Abstract
Mechanisms of colorectal cancer (CRC) development can be roughly divided into three categories: genetic, epigenetic, and aberrant immunologic signaling pathways, all of which may be triggered by an imbalanced intestinal microbiota. Aberrant gut microbial composition, termed “dysbiosis”, has been reported in inflammatory bowel disease patients who are at increased risk for CRC development. Recent studies indicate that it is feasible to rescue experimental models of colonic cancer by oral treatment with genetically engineered beneficial bacteria and/or their immune-regulating gene products. Here, we review the mechanisms of epigenetic modulation implicated in the development and progression of CRC, which may be the result of dysbiosis, and therefore, may be amenable to therapeutic intervention.
Keywords: colorectal cancer, commensal bacteria, epigenetic regulation, inflammatory bowel disease, microbiota
A role for the microbiota in colorectal cancer development
Colorectal cancer (CRC) is one of the leading contributors to cancer death and morbidity in the United States, and is the fourth most commonly diagnosed cancer in the world, with more than one million new cases diagnosed annually [1, 2]. CRC begins with the formation of polyps, masses of cells protruding from the bowel wall, which subsequently progress to dysplastic adenomas, and ultimately, to colonic carcinoma [3]. The mechanisms of CRC pathogenesis have been widely investigated and current evidence indicates that genetic mutations, epigenetic changes, and aberrant immunologic signaling pathways are major contributors to CRC [4]. Mutations in tumor suppressor genes or oncogenes of colonic epithelial cells lead to dysregulated signaling networks that control cellular metabolism, differentiation, proliferation, and survival, resulting in the transformation of normal cells into cancer cells [5]. In addition to genetic mutations, epigenetic modulation of tumor suppressor genes, oncogenes, or pro-inflammatory mediators represent other important mechanisms whereby homeostatic balance is lost. Host chromatin can be altered epigenetically by covalent modification of histones (e.g., acetylation, methylation, phosphorylation, and ubiquitylation), or of DNA (e.g., cytosine methylation); additionally, chromatin can be regulated by nucleosome positioning that governs accessibility of trans-acting factors to DNA and the presence of non-coding RNAs [6]. Various epigenetic profile analyses comparing healthy and neoplastic cells indicate that epigenetic regulation plays a key role in tumor initiation and progression in CRC [5].
Cancer development and progression is a complex process involving a myriad of host-tumor interactions and countless molecular and cellular elements in the tumor microenvironment [7]. Recently, data have established a link between alterations in the commensal bacteria of the gut, termed the “microbiota”, and the pathogeneses of inflammatory bowel disease (IBD) and colitis-associated colorectal carcinoma (CAC) [8]. This is not surprising, as the gut microbiota is a pivotal component of the gastrointestinal ecosystem, wherein the microbes maintain complex interactions with host mucosal epithelial and immune cells. Microbial surface antigens and bacterial metabolic end-products can trigger downstream signaling pathways resulting in the activation of immune cells and the production of cytokines. Thus, a balance between immunostimulation against potential pathogens and immunosuppression against gut commensal microbes and dietary antigens is required for intestinal homeostasis [9].
This homeostasis is lost in IBD, a chronic autoinflammatory syndrome manifesting as Crohn's disease (CD) or ulcerative colitis (UC). CD is characterized by discontinuous, transmural lesions in the intestinal wall [10], while UC presents with diffuse, continuous, superficial inflammation in the colon [11]. Gradually, this chronic inflammation in the gut may provide a suitable microenvironment for aberrant molecular events [12]. For example, patients with IBD, especially UC, have an increased risk for developing CRC, and this risk is directly proportional to the extent of the colonic inflammation [9, 13]. Furthermore, a pathologic role for gut luminal bacteria in mucosal inflammation has been demonstrated in interleukin (IL)-2 and IL-10 gene knockout models of colitis [14]; and mice that are genetically susceptible to developing CRC have fewer tumors when living in germ-free conditions than when housed conventionally [2]. Thus, the importance of the intestinal microbiota during CRC development and progression is gaining significant attention.
It is thought that, in most patients, the progression of normal colonic mucosa to CRC requires a stepwise accumulation of genetic and epigenetic alterations [15]. The approximate amount of time for the malignant transformation of normal mucosa into adenomatous polyps and, finally, into invasive carcinoma is thought to be 5-15 years [16, 17]. This window of time provides an opportunity for early detection and therapeutic intervention [15]. Here, we review evidence for the role of epigenetic regulation in the development and progression of CRC and consider how an altered microbiota plays a role in these processes and may even represent a platform for intervention.
Intestinal microbiota are necessary for a healthy immune system
As most pathogens gain entry by breaching a mucosal surface, the mucosal epithelia and resident immune cells of the gastrointestinal, respiratory, and urogenital tracts play critical roles in the protection of the host against infection [18]. Within the healthy intestine, the normal commensal microbiota and the cells of the intestinal mucosa co-exist in a mutualistic relationship, whereby the commensal microorganisms help to shape the local and systemic immune response, and the intestinal mucosal cells modulate the habitat and composition of the microbiota [9]. In particular, the colon is the most heavily colonized tissue in the body, containing an estimated 1014 bacteria and over 70% of all the microorganisms that reside in the host [19]. Obligate anaerobic bacteria belonging to the two phyla Bacteroidetes and Firmicutes dominate the human digestive tract where more than 50 different phyla reside [20]. Other phyla, including Proteobacteria, Verrucomicrobia, and Actinobacteria, are present in the gut to a much lesser extent. An imbalance in this composition of commensal microbes, or “dysbiosis”, is characterized by a considerable decrease in the resident obligate anaerobic bacteria, and an increase in facultative anaerobes such as Enterobacteriaceae [21]. For example, adherent-invasive Escherichia coli (AIEC), which are Enterobacteriaceae, are isolated more frequently from the intestinal mucosa of patients with CD than from healthy individuals [22]. Interestingly, the endoplasmic reticulum-localized stress response chaperone, Gp96, is abnormally expressed on the apical surface of epithelial cells in the ileum of patients with CD, and acts as a receptor for the OmpA of AIEC to promote this bacterium's pathogenic invasion [23].
