have generally been assumed to be proportional to the more traditional oxidation-reduction ratio of NAD+/NADH13. However, this assumption of a direct relationship between any optical redox ratio and a traditional ratio of cofactor concentrations has not been robustly tested. Furthermore, because NADH and FAD are ubiquitously utilized throughout metabolic pathways, understanding the specific mechanisms that contribute to a change in the optical redox ratio remains unclear for many applications, such as monitoring stem cell status.
A number of studies have identified changes in stem cell autofluorescence characteristics during differentiation4,14,18,19. By defining a normalized fluorescence-based redox ratio of FAD/(NADH + FAD), putative metabolic changes have been detected in mesenchymal stem cell cultures upon the induction of adipogenic and osteogenic differentiation18. Similarly, by defining individual cell redox ratios within 3D adipose cultures, a transient decrease in redox ratio was identified during periods of lipogenesis4. Recent studies have also provided complementary information to traditional ratiometric fluorescence intensity imaging, by identifying characteristic changes in the fluorescence lifetime of NADH in differentiating stem cells14,20,21. These findings suggest changes in NADH fluorescence intensity may not be solely related to changes in cofactor concentration. Collectively, both intensity-based and lifetime-based studies provide phenomenological evidence of an optical sensitivity to stem cell differentiation, but no study has elucidated the underlying mechanisms of this putative sensitivity.
The goal of this study was to establish whether quantitative readouts of NADH and FAD autofluorescence relate to biologically-relevant changes in cell redox state and then to elucidate the metabolic pathways that produce such changes during stem cell differentiation. To this end, we (a) determined how changes in the normalized optical redox ratio of FAD/(NADH + FAD) fluorescence intensities are related to changes in the ratio(s) of intracellular cofactor concentrations using destructive mass spectrometry methods, (b) evaluated whether the optical redox ratio is associated with qPCR expression of traditional differentiation markers, and (c) assessed how the biosynthetic demands of adipogenic differentiation are reflected in both mitochondrial organization and redox state. By understanding how the optical redox ratio can be related to specific metabolic pathways, this study provides a framework to explore the utility of these non-invasive imaging methods for a wide range of applications.
Results
Optical redox ratio is associated with intracellular NAD+/(NADH + NAD+) concentrations
To isolate NADH and FAD fluorescence, bandpass emission filters centered at 460 nm and 525 nm (ET460/40M-2P and ET525/50M-2P, respectively; Chroma, Bellows Falls, VT) were selected for non-descanned detectors based on non-negative matrix factorization unmixing of stem cell emission spectra18. Due to the wide variation in fluorescence intensities among stem cell populations at different stages of differentiation, the relationships between detector gain, laser power, and fluorescence intensity had to be established to facilitate a quantitative analysis of fluorescence data within the dynamic range of our system (Supplementary Fig. S1). The normalized fluorescence intensities at each pixel of our images were used to produce maps of the optical redox ratio of FAD/(NADH + FAD) with subcellular resolution (Fig. 1). These pixel-wise normalized fluorescence data can also be averaged on a cell-by-cell or field-by-field basis to provide average redox ratios that are representative of individual cells or cultures.
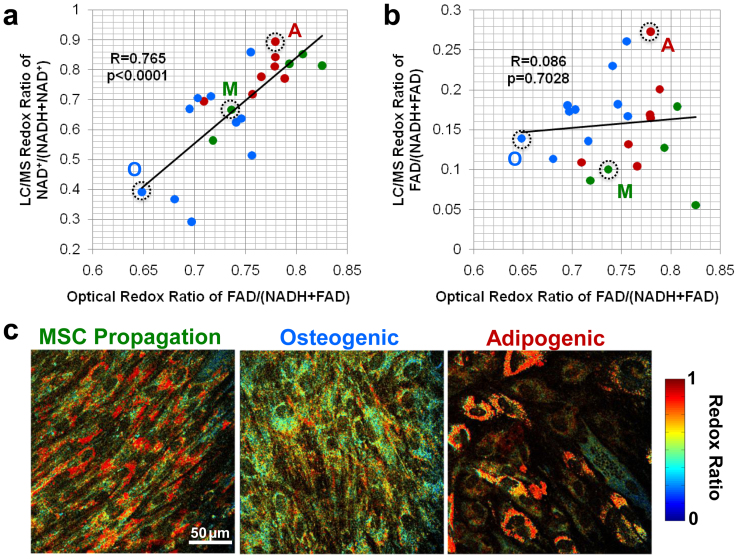
Figure 1. Comparison of optical redox ratio with intracellular cofactor concentration ratio measured through LC/MS-MS.
(a) The optical redox ratio is significantly correlated (R = 0.765, p < 0.0001) with the LC/MS-MS ratio of NAD+/(NADH + NAD+). (b) However, the optical redox ratio is not associated (R = 0.086, p = 0.7028) with the LC/MS-MS ratio of FAD/(NADH + FAD). Green points represent individual MSC propagation cultures, blue points represent osteogenic cultures, and red points represent adipogenic cultures. (c) Representative images from the cultures circled in (a) and (b) demonstrate overall differences in the average redox ratio among cultures, but also inter- and intra-cellular variability in the optical redox ratio, which is not measurable with LC/MS-MS.
To evaluate whether this optical redox ratio is associated with intracellular changes in NADH and FAD concentrations within differentiating stem cell cultures, the average optical redox ratio was compared with the ratio of intracellular cofactor concentrations extracted and measured using liquid chromatography/tandem mass spectrometry (LC/MS-MS) (Fig. 1). Human bone marrow-derived mesenchymal stem cell (MSC) cultures (n = 22) undergoing adipogenic differentiation, osteogenic differentiation, or MSC propagation at different post-induction time points were monitored through TPEF imaging and immediately processed to extract intracellular metabolites. The average optical redox ratio of each culture prior to metabolite extraction exhibited a significant correlation (R = 0.765, p < 0.0001) with the intracellular ratio of NAD+/(NADH + NAD+) across all treatment groups (Fig. 1a). Interestingly, the optical redox ratio was not significantly correlated (R = 0.086, p = 0.7028) with LC/MS-MS measurements of FAD/(NADH + FAD) (Fig. 1b), suggesting changes in FAD fluorescence measurements may not be directly related to changes in the total intracellular FAD concentrations due to the different fluorescence quantum yields among flavoproteins15. Yet the significant correlation of the optical redox ratio with LC/MS-MS measurements of NAD+/(NADH + NAD+) suggests those specific flavoproteins that are highly fluorescent with a high quantum yield may be directly reducing NAD+ to NADH.
