Abstract
The transcription factor Krüppel-like factor 4 (KLF4) can activate or repress gene expression in a cell-context dependent manner. We have previously shown that KLF4 inhibits the proliferation of naïve CD8+ T cells in vitro downstream of the transcription factor ELF4. In this work, we describe a novel role of KLF4 in the differentiation of CD8+ T cells upon infection. Loss of KLF4 had minimal effect on thymic T cell development and distribution of mature T cells in the spleen, blood, and lymph nodes. KLF4-deficient naïve CD8+ T cells also displayed normal homeostatic proliferation upon adoptive transfer into lymphopenic hosts. However, activation of KLF4-deficient naïve CD8+ T cells by in vitro TCR crosslink and co-stimulation resulted in increased proliferation. Furthermore, naïve KLF4-deficient OT-I CD8+ T cells generated increased numbers of functional memory CD8+ T cells compared to wild type OT-I CD8+ T cells co-injected in the same recipient in both primary and recall responses to Listeria monocytogenes-OVA. Collectively, our data demonstrate that KLF4 regulates differentiation of functional memory CD8+ T cells while sparing development and homeostasis of naïve CD8+ T cells.
Keywords: KLF4, CD8+ T cells, proliferation, differentiation, memory
1. Introduction
The Krüppel-like factor family is comprised of highly conserved transcription factors that control proliferation, development, and differentiation [1]. The Krüppel-like factor 4 (KLF4) promotes re-programming of somatic cells into pluripotent stem cells [2,3]. Depending on the cellular context, KLF4 can act as a tumor suppressor or an oncogene [4–7] by either enhancing or inhibiting proliferation [8–10]. This duality in function can be attributed to the presence of both activating and inhibitory regulatory domains within the KLF4 protein [11,12]. In the immune system, KLF4 regulates the development of monocytes, maintenance of splenic dendritic cells, and differentiation of T cells [10,13–16]. In addition, KLF4 inhibits proliferation of B cells downstream of FOXO proteins [17]. We have previously shown that in vitro TCR stimulation of CD8+ T cells leads to a rapid downregulation of the transcription factor E74-like Factor 4(ELF4) followed by suppression of KLF4, a direct target of ELF4 [9,18]. In addition to the linear model ELF4→KLF4, differential target genes may lead to unique functions of KLF4 in lymphocytes. Increased in vitro proliferation of KLF4-deficient CD8+ T cells suggested a role of KLF4 in the quiescence of lymphocytes [9]. However, the role of KLF4 in the differentiation of CD8+ T cells has not been studied in vivo.
Naïve CD8+ T cells originate from bone marrow derived progenitor cells, which undergo sequential expansion, differentiation, and selection processes in the thymus to become functional and mono-specific T cells [19,20]. Once in peripheral tissues, naïve CD8+ T cells rely on TCR signaling for their maintenance and differentiation [21]. Low affinity self peptide–MHC recognitions, in synergy with IL-7 and IL-15, trigger rare events of cell divisions in naïve CD8+ T cells that result in a slow accumulation of CD8+ T cells with a memory phenotype [21]. On the opposite end of the spectrum, acute infections cause vigorous activation and differentiation of pathogen-specific CD8+ T cells by providing strong TCR stimulation and co-stimulation (signals 1 and 2) and eliciting high levels of inflammatory cytokines (signal 3) [22,23]. Immediate and long-term protection of the host is ensured by the generation of short-lived effector and longlived memory CD8+ T cells. Even though the conventional in vitro activation of naïve CD8+ T cells by plate bound anti-CD3 and anti-CD28 antibodies also efficiently triggers proliferation, this system does not accurately resemble the strength of stimulation and effect of the microenvironment to ensure a physiological differentiation program. Therefore, we decided to investigate the role of KLF4 in homeostatic processes of development and differentiation of CD8+ T cells in response to infectious agents because of the capacity of KLF4 to inhibit in vitro proliferation of CD8+ T cells [9].
In the present study, we show that KLF4 does not have a significant role in the homeostatic proliferation of CD8+ T cells and thymic differentiation but lessens the formation of memory CD8+ T cells after infection with Listeria monocytogenes. Our findings revealed a novel function of KLF4 in the regulation of memory CD8+ T cell development in response to infection.
2. Material and Methods
2.1. Mice
Klf4fl/fl mice were obtained from Dr. K. Kaestner and bred with Vav-Cre and CD4-Cre mice as previously described [24,25]. For experiments with Lm-OVA, Klf4fl/fl CD4-Cre and B6.SJL mice were bred with the OT-I transgenic mice to obtain Klf4fl/fl CD4-Cre OT-I (CD45.2+) and B6.SJL OT-I (CD45.1+) mice, respectively. C57BL/6 mice were crossed with B6.SJL to obtain B6xB6.SJL (CD45.1+ CD45.2+) recipients. Mice were maintained in specific pathogen-free conditions at Baylor College of Medicine. All experiments and procedures were done in compliance with the Institutional Animal Care and Usage Committee of Baylor College of Medicine.
2.2. T cell proliferation
Naïve CD44low CD8+ T cells were purified from the spleen of Klf4fl/fl and Klf4fl/fl Vav-Cre mice and labeled with 4 µM CFSE (Invitrogen) according to the manufacturer’s instructions. For in vitro proliferation assay, CD8+ T cells were then cultured in 96-well plates (105 cells per well) coated with 2 µg/ml or 10 µg/ml anti-CD3ε (Bio × Cell) in RPMI media (Lonza) containing 10% FBS in the presence of 2 µg/ml anti-CD28 (BD Biosciences). Dilution of the CFSE dye was analyzed by flow cytometry on days 2.5 and 3 after plating. For in vivo homeostatic proliferation assay, purified naïve CD8+ T cells were labeled with CFSE and adoptively transferred (4×105 per mouse i.v.) into congenic B6.SJL recipient mice sub-lethally irradiated (4.5 Gy) 15 h before to induce lymphopenia. CFSE dilution was analyzed by flow cytometry in donor-derived CD8+ T cells isolated from the spleen on day 9 after transfer. For thymocyte proliferation assay, BrdU (10mg/ml) was injected i.p. to 3 month-old mice and thymi were collected 3 h later. BrdU detection was performed using BrdU Detection Kit (BD Biosciences) by flow cytometry.