It is important to note that the composition of the microbiome of the murine gut is very similar to that of humans [24]; therefore, mouse experimental models of gastrointestinal disease likely have translational relevance [25]. The gut microbiota of both species can further be divided according to their location in the gut, namely whether the microbes are present in the intestinal lumen or penetrate the mucus layer overlying the intestinal epithelium [26]. A thick mucin layer protects mucosal enterocytes from excessive exposure to bacteria and dietary antigens throughout the length of the intestines, particularly in the colon, thus preventing immune hypersensitivity responses [27].
Microbial colonization of the human gut is thought to begin at birth via commensal microbes from the maternal skin, vagina, and feces, as well as the external environment of the neonate [28, 29]. Emerging data also suggest that the environment in utero is not sterile, as Enterococcus fecalis, Staphylococcus epidermidis, and E. coli have been isolated from the meconium of healthy neonates [30]. The establishment of a diverse and balanced microbiota plays a critical role in the development and maturation of a healthy immune system. This has been demonstrated using germ-free animals raised in bacteria-free conditions that exhibit physical and functional immune abnormalities, including defects in the formation of the spleen, lymph nodes, and Peyer's patches [18]. Additionally, there are significant reductions in the number of IgA-producing plasma cells and CD4+ T cells, as well as decreased immunoglobulins and imbalanced cytokine levels [18]. Germ-free mice also have few, if any, pro-inflammatory Th17 cells in the intestinal lamina propria [31]. Thus, exposure to commensal bacteria greatly affects gut health and immunity. This microbial exposure is tightly regulated, as limited or no bacterial interaction significantly impairs development of the host immune system, while excessive bacterial contact can result in exaggerated immune responses and autoinflammatory disease [27]. The critical role of the microbiota in overall gut health and the extent of the deleterious consequences of dysbiosis speak to the importance of understanding the molecular interactions between the microbiota and the cells of the host.
Control of intestinal inflammation and tolerance by the gut microbiota
While a basal level of inflammation is needed to recognize and eliminate invading enteropathogens, the gastrointestinal tract must also coexist with the diverse and abundant commensal bacteria and the daily estimated 130-190 grams of ingested protein replete with dietary antigens [32]. To maintain intestinal homeostasis both locally and systemically, tolerance must be achieved by the induction of anti-inflammatory molecules. Commensal microbes are critical regulators of the immune system, and can maintain homeostasis by stimulating antibody production and activating immune cells, as well as inducing anti-inflammatory cytokine responses (Figure 1A). The microbial pattern sensors, Toll-like receptors (TLRs), play particularly important roles in microbe recognition, homeostasis, and immune defense in the gut [33]. In the healthy gut, TLR3 and TLR5 seem to be constitutively expressed. In contrast, TLR2 and TLR4 are expressed at very low levels, suggesting that the expression of these two receptors is regulated to avoid autoinflammatory immune activation in response to commensal microbes [27]. However, current data reveal that microbiota-mediated TLR signal transduction is complex. For example, constitutive TLR4 signaling in intestinal epithelium reduced tumor burden by increasing apoptosis in Apc Min/+ mice, an experimental model of CRC in which Min (multiple intestinal neoplasia) is a mutant allele of the mouse ortholog of the tumor suppressor gene, APC (adenomatous polyposis coli) [34].
Figure 1. Mechanisms for maintaining basal immune activation and tolerance.
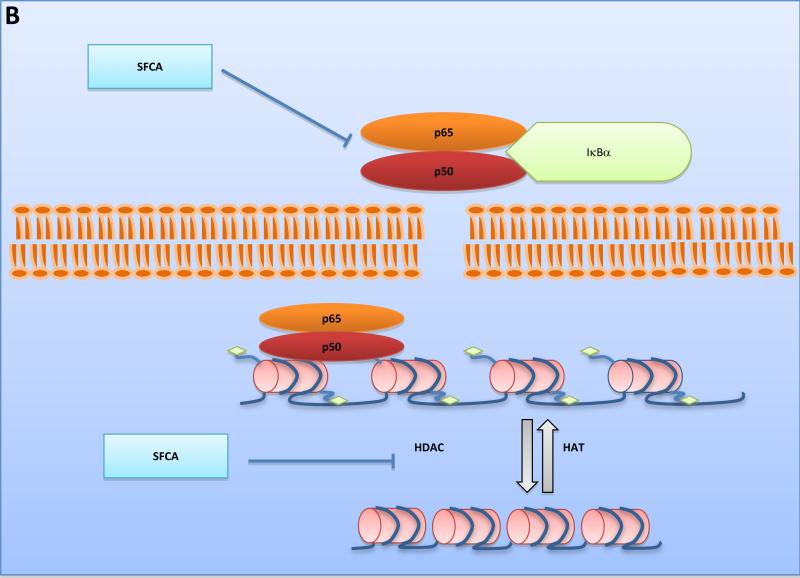
A. Commensal bacteria help to preserve a healthy gut environment. Abundant beneficial microbes exist at the mucosal layer where epithelial cells can be stimulated by bacterial antigens such as surface layer proteins and metabolic products. The release of signaling molecules or the presentation of antigens by dendritic cells is able to induce the activation of T and B lymphocytes, meanwhile recruiting more dendritic cells and macrophages. B. Bacterial fermentation products such as SFCAs inhibit the NF-κB signaling pathway and decrease HDAC activity to regulate homeostasis. HATs, acting as transcription co-activators, can open chromosome structure by adding acetyl residues to histones, while HDACs do the opposite. Inhibition of HDACs may lead to accessible chromatin. The transcription of cell cycle arrest and apoptosis-related genes can result in suppression of tumor development. Inhibition of the NF-κB signaling pathway prevents its contribution to pathogenic inflammation as well. SlpA, Surface Layer Protein A; SFCA, Short Fatty Chain Acid; HAT, Histone Acetyltransferase; HDAC, Histone Deacetylase
Another way in which intestinal bacteria affect the host is via the end-products of their unique metabolism. For example, the short chain fatty acids (SCFAs), acetate, proprionate, and butyrate, are products of microbial fermentation of dietary fiber in the colon and have been reported to be effective in immune regulation [10]. These metabolites are important sources of energy for colonic epithelial cells and are reportedly decreased in patients with IBD [10]. While acetate is the most abundant SCFA found in the colon, butyrate has been the subject of intense study because of its role in the preservation of mucosal health and its anti-inflammatory and anti-tumorigenic properties [35]. The most widely studied function of butyrate is as a histone deacetylase (HDAC) inhibitor that results in hyper-acetylation, maintenance of an open euchromatic state, and the activation of genes involved in cellular differentiation, apoptosis, and cell cycle arrest in tumor cells [36]. Butyrate has disparate effects in normal colonocytes versus malignant cells because most of this SCFA is oxidized to produce ATP in normal cells, resulting in cellular proliferation. Conversely, butyrate is inefficiently oxidized in tumor cells, which rely on anaerobic metabolism, and is able to reach the nucleus and function as a HDAC inhibitor [36]. Apart from this epigenetic regulation, butyrate is also capable of suppressing the activation of the pro-inflammatory transcription factor, nuclear factor-kappa B (NF-κB), which plays a central role in the host immune responses to infection. Activated NF-κB translocates into the nucleus where it binds to specific sequences of target genes and promotes the expression of a series of pro-inflammatory factors [37]. Activation of NF-κB is one of the principal contributing factors to the development of inflammation-associated CRC and its constitutive activation was reported in 40% of excised CRC tissues and 67% of human CRC cell lines [35, 38]. These mechanisms of butyrate action are important to limit the inflammatory response and to maintain intestinal homeostasis (Figure 1B).