Decrease in redox ratio coincides with the onset of differentiation
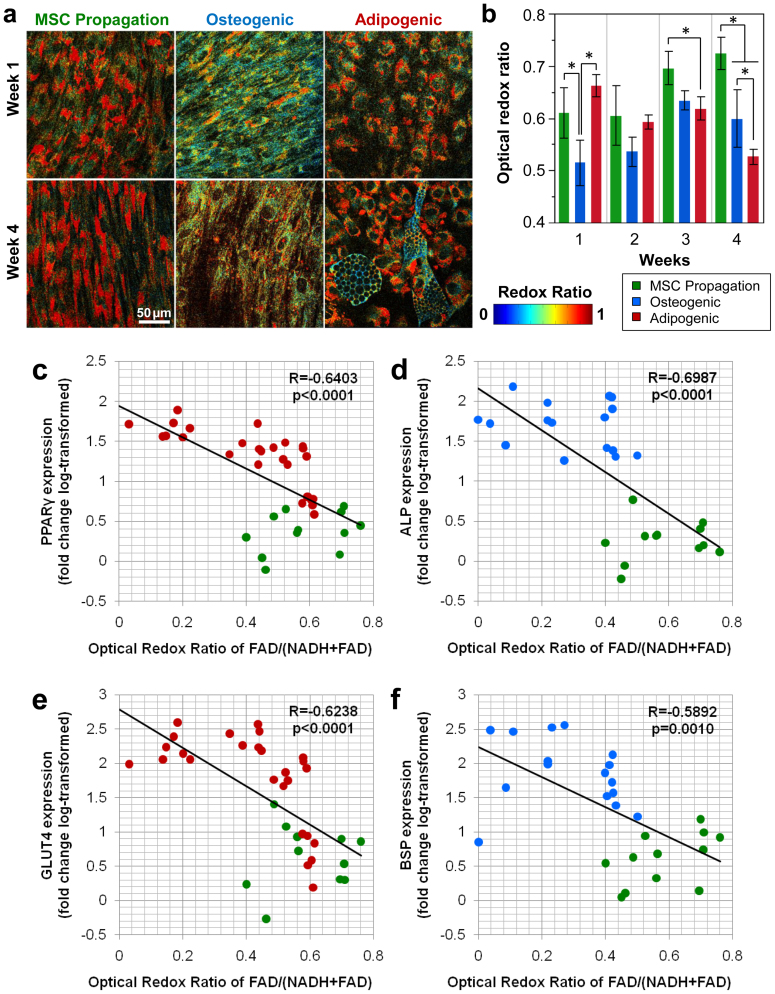
With the verification that the optical redox ratio during stem cell differentiation is correlated with the more traditional ratio of NAD+/(NADH + NAD+) concentrations, we sought to characterize the dynamic changes in redox ratio that occur upon differentiation during 4 weeks of culture in induction media (n = 4 cultures per group and time point). We found that the average optical redox ratio initially decreases on week 1 in osteogenic cultures (p = 0.0016) relative to the undifferentiated MSC group (Fig. 2b). Following this initial decrease, osteogenic cultures demonstrated a slight increase in redox ratio over time; however, a lower redox ratio (p < 0.0001) was still detected at week 4 relative to the MSC propagation group (Fig. 2b). Adipogenic cultures did not exhibit an immediate decrease in the optical redox ratio compared to MSC groups and were significantly higher than osteogenic cultures at Week 1 (p < 0.0001). However, the average optical redox ratio was lower on week 3 (p = 0.0257) and week 4 (p < 0.0001), than the corresponding MSC control groups. In fact, at Week 4, the average redox ratio of the adipogenic cultures was also significantly lower than osteogenic cultures (p = 0.0456). This decrease in the field-average redox ratios of adipogenic cultures was driven by a substantially lower redox ratio in cells demonstrating lipid droplet formation (Fig. 2a).
Figure 2. Decrease in optical redox ratio coincides with differentiation.
(a) Representative redox ratio maps and (b) field-averaged redox ratios demonstrate a decrease in redox ratio upon differentiation. Cultures in MSC propagation media demonstrate a slight increase in redox ratio over time, while osteogenic differentiation produces an immediate decrease in redox ratio at week 1 (p = 0.0016). By week 4 both osteogenic and adipogenic cultures exhibit lower redox ratios compared to MSC propagation cultures (p ≤ 0.0001). Error bars in (b) represent standard deviation. (c) PPAR-γ expression was significantly correlated (R = −0.6403, p < 0.0001) with the average optical redox ratio of adipogenic and MSC propagation cultures. (d) ALP expression in osteogenic and MSC propagation cultures was also significantly correlated (R = −0.6987; p < 0.0001) with average redox ratio. (e) GLUT4 expression (R = −0.6238, p < 0.0001) and (f) BSP expression (R = −0.5892, p = 0.0010) exhibited significant correlations with redox ratio as well.
Each week, cultures (n = 4 per group) were sacrificed immediately following imaging for analysis by qPCR for standard gene expression markers of adipogenic and osteogenic differentiation. Peroxisome proliferator-activated receptor gamma (PPAR-γ) and glucose transporter type 4 (GLUT4) expressions were quantified to assess adipogenic differentiation, whereas alkaline phosphatase (ALP) and bone sialoprotein (BSP) expression were quantified to assess osteogenic differentiation. The optical redox ratio significantly and negatively correlated with the gene expression markers (Fig. 2c–f). The strongest correlations between redox ratio and adipogenic and osteogenic differentiation were found with PPAR-γ (R = −0.6403, p < 0.0001) and ALP (R = −0.6987, p < 0.0001), respectively (Fig. 2c and 2d). In addition, GLUT4 (R = −0.6238, p < 0.0001), and BSP (R = 0.5892, p = 0.0010) demonstrated a correlation with decreased redox ratio (Fig. 2e and 2f).