2.3. Infection with Lm-OVA
Naïve splenic OT-I CD8+ T cells from wild type (WT) B6.SJL OT-I or Klf4fl/fl CD4-Cre OT-I mice were negatively purified with BD-Imag magnetic separation system (BD Biosciences). Biotinylated anti-CD44 antibodies (BD Biosciences) were added to the antibody cocktail to enrich for CD44low CD8+ T cells (90–95% purity). CD8+ T cells were adoptively transferred by i.v. injection into sex- and age-matched B6xB6.SJL recipients (1×103 OT-I cells per mouse). Recipient mice were infected 24 h later with a recombinant OVA-expressing Listeria monocytogenes strain (Lm-OVA, 4×103 CFU i.v.). To study secondary responses, mice were re-challenged with Lm-OVA (200×103 CFU, i.v.) 60 d after initial infection. The expansion of OT-I T cells was monitored in peripheral blood by serial tail vein bleeding.
2.4. Flow cytometry and cell sorting
Cells were pre-incubated with anti-CD16/CD32 (BioLegend) to block Fc receptors prior to cell surface staining. The following antibodies were used for surface staining: allophycocyanin-labeled anti-CD45.1, PerCP-Cy5.5-labeled anti-CD45.2, FITC-labeled anti-CD62L, Alexa Fluor® 488 labeled anti-CD44, PE-Cy7-labeled anti-CD8, PerCP-Cy5.5-labeled anti-CD4, PerCP-Cy5.5-labeled anti-CD8, PE-Cy7-labeled anti-c-kit, Alexa Fluor® 647-labeled anti-CD25 (BioLegend); PE-labeled anti-CD4, PerCP-Cy5.5-labeled streptavidin, FITC-labeled anti-Sca1, APC-labeled anti-c-kit, and PE-labeled anti-IL7Rα (BD Biosciences); PerCP-Cy5.5-labeled anti-CD25, APC-labeled anti-CD8, and APC-eFluor® 780-labeled anti-CD24 (eBioscience). For lineage depletion we used the hematopoietic progenitor cell enrichment set (BD Biosciences). Thymocyte subsets were sorted on a MoFlo cell sorter (Cytomation, 97–99% purity). To study apoptosis, thymocytes were first stained with surface marker and then stained for Annexin V by using the Apoptosis Detection Kit from BD Biosciences. Effector functions of OT-I CD8+ T cells were evaluated by activating splenocytes for 5 hours with 1 µg/ml OVA(257–264) peptide (AnaSpec) in RPMI medium containing 10% FBS at 37°C in the presence of 1 µl/ml Golgi Plug and 0.8 µl/ml Golgi Stop (BD Biosciences). Cells were then stained for CD8, CD45.1 and CD45.2, fixed and permeabilized using BD Cytofix/Cytoperm buffer (BD Biosciences), and finally stained with FITC-labeled anti-IFNγ, allophycocyanin-labeled anti-TNFα (BD Biosciences), PE-labeled anti-perforin, or FITC-labeled anti-granzyme B (eBioscience).
2.5. Quantitative real-time and genomic PCR
Total RNA was extracted using RNeasy Micro kit (Qiagen) and cDNA was synthesized using a SuperScript III kit (Invitrogen) with random hexamer primers. Real-time PCR was performed on the Mx3005P QPCR System (Agilent Technologies) using LightCycler FastStart DNA Master SYBR Green I kit (Roche). The following primers were used: Actin forward, 5′-GTGGGCCGCTCTAGGCACCA-3′, and reverse, 5′-CGGTTGGCCTTAG GGTTCAGGGG-3′; KLF4 forward, 5′- CTGAACAGCAGGGACTGTCA -3′, and reverse, 5′- GTGTGGGTGGCTGTTCTTTT -3′. To assess deletion of the KLF4 gene, genomic DNA was extracted from bone marrow, thymus and purified splenic CD8+ T cells using DNeasy Blood and Tissue kit (QIAGEN). The following PCR primers were used: forward-1, 5’- CTGGGCCCCCACATTAATGAG- 3’; forward-2, 5’-CGCTGACAGCCATGTCAGACT-3’; reverse, 5’-CAGAGCCGTTCTGGCTGTTTT-3’.
2.6. Statistical analysis
Statistical significance was determined by two-tailed Student t-tests calculated with GraphPad Prism 5 software as indicated in the legend to figures.