Intestinal pathogenesis elicited by altered microbiota composition
Discussions about whether the altered composition of the microbiota is a cause or consequence of IBD and CAC are ongoing. A possibility is that alteration of either the microbiome composition or the number of inflammatory stimuli, whichever occurs in excess first, will tip the balance of homeostasis (Figure 2). As a consequence, changes in barrier function of the epithelium, activation of the innate immune system via TLRs and other microbial sensors, and polarization of T lymphocytes, will result in the release of inflammatory mediators that lead to pathologic autoinflammation. A decreased diversity of species in gut microbial communities is often associated with a high density of mucosal surface colonization and epithelial invasion in areas with active disease [39]. Further investigations have demonstrated that it is not only the alteration of microbial composition, but also aberrant molecular signaling induced by metabolites of altered bacteria that are involved during the transformation from acute to chronic inflammation, IBD, or even colon cancer [40]. This is not surprising, as an estimated 20-30% of cancers are associated with chronic microbial infections, including gastric cancer (Helicobacter pylori), liver cancer (hepatitis B and C viruses), urinary bladder cancer (Schistosoma hematobium), cholangiocellular neoplasia (Opisthorchis viverrini), and cervical cancer (human papilloma viruses) [41].
Figure 2. Interactions between the gut microbiota and the immune system maintain healthy homeostasis.
Either alteration of the gut microbiota composition or overactivation of the immune system may disrupt the balance, and therefore, induce inflammation and even IBD or CRC. IBD, Inflammatory Bowel Disease; UC, Ulcerative Colitis; CRC, Colorectal Cancer
The connection between infection, inflammation, and carcinogenesis is explained, in part, by oxidative and nitrative stress and the resulting pro-mutagenic DNA lesions: DNA-protein crosslinks, depurination/depyrimidination, base and sugar modifications, and single- and double-stranded breaks, all of which have been reported to play key roles in inflammation-related carcinogenesis [3, 42]. For example, nitrative- and oxidative-induced DNA damage have been identified at sites of carcinogenesis in animal models of chronic inflammation induced by infection with the liver fluke, Opisthorchis viverrini [43]. The reactive oxygen, nitrogen, and halogen species caused profound damage to cellular nucleic acids, proteins, lipids, and carbohydrates, and resulted in the induction of pro-inflammatory molecules, including IL-1, IL-6, tumor necrosis factor (TNF)-α, cyclooxygenase-2, and inducible nitric oxide synthase (iNOS) [43].
Molecular mechanisms underlying infection-induced inflammation and colon cancer were recently described using Helicobacter hepaticus-infected Rag2−/− mice [44]. This bacterium colonizes the liver and colon of various mouse strains, but does not typically cause disease in immunocompetent mice. However, it is linked with hepatitis and chronic colitis in several immunodeficient murine models; the lack of Tregs in Rag2−/− mice renders them unable to achieve immune tolerance to this microbe [45]. In this study, liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) was used to quantify DNA and RNA damage products, revealing chlorinated nucleic acid (5-chlorocytosine, 5-Cl-C) in the colonic epithelium due to neutrophil-derived HOCl [44]. Lao et al. [46] reported that the 2-deoxyribo-halogenated product, 5-Cl-dC, mimics 5-methyl-dC and induces inappropriate, increased CpG methylation that can silence tumor suppressor genes and initiate carcinogenesis [44].
Another role for inflammatory cell-derived nitrogen species in the disruption of intestinal homeostasis is the ability of certain microbes to use host inflammation-derived nitrate as a terminal electron acceptor for anaerobic respiration. This gives the facultative anaerobe, E. coli, a distinct growth advantage in the lumen of the inflamed intestine over commensal obligate anaerobes, which cannot utilize nitrate and must rely on fermentation for energy [47].
