Abstract
[11C]AFM, or [11C]2-[2-(dimethylaminomethyl)phenylthio]-5-fluoromethylphenylamine, is a new positron emission tomography (PET) radioligand with high affinity and selectivity for the serotonin transporter (SERT). The purpose of this study was to determine the most appropriate kinetic model to quantify [11C]AFM binding in the healthy human brain. Positron emission tomography data and arterial input functions were acquired from 10 subjects. Compartmental modeling and the multilinear analysis-1(MA1) method were tested using the arterial input functions. The one-tissue model showed a lack of fit in low-binding regions, and the two-tissue model failed to estimate parameters reliably. Regional time–activity curves were well described by MA1. The rank order of [11C]AFM binding potential (BPND) matched well with the known regional SERT densities. For routine use of [11C]AFM, several noninvasive methods for quantification of regional binding were evaluated, including simplified reference tissue models (SRTM and SRTM2), and multilinear reference tissue models (MRTM and MRTM2). The best methods for region of interest (ROI) analysis were MA1, MRTM2, and SRTM2, with fixed population kinetic values (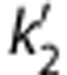 or b′) for the reference methods. The MA1 and MRTM2 methods were best for parametric imaging. These results showed that [11C]AFM is a suitable PET radioligand to image and quantify SERT in humans.
or b′) for the reference methods. The MA1 and MRTM2 methods were best for parametric imaging. These results showed that [11C]AFM is a suitable PET radioligand to image and quantify SERT in humans.
Keywords: brain imaging, dopamine, 5-HT, kinetic modeling, positron emission tomography, receptor imaging
Introduction
The serotonin transporter (SERT) is located on the presynaptic terminals of neurons, and is responsible for the reuptake of serotonin, a neurotransmitter involved in the regulation of brain functions such as mood, appetite, and sleep.1 Serotonin transporter is a primary target of action for selective serotonin reuptake inhibitors, which are effective therapeutic agents for the treatment of major depressive disorder, obsessive-compulsive disorder, posttraumatic stress disorder, and other mood and anxiety disorders. Because the serotonin system has such an important role in the pathophysiology of these diseases, there has been considerable interest in developing in vivo positron emission tomography (PET) radioligands for imaging SERT distribution. In the human brain, highest concentrations of SERT are found in the raphe nucleus and midbrain, followed by thalamus, striatum, and amygdala. Intermediate levels are in the limbic system (hippocampus and cingulate cortex), and lower levels in the neocortex regions, with cerebellum displaying lowest to negligible SERT density.2, 3, 4, 5
Over the years, a number of high-affinity ligands have been developed for SERT imaging with PET. [11C]McN5652 [11C]McN5652 (trans-1,2,3,5,6,10β-hexahydro-6-[4-[11C](methylthio)phenyl]-pyrrolo-[2,1-a]-isoquinoline)6 was the first successful and widely used tracer. However, [11C]McN5652 has many limitations, which include high nonspecific binding, nonmeasurable free fraction in the plasma (<1%), and long scanning time (120 minutes). Because of high-nonspecific binding, this tracer does not provide reliable quantification in brain regions with moderate and low SERT densities. Given these limitations and the need for SERT tracers with improved imaging properties, a number of ligands derived from diphenyl sulfides were developed [11C]DASB ([11C]N,N-dimethyl-2-(2-amino-4-cyanophenylthio)benzylamine),7 [11C]AFM ([11C]2-[2-(dimethylaminomethyl)phenylthio]-5-fluoromethylphenylamine),8 [11C]ADAM ([11C]2-((2-((dimethylamino)methyl)phenyl)thio)-5-iodophenylamine),9 [11C]DAPA ([11C]-5-bromo-2-[2-(dimethylaminomethylphenylsulfanyl)]phenylamine))10. In a comparative evaluation of these tracers and [11C]McN5652, both [11C]DASB and [11C]AFM were shown to be superior radioligands to image SERT in baboons, with [11C]DASB displaying fastest brain kinetics that enabled the measurement of SERT availability in a shorter scanning time, and [11C]AFM having the highest specific-to-nonspecific binding ratios (BPND).11 The plasma free fraction for [11C]DASB and [11C]AFM was high enough to be reliably measured. Since then, [11C]DASB has become the most widely used ‘standard' tracer. Subsequently, [11C]MADAM ([11C]N,N-dimethyl-2-(2'-amino-4'-methylphenylthio)benzylamine)12 and [11C]HOMADAM ([11C]N,N-dimethyl-2-(2′-amino-4′-hydroxymethylphenylthio)benzylamine)13 were also developed and evaluated in humans.14, 15 Like [11C]AFM in baboons, [11C]MADAM and [11C]HOMADAM appeared to have higher BPND than [11C]DASB in humans, even though these comparisons were not made within subjects. High BPND might allow more reliable measurement of SERT availability in brain regions, especially cortical regions, where SERT concentrations are low, but SERT abnormalities have been observed in several postmortem studies of patients with psychiatric disorders.16 Thus, to investigate SERT in cortical regions, tracers with better imaging characteristics than [11C]DASB are needed.
[11C]AFM is a potential PET radioligand that could measure the SERT in both high- and low-density regions. Here we report the first human PET study with [11C]AFM to evaluate its pharmacokinetics and brain distribution and to develop appropriate kinetic modeling techniques for the quantitative analysis of its binding parameters.
Materials and Methods
Human Subjects
Positron emission tomography scans were performed in 10 healthy volunteers whose age ranged from 20 to 45 years (6 men, 4 women). This study was performed under a protocol approved by the Human Investigation Committee, Yale University School of Medicine, and the Yale–Haven Hospital Radiation Safety. Written informed consent was obtained from all subjects. This study was performed according to the Ethical Principles and Guidelines for the Protection of Human Subjects of Research (Belmont Report).
As part of the subject evaluation, magnetic resonance (MR) images were acquired on all subjects to eliminate those with structural brain abnormalities and for image registration. Magnetic resonance imaging was performed on a 3 T whole-body scanner (Trio; Siemens Medial Systems, Erlangen, Germany) with a circularly polarized head coil. The dimension and pixel size of MR images were 256 × 256 × 176 and 0.98 × 0.98 × 1.0 mm3, respectively.
Radiochemistry
[11C]AFM was prepared using the TRACERLabTM FXC automated synthesizer (GE Healthcare, Chalfont St Giles, UK) by N-methylation of the desmethyl precursor with [11C]methyl triflate instead of [11C]methyl iodide as previously reported using a manual method.8 Briefly, [11C]methyl triflate was trapped at 0°C in a solution of 0.5 to 1 mg precursor in 400 μL of anhydrous methyl ethyl ketone. The mixture was then heated for 1 minute at 60°C, and then purified by reverse-phase semipreparative high-performance liquid chromatography (HPLC). The HPLC fraction containing the product was further purified by solid-phase extraction and formulated in ethanol and saline into a sterile solution ready for administration. Radiochemical purity of the final product was 93.0±1.5%. (see Supplementary Data and Supplementary Figure S1 for further details).
Positron Emission Tomography Scanning Procedure
Positron emission tomography scans were performed on the high-resolution research tomography (HRRT) (Siemens Medical Solutions, Knoxville, TN, USA), which acquires 207 slices (1.2-mm slice separation) with a reconstructed image resolution of ∼3 mm. Before tracer administration, a 6-minute transmission scan was conducted for attenuation correction. [11C]AFM (dose: 712±47 MBq; specific activity at the time of injection: 193±127 GBq/μmoL) was administered intravenously over 1 minute by an automatic pump (Harvard PHD 22/2000; Harvard Apparatus, Holliston, MA, USA), and a 120-minute PET scan was acquired in list mode. Dynamic scan data were reconstructed in 33 frames (6 × 0.5 minutes, 3 × 1 minutes, 2 × 2 minutes, and 22 × 5 minutes) with all corrections (attenuation, normalization, scatter, randoms, and deadtime) using the MOLAR algorithm.17 Motion correction was included in the reconstruction based on measurements with the Polaris Vicra sensor (NDI Systems, Waterloo, ON, Canada) with reflectors mounted on a swim cap worn by the subject.
Input Function Measurement
For each study, a catheter was inserted into the subject's radial artery for blood sampling. An automated blood counting system (PBS-101; Veenstra Instruments, Joure, The Netherlands) was used to measure the radioactivity in whole blood during the first 7 minutes, with blood withdrawn continuously at 4 mL/min. Thereafter, 15 samples (1 to 6 mL) were collected manually at 3, 5, 7, 10, 15, 20, 25, 30, 40, 50, 60, 75, 90, 105, and 120 minutes after tracer administration. For each sample, plasma was obtained by refrigerated centrifugation (2930 g at 4°C for 5 minutes). Whole blood and plasma were counted in cross-calibrated gamma counters (1480 WIZARD; Perkin-Elmer, Waltham, MA, USA).
To determine radioactivity in the plasma for the first 7 minutes, the whole blood-to-plasma ratios (r(t)) were calculated from the hand-drawn samples. The ratios between 3 and 120 minutes were fitted to the following equation: r(t)=a exp(bt)+c, and the plasma time–activity curve (TAC) in the first 7 minutes was calculated from the measured whole blood TAC and the extrapolated ratio. These data were combined with the plasma samples to produce the final curve of total radioactivity in the plasma. To reduce noise in these data, the total plasma curve from ∼6 minutes onward was fitted to a sum of exponentials.