3. Results
3.1. T cell development in Klf4-deficient mice
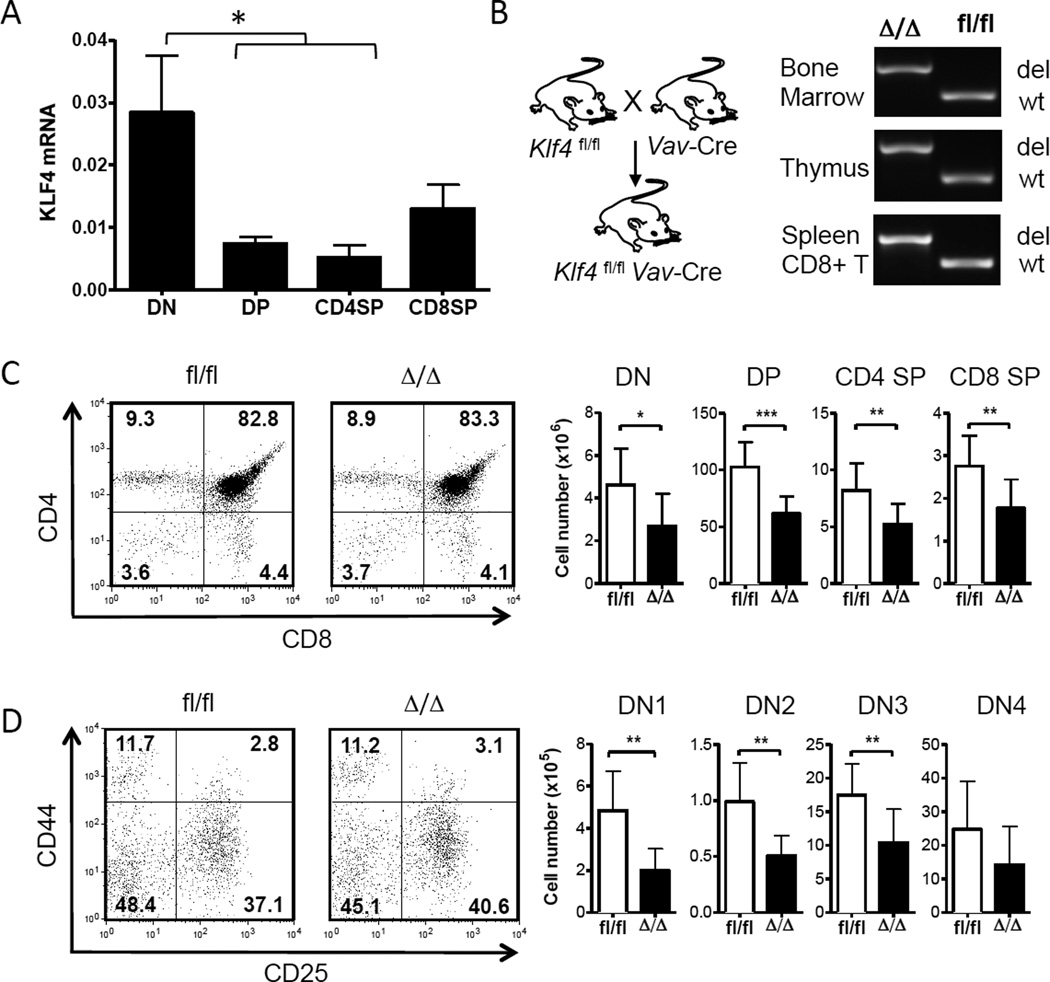
We first assessed the relative expression of KLF4 at different stages of thymic development before studying immune response of mature CD8+ T cells. Double-negative (DN; CD4−CD8−), double-positive (DP; CD4+CD8+) and single-positive (SP) CD4+ and CD8+ T cells were purified from the thymi of wild-type CD57BL/6 mice by cell sorting. KLF4 transcripts were elevated in DN thymocytes and decreased in DP thymocytes and mature SP T cells (Fig. 1A), suggesting that KLF4 may play a role in early stages of T cell development. We used Klf4fl/fl Vav-Cre+ mice to delineate the role of KLF4 in thymocytes [24]. In this model, KLF4 was deleted in all hematopoietic cells including the bone marrow-derived T cell progenitor cells that emigrate to thymus for lineage commitment and development of functional mono-specific T cells. KLF4 deletion was confirmed by PCR with genomic DNA from bone marrow cells, thymocytes, and splenic CD8+ T cells (Fig. 1B). Loss of KLF4 did not alter frequency of major subsets based on the expression of CD4 and CD8 in Klf4fl/fl Vav-Cre (Δ/Δ) and Klf4fl/fl (fl/fl) thymocytes (Fig. 1C). The frequencies of DN subsets defined by the expression of CD44 and CD25 (DN1- DN4) were also similar to controls (Fig. 1D). However, a reduction in the thymic cellularity of KLF4-deficient mice resulted in lower numbers of DN1-DN4, DP, and SP cells (Fig. 1C, D). A previous report demonstrated that deletion of KLF4 at the DN3-DN4 stage with CD4-Cre lowered thymic cellularity by reducing proliferation of DN thymocytes [10].
Fig. 1. Redundant role of KLF4 in T cell development.
(A) Relative expression of KLF4 transcripts was measured in double-negative (DN; CD4− CD8−), double-positive (DP; CD4+ CD8+), and CD4+ and CD8+ single-positive (SP) thymocytes purified from WT thymi by cell sorting (mean ± SD, n=3). (B) Diagram depicting deletion of KLF4 gene in genomic DNA extracted from bone marrow, thymus, and purified splenic CD8+ T cells of KLF4fl/fl Vav-Cre (Δ/Δ) and Cre-negative control (fl/fl) CD8+ T cells was assessed by PCR (del = deleted allele; wt = wild-type allele). (C) Representative dot plots showing expression of CD4 and CD8 on thymocytes. Numbers indicate percentage of each population. Corresponding cell counts are shown as bar graphs (mean ± SD, n=9). (D) Expression of CD25 and CD44 on DN thymocytes with indicated relative frequency of each sub-population. Total cell counts of DN1 (CD44high CD25low), DN2 (CD44high CD25high), DN3 (CD44low CD25high) and DN4 (CD44low CD25low) sub-populations are shown as bar graphs (mean ± SD, n=9). *, P<0.05; **, P<0.01; ***, P<0.005 (unpaired two-tailed Student t-test).