The means by which colonization with the gut commensal, E. coli NC101, promotes invasive colorectal carcinoma in germ-free, colitis-susceptible Il-10−/− mice treated with the colon-specific carcinogen, azoxymethane (AOM) was also recently investigated [48]. The promotion of tumorigenesis was related to the DNA-damaging, polyketide synthase (pks) pathogenicity island of the bacteria; and mucosal pks+ E. coli were found in a high percentage of IBD and CRC patients. These data suggest that colitis can promote tumorigenesis by altering microbial composition and inducing the expansion of microorganisms with the ability to damage DNA [48]. Additional evidence linking microbial infection and CRC includes the capability of enterotoxigenic Bacillus fragilis (ETBF) to trigger colitis and induce colonic hyperplasia and tumor initiation in Min mice via induction of signaling transducer and activator of transcription (STAT)-3 activation and a pro-inflammatory Th17 response [49]. Conversely, while non-toxigenic B. fragilis (NTBF) also chronically colonized mice, it did not induce STAT-3 activation or colitis; importantly, these data reveal distinct roles for a commensal bacterium and adaptive immunity during CRC pathogenesis [49].
It would stand to reason that in a mutualistic relationship, the gut microbiome could impact the host's genetic and epigenetic structures. A supportive example of the role of diet and gut commensals in host mucosal epigenetic and microbiomic changes came from Richard Kellermayer's laboratory, where they demonstrated that a maternal diet supplemented with methyl-donors (e.g., folic acid, vitamin B12, and choline) resulted in offspring with increased susceptibility to dextran sodium sulfate (DSS)-induced colitis, due in part, to aberrant methylation of anti-inflammatory genes [50].
Genetic regulation of colorectal carcinogenesis
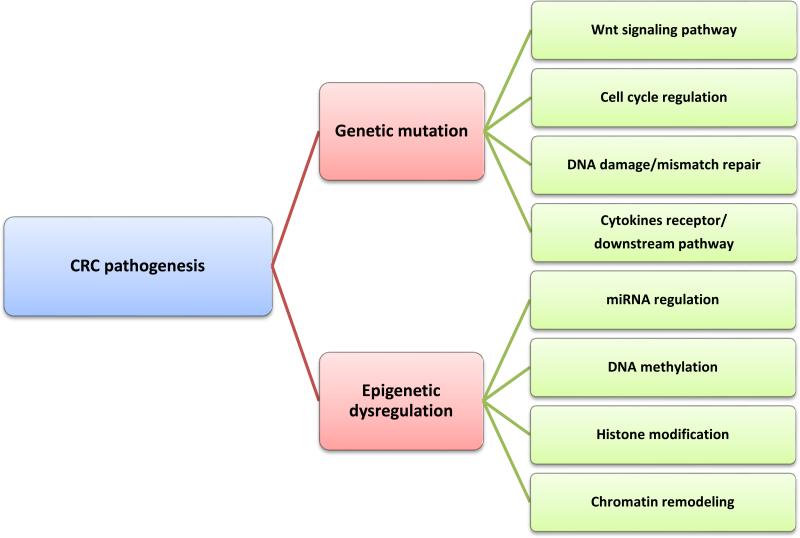
For decades, genetic instability has been considered to underlie the multistep process of tumor growth and metastatic dissemination [51]. An array of genetic mutations is found in most, if not all, cancer subtypes (Figure 3). The most common gene mutation in CRC is the inactivating mutation of the APC gene [52]. Approximately 70–80% of sporadic colorectal adenomas and carcinomas have somatic mutations of APC, and nearly all of these mutations result in premature truncation of the APC protein [5]. Inactivation of APC leads to inappropriate and constitutive activation of the Wnt signaling pathway, a general mechanism of CRC development [53]. APC mutations are found in even the earliest lesions, such as microscopic adenomas consisting of a few dysplastic glands [5]. Several non-functional Apc mutant mouse strains show similar CRC phenotypes, with polyps arising within the small intestine or colon [54, 55]. Multiple mechanisms of APC inactivation have been reported, including hypermethylation of CpG sites in the APC promoter and decreased translation due to inhibition by microRNAs [52]. Interestingly, it was recently demonstrated that chronic NF-κB activation within intestinal epithelial cells accelerates loss of heterozygosity at the APC locus via increased nitrosative DNA damage, leading to spontaneous adenomas within the small intestine and the colon [56]. TP53, the gene encoding the tumor suppressor protein, p53, is also mutated with high incidence in CRC [57]. p53 is a cell cycle regulator that induces cell growth arrest by holding the cell cycle at the G1/S regulation checkpoint. TP53 mutations can lead to the inactivation of p53 or a 17p chromosomal deletion that eliminates the second TP53 allele [58]. Not surprisingly, this abnormality often correlates with colorectal tumorigenesis [59]. It has also been reported that the K-ras oncogene is mutated in 30-60% of large adenomas and CRC; the mutated, constitutively active form of this protein affects signaling pathways controlling cellular differentiation, motility, proliferation, and apoptosis [17].
Figure 3. Genetic and epigenetic mechanisms of CRC pathogenesis.
Mutation of genes (e.g., APC) involved in the Wnt signaling pathway plays a dominant role in CRC pathogenesis. Genes that are related to cell cycle progression, DNA repair, and cytokine signaling have also been shown to be crucial for CRC pathogenesis. DNA hypermethylation of tumor suppressor gene promoter regions has been intensively studied to demonstrate its pivotal role in gene silencing. Histone modification includes histone methylation and deacetylation, both of which have been shown to be associated with DNA methylation.
Epidemiologic data reveal that approximately 15% of all colorectal tumors demonstrate a defect in DNA mismatch repair proteins resulting in insertions/deletions in repetitive sequences (e.g., microsatellites) and microsatellite instability [60]. The most commonly mutated DNA repair enzyme genes in CRC are MSH2 (bacterial mutS homolog 2), MLH1 (bacterial mutL homolog 1), and MSH6 [61]. The subsequent colorectal tumors tend to occur in the proximal or “right-sided colon,” are less invasive, occur in younger patients, and are rather undifferentiated with fewer mutations in K-ras or TP53 [60]. Conversely, 55-70% of colorectal tumors develop in the distal or “left-sided colon” [61]. Regardless of location, most of these tumors demonstrate mutations that result in over-activation of the Wnt/β-catenin signaling pathway [52].