Plasma Metabolite and Protein-Binding Analysis
Metabolism of the radiotracer was analyzed with arterial blood samples collected at 3, 7, 15, 30, 60, and 90 minutes after tracer injection, using the column-switching HPLC method.18 Plasma samples were first treated with urea (8 mol/L) and citric acid (50 mmol/L), and then loaded onto a capture column (19 × 4.6 mm2) packed with Phenomenex SPE C18 Strata-X sorbent and eluting with 1% acetonitrile in water at a flow rate of 2 mL/min. At 4 minutes after sample injection, the activity on the capture column was back flushed onto an analytical column (Phenomenex Luna Phenyl hexyl, Torrance, CA, USA; 5 μm, 250 × 4.6 mm2) eluting with 30% acetonitrile in 0.1 mol/L ammonium formate at a flow rate of 1.50 mL/min. The HPLC eluent was collected with an automated fraction collector and the fractions were counted with an automatic gamma well counter (Wizard 1480; Perkin-Elmer). The unmetabolized parent fraction was determined as the ratio of the sum of radioactivity in fractions containing the parent tracer to the total amount of radioactivity collected, and fitted with bounded sum of exponentials. The final plasma input function was calculated as the product of the total plasma curve and the parent fraction curve.
Ultrafiltration-based method was used for measuring the unbound portion (free fraction, fP) of [11C]AFM in the plasma. [11C]AFM (∼6 MBq) was mixed with arterial blood (6 mL) taken immediately before tracer injection. After standing for 10 minutes at room temperature, the spiked blood sample was centrifuged at 2930 g for 5 minutes to separate the plasma. Plasma aliquots (0.3 mL) were then taken and loaded onto the reservoir of the Millipore Centrifree micropartition device in triplicate and centrifuged at 1228 g for 20 minutes. The free fraction fP was determined from the activity ratio of ultrafiltrate to plasma. Similarly, the amount of nonspecific binding of [11C]AFM to the filter was also determined as described above by spiking a sample of saline with [11C]AFM. The ultrafiltrate to spiked saline ratio was 0.96±0.03 (n=10), indicating negligible tracer retention on the filter.
Image Registration and Regions of Interest
Regions of interest (ROI) were taken from the automated anatomic labeling (AAL) for SPM219 in Montreal Neurological Institute (MNI) space.20 For each subject, the dynamic PET images after hardware motion correction were coregistered to the early summed PET images (0 to 10 minutes after injection) using a six-parameter mutual information algorithm21 (FMRIB's Linear Image Registration Tool, FMRIB Software Library) to eliminate any residual uncorrected motion. The summed PET images were then coregistered to the subject's T1-weighted 3 T MR images (six-parameter affine registration), which were subsequently coregistered to the AAL template in MNI space using a 12-parameter affine transformation. Using the combined transformation from template to PET space, TACs were generated for 13 ROIs: amygdala, caudate, cerebellum, anterior cingulate, posterior cingulate, frontal lobe, hippocampus, occipital lobe, putamen, midbrain raphe, pontine raphe, temporal lobe, and thalamus. The raphe regions (0.64 mL for midbrain raphe, and 0.21 mL for pontine raphe) were added to the standard AAL template as described previously.22 In this study, we used a portion of the cerebellum region located at the inferior end, based on a blocking study with citalopram, which showed minimal blockade (<15%).23 The ROIs were a priori divided into four categories: regions with very high SERT density (raphe regions), regions of high density (thalamus, putamen, caudate, amygdala), regions of intermediate density (hippocampus, cingulate cortex), and regions of low density (neocortices).
Kinetic Modeling
Compartment models with the arterial input function
Kinetic analysis of tissue data was performed with the one-tissue (1T) and two-tissue (2T) compartment models.24 The rate constants, K1 and k2 for the 1T model and K1, k2, k3, and k4 for the 2T model, were estimated by weighted nonlinear least squares (see below). The primary outcome measure was the total volume of distribution, VT.25 Three versions of the binding potential were then computed from the VT values: BPND=VT/VND−1, BPP=VT−VND, and BPF=(VT−VND) /fP.25 Here the cerebellum gray matter was used as the reference region. Previous studies reported extremely low2, 3 or low4, 5, 26 SERT density in the cerebellum.
Graphical analysis with the arterial input function
The multilinear analysis-1 (MA1) method27 was applied for [11C]AFM quantification using the measured input function. The MA1 method is intermediate between 1T and 2T models. When the TAC is described well by the 1T model, early t* setting for MA1 provides small bias in the estimate. Conversely, when the TAC is described well by the 2T model, late t* setting will help minimize bias, although stability of parameter estimates must also be considered. The equation for MA1 is:
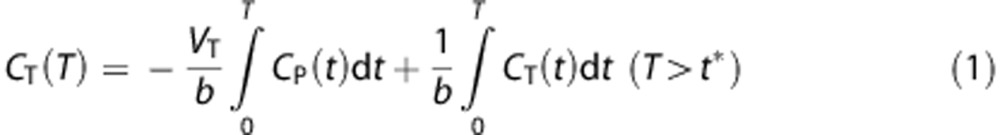 |
where CT is the tissue curve and CP is the metabolite-corrected input function.
Effect of scan duration on VT estimates
The effect of scan duration on parameter stability was calculated by shortening the fitting interval. Scan durations ranging from 70 to 120 minutes were evaluated. For each region and duration, the ratio of the estimated VT values to the 120-minute values was calculated. The following two criteria were used to select optimal scan duration28: (a) the average of this ratio was between 0.95 and 1.05; and (b) the intersubject s.d. of this ratio was <0.1.
Reference tissue compartment models
Reference tissue models use a TAC from the reference region, CREF(t), instead of the input function. The simplified reference tissue model (SRTM)29 is described with the equation:
where k2 and 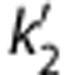 are the efflux rate constants from the target and the reference regions, respectively, and Rl is the ratio of influx constants K1 between target and reference regions. The SRTM method assumes that all regions can be described by a 1T model and has three parameters to estimate. The parameter
are the efflux rate constants from the target and the reference regions, respectively, and Rl is the ratio of influx constants K1 between target and reference regions. The SRTM method assumes that all regions can be described by a 1T model and has three parameters to estimate. The parameter 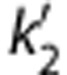 should be common among all regions or voxels. However, a different value of
should be common among all regions or voxels. However, a different value of 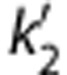 is estimated by equation (2) for each voxel. The two-parameter version of SRTM30 (SRTM2) was also applied. The two-parameter method has been shown to be effective in reducing noise from the original three-parameter models. The use of SRTM2 requires fixing k2′ with a preliminary analysis. Approaches to fix this parameter for [11C]AFM are described below.
is estimated by equation (2) for each voxel. The two-parameter version of SRTM30 (SRTM2) was also applied. The two-parameter method has been shown to be effective in reducing noise from the original three-parameter models. The use of SRTM2 requires fixing k2′ with a preliminary analysis. Approaches to fix this parameter for [11C]AFM are described below.
Graphical analyses with reference region
The multilinear reference tissue model (MRTM),31 which is based on graphical analysis, was evaluated. The equation for MRTM is described as:
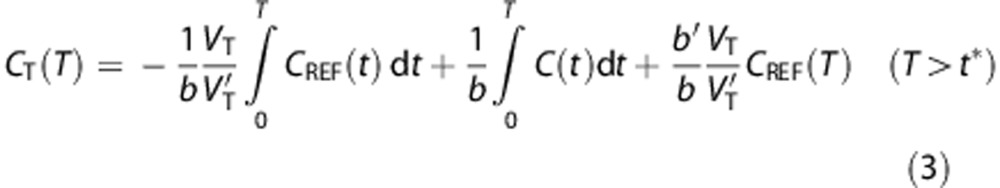 |
The MRTM method has three parameters to estimate. Like SRTM, MRTM has a two-parameter version: MRTM2.31 The use of MRTM2 requires fixing b′ (=−1/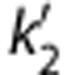 for 1T model) from a preliminary analysis. Approaches to determine the value of b′ are described below.
for 1T model) from a preliminary analysis. Approaches to determine the value of b′ are described below.
Determination of reference region parameters for simplified reference tissue model 2 and multilinear reference tissue model 2
In the reference tissue models that fix one parameter, two approaches were evaluated:
Coupled fit: All TACs were fitted simultaneously under the constraint of finding a common parameter (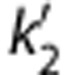 or b′) to all regions in SRTM2 and MRTM2, i.e., parameter coupling.32
or b′) to all regions in SRTM2 and MRTM2, i.e., parameter coupling.32
Population value: The value of 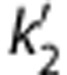 or b′ was fixed to the population average values estimated from the 1T model (SRTM2) or MA1 models (MRTM2). To differentiate from the coupled fit, a model with population value is denoted with the suffix ‘pop' (e.g., SRTM2pop).
or b′ was fixed to the population average values estimated from the 1T model (SRTM2) or MA1 models (MRTM2). To differentiate from the coupled fit, a model with population value is denoted with the suffix ‘pop' (e.g., SRTM2pop).
For all reference region fits, to reduce estimation error, the reference region TAC was fitted by a sum of exponentials (1 to 3 exponentials; the number of exponentials was selected by minimizing χ2) starting from ∼25 minutes after injection.
Selection of start of fitting period (t*) for graphical analyses
The effect of the choice of starting time (t*) for graphical analyses was evaluated by varying t* from 10 to 60 minutes after injection in 10-minute increments. The determination of t* for MA1 was conducted by visual inspection of fit quality. After selecting the best t* for MA1 (40 minutes, see Results), the optimal t* for MRTM and MRTM2 were selected by comparing the values of MRTM and MRTM2 BPND with the MA1 BPND estimates.