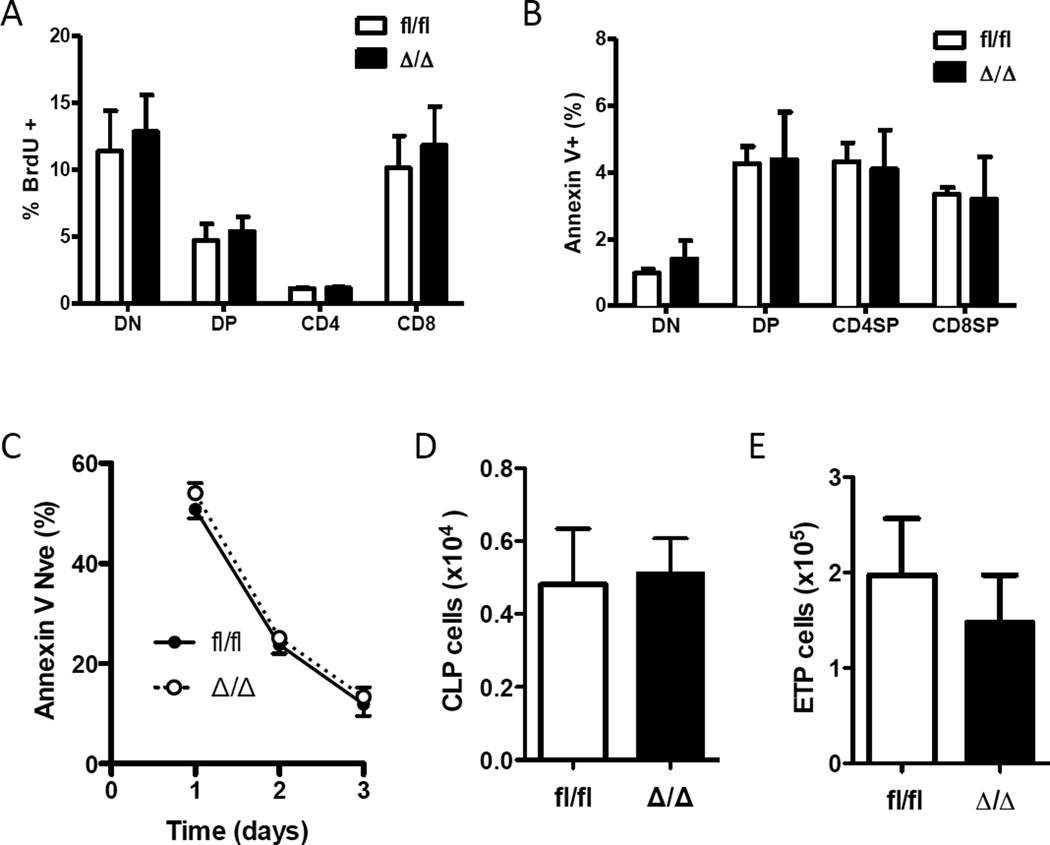
In our model, the reduction of KLF4-deficient thymocyte numbers was not due to the altered proliferation, as evidenced by similar frequencies of BrdU-positive cells compared to the control group (Fig. 2A). Of interest, we also did not observe differences in survival since the frequency of Annexin V-positive thymocytes was similar in both groups (Fig. 2B). Survival was further examined by in vitro culture of thymocytes from wild type and KLF4-deficient mice and analyzed by the expression of Annexin V. Loss of KLF4 did not affect the number of viable thymocytes (Annexin V negative), suggesting a similar cell intrinsic predisposition to undergo cell death (Fig. 2C). Therefore, it is possible that a lower thymic cellularity was caused by a paucity of thymic immigrants. However, we found a similar number of common lymphoid progenitor cells (Lin− Sca-1int c-kitint IL7Rα+) in bone marrow (Fig. 2D) and early thymic progenitor cells (Lin− CD4− CD8− CD44+ CD25− c-kit+ CD24lo) in thymus (Fig. 2E) [19,26]. Despite reduced numbers of CD4+ and CD8+ SP in KLF4-deficient thymi, we did not detect significant differences in the numbers of mature CD4+ and CD8+ T cells in the spleen and inguinal lymph nodes, and frequency in peripheral blood (Appendix).
Fig. 2. Normal seeding, proliferation, and survival of KLF4-deficient thymocytes.
(A) Percent of BrdU-positive cells in each thymic population was determined by flow cytometry 3 h after BrdU injection (mean ± SD, n=4). (B) Percentage of Annexin V-positive cells in DN, DP and single-positive thymocytes are shown as bar graphs (mean ± SD, n=3). (C) Survival of thymocytes to in vitro culture was examined by flow cytometric detection of Annexin V. Live thymocytes (Annexin V negative) were determined at each time (mean ± SD, n=4). (D) Number of common lymphoid progenitors (CLP) in bone marrow was determined by flow cytometry (Lin− Sca-1int c-kitint IL7Rα+) (mean ± SD, n=5). (E) Number of early thymic progenitor cells (Lin− CD4− CD8− CD44+ CD25− c-kit+ CD24lo) was determined by flow cytometry (mean ± SD, n=4).
3.2. Dispensable role of KLF4 in homeostatic proliferation of naïve CD8+ T cells
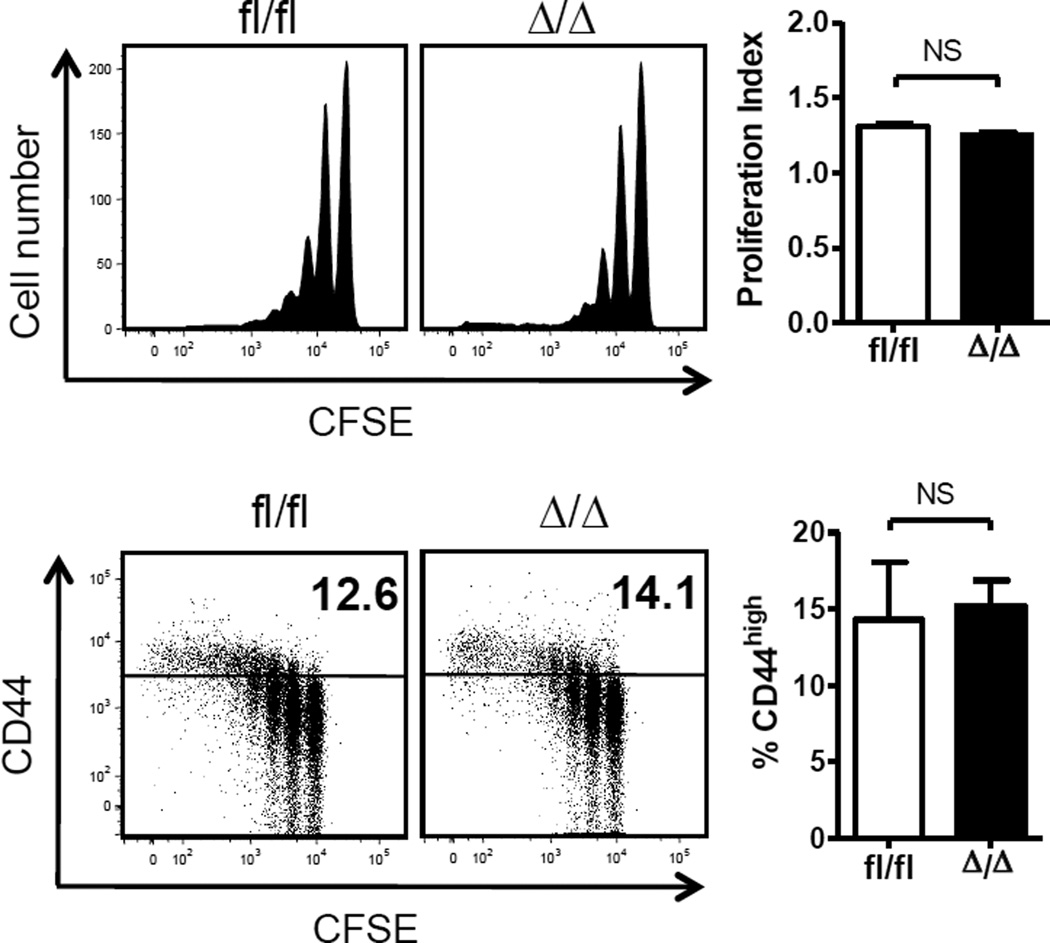
T cell development is regulated by sequential processes of proliferation and differentiation. A lower thymic cellularity with normal T cell numbers in the spleen of Klf4fl/fl Vav-Cre mice suggests compensation with mechanisms of T cell maintenance. To further dissect the role of KLF4 in homeostatic proliferation of CD8+ T cells, we analyzed lymphopenia-induced proliferation of KLF4-deficient CD8+ T cells, which is largely driven by weak interactions with self peptide-MHC complexes in concert with IL-7 [21]. We adoptively transferred naïve CFSE-labeled CD44low CD8+ T cells purified from Klf4fl/fl Vav-Cre+ or Klf4fl/fl mice into lymphopenic mice, generated by sub-lethal irradiation, and analyzed CFSE dilution in the spleen 9 days after adoptive transfer. No significant differences were observed in T cell activation, based on the expression of CD44 and proliferation of KLF4-deficient CD8+ T cells (Fig. 3). These findings suggest that KLF4 plays a redundant role in the homeostatic proliferation of CD8+ T cells and thus previously reported enhanced proliferation likely takes place under stronger stimulation [9].