Differentiation coincides with increased fractal clustering of mitochondria
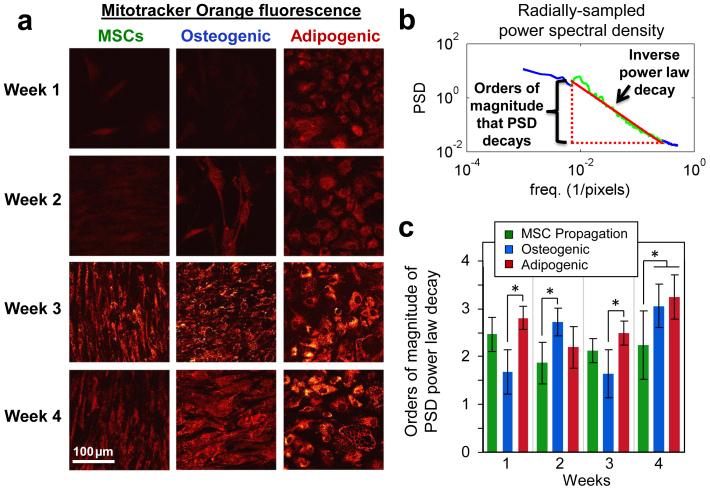
Previous studies have demonstrated that as stem cells differentiate, their mitochondria grow in response to their changing metabolic demands and elongate into organized networks throughout the cytosol22,23,24,25. To contextualize the changes in metabolism detected through the optical redox ratio, mitochondrial organization at each time point was characterized through Mitotracker Orange fluorescence images (Fig. 3a). An increase in Mitotracker Orange intensity is apparent over 4 weeks in both MSC propagation and differentiation groups, suggesting mitochondrial biogenesis (Fig. 3a). However with a number of potential factors affecting the absolute fluorescence intensity of the stain (e.g. membrane potential, mitochondrial density), we focused on quantifying changes in the spatial organization of mitochondria associated with differentiation. We have previously developed a simple, automated Fourier-based analysis method to quantify the fractal organization of mitochondria based on an inverse power law decay of power spectral density (PSD) with respect to spatial frequency26 (Fig. 3b). In this study, we measured the range of fractal clustering by calculating the orders of magnitude over which the PSD of Mitotracker images decayed according to a power law. Through this metric of mitochondrial organization, a significant increase in the range of fractal clustering was detected in adipogenic cultures at week 4 (p = 0.0037) and osteogenic cultures at weeks 2 (p = 0.0353) and 4 (p = 0.0497) compared to MSC propagation cultures at those time points. While Mitotracker staining was quantified in this study because it has traditionally been used to assess mitochondrial organization, both the intensity and fractal organization of NADH autofluorescence was correlated with the corresponding Mitotracker Orange outcomes reported above (p < 0.0001).
Figure 3. Mitochondrial organization changes upon stem cell differentiation.
(a) Mitotracker Orange fluorescence staining demonstrated an increase in intensity over time in all groups, and increased organization throughout differentiated cells. (b) The range of fractal clustering of the mitochondria was measured by identifying the spatial frequencies over which the power spectral density (PSD) decays according to an inverse power law and quantifying the orders of magnitude over which the PSD decays as a power law. (c) Osteogenic cultures on weeks 2 and 4, as well as Adipogenic cultures on week 4, exhibited a significantly higher range of fractal clustering compared to MSC propagation cultures (p ≤ 0.0497). Error bars represent standard deviation.
De novo fatty acid synthesis drives a decrease in redox ratio during adipogenic differentiation
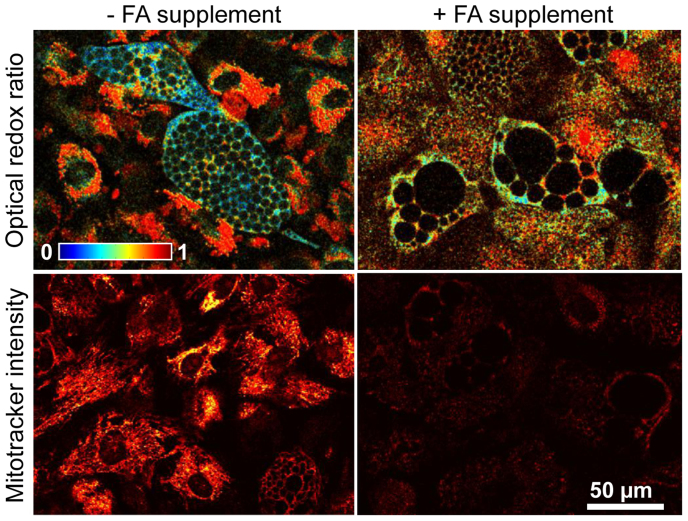
We hypothesized that the accumulation of mitochondrial NADH during adipogenic differentiation was due to the demand for fatty acids (FA) outpacing ATP demand. To test this hypothesis, exogenous FAs were added to the adipogenic induction media (1 mM oleic acid) to obviate the need for de novo FA synthesis27. Although no consistent differences in redox ratio or mitochondrial organization were detected between adipogenic cultures with and without the FA supplement during the first 3 weeks, substantial differences between the groups were observed with the increased presence of maturing adipocytes on week 4. By week 4, FA supplemented adipogenic cultures exhibited larger and substantially more lipid droplets compared to control adipogenic cultures without FA supplementation (Fig. 4). Consistent with our hypothesis, the decrease in redox ratio observed during adipogenic differentiation was significantly attenuated (p = 0.0002) in the FA supplemented cultures despite the larger lipid droplets (Fig. 4, Table 1). This attenuation of the decrease in redox ratio was not associated with reduced differentiation; PPAR-γ and GLUT4 expression were not significantly lower in the FA supplemented group at any time point (Table 1). Interestingly, FA supplemented cultures also demonstrated reduced Mitotracker Orange fluorescence intensities (p < 0.0001) and less mitochondrial organization as measured by the range of fractal clustering (p = 0.0013) on week 4 compared to normal adipogenic cultures (Table 1).
Figure 4. Exogenous fatty acids (FA) during adipogenic differentiation attenuated changes in mitochondrial function and structure.
Treatment with an oleic acid supplement attenuated the decrease in redox ratio normally observed in differentiating cells, despite an increase in lipid droplet size. Lipid droplets could easily be identified by the lack of autofluorescence in these spherical-shaped organelles. Additionally, mitotracker orange staining intensity was attenuated and the range of fractal clustering was reduced in the adipogenic group with a FA supplement.
Table 1. Summary statistics from adipogenic cultures after 4 weeks with and without the addition of a fatty acid (FA) supplement (Mean ± SD).
| Quantitative Metric | without FA supplement | with FA supplement | p value |
|---|---|---|---|
| PPAR-γ expression (fold change) | 29.6 ± 16.0 | 44.8 ± 7.8 | 0.6202 |
| GLUT4 expression (fold change) | 248.3 ± 104.1 | 109.2 ± 9.6 | 0.8880 |
| Average optical redox ratio | 0.527 ± 0.015 | 0.634 ± 0.032 | 0.0002 |
| Average mitotracker orange intensity (a.u.) | 29.17 ± 7.32 | 8.87 ± 2.25 | <0.0001 |
| Range of fractal clustering (orders of magnitude of PSD power law decay) | 3.25 ± 0.47 | 2.17 ± 0.27 | 0.0013 |
Cell-specific redox ratio measurements enable assessments of intra-culture heterogeneity
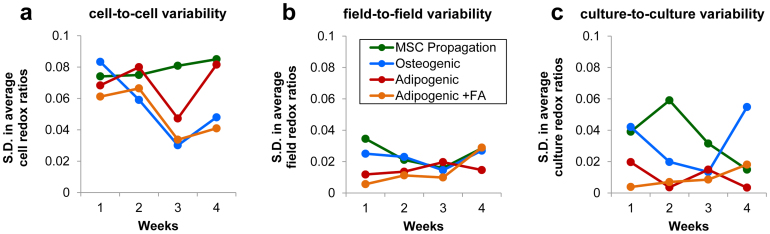
Unlike LC/MS-MS or qPCR measurements, TPEF-based optical redox ratio measurements provide the unique opportunity to calculate the inherent heterogeneity of cell metabolism and differentiation status within cell culture populations. This heterogeneity was quantified by computing cell-specific redox ratios within the individual 238 × 238 μm image fields acquired over 4 weeks of culture and measuring their standard deviation about the average redox ratio of the field (Fig. 5a). Furthermore, field-to-field variability of the redox ratio within individual cultures, as well as culture-to-culture variability within the different groups and time points can be measured (Fig. 5b and 5c). For each time point and cell culture group, variability in cell redox ratio measurements within individual fields exceeded the variability in average field measurements within individual cell cultures. This substantial cell-to-cell variability can be visualized within the representative redox ratio maps (Fig. 1c, 2a, and 4) and suggests a reasonable estimation of the percent of differentiated cells within a culture can be made from a few representative fields. To account for the inherent biological variability within these stem cell cultures during the correlative studies with LC/MS-MS and qPCR measurements, field-averaged redox ratio measurements were computed using 6–10 random fields in each culture. From the estimate of random effects in our ANOVA model of field-averaged redox ratio measurements, we find that field-to-field variability accounted for 39.7% of the total random effect variance, while culture-to-culture variability accounted for 60.3% of variance in this dataset. Collectively, these findings suggest that much of the biological variability within stem cell populations can be assessed by computing cell-specific redox ratios within a limited number of randomized image fields.