Parametric Imaging
Parametric imaging was performed using 1T, MA1, SRTM, SRTM2, MRTM, and MRTM2. A basis function method was used for parametric imaging for the compartmental methods where the range of k2 for the basis functions was 0.005≤k2≤1/min. Region of interests were applied to the parametric images and these values were compared between methods and with the values obtained by MA1 fits (t*=40 minutes) of the ROI TACs. For the SRTM2 or MRTM2 calculation, the value 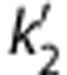 or b′ was fixed to a global value among all voxels. The SRTM (or MRTM) method was applied to estimate
or b′ was fixed to a global value among all voxels. The SRTM (or MRTM) method was applied to estimate 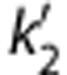 (or b′) from all brain voxels. Then, the global value was computed as the median of
(or b′) from all brain voxels. Then, the global value was computed as the median of 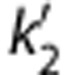 or b′ estimates from selected brain voxels (BPND>0.5). The effect of smoothing to reduce noise before estimation was investigated by applying a three-dimensional Gaussian filter with full-width at half-maximum (FWHM) of 3.6 and 6.0 mm (3 and 5 voxels) to the original image data.
or b′ estimates from selected brain voxels (BPND>0.5). The effect of smoothing to reduce noise before estimation was investigated by applying a three-dimensional Gaussian filter with full-width at half-maximum (FWHM) of 3.6 and 6.0 mm (3 and 5 voxels) to the original image data.
Implementation
For all approaches, kinetic parameters were estimated using weighted least squares, with the weights determined based on noise equivalent counts in each frame. A Marquardt–Levenberg algorithm was used for nonlinear least-squares estimation applied to ROI TACs. Estimates of relative standard error (rSE) of the parameters were derived from the covariance matrix. Time shifts between blood and brain were estimated by applying the 1T model to the whole brain TAC and these values were then used as a global correction for all analyses. All methods were implemented with IDL 8.0 (ITT Visual Information Solutions, Boulder, CO, USA).
Statistics
Goodness of fit was evaluated by visual inspection, and by calculating χ2 statistics, the Akaike information criterion,33 the Schwartz criterion,34 and the F test using the weighted residual sum of squares. Statistical significance using the F test was assessed with P<0.05.
Results
Arterial Input Function
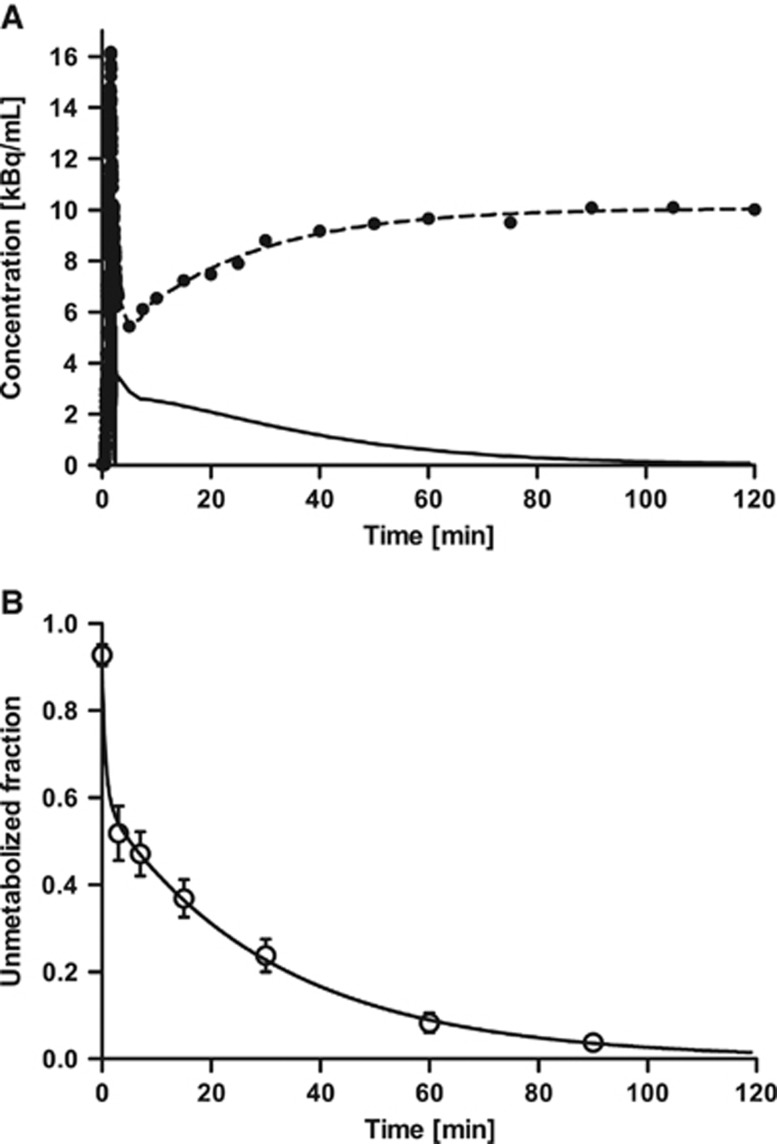
Figure 1A shows a typical example of the radioactivity concentration in the plasma over time. In most cases, the whole blood to plasma ratio was >1 for the first 20 minutes (1.22±0.14 at 5 minutes, n=10), and subsequently decreased (0.82±0.03 at 90 minutes, n=10). This pattern was similar to that of [11C]DASB.35 The plasma free fraction (fP) of [11C]AFM was 8.6±1.8% (n=10), which was similar to that in baboons (9.6±0.5%, n=4).11 [11C]AFM was rapidly metabolized, and the parent fraction was 47±5%, 37±4%, 24±4%, 8±2%, and 3.8±1.4% at 7, 15, 30, 60, 90 minutes after injection, respectively. Figure 1B shows the average parent fraction over time and the fitted curve averaged over all subjects. The rapid clearance and metabolism of [11C]AFM required careful handling to avoid low statistical quality of the metabolite data at later sampling times. AFM metabolites in arterial plasma were polar in comparison with the parent. Examples of the HPLC results are shown in the Supplementary Data (Supplementary Figure S2).
Figure 1.
(A) Typical example of time–activity curves of total radioactivity in the plasma (dots) and its fit (dashed line), and the corresponding metabolite-corrected input curve (solid line). (B) Mean±s.d. of unmetabolized [11C]AFM ([11C]2-[2-(dimethylaminomethyl)phenylthio]-5-fluoromethylphenylamine) fraction over time in 10 healthy human subjects and mean of fitted curve (solid line). The parent fraction was fitted using a sum of exponentials.
Region of Interest Data Analysis
Regional TACs
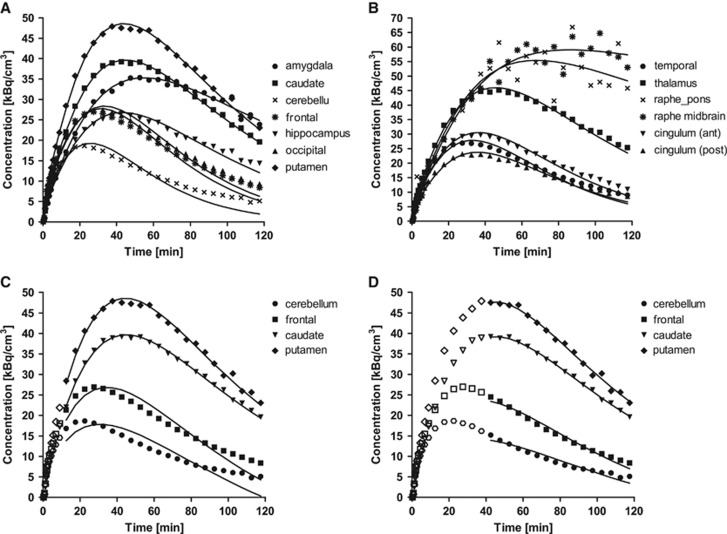
Figures 2A and 2B, shows typical TACs from the 13 ROIs. Radioactivity concentrations were highest in the raphe regions and high levels were seen in the putamen, thalamus, caudate, and amygdala. Intermediate levels of activity were observed in the hippocampus and anterior cingulate cortex, with lower levels in the posterior cingulate and other cortical regions, as well as the cerebellum. In the cerebellum region, the concentration peaked at ∼30 minutes after injection. The peak times were similar in regions with low SERT densities. Regions with higher densities peaked at later times (50 to 70 minutes). The data from raphe, a high-density, small region, were too noisy to obtain stable parameter estimates. On average, pontine raphe and midbrain raphe peaked at around 80 minutes, and then slightly decreased.
Figure 2.
Typical time activity curves in 13 region of interest (ROIs) measured after injection of 700 MBq of [11C]AFM ([11C]2-[2-(dimethylaminomethyl)phenylthio]-5-fluoromethylphenylamine). For each region, the symbols correspond to the measured activities and the lines are values fitted to one-tissue compartment model. Curves are shown in two graphs (A and B) for clarity, using different scales. Typical examples of fitting are shown for multilinear analysis with (C) t*=10 minutes and (D) t*=40 minutes. ROIs are cerebellum (circles, •), frontal lobe (triangles, ▴), caudate (downward-pointing triangles, ▾), and putamen (diamonds, ♦). Points used for the estimation are shown in closed symbols.
Compartmental modeling with the input function
In most cases, the 2T model converged and fitted the data well; however; the parameter values were often implausible (e.g., k3 <10−3/min, VT >500 mL/cm3). Such implausible parameters were seen mainly in neocortical regions across most subjects. In addition, for two subjects, questionable parameters were seen in most regions.
Results obtained from the 1T model are presented in Table 1. Figures 2A and 2B, show representative fits to the 1T model in one subject, chosen as the subject with best correlation of individual VT values to the population average values (Table 1). Convergence was achieved for all regions using the 1T model. Visual inspection of 1T fitting showed some lack of fit in regions with intermediate to low binding, with nonrandom distribution of the residuals. In those cases, the fitted curve tended to overestimate the data at intermediate time points (∼30 minutes), and underestimate it at late time points (∼90 minutes). In high-binding regions, 1T fitting was good.