Fig. 3. KLF4 deletion in CD8+ T cells does not affect homeostatic proliferation.
Homeostatic proliferation of splenic naive CD8+ T cells after adoptive transfer into lymphopenic recipient mice. Proliferation index for each group is plotted as bar graph (mean ± SD; n=3). Representative dot plots on the bottom show upregulation of CD44 in proliferating cells. Percentage of CD44high cells is shown on each dot plot and plotted as bar graph (mean ± SD; n=3). Data represent 2 independent experiments.
3.3. Loss of KLF4 increases in vitro proliferation of CD8+ T cells
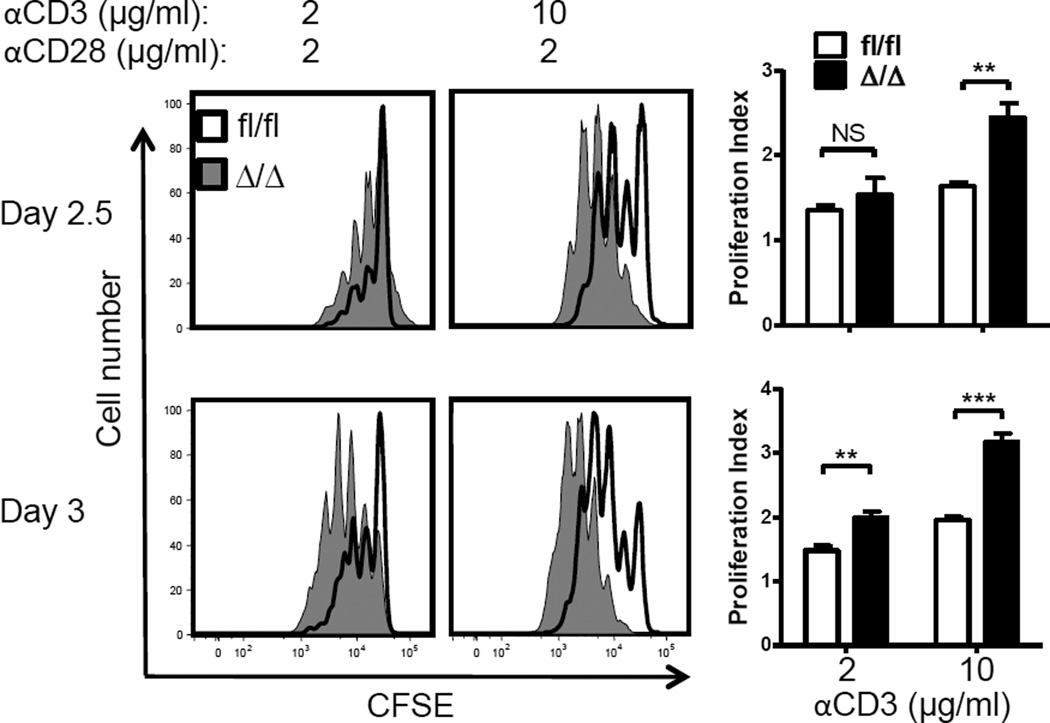
The transcription factor ELF4 controls proliferation of naïve CD8+ T cells in part by activating the KLF4 gene [9]. Somatic deletion of the KLF4 gene using the Klf4fl/fl Mx1-Cre system led to increased in vitro proliferation of CD8+ T cells [9]. However, in this model systemic deletion of KLF4 was induced by type I interferons secreted in response to poly-I:C administration and CD8+ T cells were isolated for analysis 9 months later, which may have affected the function of naïve T cells [27]. To rule out artifacts of inducible deletion, naïve CD44low CD8+ T cells isolated from the spleen of Klf4fl/fl Vav-Cre+ mice were labeled with CFSE and activated in vitro on plates coated with anti-CD3 (2 or 10 µg/ml) in the presence of anti- CD28 (2 µg/ml). Consistent with our previous observations, KLF4-deficient CD8+ T cells showed increased proliferation in both conditions (Fig. 4). By increasing the density of crosslinking antibody and degree of stimulation, we observed a more pronounced increase in the proliferative response of KLF4-deficient CD8+ T cells suggesting that KLF4 may restrict response to strong stimulations, such as bacterial infection.
Fig. 4. KLF4 restrains CD8+ T cell proliferation in response to TCR stimulation.