Figure 5. Variability in redox ratio measurements.
(a) The standard deviation (S.D.) of the cell redox ratio values within individual image fields is summarized for each group and time point. (b) The S.D. of field-averaged redox ratios within each individual culture demonstrates field-to-field variability is substantially lower than cell-to-cell variability within fields. (c) The S.D. in the average culture redox ratios within each group and time point are plotted, and demonstrate culture-to culture variability is similar to field-to-field variability.
Discussion
Although NADH and FAD autofluorescence have been used in a variety of applications over the last half century7,8,10,11,12,13 and recently been utilized to detect stem cell differentiation4,14,18,19,20,21, the biological relevance of an optically-derived redox ratio and the metabolic pathways that contribute to alterations in this ratio have not been rigorously evaluated. Using gold-standard LC/MS-MS measurements of intracellular cofactor concentrations, we have demonstrated the optical redox ratio is strongly associated with changes in cofactor concentrations (Fig. 1). Unlike LC/MS-MS measurements that require a destructive extraction protocol, the optical method described here can provide non-invasive, quantitative redox data with subcellular resolution in live cultures, which allows for the characterization of cell population heterogeneity at different length scales (Fig. 5). This approach can be adapted for use with any TPEF-capable microscope and relies solely on endogenous sources of fluorescence, which can enable immediate applications in the laboratory or the clinic. Adaptation of this technique to new stem cell applications in the lab and clinic requires an understanding of the underlying metabolic pathways that contribute to changes in the optical redox ratio. Although the optical redox ratio has frequently been summarized as inversely proportional to the metabolic rate of a cell and likely reflecting levels of oxidative phosphorylation17,18, we have identified a sensitivity specifically related to fatty acid synthesis during adipogenic differentiation in this study (Fig. 4). This suggests the need for a more nuanced interpretation of the biological significance of the optical redox ratio that considers multiple metabolic pathways. During stem cell differentiation, the optical redox ratio is sensitive to the balance between the rates of ATP consumption and glucose catabolism in a cell. Glucose catabolism produces the reduced, fluorescent form of NADH. In the absence of a proportional demand for ATP production through NADH oxidation to NAD+, this fluorescent NADH accumulates and the redox ratio decreases (Fig. 6).
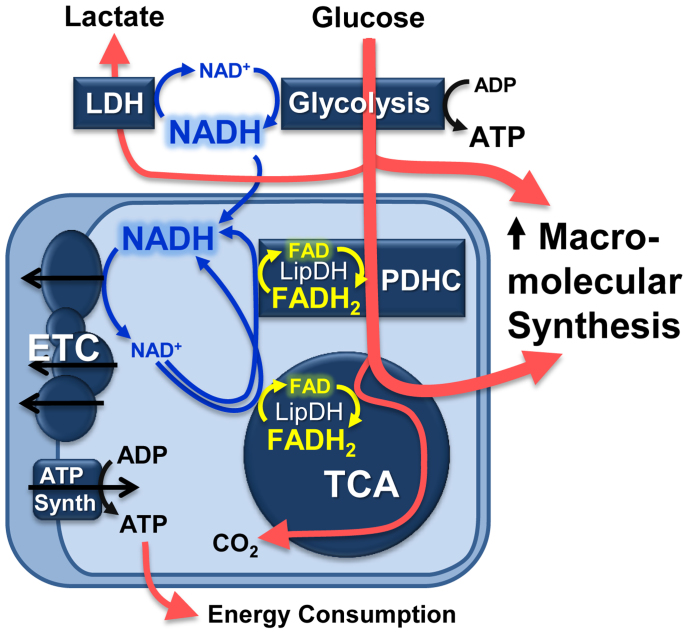
Figure 6. Schematic relating the increase in macromolecular biosynthesis during stem cell differentiation to changes in NADH and FAD concentrations.
When glucose catabolism outpaces ATP production due to the biosynthesis of macromolecules (e.g. fatty acids), NADH concentrations will increase and the concentration of FAD bound to enzyme complexes containing lipoamide dehydrogenase (LipDH) will decrease. These changes in NADH and FAD concentrations can be detected through the optical redox ratio as demonstrated in Figure 1, and result in a decrease in the optical redox ratio during the induction of differentiation in stem cell cultures. Abbreviations: LDH = lactate dehydrogenase, PDHC = pyruvate dehydrogenase complex, ETC = electron transport chain, TCA = tricarboxylic acid cycle.
The specificity of optical redox ratio measurements to the intracellular cofactor concentration ratio of NAD+/(NADH + NAD+) was quantitatively confirmed, for the first time, through LC/MS-MS correlations. Interestingly, the optical redox ratio is significantly correlated with the intracellular concentration ratio of NAD+/(NADH + NAD+) rather than FAD/(NADH + FAD) (Fig. 1). While this result is somewhat surprising, other groups have reported that when FAD is bound to most proteins, its fluorescence is substantially quenched15,28. Among these flavoproteins, lipoamide dehydrogenase (LipDH) protein complexes appear to be a major source of the overall flavin fluorescence emanating from cells15. LipDH serves as an intermediate electron carrier by transferring electrons produced by decarboxylation to local NAD+ in the mitochondria (Fig. 6). Therefore, the LipDH-containing complexes involved in glucose metabolism, such as pyruvate dehydrogenase and oxoglutarate dehydrogenase, have a flavin moiety that is in direct equilibrium with the local balance of NAD+ and NADH concentrations.