Table 1. Kinetic parameters estimated from 1T compartment model and MA1.
|
VT
(mL/cm3)a |
|||||
|---|---|---|---|---|---|
| Regions | 1T | MA1b | K1 (mL/min per cm3)a | k2 (1/min)a | Minimal scan duration (min) |
| Cerebellum | 9.7 (20%) | 10.6 (19%) | 0.52 (24%) | 0.053 (10%) | 90 |
| Amygdala | 44.2 (29%) | 42.9 (23%) | 0.42 (20%) | 0.010 (22%) | 80 |
| Putamen | 40.8 (17%) | 41.0 (17%) | 0.68 (23%) | 0.017 (13%) | 70 |
| Thalamus | 39.4 (21%) | 40.3 (21%) | 0.66 (20%) | 0.017 (11%) | 70 |
| Caudate | 33.2 (13%) | 32.9 (12%) | 0.56 (21%) | 0.017 (15%) | 70 |
| Hippocampus | 21.2 (24%) | 22.3 (24%) | 0.42 (18%) | 0.020 (13%) | 80 |
| Ant. cingulate cortex | 21.5 (18%) | 22.3 (17%) | 0.56 (26%) | 0.026 (17%) | 70 |
| Post. cingulate cortex | 17.1 (19%) | 17.8 (18%) | 0.53 (24%) | 0.031 (12%) | 80 |
| Occipital lobe | 16.9 (21%) | 17.7 (20%) | 0.58 (22%) | 0.034 (12%) | 80 |
| Temporal lobe | 16.2 (16%) | 17.0 (15%) | 0.55 (20%) | 0.034 (11%) | 80 |
| Frontal lobe | 15.7 (18%) | 16.6 (18%) | 0.58 (23%) | 0.037 (11%) | 80 |
| Raphe midbrain | 231.8 (127%) | 151.1 (47%) | 0.48 (17%) | 0.004 (46%) | |
| Raphe pons | 124.8 (91%) | 100.1 (52%) | 0.57 (21%) | 0.007 (48%) | |
COV, coefficients of variation; 1T, one-tissue; MA1, multilinear analysis-1.
Values are means (%COV, intersubject variability) in 10 subjects.
t* was set to 40 minutes after injection.
The uptake rate constant K1 ranged from 0.42 to 0.68 mL/min per cm3 reflecting moderate to high extraction of [11C]AFM. The K1 value had an rSE of <6% (mean 2%) in all regions except for the raphe. The rate constant k2 had an rSE of <8% (mean 3%), excluding the raphe. VT and BPND values were estimated with rSE of <7% (mean 2%) and <10% (mean 4%) (except for raphe regions), respectively. Excluding the raphe, the order of VT values from the 1T model was: amygdala>putamen>thalamus>caudate>hippocampus≈ anterior cingulate cortex> posterior cingulate cortex≈occipital lobe>temporal lobe>frontal lobe>cerebellum. Estimates of VT and BPND are presented in Tables 1 and 2.
Table 2. Comparison of binding potential values between the 1T compartment model and MA1.
|
BPNDa |
BPP(mL/cm3)a |
BPF(mL/cm3)a |
||
|---|---|---|---|---|
| Regions | 1T | MA1b | MA1b | MA1b |
| Amygdala | 3.60 (34%) | 3.07 (24%) | 32.29 (26%) | 393 (35%) |
| Putamen | 3.25 (11%) | 2.89 (8%) | 30.43 (17%) | 365 (19%) |
| Thalamus | 3.08 (13%) | 2.81 (12%) | 29.76 (23%) | 352 (18%) |
| Caudate | 2.49 (16%) | 2.14 (14%) | 22.31 (13%) | 270 (23%) |
| Hippocampus | 1.19 (21%) | 1.10 (20%) | 11.71 (31%) | 140 (30%) |
| Ant. cingulate cortex | 1.23 (14%) | 1.12 (12%) | 11.72 (17%) | 141 (22%) |
| Post. cingulate cortex | 0.77 (16%) | 0.69 (17%) | 7.21 (21%) | 86 (21%) |
| Occipital lobe | 0.74 (10%) | 0.67 (10%) | 7.09 (23%) | 84 (21%) |
| Temporal lobe | 0.69 (17%) | 0.62 (16%) | 6.41 (15%) | 78 (27%) |
| Frontal lobe | 0.63 (10%) | 0.57 (10%) | 6.02 (18%) | 72 (22%) |
| Raphe midbrain | 24.0 (141%) | 13.62 (55%) | 140.49 (50%) | 1769 (61%) |
| Raphe pons | 12.5 (107%) | 8.84 (67%) | 89.47 (58%) | 1143 (72%) |
COV, coefficients of variation; 1T, one tissue; MA1, multilinear analysis-1.
Values are means (%COV, intersubject variability) in 10 subjects.
t* was set to 40 minutes after injection.
The parameters in the raphe regions showed large intersubject variability, and rSE for VT and BPND were >20%. The combination of small size and slow kinetics in the raphe leads to a noisy TAC and unstable estimation. Therefore, the raphe regions were excluded from further analysis.
Graphical analyses with the input function
Representative fitting plots using MA1 with starting times for fitting t* set to 10 and 40 minutes are displayed in Figures 2C and 2D, respectively. Visual inspection of the MA1 fitting with early t* (10 to 30 minutes) showed a similar pattern of lack of fit to the 1T model. MA1 fitting was improved by increasing t* to 40 minutes, as shown in Figure 2D.
Supplementary Figure S3 shows the VT values from MA1 with different t*. When t* was increased, VT estimates increased in the cerebellum and other regions with low SERT densities. However, in regions with high densities, the estimates slightly decreased with larger t*. For example, with t*=40 minutes, VT values increased by 6% in the cerebellum and decreased by 3% in the amygdala when compared with t*=10 minutes. The values of BPND were therefore decreased with increasing t* in all regions. Based on visual inspection, t*=40 minutes was selected for use in further analysis.
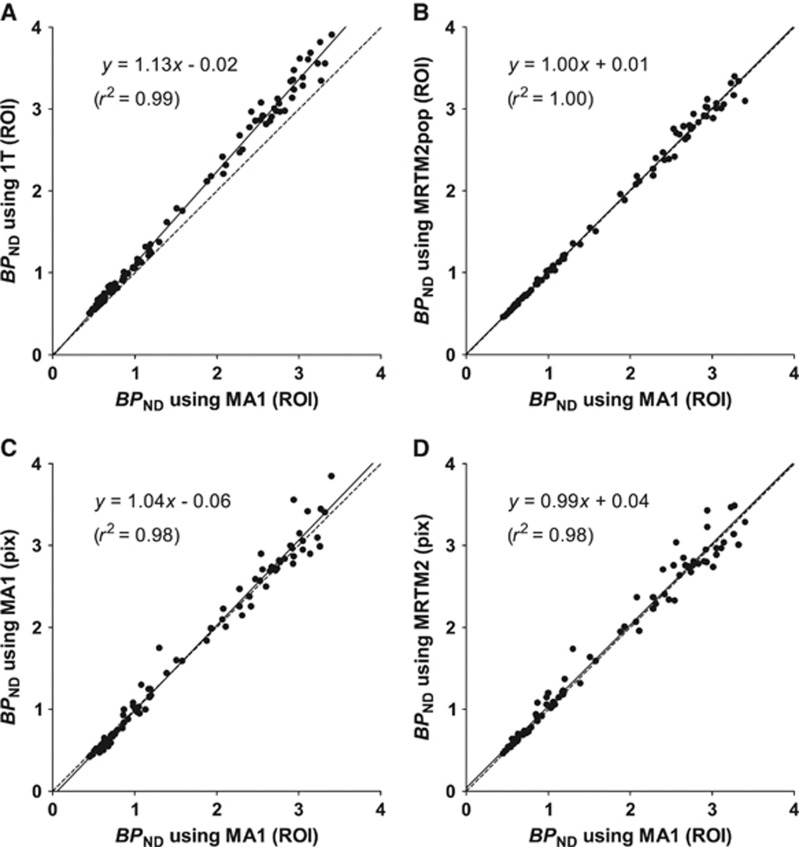
The MA1 estimates of VT and BPND are presented in Tables 1 and 2. The MA1 method provided values of VT similar to those derived from 1T model in high-binding regions, whereas in intermediate and low-binding regions, MA1 provided higher values of VT. VT estimates using MA1 matched well with VT estimates using the 2T model (VT(MA1, t*=40 minutes)=0.97 VT(2T)−0.87, r2=0.98). Note that estimates with 2T %SE >10 (54 out of 110 fits) were excluded from this comparison. Because of this stronger bias observed with 1T in the cerebellum and low-binding regions, BPND estimates from the MA1 method were smaller than those from 1T. For MA1, estimates of the intercept b ranged from −103±27 minutes in the amygdala to −28±4 minutes in the cerebellum. BPND values ranged from 0.57±0.06 in the frontal lobe to 3.1±0.7 in the amygdala. The correlation scatter plot and regression line between BPND estimates from 1T and MA1 are shown in Figure 3A.
Figure 3.
Comparison of binding potential (BPND) values between region of interest (ROI) analysis with multilinear analysis-1 (MA1) (t*=40 minutes) and other ROI analyses and parametric image methods. For parametric imaging, a three-dimensional Gaussian filter with full-width at half-maximum=3.6 mm was applied to the dynamic image data. The reference region was the cerebellum. Raphe regions were excluded from this analysis. The line of identity (dotted line) is added to each panel for reference. Region of interest analyses using (A) one-tissue (1T) compartment model and (B) two-parameter multilinear reference tissue model (MRTM2) with a population value of b′, and parametric images using (C) MA1 and MRTM2. The value of t* was set to 40 minutes after injection in panels B, C, and D.
The three binding potentials, BPND, BPP, and BPF using MA1, are presented in Table 2. These three outcome measures were well correlated with one another. Regional BPP and BPF values in general displayed larger intersubject variability than BPND. Average values for each variability were 14%, 20%, and 24% for BPND, BPP, and BPF, respectively. For MA1 with t*=40 minutes, the tissue free fraction (fND) calculated from the cerebellum VT and fP was 0.008±0.002. No significant correlation was found between the measured free fraction fP and VND (r2=0.26, n=10, Student's t-test, P=0.13), suggesting why correction for fP does not reduce intersubject variability.