CFSE dilution profiles of purified CD8+ T cells cultured on plates coated with either 2 µg/ml or 10 µg/ml of anti-CD3 antibody. Corresponding proliferation indexes were plotted as bar graphs (mean ± SD, n=3). Data represent 3 independent experiments. **, P<0.01; ***, P<0.005 (unpaired two-tailed Student t-test)
3.4. KLF4 restricts generation of memory CD8+ T cells after Lm-OVA infection
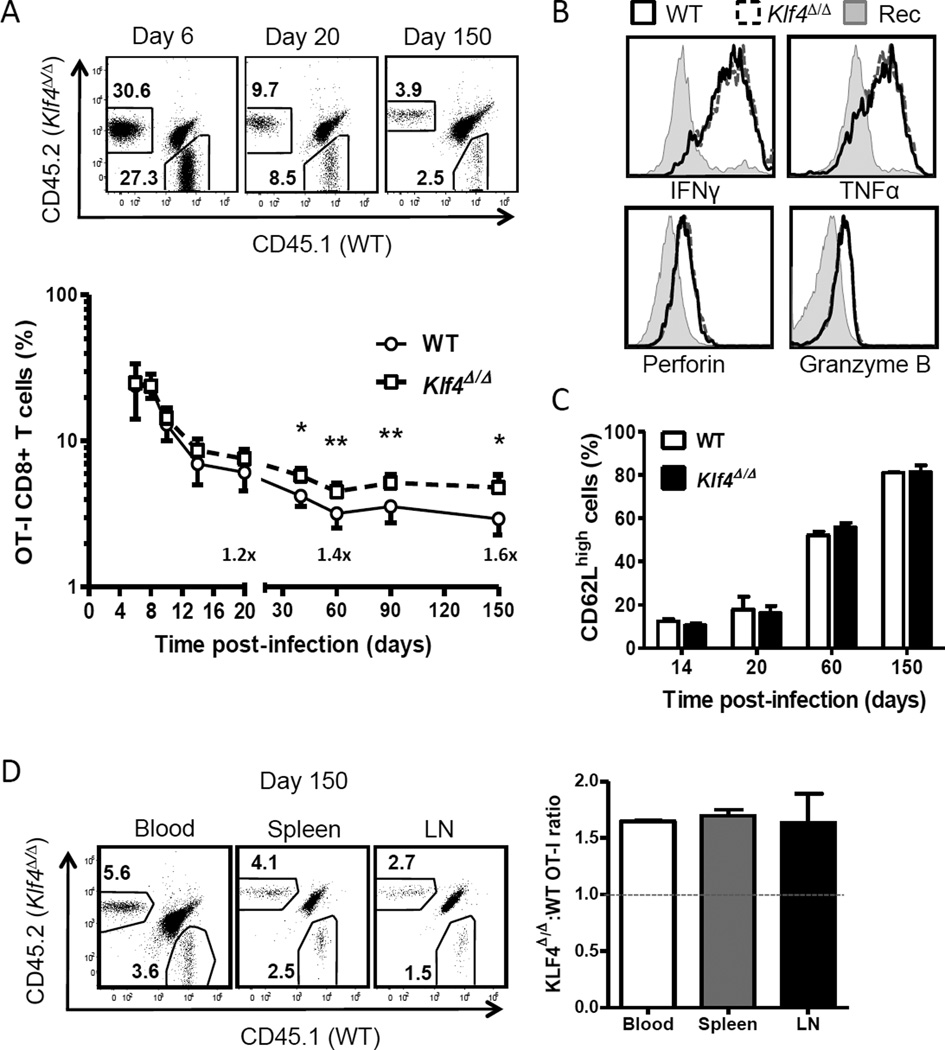
Systemic infection with Lm-OVA results in a strong activation, proliferation and differentiation of naïve CD8+ T cells to effector and memory T cells [23]. We therefore studied the response to Lm-OVA infection of KLF4-deficient CD8+ T cells expressing transgenic OVAspecific TCR (OT-I) in an adoptive co-transfer model to compare to wild type CD8+ T cells in same recipient mice. We purified naïve CD44low CD8+ T cells from Klf4fl/fl CD4-Cre OT-I and WT OT-I mice and adoptively transferred a 1:1 mixture to congenic recipient hosts followed by infection with Lm-OVA. Then, donor WT and KLF4-deficient CD8+ T cells were monitored in peripheral blood by flow cytometry. Although the expansion of KLF4-deficient CD8+ T cells was comparable with WT CD8+ T cells at the peak of response (day 6), we observed 40–60% increase of KLF4-deficient memory CD8+ T cells at the end of immune response (Fig. 5A).
Fig. 5. KLF4 regulates formation of memory CD8+ T cells upon Lm-OVA infection.
(A) Expansion of co-transferred WT (CD45.1+) and KLF4fl/fl CD4-Cre (Klf4Δ/Δ) (CD45.2+) OT-I CD8+ T cells in peripheral blood of congenic recipient mice infected with Lm-OVA. Representative dot plots and total kinetics are shown for days 6, 20 and 150 post-infection. (B) Expression of IFNγ, TNFα, perforin and granzyme B was assessed in WT and Klf4Δ/Δ effector OT-I CD8+ T cells by intracellular staining. Solid and dashed histograms represent WT and Klf4Δ/Δ OT-I effector CD8+ T cells, respectively; gray filled histograms show corresponding profiles of recipient CD8+ T cells used as a control. (C) Percentages of CD62Lhigh OT-I CD8+ T cells in each donor-derived population is plotted as bar graphs throughout the response. (D) Relative frequencies of WT and Klf4Δ/Δ OT-I memory CD8+ T cells in blood, the spleen and inguinal lymph nodes (LN) are shown in representative dot plots. The ratio of Klf4Δ/Δ to WT OT-I CD8+ T cells in each tissue was calculated and plotted as bar graphs. Dashed line indicates initial ratio upon infection. The data are representative of 2 independent experiments (mean ± SD, n=3). *, P<0.05; **, P<0.01; (paired two-tailed Student t-test).