Protein binding can also affect the fluorescence quantum yield of NADH, with up to a ten-fold increase in the fluorescence intensity of mitochondrial protein bound NADH relative to free NADH13,29,30. Given such a large putative range in the quantum yield of bound and unbound NADH and the success of fluorescence lifetime imaging (FLIM) studies in detecting stem cell differentiation14,20,21,31, it is perhaps surprising that changes in the quantum yield of NADH do not substantially affect the strong correlation between the optical redox ratio and NAD+/(NADH + NAD+) concentration ratios (Fig. 1). However, those FLIM studies report a relatively small change (150–250 ps) in mean lifetime during stem cell differentiation21,31, which would suggest a change in the quantum yield of NADH that would account for only a small fraction (<10%) of the average change in the intensity-based redox ratio measured during differentiation in this study. While NADPH fluorescence cannot be distinguished from NADH using standard TPEF or fluorescence lifetime imaging, the fluorescence correlation with LC/MS-MS-derived concentration ratios suggests the NADPH contributions are also negligible in differentiating MSCs. Furthermore, the intracellular concentration of NADH was 33.7 ± 13.1 fold higher than the concentration of NADPH, based on additional LC/MS-MS assessments performed on a subset of cell extracts analyzed in this study. These findings provide additional support for previous work suggesting the majority of fluorescence emitted in the blue range is produced by protein-bound NADH in mitochondria, rather than by cytosolic NADH or NADPH11,32.
Undifferentiated, highly proliferative stem cells have a substantial need for biosynthesis, and glucose metabolites are diverted from the glycolytic pathway and the TCA cycle to facilitate glycoprotein, proteoglycan, and nucleotide synthesis even in the presence of sufficient oxygen22,23,33. The increase in glycolysis associated with biosynthesis can lead to an accumulation of NADH in the cytosol, which can be balanced by lactate dehydrogenase activity or transport into the mitochondria for oxidation33,34 (Fig. 6). As propagating stem cells reach confluence, they reach a quiescent state, and their anabolic demands decrease, likely causing the observed trend of increasing redox ratios over time (weeks 1 to 4, MSC propagation group) (Fig. 2). Compared to stem cells cultured with propagation media, the induction of osteogenic differentiation produces a lower redox ratio at week 1 (Fig. 2). Given that this change in redox ratio upon osteogenic differentiation occurs prior to any noticeable changes in mitochondrial organization, the underlying mechanisms of the redox ratio change may be related to changes in the concentrations of cofactors in the cytosol rather than the mitochondria. In fact, it has been suggested that a reduction in the cytosolic NAD+/NADH ratio is needed to inhibit ADP-ribosylation, which in turn promotes collagen synthesis35,36. Certainly, additional work is necessary to understand the underlying causes of this initial decrease in redox ratio during osteoblastic differentiation and its suggested association with collagen synthesis.
Unlike MSC proliferation and osteogenic differentiation, adipogenic differentiation, at least in vitro, is associated with an increase in flux through metabolic pathways with enzymes expressed in the mitochondria37,38. The typical pathway to facilitate de novo fatty acid synthesis involves glucose being broken down to pyruvate, shuttled into the mitochondria, re-assembled into citrate during the first steps of the TCA cycle, and exported out of the mitochondria to serve as a carbon supply for fatty acid and triglyceride synthesis (Fig. 6). This process would require the conversion of NAD+ to NADH by the LipDH-containing pyruvate dehydrogenase complex (PDHC). Carbon storage in lipid droplets through de novo fatty acid synthesis would likely not occur when substantial ATP production is necessary, and consequently the reduced form of NADH may accumulate. Indeed, a decrease in the optical redox ratio is detected upon adipogenic differentiation in this (Fig. 2) and previous studies4. Furthermore, it has been suggested that a lower baseline ratio of NAD+/NADH concentrations in the mitochondria may facilitate citrate transport out of the mitochondria rather than continued catabolism through the TCA cycle34. Studies have also demonstrated there is coordinated counter-regulation of lipogenesis and NADH oxidation in adipocytes through regulatory pathways anchored by adenosine monophosphate-activated protein kinase (AMPK)39. Collectively, these previous studies and the attenuation of changes in the redox ratio following the addition of exogenous fatty acids in the current study (Fig. 4, Table 1), support the hypothesis that our optical sensitivity to adipogenic differentiation is related to de novo fatty acid synthesis.
In addition to detecting changes in the cell redox state during stem cell differentiation, the microscopy techniques employed here can be utilized to characterize changes in mitochondrial organization that also reflect the metabolic demands of the cell. Using a variety of techniques, increases in mitochondrial content and organization have been detected upon stem cell differentiation22,23,24,25,40,41. Previous autofluorescence studies of adipogenic differentiation have measured a 2.7-fold increase in the average NADH fluorescence and 3.4-fold increase in FAD over 6 months further supporting the potential for mitochondrial biogenesis4. In the current study, we have identified increases in the range of fractal clustering of mitochondria after 4 weeks of differentiation (Fig. 3), which match qualitative descriptions of the elongation and maturation of interconnected mitochondrial networks in previous studies22. A transition from a reliance on glycolysis during stem cell proliferation to the increased utilization of mitochondrial oxidative phosphorylation as a differentiated cell type had been thought to drive the observed changes in mitochondrial content and organization22,23,24,34,42,43. However, the reduced mitochondrial staining and a lower range of fractal clustering in adipogenic cultures with FA supplementation (Fig. 4) suggests mitochondrial biogenesis may not occur solely to prepare the cell for increased oxidative phosphorylation, but also to facilitate the biosynthetic demnands of de novo FA synthesis. By upregulating specific mitochondrial proteins, such as PDHC, and reorganizing mitochondria around lipid droplets25,44, these changes in mitochondrial content and structure could help to facilitate more efficient lipogenesis that is thought to contribute to the decrease in redox ratio observed during adipogenic differentiation.
In summary, we have demonstrated a quantitative method for measuring autofluorescence-based redox ratios that is sensitive to changes in metabolic cofactor concentration ratios (Fig. 1) and have begun elucidating the metabolic pathways that contribute to these optical changes during stem cell differentiation (Figs. 2 and 4). Given the inherent heterogeneity of differentiating stem cell cultures (Fig. 5), the ability to non-destructively measure redox state, and by extension differentiation status, on a cell-by-cell basis is particularly useful. This method can also be used in conjunction with other non-linear optical imaging techniques such as FLIM, third harmonic generation, or coherent anti-Stokes Raman spectroscopy imaging to provide additional biochemical or structural information for each cell. In particular, future work that integrates these TPEF outcomes with FLIM-based measurements of the ratio of free and protein-bound NADH14,21,31 may be able to distinguish between changes in cytosolic and mitochondrial biosynthetic pathways. These non-linear optical imaging techniques to non-destructively characterize cell metabolic status during differentiation can serve complementary roles to traditional immunohistochemistry, PCR, or proteomic analyses of tissues. The many diverging and converging pathways that can contribute to a change in the optical redox ratio demonstrates just how ubiquitous these metabolic cofactors are, and suggests applications for this imaging method beyond the detection of stem cell differentiation. Specifically, this depth-resolved imaging technique has potential utility in detecting pathologies, such as epithelial cancers17,45, and in establishing high-content drug discovery platforms using in vitro disease models. In the context of tissue engineering, the TPEF measurements detailed in this study can potentially enable rapid in vitro differentiation protocol development, non-invasive in vivo assessments of differentiation, and good manufacturing practice for engineered tissue implants derived from MSCs. As cell therapy and tissue engineering research matures and requires long-term evaluations of cell status, the methods described here to assess cell differentiation can be coupled with microstructural information obtained through non-linear microscopy to provide a non-destructive, label-free overview of tissue structure and function.