Effect of scan duration on parameter estimates
Time–activity curves were fitted for different scan durations, to define the minimum scan time required for reliable estimates of binding parameters. Using comparatively strict criteria, minimal scanning time ranged between 70 to 90 minutes (Table 1). The mean VT values in high-binding regions did not change with decreasing scan duration (see Supplementary Figure S4).
Compartmental modeling without input function
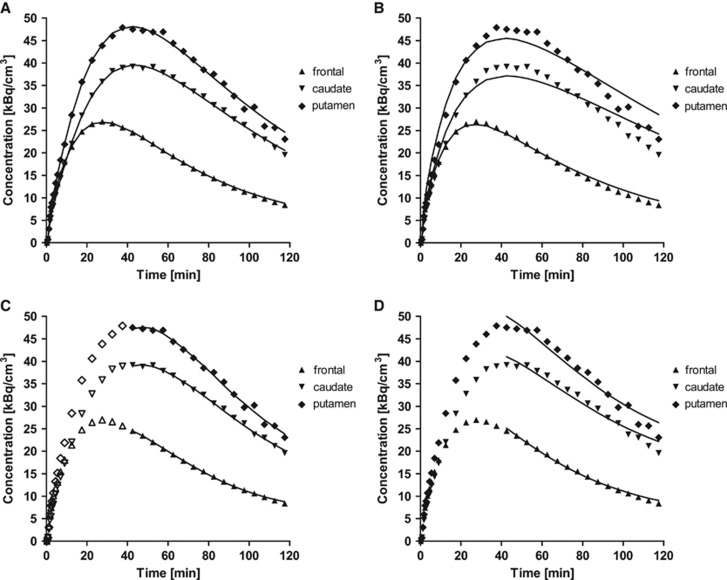
Typical fits using SRTM and SRTM2pop are presented in Figures 4A and 4B. The values of 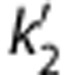 in SRTM (0.095±0.020/min) and SRTM2 (0.096±0.017/min) were larger than the population value of k2 from 1T in the cerebellum (0.053/min). Visual inspection indicates that SRTM and SRTM2 provide better quality of fit than SRTM2pop. The SRTM method was compared with SRTM2pop using the F test, which showed that weighted residual sum of squares of SRTM was significantly smaller than that of SRTM2pop in almost all fits (98 out of 100 fits). The correlation scatter plot and regression line between BPND estimates from SRTM2, and SRTM2pop and MA1 with t*=40 minutes are shown in Supplementary Figures S5B and S5C, respectively. Simplified reference tissue model BPND showed a very similar regression line to SRTM2 BPND (Supplementary Figure S5). The SRTM and SRTM2 BPND estimates were smaller than MA1 estimates. In spite of the lack of fits, BPND estimates from SRTM2pop were similar to MA1 estimates. Presumably, the lack of fit with SRTM2pop is because of the inadequacy of 1T fitting in low-binding regions (Figure 2A). Intersubject variability of BPND was similar among SRTM, SRTM2, and SRTM2pop.
in SRTM (0.095±0.020/min) and SRTM2 (0.096±0.017/min) were larger than the population value of k2 from 1T in the cerebellum (0.053/min). Visual inspection indicates that SRTM and SRTM2 provide better quality of fit than SRTM2pop. The SRTM method was compared with SRTM2pop using the F test, which showed that weighted residual sum of squares of SRTM was significantly smaller than that of SRTM2pop in almost all fits (98 out of 100 fits). The correlation scatter plot and regression line between BPND estimates from SRTM2, and SRTM2pop and MA1 with t*=40 minutes are shown in Supplementary Figures S5B and S5C, respectively. Simplified reference tissue model BPND showed a very similar regression line to SRTM2 BPND (Supplementary Figure S5). The SRTM and SRTM2 BPND estimates were smaller than MA1 estimates. In spite of the lack of fits, BPND estimates from SRTM2pop were similar to MA1 estimates. Presumably, the lack of fit with SRTM2pop is because of the inadequacy of 1T fitting in low-binding regions (Figure 2A). Intersubject variability of BPND was similar among SRTM, SRTM2, and SRTM2pop.
Figure 4.
Typical examples of fitting methods based on a reference region (cerebellum) (A) simplified reference tissue model (SRTM), (B) SRTM2, (C) multilinear reference tissue model (MRTM), and (D) MRTM2. The values of 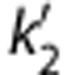 in SRTM2 and MRTM2 were set to the population average. Points are concentrations measured in the frontal lobe (triangles, ▴), caudate (downward-pointing triangles, ▾), and putamen (diamonds, ♦). The starting point for fitting, t*, was set to 40 minutes for panels C and D; points used for the regression are shown in closed symbols.
in SRTM2 and MRTM2 were set to the population average. Points are concentrations measured in the frontal lobe (triangles, ▴), caudate (downward-pointing triangles, ▾), and putamen (diamonds, ♦). The starting point for fitting, t*, was set to 40 minutes for panels C and D; points used for the regression are shown in closed symbols.
Graphical analyses without input function
Typical fits using MRTM and MRTM2pop are presented in Figures 4C and 4D, respectively. The value of t* was set to 40 minutes after injection. The comparison between BPND values obtained from MRTM2, and MRTM2pop, and MA1 is presented in Supplementary Figure S5I and Figure 3B, respectively. Multilinear reference tissue model BPND showed a very similar regression line to SRTM BPND (Supplementary Figure S5). The value of b′ was not constant among regions in MRTM, but the interregion average (−26±13 minutes) was similar to the population value of b from MA1 model in the cerebellum (−28 minutes). As seen with SRTM and SRTM2, the MRTM and MRTM2 BPND estimates were smaller than MA1 estimates (Supplementary Figure S5). The MRTM2pop method provided BPND estimates similar to those from MA1, as well as good fits. Intersubject variability of BPND estimates was similar among MRTM, MRTM2, and MRTM2pop.
Parametric Imaging
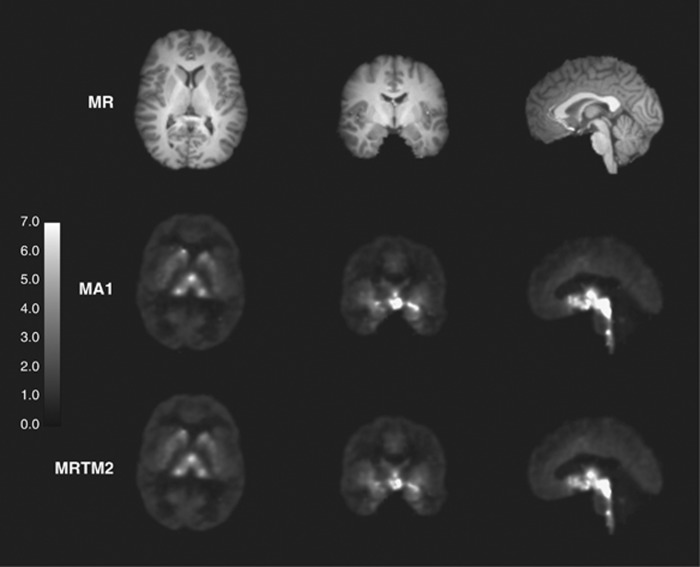
Parametric images of BPND (MA1 and MRTM2) are shown in Figure 5. Highest binding was seen in the thalamus, striatum, and raphe regions. Given the smoothing parameters chosen, the parametric images had low noise, and the statistical quality of MA1 and MRTM2 images was similar. Region of interests were applied to the parametric images, and BPND values were compared with those from the MA1 ROI analysis.
Figure 5.
Typical coregistered magnetic resonance (MR) anatomic image (top) and parametric binding potential (BPND) images derived from multilinear analysis-1 (MA1) (middle) and two-parameter multilinear reference tissue model (MRTM2) (bottom). Images were generated with a pre-smoothing filter (three-dimensional Gaussian filter, full-width at half-maximum=3.6 mm). The BPND color scale ranged from 0 to 7.
For all smoothing levels and models, regional BPND values correlated well with those from MA1 ROI analysis. Because of the noise included in the unsmoothed image, the parametric VT images using MA1 included some voxels with negative values or unreasonably high values (>50). In all models, regional BPND values decreased with larger FWHM (except for MRTM, between no smoothing and FWHM=3.6 mm). The value of b′ (MRTM2) increased with larger FWHM, whereas the value of 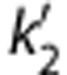 (SRTM2) decreased (b′=−23±2 minutes,
(SRTM2) decreased (b′=−23±2 minutes, 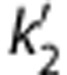 =0.099±0.014/min at FWHM=3.6 mm). With a smoothing filter of 3.6 mm, the MA1 BPND estimates from parametric images matched well with those from ROI analysis (BPND(pix,MA1)=1.04 BPND(ROI,MA1)−0.06, r2=0.98). The MRTM2 BPND estimates were also in excellent agreement with those from MA1 (BPND(pix,MRTM2)=0.99 BPND(ROI,MA1)+0.04, r2=0.98). The comparison of the parametric and regional BPND values is shown in Figures 3C and 3D, for MA1 and MRTM2, respectively, and in Supplementary Figure S5 for SRTM, SRTM2, and MRTM.
=0.099±0.014/min at FWHM=3.6 mm). With a smoothing filter of 3.6 mm, the MA1 BPND estimates from parametric images matched well with those from ROI analysis (BPND(pix,MA1)=1.04 BPND(ROI,MA1)−0.06, r2=0.98). The MRTM2 BPND estimates were also in excellent agreement with those from MA1 (BPND(pix,MRTM2)=0.99 BPND(ROI,MA1)+0.04, r2=0.98). The comparison of the parametric and regional BPND values is shown in Figures 3C and 3D, for MA1 and MRTM2, respectively, and in Supplementary Figure S5 for SRTM, SRTM2, and MRTM.