Epigenetic regulation of colorectal carcinogenesis
The link between epigenetic modifications and cancer was first made in 1983 [62]. However, until recently, appreciation of epigenetic alterations has lagged behind genetic mutations in regard to their contributions to human cancer development. With the rapidly increasing understanding of specific epigenetic mechanisms involved in gene expression regulation, and the advent of techniques to study these changes, epigenetics has become a favored area of research in the field of cancer biology. Epigenetic modifications are heritable, potentially reversible, and regulate gene expression through post-replicative DNA modification, post-translational histone modification, and chromatin remodeling without affecting the sequence of nucleotides [63] (Figure 3). As epigenetic changes are reversible, they are appealing targets for cancer therapy [64].
miRNAs
Discovered over 20 years ago, microRNAs (miRNAs) are small, non-coding RNAs composed of approximately 18-24 nucleotides. miRNAs post-transcriptionally regulate target gene expression by inhibiting translation or inducing the degradation of messenger RNA (mRNA), depending on the amount of complementarity with the target sequence [65]. Mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) where they are able bind to the 3’ untranslated region of the target mRNA. [66]. miRNAs are frequently dysregulated in tumors either by genetic or epigenetic factors, and are currently being investigated for their potential as cancer biomarkers [67]. For example, miR-21 and miR-106 are upregulated in stool samples from patients with CRC compared to normal individuals, and higher levels of miR-144 are found in the stool of CRC patients with a sensitivity of 74% and specificity of 87% [68]. Because of their small size, miRNA levels are very stable in biologic samples [15], and investigators have recently demonstrated that extracellular miRNAs are stable for at least one month, even in stool samples [69]. As there is currently no FDA-approved blood screening test for CRC, there is increasing interest in detectable, circulating markers of these tumors such as miRNA [15]. miR-21 is considered to be an oncogenic miRNA or “oncomir” that is upregulated in many solid and hematological cancers, including CRC [68].
Recently, it has been demonstrated that miR-18a reduces the repair of DNA double-strand breaks by directly suppressing the translation of the multimeric Ataxia telangiectasia mutated (ATM) repair enzyme [70]. Double-strand DNA breaks are an example of genetic instability, are extremely cytotoxic, and result from endogenous, oxidative stress (e.g., inflammation) or exogenous sources such as ionizing radiation or genotoxic agents [70]. Once double-strand DNA breaks are detected by the Mre11 sensor complex, ATM undergoes autophosphorylation, which promotes its monomerization and kinase activity [71]. This causes G1 cell cycle arrest by activating p53, with subsequent upregulation of the cyclin-dependent kinase inhibitor, p21 [72]. Furthermore, there was a significant inverse correlation between levels of miR-18a and ATM in 45 pairs of human rectal tumors and adjacent normal tissues; this down-regulation of ATM was also observed in CRC cell lines when compared to normal colon biopsies [70].
DNA methylation
DNA methylation is an important regulator of gene expression and was the first epigenetic mechanism identified in cancer development [62]. In this process, a methyl group (CH3) is covalently added by DNA methyltransferase (DNMT) enzymes to carbon 5 of the cytosine base, usually in CpG dinucleotides [73]. Aberrant DNA hypermethylation at CpG-rich regions near gene promoters has consistently been associated with the inactivation of target genes. Since its postulation by Toyota et al. in 1999 [74], current investigations have increasingly focused on what is known as the CpG island methylator phenotype (CIMP) of tumor suppressor genes involved in CRC. More recent studies have revealed that numerous genes are hypermethylated at their promoter regions and silenced in CRC [75-78]. These include regulators of DNA mismatch repair such as MLH1 and MGMT (O6-methylguanine-DNA methyltransferase), and negative regulators of Wnt signaling such as Wnt inhibitory factor 1 (WIF1) [79]. MGMT encodes a DNA repair protein that demethylates lesions that result from alkylation at the O6 position of guanine that are induced by dietary nitrosamines or chemotherapeutic agents [80].
Hypermethylation of DNA can also lead to the silencing of genes encoding tumor suppressors and transcription factors required for cellular homeostasis. Among these is RUNX3 that codes for runt-related transcription factor 3, which modulates both the transforming growth factor (TGF)-β [81] and the Wnt/β-catenin signaling pathways [82]. Runx3–/– mice exhibit gastric epithelial proliferation due to decreased apoptosis and reduced sensitivity to the growth inhibitory cytokine, TGF-β. They also demonstrate intestinal hyperplasia due to enhanced Wnt signaling activity and upregulation of the Wnt target genes, CMYC and cyclinD1 [83]. Thus, suppression of RUNX3 expression leads to aberrant signaling of both pathways, which are commonly dysregulated in colonic carcinogenesis. Loss of function of RUNX3 has significant and constitutive effects on the expression of its target genes, which may then contribute to CRC development [76, 82]. Decreased expression of RUNX3 mRNA due to epigenetic hypermethylation of the RUNX3 promoter occurs in a range of human CRC cell lines [84]. Treatment of CRC cell lines with the DNMT inhibitor, 5-aza-2′ -deoxycytidine, restored the expression of RUNX3 mRNA, suggesting that epigenetic inactivation of RUNX3 is due to aberrant promoter methylation [84].
Moreover, the DNA mismatch repair enzyme gene, MLH1, has also been found to be inactivated by the hypermethylation of its promoter region. Decreased expression of MLH1 results in a larger number of DNA mismatches and increased microsatellite instability, and ultimately, increased carcinogenesis [85]. As discussed above, mutations of the APC gene promote CRC onset by failing to negatively regulate the Wnt/β-catenin pathway. However, it has also been suggested that functional but reduced expression of APC caused by hypermethylation of its promoter can lead to CRC associated with over-expression of the oncogene, CMYC, which is a downstream effector of the Wnt/β-catenin pathway [86].