Since KLF2 regulates the expression of CD62L and trafficking of T cells, a member of the KLF family with the high homology to KLF4 [1,28], we next investigated differentiation and tissue distribution of KLF4-deficient CD8+ T cells. In contrast to KLF2, loss of KLF4 did not affect acquisition of effector functions and polarization to CD62Lhigh central and CD62Llow effector memory CD8+ T cells (Fig. 5B,C). At day 150 post-infection, we detected a similar distribution of KLF4-deficient and WT OT-I CD8+ T cells in the spleen, blood and lymph nodes, indicating that loss of KLF4 enhanced memory development without altering their homing capacity to these tissues (Fig. 5D).
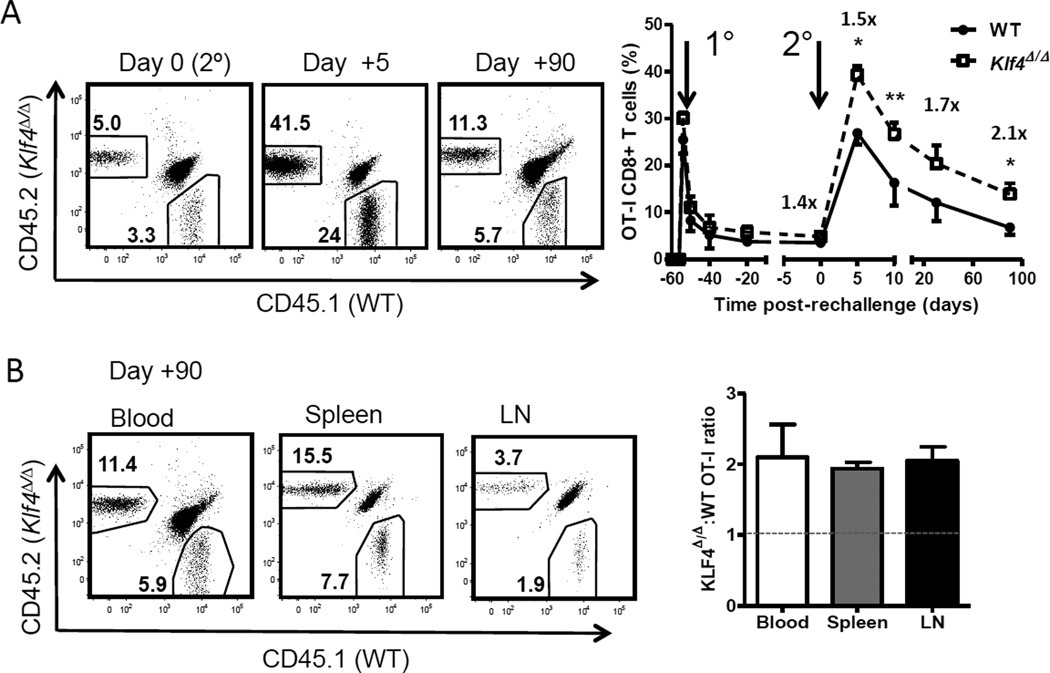
An important feature of memory T cells is their capacity to rapidly respond to a secondary encounter with same antigen/pathogen. Thus, we re-challenged mice that received 60 days earlier a 1:1 mixture of naïve WT and KLF4-deficient OT-I CD8+ T cells followed by infection with Lm-OVA to test the role of KLF4 in the function of memory CD8+ T cells. Wild type and KLF4-deficient memory CD8+ T cells showed a comparable expansion 5 days after rechallenge whereas the number of KLF4-deficient memory T cells left at the end of the recall response was significantly increased (Fig. 6A). Similar to primary response, the ratio of WT and KLF4-deficient memory CD8+ T cells increased uniformly in blood, spleen and lymph nodes and reached 2:1 on day 90 after re-challenge (Fig. 6B). Altogether, our findings show that KLF4 regulates the formation of memory CD8+ T cells following Lm-OVA infection.
Fig. 6. Loss of KLF4 augments secondary response of memory CD8+ T cells.
(A) Secondary expansion of WT and Klf4Δ/Δ OT-I memory CD8+ T cells in blood. Naive WT and Klf4Δ/Δ OT-I CD8+ T cells were mixed at 1:1 ratio, adoptively transferred into congenic recipients and activated with Lm-OVA infection as shown in Fig. 4. Sixty days later, recipient mice were re-challenged with a lethal dose of Lm-OVA. Representative dot plots show total blood CD8+ T cells with relative frequencies of WT and Klf4Δ/Δ OT-I CD8+ T cells before re-challenge (day 0), at the peak of secondary response (day +5) and 90 days after re-challenge (day +90). Total kinetics of primary and secondary response is plotted as percentage of total CD8+ T cells in blood. (B) Relative frequencies of secondary memory WT and Klf4Δ/Δ OT-I CD8+ T cells in the spleen, blood and inguinal lymph nodes (LN) are shown in representative dot plots. The ratio of Klf4Δ/Δ to WT OT-I CD8+ T cells in each tissue plotted as bar graphs. Dashed line indicates the ratio in blood before rechallenge. The data are representative of 2 independent experiments (mean ± SD, n=3). *, P<0.05; **, P<0.01; (paired two-tailed Student t-test).
4. Discussion
The transcriptional programs that regulate the development of effector and memory T cells to efficiently eliminate pathogens and ensure long-lasting protection have not been completely elucidated. In this work, we show that loss of KLF4 increased formation of memory CD8+ T cells in both primary and secondary responses to Lm-OVA in addition to restrain proliferation of CD8+ T cells activated by in vitro TCR stimulation. KLF4 had a minimal role in regulating differentiation of thymocytes and homeostatic proliferation of mature CD8+ T cells.
The total number of cells in the KLF4-deficient thymus was significantly reduced despite a normal phenotypic distribution of thymocytes. Loss of KLF4 did not affect survival and proliferation of thymocytes and the numbers of bone marrow-derived early thymic precursors (ETP). The numbers of common lymphoid progenitors –direct precursors of DN1 thymocytes– were also normal in KLF4-deficient bone marrow. Therefore, it is possible that KLF4 could modulate function of ETPs or control hematopoietic derived “niches” in the thymus for early immigrants. Despite a reduced thymic cellularity, peripheral CD4+ and CD8+ T cell counts were normal, suggesting that KLF4 is not required for the maintenance of mature T cell subsets.