Discussion
The primary aim of this study was to evaluate kinetic modeling methods and to determine the most appropriate method to obtain estimates of regional SERT densities with [11C]AFM in the human brain. Because the 1T model did not adequately describe low-binding regions, the MA1 model was found to be quite appropriate for this tracer. The binding potential measure BPND was found to have the smallest intersubject coefficients of variation. Conventional reference tissue models (SRTM and MRTM) showed some underestimation of BPND, with best agreement to MA1 found for SRTM2 and MRTM2 with population 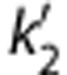 (or b').
(or b').
Parent Fraction and Free Fraction in the Input Function
Like [11C]DASB, metabolism of [11C]AFM was rapid. For [11C]DASB, an increase in parent fraction from 2 to 12 minutes after injection was reported by Parsey et al.36 This pattern was absent for other SERT PET ligands in humans, such as [11C]McN5652 and [11C]MADAM. We did not observe this pattern in our study either. Comparing parent fractions at 3, 7, and 15 minutes in each subject, the fraction decreased monotonically in most cases. In one subject, the parent fraction at 7 minutes was higher than that at 3 minutes by 2%. The initial metabolism rate of [11C]AFM was faster or similar to that of [11C]DASB.28, 35, 37
The effect of interpolation/extrapolation of parent fraction curves was investigated: (1) Assuming very slow metabolism at early time, i.e., the first 3 minutes parent fraction was set to one. (2) Avoiding the potential extrapolation errors, the parent fraction at 90 to 120 minutes was set to be the (constant) 90-minute value. Both cases are not physiologic, but they would be potentially large effects given the use of the exponential function. The VT estimates using MA1 decreased by ∼3% in both cases. These results indicate that interpolation/extrapolation errors effect is small.
We adopted the column-switching method of Hilton et al18 for metabolite analysis. This method permits the use of larger volumes of plasma producing higher radioactivity levels and lower statistical noise in the final results compared with protein precipitation method previously used in our lab. Application of this method was important because statistical errors in the low parent fraction values at later time points introduced variability into the fitting results.
All metabolites and radiochemical impurities were more polar than the parent compound, i.e., they eluted earlier in the analytical HPLC chromatogram. It is not known if the metabolites and/or impurities can enter the brain in humans, but in rat studies, HPLC analysis of rat brains showed that the parent compound accounted for >95% of the total radioactivity at 10 and 30 minutes after the injection, whereas at the same time points, the parent compound accounted for only 20% and 10% of the radioactivity in the plasma,8 respectively. These results in rat brain are suggestive that the metabolites and impurities do not enter the brain in humans.
The plasma free fraction of [11C]McN5652 is not measurable. In contrast, [11C]AFM plasma free fraction was low (8.6±1.8%), but high enough to be measurable, and similar to that of [11C]DASB (8.9±1.6%).28 The plasma free fraction for other SERT ligands, [11C]MADAM and [11C]HOMADAM, was not reported.
Compartmental Modeling and Graphical Analysis
Kinetic analyses indicated that both 1T and 2T compartment models were not sufficient to represent regional brain uptake curves for the following reasons. First, the 1T model showed lack of fit in intermediate and low-binding regions. The 1T fitted curves were above the actual activity levels at intermediate time points and below at late time points. Frankle et al28 reported a similar pattern in a study with [11C]DASB. Second, the 2T model generated implausible parameter estimates in most cases, although it did converge in all cases. VT is determined by the model prediction of the shape of the curves after the end of the scan as well as by the height of the tails of the curves. For a bolus injection, VT is 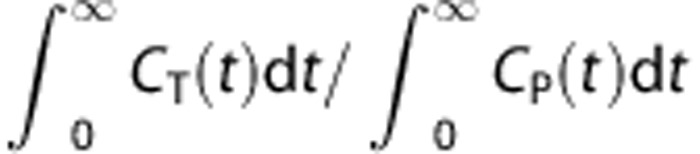 .38 In real situation, PET data are not acquired to infinity. The model provides an extrapolation of the PET data. In the 1T model, the extrapolation is performed by estimating only one exponential rate (k2) and one scale factor (K1). In the 2T model, there are two exponential rates and two scale factors, which cause the model-predicted extrapolation to become more unstable. There are many tracers for which the unconstrained 2T model does not provide stable VT estimates, including [11C]DASB.37, 39 Fit quality, Akaike information criterion, and Schwartz criterion were not improved by the 2T model in SERT-rich regions. This is because of the greater difficulty in determining the 2T parameters in regions with a high fraction of specific binding as reflected in larger BPND.40
.38 In real situation, PET data are not acquired to infinity. The model provides an extrapolation of the PET data. In the 1T model, the extrapolation is performed by estimating only one exponential rate (k2) and one scale factor (K1). In the 2T model, there are two exponential rates and two scale factors, which cause the model-predicted extrapolation to become more unstable. There are many tracers for which the unconstrained 2T model does not provide stable VT estimates, including [11C]DASB.37, 39 Fit quality, Akaike information criterion, and Schwartz criterion were not improved by the 2T model in SERT-rich regions. This is because of the greater difficulty in determining the 2T parameters in regions with a high fraction of specific binding as reflected in larger BPND.40
The multilinear analysis MA1, which is based on the graphical analysis approach, constitutes a trade-off between the lower quality of fit of the 1T model and the greater instability of the 2T model. Similar types of lack of fit were observed with the 1T model or MA1 method with early t*. When a later t* setting was used, MA1 provided good fit quality in most regions, although some lack of fit could still be seen in the cerebellum and other low-binding regions, which were better fitted by the 2T model. Here, based on curve fitting and time stability of estimates, the MA1 model with later t* was chosen as the method of choice for analysis. An early t* results in poor quality of fit and bias in VT estimate. Conversely, a late t* results in greater variability in VT estimates because too few points are available for the fitting. It is common to select the value of t* based on visual inspection in a graphical analysis.41, 42, 43, 44 The chosen t* value (40 minutes) is a compromise to balance the quality of fit (and thus bias) and variability in VT estimates. By setting t* to 40 minutes, we minimize both the bias and instability in VT estimates.
Different models have been proposed as appropriate for the analysis of [11C]DASB data: the 1T model,28 constrained 2T model,37 and likelihood estimation in graphical analysis.42 For [11C]MADAM and [11C]HOMADAM, the 2T model failed to provide stable rate constants.14, 15 Appropriate models for [11C]MADAM are constrained 2T model and Logan graphical analysis. For [11C]HOMADAM, the 1T model was chosen. Here, based on curve fitting and time stability of estimates, the MA1 model with later t* was chosen as the method of choice for analysis. However, model choice may evolve with the addition of data from more subjects.
From the correlation with the BPND estimates by MA1 model (Figure 3 and Supplementary Figure S5), SRTM, SRTM2, MRTM, and MRTM2 produced lower BPND estimates in high-binding regions. Because the 1T model does not fit well the [11C]AFM data, the assumptions of SRTM and SRTM2 are violated. However, there are examples where SRTM can provide good quality fits for tracers whose TACs are not well fitted by the 1T model, e.g., [11C]PHNO ([11C](+)-4-propyl-3,4,4a,5,6,10b-hexahydro-2H-naphtho[1,2-b][1,4]oxazin-9-ol),45 [11C]PE2I (N-3-isodoprop-2E-ethyl)-2β-carbo[11C]methoxy-3β-(4-methylphenyl)nortropane),46 or [11C]P943.47 For such tracers, SRTM BPND estimates can match those obtained with arterial input function data (e.g., [11C]PHNO), or be biased (e.g., [11C]PE2I and [11C]P943). [11C]AFM belongs to the second category, where TACs are well fitted with SRTM, but SRTM BPND estimates are biased. Therefore, quality of fit can be deceptive for reference tissue models, and it is important to determine a gold standard model using an input function before reference tissue methods are investigated. The fits with SRTM2pop and MRTM2pop are visually not optimal. However, considering that MA1 VT estimates matched well with 2T VT estimates and that SRTM2pop and MRTM2pop provides comparable results to MA1, SRTM2pop and MRTM2pop are acceptable methods.
The value of kinetic parameter, K1, was moderate to high, indicating that [11C]AFM is well extracted from the plasma. This K1 was higher than that of [11C]MADAM, similar to [11C]DASB, and lower than that of [11C]HOMADAM. Similar to [11C]DASB and many other tracers, [11C]AFM K1 values are high compared with the theoretical maximum value, the cerebral blood flow, even though the plasma free fraction fP is low (<10%). This combination of K1 and fP values can be explained if the [11C]AFM–plasma protein association and dissociation rates are fast, and that within one capillary transit, all/most of the tracer in the plasma is effectively available for extraction into tissue.
Using fairly strict criteria, the minimal scan duration for [11C]AFM was determined based on MA1 analysis (t*=40 minutes) and found to be 70 to 90 minutes. In raphe regions, the optimal minimal scan duration could not be determined. Raphe regions and amygdala have slow kinetics and thus require longer scanning time. Except for raphe, a 90-minute scan is sufficient for reliable estimates of [11C]AFM binding parameters with MA1 analysis.
Among the three definitions of binding potential, BPND showed the smallest intersubject variability. Calculation of BPP assumes that fP is similar across subjects, and BPND assumes a similar fND across subjects. In this study, the intersubject variation in free fraction was similar between the plasma fP (∼20%) and the nondisplaceable compartment fND (∼20%); however, fND is derived from fP, so this agreement may not be surprising. This suggests that variability in the measurement of the input function is a substantial contributor to the larger intersubject variability in BPF and BPP. Interestingly, [11C]DASB showed the opposite results: BPP displayed smaller intersubject variability (coefficients of variation) than BPND when the 1T model was used (n=6).28
Comparison of Various Serotonin Transporter Tracers
All the four diphenyl sulfide-derived SERT tracers ([11C]DASB, [11C]MADAM, [11C]HOMADAM, and [11C]AFM) show a high selectivity to SERT. Among the four tracers, [11C]HOMADAM displays the fastest kinetics, with early regional peak uptake times of 10 to 40 minutes.15 Regional peak uptake times for [11C]MADAM range from 25 to 70 minutes.14 [11C]DASB and [11C]AFM have slightly slower uptake, with regional peak uptake times of 30 to 70 minutes.28 Correspondingly, the minimal scanning time to estimate stable outcome measures is the shortest for [11C]HOMADAM (65 minutes) and longer for [11C]DASB and [11C]AFM (90 minutes).