In the case of genes lacking CpG-rich promoter regions, hypomethylation of the gene body may contribute to cancer initiation [87]. Genome-wide DNA hypomethylation has been detected in many human cancers. Unlike inactivation of tumor suppressor genes by hypermethylation in CRC, global genomic hypomethylation plays a causal role in tumor formation through the activation of proto-oncogenes [74] or by promoting chromosomal instability [87, 88]. For instance, immunohistochemistry analyses indicated that the c-myc oncoprotein was highly disseminated throughout the adenomatous polyps of the colonic mucosa compared to normal colonic mucosa [89, 90], where c-myc was found to be solely localized in the nuclei. Elevated levels of the c-myc protein correlated with the loss of methylation of the third exon of the CMYC gene. In addition, parallel analyses of paired normal-CRC human tissues indicated that global DNA hypomethylation occurs early in carcinogenesis and that progression of this event is followed by several causative alterations, including increased activity of DNMTs and the appearance of hypermethylated sites [62, 63].
Histone modifications
Unlike the intensive studies aimed at understanding DNA methylation in tumor initiation, much less is known regarding the potential contribution of aberrant histone post-translational modifications and nucleosome positioning to the initiation and progression CRC. The basic unit of chromatin is the nucleosome, which comprises a histone octamer (2 copies each of H2A, H2B, H3, and H4) around which 147 base pairs of DNA are wrapped [91]. Evidence suggests that accumulated loss of histone H4 lysine monoacetylation, which is associated with loss of H4K16 and H4K20 trimethylation, may be a hallmark in human tumor cells, including CRC [92]. Distinct post-translational methylation of the N-termini of histones can play opposing roles in the expression of target genes. For example, H3K4 mono- and tri-methylation are generally considered to be markers for activation. However, di- or tri-methylation of H3K9 or H3K27 are associated with repression [93]. The Polycomb Repressive Complexes (PRC1 and PRC2) are histone-modifying complexes that set the repressive mark tri-methylation of H3K27 [94]. Investigators have recently determined a “DNA methylome” using murine ApcMin adenoma as a model for cancer initiation. In this study, a core panel of differentially methylated regions (DMRs) was conserved between mouse adenomas and excised tissues from patients with advanced CRC. These patterns were also distinct from the surrounding the intestinal epithelia, including intestinal stem cells, indicating that the patterns were not pre-existing, but formed de novo [95]. Hypermethylated DMRs were prevalent at PRC2 binding sites in both mice and humans tumors, suggesting that increased PRC2 activity may be critical in the intestinal tumor-specific pattern of DNA hypermethylation following the loss of APC [95].
Unlike histone methylation, acetylation of histone tails by histone acetyltransferases (HATs) predominantly demarcates an active state. Addition of the acetyl group to lysines neutralizes the positive charge of the histone. This disrupts interactions between the histone N-terminal tails and the negatively charged DNA as well as interferes with higher-order compaction of the chromatin fiber [96, 97]. In addition, histone acetylation recruits chromatin remodeling activities that contain subunits with bromodomains that bind preferentially to acetylated nucleosomes [98]. As a consequence, a more open chromatin structure is formed, often accompanied by histone depletion that leads to higher accessibility to the DNA by transcription factors [99]. Histone deacetylases (HDACs) are a class of enzymes that remove acetyl groups from acetylated histones (as well as from some non-histone proteins such as the transcription factors, Sp1 and Sp3) [100], reversing the open chromatin structure, and resulting a condensed heterochromatic state and inactivation of transcription [101]. Consequently, HATs and HDACs collaboratively maintain the balance of histone acetylation in vivo to achieve homeostasis [102]. In 2002, a link between DNA methylation and histone modification in cancer was reported [103]. Genes that become hypermethylated at their promoter CpG islands are often associated with histone hypoacetylation, loss of active histone methylation marks, and gain of repressive marks [99]. Accumulating evidence indicates that bacteria can directly interfere with DNA replication and repair, RNA splicing, transcription, and chromatin remodeling [6]. Helicobacter pylori, which was first discovered and identified in a chronic gastritis patient in 1982, was shown to be a potential causative agent of gastric carcinogenesis due to altered DNA methylation [104] and histone acetylation patterns [105].
In a recent study that examined the dynamic interplay between host and gut microbiota [106], wild-type and Tlr2–/– mice were employed to evaluate epigenomic and metagenomic differences in the colonic mucosa. Significantly modified DNA methylation patterns of host genes involved in immune processes were identified. Alterations in the number and composition of the mucosal microbiome were also shown with the use of next-generation pyrosequencing of bacterial 16S rRNA. Collectively, these consequences of gut microbial imbalance may result in epigenetic abnormalities that contribute to differential gene expression and potentially trigger the onset and progression of inflammation-associated carcinomas [106].
Tumor therapy and protection by genetically modified bacteria
There are different causative mechanisms responsible for the generation and progression of CRC. Based on our current knowledge, drugs approved by the Food and Drug Administration (FDA) for use in CRC treatment, include Bevacizumab (an angiogenesis inhibitor); Cetuximab (a monoclonal antibody against the epidermal growth factor receptor); and Capecitabine (a drug precursor of the DNA synthesis inhibitor, 5-fluorouracil). These drugs are designed for inhibiting and/or eliminating aberrant tumor cells. However, the regular metabolism of normal cells is also disrupted by these drugs, which may result in unwanted side effects. Although inhibitors with therapeutic potential are being designed based on HDACs, HATs, and DNMTs [107], there will always be a major concern of how to enhance the drug specificity in vivo while diminishing side effects.
Changes in the composition of the microbiota can lead to altered immunologic signaling; thus, it is reasonable to speculate that oral administration of gut commensal bacteria could enhance anti-inflammatory responses at the genetic, epigenetic, and/or immune cytokine levels to rebalance disrupted homeostasis. Recently, our laboratory and other investigators have provided supportive evidence that the gut microbiota plays an essential role in intestinal epigenomic mechanisms of the host [108, 109]. We demonstrated that deletion of lipoteichoic acid (LTA), a TLR2 ligand, normalizes innate and adaptive pathogenic immune responses and causes regression of established colonic polyps in a novel transgenic mouse model that develops multiple polyps in the colon and distal ileum as early as 5-6 months of age. This study revealed the pro-inflammatory role of LTA and the ability of LTA-deficient Lactobacillus acidophilus to suppress inflammation and protect against colon cancer [55]. Furthermore, this genetically modified gut commensal bacterium, LTA-deficient L. acidophilus, could prevent and/or ablate established colitis [110] and polyposis [111], downregulate downstream signalling [112], and upregulate tumor suppressor genes in CRC cell lines [113] (Figure 4). These observations suggest that overt inflammation can be reshaped by immune regulatory agents or even by natural microbial gene products, including the bacterial surface layer proteins upon which our laboratory's focus is currently centered.