Methods
Stem cell culture
Human mesenchymal stem cells (MSCs) were isolated from bone marrow aspirate (Lonza, Walkersville, MD) as described previously46. Osteogenic and adipogenic differentiation were induced through the addition of soluble factors to the media. To supplement adipogenic medium with fatty acids, 1 mM oleic acid and 0.5 mM bovine serum albumin (Sigma-Alrich, St. Louis, MO) were added to the standard adipogenic medium. Additional details regarding the stem cell isolation and differentiation protocols can be found in the SI Methods.
Two-photon excited fluorescence image acquisition protocol
For TPEF imaging, cell cultures were placed in microscope-compatible micro-incubator system (OkoLab; Naples, Italy), which maintained 37°C and 5% CO2 within a humidified environment throughout the imaging session. Images were obtained using a Leica TCS SP2 confocal microscope equipped with a tunable (710–920 nm) titanium-sapphire laser (Mai Tai; Spectra Physics; Mountain View, CA). Images (512 × 512 pixels; 238 × 238 μm; 12-bit depth) were acquired using water-immersion 63× objective (NA 1.2; 220 μm working distance), with simultaneous collection by two non-descanned photomultiplier tube (PMT) detectors using a filter cube containing filters from Chroma (Bellows Falls, VT), including a 700 nm short pass filter (ET700SP-2P) and a 495 nm dichroic mirror (495DCXR). To isolate NADH fluorescence, a 460(±20) nm emission filter (Chroma, ET460/40M-2P), corresponding to the NADH emission peak, was placed before one of the non-descanned detectors. NADH fluorescence images were acquired from this 460 nm channel using 755 nm excitation. FAD fluorescence was isolated using a 525(±25) nm emission filter (Chroma, ET525/50M-2P) for the other non-descanned detector and 860 nm excitation. The two-photon action cross section of NADH decreases by several orders of magnitude between 755 nm and 860 nm excitation12,47, enabling an efficient isolation of FAD at longer wavelengths. For each randomly-selected field, images were acquired at 755 nm and 860 nm excitation within 30 seconds of each other. PMT gain was adjusted for each image to maximize contrast while preventing a saturated pixel intensity value. The PMT gain and laser power were recorded for each image and used to normalize fluorescence intensity.
Optical redox ratio calculation
Prior to calculation of an optical redox ratio, fluorescence intensities were normalized by the PMT gain and the laser power used during acquisition, and automated cell segmentation was performed for each field. Additional details regarding these analysis steps can be found in the Supplementary Methods. Previous work has suggested lipofuscin autofluorescence may affect NADH and FAD measurements18. Pixels with a fluorescence intensity exceeding a normalized value of 2 in the 460 nm emission channel at 860 nm excitation were assumed to be lipofuscin. With this combination of emission and excitation wavelengths, minimal NADH or FAD fluorescence would be detected7,12,47, so any such lipofuscin-containing pixels were removed from the segmented cell masks and subsequent redox ratio calculations. For visualization purposes, pixel-wise redox ratio maps were computed from the normalized fluorescence intensities as FAD/(NADH + FAD). These redox ratio maps were assigned to the jet color map in MATLAB and multiplied by the merged grayscale image of the unmasked NADH and FAD fluorescence intensity images to produce the images displayed in Fig. 1, 2, and 4. To obtain an average redox ratio of an entire field for correlations with LC/MS-MS and qPCR outcomes, the average fluorescence intensities were computed from the masked cell regions of the field and a redox ratio of FAD/(NADH + FAD) was computed from the average field intensities of the fluorophores. To compute cell-to-cell variability in the redox ratios within each field, individual cell objects within the cell mask were identified through pixel connectivity and the average fluorescence intensities within each cell object were calculated and used to compute a redox ratio for each contiguous cell object. The standard deviation of the cell measurements within each field was computed, and the redox ratio standard deviation from each field was averaged for every combination of culture group and time point. Similarly, the standard deviation of field-averaged redox ratios was computed for the culture in each well to characterize field-to-field variability, and the standard deviation of the average redox ratio of each culture was computed within each combination of group and time point to characterize culture-to-culture variability.
LC/MS-MS correlations with optical redox ratio
To validate our optical sensitivity to the cofactors NADH and FAD, intracellular metabolites were extracted and measured through liquid chromatography/mass spectrometry (LC/MS-MS). In order to achieve sufficient signal intensity from the mass spectrometer (Supplementary Fig. S2) using a single culture of cells, confluent T75 flasks of differentiating MSCs were used. To facilitate optical imaging of each T75 flask with our high NA objective, a 3/8 inch hole was drilled in a sterile environment and a 25 mm glass coverslip (No 1.5) was adhered to the flask using an aquarium-safe silicone sealant. MSC cultures undergoing adipogenic differentiation, osteogenic differentiation, or MSC propagation at a range of post-induction time points up to 2 weeks (n = 22 cultures) were assessed through TPEF imaging (n = 10 fields per culture) and immediately sacrificed for metabolite extraction.
A previously established extraction protocol was modified to extract intracellular metabolites from each cell culture after imaging48. The LC/MS-MS analysis was performed using a 3200 QTRAP Hybrid Triple Quadrupole Linear Ion Trap mass spectrometer (AB SCIEX, Foster City, CA) coupled to a 1200 Series Binary LC System (Agilent Technologies, Santa Clara, CA)52. Standard curves for NAD+, NADH, and FAD were established by dissolving 0.01 g of each compound in 10 mL of 50:50 v% methanol:water to obtain a 1 g/L stock solution. The stock solutions were diluted 100-fold, and subsequently serially diluted to obtain a set of standards ranging from 1–10 mg/L. All compounds and solvents were purchased from Sigma-Aldrich (St. Louis, MO). For both the standards and cell extracts, the areas under the NAD+, NADH, and FAD peaks were quantified using Analyst software (Version 1.5, AB Sciex) (Supplementary Fig. S2). Standard curves exceeding R2 = 0.995 were achieved, and used to calculate the concentration of each cofactor. Based on the measured concentrations, redox ratios of FAD/(NADH + FAD) and NAD+/(NADH + NAD+) were computed for each culture, and a linear regression with the optical redox ratio was performed. Additional details on the LC/MS-MS extraction, data acquisition, and analysis can be found in the Supplementary Methods.