[11C]AFM VT and BPND values were compared with those of [11C]DASB, [11C]MADAM, and [11C]HOMADAM. Note that the values for [11C]DASB were variable in the literature. For this comparison purpose, values from Frankle et al39 were used for [11C]DASB. [11C]AFM VT and BPND values were highest among the four tracers across regions. Nonspecific binding, as measured by VT in the cerebellum, is lowest for [11C]HOMADAM, followed by [11C]MADAM, and higher for [11C]DASB and [11C]AFM.
BPND is the most critical parameter to evaluate tracers, although other factors (e.g., peripheral metabolism rate) also play a role. [11C]AFM BPND values in SERT-rich regions was 2.6, 2.1, and 1.6 times higher than those of [11C]DASB, [11C]MADAM, and [11C]HOMADAM, respectively. In neocortical regions, [11C]AFM BPND values were 6.7, 1.4, and 1.4 times higher than those of [11C]DASB, [11C]MADAM, and [11C]HOMADAM, respectively. Note that these parameters were obtained from scanners with different spatial resolution (EXACT HR+ scanner for [11C]DASB, EXACT HR47 scanner for [11C]MADAM, and HRRT scanner for [11C]HOMADAM). According to Schain et al,48 SRTM BPND values for [11C]MADAM were higher by ∼20% on average with HRRT than the HR+.
Cerebellum as a Reference Region
According to previous postmortem studies using [3H]paroxetine binding,2, 3, 4, 5, 26 nondetectable or extremely low SERT density was detected in the cerebellum. However, several blocking studies showed that there were changes in radioligand binding in the cerebellum, especially in the white matter and vermis.35, 49 In this study, white matter, vermis, and cerebellar subregions with confirmed blocking effect by citalopram were excluded from the cerebellum region used in our analysis.
To estimate the fraction of specific binding in the cerebellum, we used information from semiquantitative Western blotting study by Kish et al,26 which showed that the relative density of SERT in the frontal lobe as compared with cerebellum was 5.3. The VT values in the cerebellum and frontal lobe were 10.6 and 16.6 (mL/cm3), respectively, in our measurement (Table 1). Assuming that nonspecific binding is equal in both regions, 13% of the VT in the cerebellum corresponds to specific binding (VT in cerebellum=VND+VS=10.6, VT in frontal lobe=VND+5.3VS=16.6, VS=1.40). According to a similar estimate between other ROIs and cerebellum, the specific binding distribution volume VS was 13±6% of the VT in the cerebellum, which would lead to a region-dependent underestimation of BPND: 13% in the amygdala, 15% in the hippocampus, 35% in the occipital lobe, and 42% in the frontal lobe. Thus, the effect of this bias must be carefully considered before using reference region analysis methods for [11C]AFM, and potentially for other SERT tracers as well.
Quantification in the Raphe Nuclei
The raphe nuclei can be easily visualized in [11C]AFM parametric images (Figure 5), especially with a high-resolution scanner. The high levels of SERT in the raphe nuclei lead to very high VT values and extremely slow kinetics. When combined with the small size of the raphe regions and the short half-life of C-11, there is insufficient statistical quality in the late PET data to provide reliable estimates of VT or BPND by any kinetic method. Compared to previous studies with [11C]DASB, this effect is more pronounced because of the high resolution of the HRRT used in this study and the higher binding affinity of AFM relative to DASB. Note that it is not unusual for tracers to be suitable for proper quantification only in certain regions of the brain, with kinetics matched to their radioisotope half-lives, especially when the density of targeted receptor/transporter varies greatly across regions.
Binding Potential Parametric Imaging
In general, image smoothing and/or limits in the permitted range of k2 values for the basis functions are required to calculate stable parametric images. The 1T, SRTM, and SRTM2 methods were implemented using a basis function method with specified range of k2 values. We set a minimum k2 value of 0.005/min; however, the k2 value is <0.005/min in some voxels in the SERT-rich regions. As a result, the estimated VT or BPND values in such regions are somewhat biased. However, k2 values are not limited in the multilinear methods MA1, MRTM, and MRTM2. To reduce the noise of image data, spatial smoothing or other noise reduction methods is recommended as a preprocessing procedure for these methods. As a result of image smoothing, spatial resolution is reduced and the voxel values in the small regions such as raphe and amygdala were biased by the partial volume effect. Although the starting time for MA1, MRTM, and MRTM2 was selected based on ROI analysis, it might not be early enough to obtain stable fitting in parametric imaging.
The MRTM2 method overestimated BPND values without image smoothing, although the value of b′ was closest to that of MA1 ROI analysis. This bias was presumably caused by noise sensitivity of MRTM2. With a 3.6-mm image smoothing, the b′ value changed substantially from that of MA1 ROI analysis. However, BPND values derived from parametric imaging with MRTM2 were close to those from MA1 ROI analysis. Thus, the two bias effects (difference in b′ and noise) tend to cancel each other out.
[11C]AFM can be used to visualize and quantify SERT binding in the human brain. Compared with previous SERT tracers, [11C]AFM showed higher specific binding in areas with low SERT densities. The MA1 method is the most suitable kinetic analysis approach with arterial input functions, both for ROI analysis and for parametric images. Among the reference region methods, MRTM2pop and SRTM2pop produced estimates comparable to those from MA1 ROI analysis and MRTM2 appears to be the most appropriate method for parametric imaging.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
This work was supported by NIMH (MH066624) and Grant-in-Aid for Young Scientists (B) (21791232). This publication was also made possible by CTSA Grant Number UL1 RR024139 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Supplementary Material
References
- Lucki I. The spectrum of behaviors influenced by serotonin. Biol Psychiatry. 1998;44:151–162. doi: 10.1016/s0006-3223(98)00139-5. [DOI] [PubMed] [Google Scholar]
- Cortés R, Soriano E, Pazos A, Probst A, Palacios JM. Autoradiography of antidepressant binding sites in the human brain: localization using [3H]imipramine and [3H]paroxetine. Neuroscience. 1988;27:473–496. doi: 10.1016/0306-4522(88)90282-5. [DOI] [PubMed] [Google Scholar]
- Laruelle M, Vanisberg MA, Maloteaux JM. Regional and subcellular localization in human brain of [3H]paroxetine binding, a marker of serotonin uptake sites. Biol Psychiatry. 1988;24:299–309. doi: 10.1016/0006-3223(88)90198-9. [DOI] [PubMed] [Google Scholar]
- Backström I, Bergström M, Marcusson J. High affinity [3H]paroxetine binding to serotonin uptake sites in human brain tissue. Brain Res. 1989;486:261–268. doi: 10.1016/0006-8993(89)90511-8. [DOI] [PubMed] [Google Scholar]
- Rosel P, Menchon JM, Oros M, Vallejo J, Cortadellas T, Arranz B, et al. Regional distribution of specific high affinity binding sites for 3H-imipramine and 3H-paroxetine in human brain. J Neural Transm. 1997;104:89–96. doi: 10.1007/BF01271297. [DOI] [PubMed] [Google Scholar]
- Suehiro M, Scheffel U, Ravert HT, Dannals RF, Wagner HN., Jr. [11C](+)McN5652 as a radiotracer for imaging serotonin uptake sites with PET. Life Sci. 1993;53:883–892. doi: 10.1016/0024-3205(93)90440-e. [DOI] [PubMed] [Google Scholar]
- Wilson AA, Ginovart N, Schmidt M, Meyer JH, Threlkeld PG, Houle S. Novel radiotracers for imaging the serotonin transporter by positron emission tomography: synthesis, radiosynthesis, and in vitro and ex vivo evaluation of 11C-labeled 2-(phenylthio)araalkylamines. J Med Chem. 2000;43:3103–3110. doi: 10.1021/jm000079i. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hwang DR, Bae SA, Sudo Y, Guo N, Zhu Z, et al. A new positron emission tomography imaging agent for the serotonin transporter: synthesis, pharmacological characterization, and kinetic analysis of [11C]2-[2-(dimethylaminomethyl)phenylthio]-5-fluoromethylphenylamine ([11C]AFM) Nucl Med Biol. 2004;31:543–556. doi: 10.1016/j.nucmedbio.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Vercouillie J, Tarkiainen S, Halldin C, Emond P, Chalon S, Sandell J, et al. Precursor synthesis and radiolabelling of [11C]ADAM: a potential radioligand for the serotonin transporter exploration by PET. J Labelled CompRad. 2001;44:113–120. [Google Scholar]
- Huang Y, Hwang DR, Zhu Z, Bae SA, Guo N, Sudo Y, et al. Synthesis and pharmacological characterization of a new PET ligand for the serotonin transporter: [11C]5-bromo-2-[2-(dimethylaminomethylphenylsulfanyl)]phenylamine ([11C]DAPA) Nucl Med Biol. 2002;29:741–751. doi: 10.1016/s0969-8051(02)00337-2. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hwang DR, Narendran R, Sudo Y, Chatterjee R, Bae SA, et al. Comparative evaluation in nonhuman primates of five PET radiotracers for imaging the serotonin transporters: [11C]McN 5652, [11C]ADAM, [11C]DASB, [11C]DAPA, and [11C]AFM. J Cereb Blood Flow Metab. 2002;22:1377–1398. doi: 10.1097/01.WCB.0000040948.67415.05. [DOI] [PubMed] [Google Scholar]