Figure 4. Cascade of molecular reactions initiated by LTA-deficient L. acidophilus.
Either component proteins or metabolic products from LTA-deficient L. acidophilus are potential antigens that can be recognized by dendritic cells. Presentation of antigen to recipient CD4+ T cells induced the genesis of a population of Tregs that could further differentiate into Th17 or Th1 cells, depending on the microenvironment. Meanwhile, several signaling pathways were triggered in dendritic cells by the recognition of antigens. The MAPK-Erk1/2 pathway was shown to be crucial for the expression of the anti-inflammatory cytokine IL-10, and tumor suppressors, such as RUNX3 and TIMP3, were also upregulated by the treatment of LTA-deficient L. acidophilus both in vitro and in vivo. RNAP II, RNA Polymerase II; TIC, Transcription Initiation Complex.
Although the beneficial effects of gut commensal bacteria have been demonstrated in a number of experimental models [114, 115], little is known about the effect of these bacteria on genomic stability or epigenetic regulation. Using an in vitro 3-dimensional intestinal mucosal model consisting of human intestinal HT-29/B6 or T84 cell lines and peripheral blood mononuclear cells (PBMC) from human healthy subjects, researchers demonstrated that the commensal bacteria, Bifidobacterium breve and Lactobacillus rhamnosus GG, inhibited the production of the pro-inflammatory cytokines, IL-17 and IL-23. Global DNA methylation and histone acetylation were also evaluated to demonstrate that commensal microbial treatment resulted in increased host DNA methylation and decreased histone acetylation [116]. It is tempting to speculate about the status of DNA methylation and histone acetylation at the promoter regions of IL-17 and IL-23 in these experiments. Using the commensal bacteria Lactobacillus and Bifidobacterium, as examples, the potential strategy to restore the composition of the intestinal microbiota with genetically modified bacteria appears feasible and promising. To this end, monoassociation and fecal flora transfer studies using germ-free animal models of CRC may provide answers to several unresolved questions (Box 1).
Concluding remarks
The mutualistic relationship between the host and the gut microbiome benefits the host in many ways. With that said, any perturbation of this delicate balance caused by altered microbial composition may induce aberrant intestinal signaling pathways and epigenetic modifications. The combinatorial impact of bacterial surface toxins and metabolic end-products may stimulate severe inflammation that can progress to cancer due to disrupted homeostasis. Instead of turning to drugs with obvious side effects for the treatment of inflammation-associated disease, genetically modified bacteria or bacterial products may be considered for a more specific correction of the original dysbiosis. At present, experimental data using genetically modified bacteria and their products are promising, and some beneficial commensal strains are currently being tested in clinical IBD cases. Although progress has been made in understanding and utilizing gut commensal microbes for these purposes, we are still at an early stage in the battle against CRC development. Additional research is necessary to better understand the association between microorganisms and carcinogenesis. Further investigation of the epigenetic regulation by natural and biologically engineered bacteria or their immune regulatory gene products in the gut will lead to the development of safer, targeted therapeutics for colorectal cancer.
Highlights.
We examine the role of the intestinal microbiota in colorectal cancer
We thoroughly discuss the epigenetic regulation of colorectal carcinogenesis
We propose the use of engineered bacteria to rebalance inflammation in the gut
Box 1. Outstanding questions.
- Is dysbiosis a cause or consequence of CAC?
- Recent studies have demonstrated that dysbiosis induces a dysregulated microenvironment, which may in turn, elicit pro-tumorigenic inflammation.
- However, clinical data show changes in the composition of the gut microbiota in patients with CAC, indicating that bacterial dysbiosis may play a critical role in the developmental process of CAC. Studies are ongoing to re-balance the condition of bacterial dysbiosis in order to elicit healthy homeostasis to mitigate cancer development and/or progression.
- What are the potential epigenetic consequences of beneficial bacteria?
- The epigenetic consequences may include DNA methylation, histone modification in target gene regions (e.g., enhancer, promoter, etc.) and dynamic chromosome remodeling across all of the gene elements. Basically, the metabolites (e.g, small peptides and chemicals) and protein components of beneficial bacteria are the major functional molecules. The targets of these molecules in the gut are enzymes with epigenetic modification activities. Acting naturally, as either antagonists or agonists of those enzymatic activities, functional molecules can control the physiological dynamics in vivo.
- What are the advantages of the administration of engineered bacteria to CRC patients?
- Most commercial drugs available for CRC treatment are chemicals that were originally designed to function as inhibitors of certain normal physiological processes to inhibit tumor cell growth or to eliminate neoplastic cells. But many of those drugs may initiate severe side effects. Therefore, the continuous use of chemical drugs raises concerns about their cell specificity in vivo. Beneficial bacteria have been widely accepted as healthy supplements. Engineered beneficial bacteria may show more anti-inflammatory properties and would restore gut homeostasis due to the removal of their pro-inflammatory gene products. Because these bacteria naturally reside in gut, their potential to cause severe side effects is much less than that of drugs.
Acknowledgements
This work was supported in part by NIH Grant 1R01AI098833-01, DoD Grant CA111002, Gatorade Seed Funds from the University of Florida, and NIH/NCRR Clinical and Translational Science Award to the University of Florida (UL1 RR 029890).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors have no conflicts of interest to disclose.