Quantitative PCR correlations with optical redox ratio
In order to evaluate a relationship between cell differentiation status and optical redox ratio, quantitative PCR analysis was employed to measure the expression of traditional markers of adipogenic and osteogenic differentiation49,50. MSCs were seeded onto uncoated glass-bottom 12 well plates (MatTek Corp., Ashland, MA) and cultured under different differentiation media conditions over 4 weeks. Each week, cultures from each differentiation group (n = 5 cultures per group) underwent TPEF imaging (n = 6 redox fields per culture). After imaging each week, cells in the imaged cultures were rinsed in PBS, trypsinized, and centrifuged to form pellets. Cell pellets were stored in Trizol (Life Technologies, Grand Island, NY) at −80°C prior to qPCR analysis.
RNA was isolated following the single step acid-phenol guanidinium method51. RNA was purified using a Qiagen RNEasy kit (Qiagen, Valencia, CA). Reverse transcription was performed on RNA using the High Capacity cDNA Reverse Transcription kit (Life Technologies, Grand Island, NY). Assays on demand (Life Technologies, Grand Island, NY) were used for the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH), ALP (Hs001029144_m1), BSP (Hs00173720_m1), GLUT4 (Hs00168966_m1) and PPAR-γ (Hs00234592_m1). Expression levels were quantified with a Stratagene Mx3000P QPCR System (Stratagene, La Jolla, CA) and normalized to GAPDH. Fold changes with respect to the average MSC expression at week 1 were computed using the delta delta Ct method, and the log-normalized fold change in expression was linear regressed against the average redox ratio of each culture.
Quantifying mitochondrial organization during differentiation
Each week, a culture from every differentiation group was stained with Mitotracker Orange (Life Technologies, Grand Island, NY) according to the manufacturer's instructions to quantify mitochondrial organization. Confocal images of Mitotracker staining were obtained through 543 nm excitation and emission collection between 550–625 nm at the same field location as NADH autofluorescence image acquisition (n = 6 fields per culture). Mitotracker intensities were normalized for PMT gain similar to other channels, and the average intensity was computed for each field.
To assess mitochondrial organization, a previously-established Fourier technique was used to obtain power spectral density (PSD) curves from each image26. Briefly, the image intensity patterns within the cell foreground regions defined by automated segmentation were cloned and randomly positioned in the image background to create a new image without distinct cell borders and only cell mitochondrial patterns spanning the entire image26. As in previous work, a 2D PSD was obtained through a discrete Fourier transform and radially sampled to produce a single PSD-frequency curve for each image. Because fractal organization is characterized by an inverse power law decay of the PSD with respect to increasing frequency, the linear portion of the PSD-frequency curve in log-log space was identified. To identify this linear region in log-log space, a linear regression of the PSD and frequency was performed following the removal of either the lowest frequency component or an equal portion of the highest frequency components (whichever removal produced the greatest increase in the R2 value). This stepwise removal of low and/or high frequency data points continued until the R2 value of the linear regression exceeded 0.99, suggesting a linear set of data. The slope of this linear portion of the PSD curve corresponds to the power law exponent, and the orders of magnitude over which the PSD decayed according to an inverse power law were computed as a measure of the range of fractal clustering.
Statistical analysis
To assess changes in redox ratio, a two-factor ANOVA (culture group, time point) of field average redox ratios was used to assess significant differences using JMP 8. The ANOVA design considered individual culture dishes as a random effect nested within each group, and all possible effect interactions were included. Post-hoc Tukey HSD tests were used to evaluate differences within each effect. A similar ANOVA design was used to compare the range of mitochondrial fractal clustering. Correlations between redox ratio and both qPCR and LC/MS-MS outcomes were computed, and significance was determined based on the null hypothesis that R = 0. Significance was defined as α = 0.05.
Author Contributions
K.P.Q., R.S.H., G.V.S., K.L., D.L.K. and I.G. designed the study. K.P.Q. conducted the imaging experiments and analyzed the fluorescence data. K.P.Q. and R.S.H. prepared media and maintained the cell cultures. R.S.H. isolated the cells and performed qPCR experiments. G.V.S. optimized and performed the LC/MS-MS experiments. K.L., I.G. and D.L.K. provided resources and guidance. K.P.Q., R.S.H., G.V.S., K.L., D.L.K. and I.G. wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This research was supported by Grant Numbers R01EB007542 from NIBIB/NIH and F32AR061933 from NIAMS/NIH, as well as an LC/MS major instrumentation grant from NSF (Grant Number 0821381) and the Tissue Engineering Resource Center (TERC) through Grant Number P41EB002520 from NIBIB/NIH. The authors would like to thank Moritz Vollrath for his technical assistance in the laboratory.
References
- Berthiaume F., Maguire T. J. & Yarmush M. L. Tissue engineering and regenerative medicine: history, progress, and challenges. Annu Rev Chem Biomol Eng 2, 403 (2011). [DOI] [PubMed] [Google Scholar]
- Guilak F. et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5 (1), 17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Kim U. J., Vunjak-Novakovic G., Min B. H. & Kaplan D. L. Influence of macroporous protein scaffolds on bone tissue engineering from bone marrow stem cells. Biomaterials 26 (21), 4442 (2005). [DOI] [PubMed] [Google Scholar]
- Quinn K. P. et al. Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios. Biomaterials 33 (21), 5341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stastna M., Chimenti I., Marban E. & Van Eyk J. E. Identification and functionality of proteomes secreted by rat cardiac stem cells and neonatal cardiomyocytes. Proteomics 10 (2), 245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W., Strickler J. H. & Webb W. W. Two-photon laser scanning fluorescence microscopy. Science 248 (4951), 73 (1990). [DOI] [PubMed] [Google Scholar]
- Georgakoudi I. & Quinn K. P. Optical Imaging Using Endogenous Contrast to Assess Metabolic State. Annu Rev Biomed Eng 14, 351 (2012). [DOI] [PubMed] [Google Scholar]
- Chance B. & Thorell B. Localization and kinetics of reduced pyridine nucleotide in living cells by microfluorometry. J Biol Chem 234, 3044 (1959). [PubMed] [Google Scholar]
- Eng J., Lynch R. M. & Balaban R. S. Nicotinamide adenine dinucleotide fluorescence spectroscopy and imaging of isolated cardiac myocytes. Biophys J 55 (4), 621 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayevsky A. & Rogatsky G. G. Mitochondrial function in vivo evaluated by NADH fluorescence: from animal models to human studies. Am J Physiol Cell Physiol 292 (2), C615 (2007). [DOI] [PubMed] [Google Scholar]
- Rocheleau J. V., Head W. S. & Piston D. W. Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response. J Biol Chem 279 (30), 31780 (2004). [DOI] [PubMed] [Google Scholar]