We previously demonstrated that KLF4 is directly activated by ELF4 in CD8+ T cells [9]. KLF4-deficient CD8+ T cells shared some functional traits with ELF4-null CD8+ T cells including increased in vitro proliferation and homeostatic accumulation of memory-like CD8+ T cells [9]. Since ELF4 also regulates T cell activation by maintaining the pool of DUSP1 and DUSP5 [18], which are not regulated by KLF4, deletion of either ELF4 or KLF4 would likely reveal overlapping and unique functions. While deletion of ELF4 decreased threshold of TCR activation triggered by suboptimal stimulation, KLF4-deficient CD8+ T cells responded better to a stronger TCR stimulation. In contrast to ELF4, loss of KLF4 did not affect activation (upregulation of CD44) and rate of in vivo homeostatic proliferation of CD8+ T cells, indicating that the accumulation of CD44high memory-like CD8+ T cells in KLF4-deficient mice was likely driven by cell-extrinsic mechanisms [9].
Krüppel-like factors have been reported to control cellular quiescence, a reversible cell-cycle arrest actively regulated by cell extrinsic and intrinsic factors. Early work largely based on in vitro studies with Jurkat cells suggested that KLF2 induces quiescence in lymphocytes by activating p21 [29]. However, it was later shown that KLF2 is dispensable during in vivo responses to antigen, demonstrating a disparity between in vitro and in vivo assays [30]. Similarly, we found KLF4 controls proliferation of CD8+ T cells in a context dependent manner, which depended on the intensity and quality of signals. The expansion of KLF4-deficient effector CD8+ T cells following infection with Lm-OVA was similar to wild type, indicating that additional co-stimulation and cytokine signals present at the priming site likely overruled the cell cycle regulation observed in vitro. However, KLF4 may alternatively promote expansion or survival of a small number of memory precursor cells, which would be undetectable in a bulk population.
T cell trafficking is in part regulated by the transcription factor FoxO1 that directly activates KLF2, which in turn controls expression of homing molecules CD62L, CCR7 and S1P1 [28,31]. KLF4 is downregulated in FoxO1-null memory CD8+ T cells that displayed defective differentiation and low expression of CD62L and CCR7 [32,33]. These observations prompted us to examine the role of KLF4 in regulating differentiation and tissue distribution of CD8+ T cells after infection. No significant alterations were observed in the expression of CD62L (L-selectin) and tissue distribution of KLF4 deficient CD8+ T cells. However, KLF4-deficient CD8+ T cells produced more memory cells without affecting the central-to-effector memory ratio and tissue distribution. A larger pool of memory CD8+ T cells from primary response was further enhanced upon secondary challenge with same pathogen leading to 2-fold increase of memory CD8+ T cells, suggesting that KLF4-deficient memory T cells are functional and have increased proliferative capacity than controls. Collectively, this study described a new role for KLF4 in the proliferation of CD8+ T cells and their differentiation to immunological memory in response to infection.
Supplementary Material
HIGHLIGHTS.
KLF4 has minimal role in the development and homeostatic proliferation of CD8+ T cells.
KLF4 restrains proliferation of CD8+ T cells in response to in vitro TCR stimulation.
Loss of KLF4 augments development of CD8+ T cell memory after Listeria infection.
Acknowledgements
Authors acknowledge T. Goltsova and C. Threeton for cell sorting assistance, Drs. K. Schluns and V. Badovinac for providing Lm-OVA. This work was supported by National Institutes of Health Grants R01-AI077536 and R01-AI077536-02S1 (to H.D.L).
Abbreviations
- ELF4
E74-like Factor 4
- KLF4
Krüppel-like factor 4
- Lm-OVA
Listeria monocytogenes-OVA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nature reviews Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA, Huettner CS, et al. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood. 2007;109:747–755. doi: 10.1182/blood-2006-03-011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–2064. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 7.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nature cell biology. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 8.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat Immunol. 2009;10:618–626. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol Rev. 2006;211:154–163. doi: 10.1111/j.0105-2896.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 11.Evans PM, Liu C. Roles of Krupel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin (Shanghai) 2008;40:554–564. doi: 10.1111/j.1745-7270.2008.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. The EMBO journal. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alder JK, Georgantas RW, 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, et al. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008;180:5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park CS, Lee PH, Yamada T, Burns A, Shen Y, Puppi M, et al. Kruppel-like factor 4 (KLF4) promotes the survival of natural killer cells and maintains the number of conventional dendritic cells in the spleen. J Leukoc Biol. 91:739–750. doi: 10.1189/jlb.0811413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen X, Liu H, Xiao G, Liu X. Downregulation of the transcription factor KLF4 is required for the lineage commitment of T cells. Cell Res. 21:1701–1710. doi: 10.1038/cr.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, Sareen P, et al. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–681. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 18.Yamada T, Gierach K, Lee PH, Wang X, Lacorazza HD. Cutting edge: Expression of the transcription factor E74-like factor 4 is regulated by the mammalian target of rapamycin pathway in CD8+ T cells. J Immunol. 185:3824–3828. doi: 10.4049/jimmunol.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 21.Sprent J, Surh CD. Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat Immunol. 12:478–484. doi: 10.1038/ni.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtulus S, Tripathi P, Hildeman DA. Protecting and rescuing the effectors: roles of differentiation and survival in the control of memory T cell development. Front Immunol. 3:404. doi: 10.3389/fimmu.2012.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 24.Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- 25.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 26.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Ngoi SM, Tovey MG, Vella AT. Targeting poly(I:C) to the TLR3-independent pathway boosts effector CD8 T cell differentiation through IFN-alpha/beta. J Immunol. 2008;181:7670–7680. doi: 10.4049/jimmunol.181.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 29.Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23:8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- 30.Takada K, Wang X, Hart GT, Odumade OA, Weinreich MA, Hogquist KA, et al. Kruppel-like factor 2 is required for trafficking but not quiescence in postactivated T cells. J Immunol. 186:775–783. doi: 10.4049/jimmunol.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tejera MM, Kim EH, Sullivan JA, Plisch EH, Suresh M. FoxO1 Controls Effector-to-Memory Transition and Maintenance of Functional CD8 T Cell Memory. J Immunol. doi: 10.4049/jimmunol.1300331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med. 210:1189–1200. doi: 10.1084/jem.20130392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.