- Tarkiainen J, Vercouille J, Emond P, Sandell J, Hiltunen J, Frangin Y, et al. Carbon-11 labelling of MADAM in two different positions: a highly selective PET radioligand for the serotonin transporter. J Labelled Compd Rad. 2001;44:1013–1023. [Google Scholar]
- Jarkas N, Votaw JR, Voll RJ, Williams L, Camp VM, Owens MJ, et al. Carbon-11 HOMADAM: a novel PET radiotracer for imaging serotonin transporters. Nucl Med Biol. 2005;32:211–224. doi: 10.1016/j.nucmedbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lundberg J, Odano I, Olsson H, Halldin C, Farde L. Quantification of 11C-MADAM binding to the serotonin transporter in the human brain. J Nucl Med. 2005;46:1505–1515. [PubMed] [Google Scholar]
- Nye JA, Votaw JR, Jarkas N, Purselle D, Camp V, Bremner JD, et al. Compartmental modeling of 11C-HOMADAM binding to the serotonin transporter in the healthy human brain. J Nucl Med. 2008;49:2018–2025. doi: 10.2967/jnumed.108.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmeier CA. Involvement of serotonin in depression: evidence from postmortem and imaging studies of serotonin receptors and the serotonin transporter. J Psychiatr Res. 2003;37:357–373. doi: 10.1016/s0022-3956(03)00050-5. [DOI] [PubMed] [Google Scholar]
- Carson RE, Barker WC, Liow JS, Johnson CA. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. IEEE 2003 Nuclear Sci Symp Conf Rec. 2003;5:3281–3285. [Google Scholar]
- Hilton J, Yokoi F, Dannals RF, Ravert HT, Szabo Z, Wong DF. Column-switching HPLC for the analysis of plasma in PET imaging studies. Nucl Med Biol. 2000;27:627–630. doi: 10.1016/s0969-8051(00)00125-6. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Viola P, Wells WM., III Alignment by maximization of mutual information. Int J Comput Vision. 1997;24:137–154. [Google Scholar]
- Ding YS, Singhal T, Planeta-Wilson B, Gallezot JD, Nabulsi N, Labaree D, et al. PET imaging of the effects of age and cocaine on the norepinephrine transporter in the human brain using (S,S)-[11C]O-methylreboxetine and HRRT. Synapse. 2010;64:30–38. doi: 10.1002/syn.20696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Neumeister A, Williams W, Ropchan J, Labaree D, Lin S, et al. Comparative evaluation of the serotonin transporter tracers [C-11]AFM and [C-11]DASB in humans J Nucl Med 2009501205abstract.19617339 [Google Scholar]
- Gunn RN, Gunn SR, Cunningham VJ. Positron emission tomography compartmental models. J Cereb Blood Flow Metab. 2001;21:635–652. doi: 10.1097/00004647-200106000-00002. [DOI] [PubMed] [Google Scholar]
- Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- Kish SJ, Furukawa Y, Chang LJ, Tong J, Ginovart N, Wilson A, et al. Regional distribution of serotonin transporter protein in postmortem human brain: is the cerebellum a SERT-free brain region. Nucl Med Biol. 2005;32:123–128. doi: 10.1016/j.nucmedbio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Ichise M, Toyama H, Innis RB, Carson RE. Strategies to improve neuroreceptor parameter estimation by linear regression analysis. J Cereb Blood Flow Metab. 2002;22:1271–1281. doi: 10.1097/01.WCB.0000038000.34930.4E. [DOI] [PubMed] [Google Scholar]
- Frankle WG, Huang Y, Hwang DR, Talbot PS, Slifstein M, Van Heertum R, et al. Comparative evaluation of serotonin transporter radioligands 11C-DASB and 11C-McN 5652 in healthy humans. J Nucl Med. 2004;45:682–694. [PubMed] [Google Scholar]
- Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. NeuroImage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- Wu Y, Carson RE. Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab. 2002;22:1440–1452. doi: 10.1097/01.WCB.0000033967.83623.34. [DOI] [PubMed] [Google Scholar]
- Ichise M, Liow JS, Lu JQ, Takano A, Model K, Toyama H, et al. Linearized reference tissue parametric imaging methods: application to [11C]DASB positron emission tomography studies of the serotonin transporter in human brain. J Cereb Blood Flow Metab. 2003;23:1096–1112. doi: 10.1097/01.WCB.0000085441.37552.CA. [DOI] [PubMed] [Google Scholar]
- Buck A, Westera G, vonSchulthess GK, Burger C. Modeling alternatives for cerebral carbon-11-iomazenil kinetics. J Nucl Med. 1996;37:699–705. [PubMed] [Google Scholar]
- Akaike H. New look at statistical-model identification. IEEE Trans Autom Control. 1974;Ac19:716–723. [Google Scholar]
- Schwarz G. Estimating dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- Hinz R, Selvaraj S, Murthy NV, Bhagwagar Z, Taylor M, Cowen PJ, et al. Effects of citalopram infusion on the serotonin transporter binding of [11C]DASB in healthy controls. J Cereb Blood Flow Metab. 2008;28:1478–1490. doi: 10.1038/jcbfm.2008.41. [DOI] [PubMed] [Google Scholar]
- Parsey RV, Ojha A, Ogden RT, Erlandsson K, Kumar D, Landgrebe M, et al. Metabolite considerations in the in vivo quantification of serotonin transporters using 11C-DASB and PET in humans. J Nucl Med. 2006;47:1796–1802. [PubMed] [Google Scholar]
- Ginovart N, Wilson AA, Meyer JH, Hussey D, Houle S. Positron emission tomography quantification of [11C]-DASB binding to the human serotonin transporter: modeling strategies. J Cereb Blood Flow Metab. 2001;21:1342–1353. doi: 10.1097/00004647-200111000-00010. [DOI] [PubMed] [Google Scholar]
- Lassen NA. Neuroreceptor quantitation in vivo by the steady-state principle using constant infusion or bolus injection of radioactive tracers. J Cereb Blood Flow Metab. 1992;12:709–716. doi: 10.1038/jcbfm.1992.101. [DOI] [PubMed] [Google Scholar]
- Frankle WG, Slifstein M, Gunn RN, Huang Y, Hwang DR, Darr EA, et al. Estimation of serotonin transporter parameters with 11C-DASB in healthy humans: reproducibility and comparison of methods. J Nucl Med. 2006;47:815–826. [PubMed] [Google Scholar]
- Carson RE, Kiesewetter DO, Jagoda E, Der MG, Herscovitch P, Eckelman WC. Muscarinic cholinergic receptor measurements with [18F]FP-TZTP: control and competition studies. J Cereb Blood Flow Metab. 1998;18:1130–1142. doi: 10.1097/00004647-199810000-00010. [DOI] [PubMed] [Google Scholar]
- Parsey RV, Ogden RT, Mann JJ. Determination of volume of distribution using likelihood estimation in graphical analysis: elimination of estimation bias. J Cereb Blood Flow Metab. 2003;23:1471–1478. doi: 10.1097/01.WCB.0000099460.85708.E1. [DOI] [PubMed] [Google Scholar]
- Ogden RT, Ojha A, Erlandsson K, Oquendo MA, Mann JJ, Parsey RV. In vivo quantification of serotonin transporters using [11C]DASB and positron emission tomography in humans: modeling considerations. J Cereb Blood Flow Metab. 2007;27:205–217. doi: 10.1038/sj.jcbfm.9600329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmenhorst D, Meyer PT, Matusch A, Winz OH, Zilles K, Bauer A. Test–retest stability of cerebral A1 adenosine receptor quantification using [18F]CPFPX and PET. Eur J Nucl Med Mol Imag. 2007;34:1061–1070. doi: 10.1007/s00259-006-0309-x. [DOI] [PubMed] [Google Scholar]
- Zanderigo F, Ogden RT, Bertoldo A, Cobelli C, Mann JJ, Parsey RV. Empirical Bayesian estimation in graphical analysis: a voxel-based approach for the determination of the volume of distribution in PET studies. Nucl Med Biol. 2010;37:443–451. doi: 10.1016/j.nucmedbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginovart N, Willeit M, Rusjan P, Graff A, Bloomfield PM, Houle S, et al. Positron emission tomography quantification of [11C]-(+)-PHNO binding in the human brain. J Cereb Blood Flow Metab. 2007;27:857–871. doi: 10.1038/sj.jcbfm.9600411. [DOI] [PubMed] [Google Scholar]
- Seki C, Ito H, Ichimiya T, Arakawa R, Ikoma Y, Shidahara M, et al. Quantitative analysis of dopamine transporters in human brain using [11C]PE2I and positron emission tomography: evaluation of reference tissue models. Ann Nucl Med. 2010;24:249–260. doi: 10.1007/s12149-010-0364-z. [DOI] [PubMed] [Google Scholar]
- Gallezot JD, Nabulsi N, Neumeister A, Planeta-Wilson B, Williams WA, Singhal T, et al. Kinetic modeling of the serotonin 5-HT1B receptor radioligand [11C]P943 in humans. J Cereb Blood Flow Metab. 2010;30:196–210. doi: 10.1038/jcbfm.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schain M, Toth M, Cselenyi Z, Stenkrona P, Halldin C, Farde L, et al. Quantification of serotonin transporter availability with [11C]MADAM—a comparison between the ECAT HRRT and HR systems. NeuroImage. 2012;60:800–807. doi: 10.1016/j.neuroimage.2011.12.047. [DOI] [PubMed] [Google Scholar]
- Parsey RV, Kent JM, Oquendo MA, Richards MC, Pratap M, Cooper TB, et al. Acute occupancy of brain serotonin transporter by sertraline as measured by [11C]DASB and positron emission tomography. Biol Psychiatry. 2006;59:821–828. doi: 10.1016/j.biopsych.2005.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.