- Huang S., Heikal A. A. & Webb W. W. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys J 82 (5), 2811 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B., Schoener B., Oshino R., Itshak F. & Nakase Y. Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem 254 (11), 4764 (1979). [PubMed] [Google Scholar]
- Stringari C., Sierra R., Donovan P. J. & Gratton E. Label-free separation of human embryonic stem cells and their differentiating progenies by phasor fluorescence lifetime microscopy. J Biomed Opt 17 (4), 046012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz W. S. & Kunz W. Contribution of different enzymes to flavoprotein fluorescence of isolated rat liver mitochondria. Biochim Biophys Acta 841 (3), 237 (1985). [DOI] [PubMed] [Google Scholar]
- Chance B., Mayevsky A., Goodwin C. & Mela L. Factors in oxygen delivery to tissue. Microvasc Res 8 (3), 276 (1974). [DOI] [PubMed] [Google Scholar]
- Skala M. C. et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A 104 (49), 19494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. L., Kaplan D. L. & Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One 5 (4), e10075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschke D. G., Squirrell J. M., Fong J. J., Eliceiri K. W. & Ogle B. M. Cell death, non-invasively assessed by intrinsic fluorescence intensity of NADH, is a predictive indicator of functional differentiation of embryonic stem cells. Biol Cell 104 (6), 352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C. et al. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc Natl Acad Sci U S A 108 (33), 13582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig K., Uchugonova A. & Gorjup E. Multiphoton fluorescence lifetime imaging of 3D-stem cell spheroids during differentiation. Microsc Res Tech 74 (1), 9 (2011). [DOI] [PubMed] [Google Scholar]
- Folmes C. D., Dzeja P. P., Nelson T. J. & Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11 (5), 596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S. et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 6 (6), e20914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. T., Shih Y. R., Kuo T. K., Lee O. K. & Wei Y. H. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26 (4), 960 (2008). [DOI] [PubMed] [Google Scholar]
- Wilson-Fritch L. et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol 23 (3), 1085 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xylas J., Quinn K. P., Hunter M. & Georgakoudi I. Improved Fourier-based characterization of intracellular fractal features. Optics Express 20 (21), 23442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y., Shi H. & Lee K. Impact of perturbed pyruvate metabolism on adipocyte triglyceride accumulation. Metab Eng 11 (6), 382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks V. A. et al. Permeabilized cell and skinned fiber techniques in studies of mitochondrial function in vivo. Mol Cell Biochem 184 (1–2), 81 (1998). [PubMed] [Google Scholar]
- Vishwasrao H. D., Heikal A. A., Kasischke K. A. & Webb W. W. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. J Biol Chem 280 (26), 25119 (2005). [DOI] [PubMed] [Google Scholar]
- Blinova K. et al. Distribution of mitochondrial NADH fluorescence lifetimes: steady-state kinetics of matrix NADH interactions. Biochemistry 44 (7), 2585 (2005). [DOI] [PubMed] [Google Scholar]
- Guo H. W. et al. Reduced nicotinamide adenine dinucleotide fluorescence lifetime separates human mesenchymal stem cells from differentiated progenies. J Biomed Opt 13 (5), 050505 (2008). [DOI] [PubMed] [Google Scholar]
- Blinova K. et al. Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry 47 (36), 9636 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacanti N. M. & Metallo C. M. Exploring metabolic pathways that contribute to the stem cell phenotype. Biochim Biophys Acta (2012). [DOI] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C. & Thompson C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324 (5930), 1029 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabold O. et al. Lactate and oxygen constitute a fundamental regulatory mechanism in wound healing. Wound Repair Regen 11 (6), 504 (2003). [DOI] [PubMed] [Google Scholar]
- Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 10 (2), 179 (2008). [DOI] [PubMed] [Google Scholar]
- De Pauw A., Tejerina S., Raes M., Keijer J. & Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol 175 (3), 927 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y., Yoon J. & Lee K. Flux profile and modularity analysis of time-dependent metabolic changes of de novo adipocyte formation. Am J Physiol Endocrinol Metab 292 (6), E1637 (2007). [DOI] [PubMed] [Google Scholar]
- Manteiga S., Choi K., Jayaraman A. & Lee K. Systems biology of adipose tissue metabolism: regulation of growth, signaling and inflammation. Wiley Interdiscip Rev Syst Biol Med (2013). [DOI] [PubMed] [Google Scholar]
- Mitra K., Rikhy R., Lilly M. & Lippincott-Schwartz J. DRP1-dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis. J Cell Biol 197 (4), 487 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguez S., Feasson L., Denis C. & Freyssenet D. Mitochondrial biogenesis during skeletal muscle regeneration. Am J Physiol Endocrinol Metab 282 (4), E802 (2002). [DOI] [PubMed] [Google Scholar]
- Rehman J. Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J Mol Med (Berl) 88 (10), 981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. T., Hsu S. H. & Wei Y. H. Mitochondrial bioenergetic function and metabolic plasticity in stem cell differentiation and cellular reprogramming. Biochim Biophys Acta 1820 (5), 571 (2012). [DOI] [PubMed] [Google Scholar]
- Ducluzeau P. H. et al. Dynamic regulation of mitochondrial network and oxidative functions during 3T3-L1 fat cell differentiation. J Physiol Biochem 67 (3), 285 (2011). [DOI] [PubMed] [Google Scholar]
- Levitt J. M., McLaughlin-Drubin M. E., Munger K. & Georgakoudi I. Automated biochemical, morphological, and organizational assessment of precancerous changes from endogenous two-photon fluorescence images. PLoS One 6 (9), e24765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman G. H. et al. Cell differentiation by mechanical stress. FASEB J 16 (2), 270 (2002). [DOI] [PubMed] [Google Scholar]
- Zipfel W. R. et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A 100 (12), 7075 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick C. A. et al. Evaluation of extraction processes for intracellular metabolite profiling of mammalian cells: matching extraction approaches to cell type and metabolite targets. Metabolomics 6 (3), 427 (2010). [Google Scholar]
- Mauney J. R. et al. In vitro and in vivo evaluation of differentially demineralized cancellous bone scaffolds combined with human bone marrow stromal cells for tissue engineering. Biomaterials 26 (16), 3173 (2005). [DOI] [PubMed] [Google Scholar]
- Mauney J. R. et al. Engineering adipose-like tissue in vitro and in vivo utilizing human bone marrow and adipose-derived mesenchymal stem cells with silk fibroin 3D scaffolds. Biomaterials 28 (35), 5280 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. & Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 1 (2), 581 (2006). [DOI] [PubMed] [Google Scholar]
- Schroer K., Zelic B., Oldiges M. & Lutz S. Metabolomics for Biotransformations: Intracellular Redox Cofactor Analysis and Enzyme Kinetics Offer Insight Into Whole Cell Processes. Biotechnology and Bioengineering 104 (2), 251 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information