References
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Tjalsma H, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 3.Tudek B, Speina E. Oxidatively damaged DNA and its repair in colon carcinogenesis. Mutat Res. 2012;736:82–92. doi: 10.1016/j.mrfmmm.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Arends MJ. Pathways of colorectal carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:97–102. doi: 10.1097/PAI.0b013e31827ea79e. [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 6.Bierne H, Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell Microbiol. 2012;14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- 7.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 8.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013 doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cario E. Microbiota and innate immunity in intestinal inflammation and neoplasia. Curr Opin Gastroenterol. 2013;29:85–91. doi: 10.1097/MOG.0b013e32835a670e. [DOI] [PubMed] [Google Scholar]
- 10.Viladomiu M, et al. Nutritional protective mechanisms against gut inflammation. J Nutr Biochem. 2013 doi: 10.1016/j.jnutbio.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford AC, et al. Ulcerative colitis. Bmj. 2013;346:f432. doi: 10.1136/bmj.f432. [DOI] [PubMed] [Google Scholar]
- 12.Sansone P, Bromberg J. Environment, inflammation, and cancer. Curr Opin Genet Dev. 2011;21:80–85. doi: 10.1016/j.gde.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Jess T, et al. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10:639–645. doi: 10.1016/j.cgh.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Okayasu I. Development of ulcerative colitis and its associated colorectal neoplasia as a model of the organ-specific chronic inflammation-carcinoma sequence. Pathol Int. 2012;62:368–380. doi: 10.1111/j.1440-1827.2012.02807.x. [DOI] [PubMed] [Google Scholar]
- 15.Mazeh H, et al. The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer - a Comprehensive review. J Cancer. 2013;4:281–295. doi: 10.7150/jca.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 17.Al-Sohaily S, et al. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol. 2012;27:1423–1431. doi: 10.1111/j.1440-1746.2012.07200.x. [DOI] [PubMed] [Google Scholar]
- 18.Dongarra ML, et al. Mucosal immunology and probiotics. Curr Allergy Asthma Rep. 2013;13:19–26. doi: 10.1007/s11882-012-0313-0. [DOI] [PubMed] [Google Scholar]
- 19.Sekirov I, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 20.Barengolts E. Vitamin D and Prebiotics May Benefit the Intestinal Microbacteria and Improve Glucose Homeostasis in Prediabetes and Type 2 Diabetes. Endocr Pract. 2013:1–40. doi: 10.4158/EP12263.RA. [DOI] [PubMed] [Google Scholar]
- 21.Winter SE, et al. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 23.Rolhion N, et al. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355–1362. doi: 10.1136/gut.2010.207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arthur JC, Jobin C. The struggle within: microbial influences on colorectal cancer. Inflamm Bowel Dis. 2011;17:396–409. doi: 10.1002/ibd.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekirov I, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 27.Kelly D, Mulder IE. Microbiome and immunological interactions. Nutr Rev. 2012;70(Suppl 1):S18–30. doi: 10.1111/j.1753-4887.2012.00498.x. [DOI] [PubMed] [Google Scholar]
- 28.Dominguez-Bello MG, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulder IE, et al. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS One. 2011;6:e28279. doi: 10.1371/journal.pone.0028279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadis U, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Capelluto DG. Tollip: a multitasking protein in innate immunity and protein trafficking. Microbes Infect. 2012;14:140–147. doi: 10.1016/j.micinf.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, et al. Constitutive TLR4 signalling in intestinal epithelium reduces tumor load by increasing apoptosis in APC(Min/+) mice. Oncogene. 2013 doi: 10.1038/onc.2012.581. [DOI] [PubMed] [Google Scholar]
- 35.Fung KY, et al. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br J Nutr. 2012;108:820–831. doi: 10.1017/S0007114512001948. [DOI] [PubMed] [Google Scholar]
- 36.Leonel AJ, Alvarez-Leite JI. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care. 2012;15:474–479. doi: 10.1097/MCO.0b013e32835665fa. [DOI] [PubMed] [Google Scholar]
- 37.Iwai K. Diverse ubiquitin signaling in NF-kappaB activation. Trends Cell Biol. 2012;22:355–364. doi: 10.1016/j.tcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto K, et al. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 2009;15:2248–2258. doi: 10.1158/1078-0432.CCR-08-1383. [DOI] [PubMed] [Google Scholar]
- 39.Manichanh C, et al. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 40.Kipanyula MJ, et al. Signaling pathways bridging microbial-triggered inflammation and cancer. Cell Signal. 2013;25:403–416. doi: 10.1016/j.cellsig.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203:306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azcarate-Peril MA, et al. The intestinal microbiota, gastrointestinal environment and colorectal cancer: a putative role for probiotics in prevention of colorectal cancer? Am J Physiol Gastrointest Liver Physiol. 2011;301:G401–424. doi: 10.1152/ajpgi.00110.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yongvanit P, et al. Oxidative and nitrative DNA damage: key events in opisthorchiasis-induced carcinogenesis. Parasitol Int. 2012;61:130–135. doi: 10.1016/j.parint.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Mangerich A, et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci U S A. 2012;109:E1820–1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fox JG, et al. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4:22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lao VV, et al. Incorporation of 5-chlorocytosine into mammalian DNA results in heritable gene silencing and altered cytosine methylation patterns. Carcinogenesis. 2009;30:886–893. doi: 10.1093/carcin/bgp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaible TD, et al. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum Mol Genet. 2011;20:1687–1696. doi: 10.1093/hmg/ddr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Wu WK, et al. Dysregulation and crosstalk of cellular signaling pathways in colon carcinogenesis. Crit Rev Oncol Hematol. 2012 doi: 10.1016/j.critrevonc.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 53.Christie M, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/beta-catenin signalling thresholds for tumourigenesis. Oncogene. 2012 doi: 10.1038/onc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robanus-Maandag EC, et al. A new conditional Apc-mutant mouse model for colorectal cancer. Carcinogenesis. 2010;31:946–952. doi: 10.1093/carcin/bgq046. [DOI] [PubMed] [Google Scholar]
- 55.Khazaie K, et al. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2012;109:10462–10467. doi: 10.1073/pnas.1207230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaked H, et al. Chronic epithelial NF-kappaB activation accelerates APC loss and intestinal tumor initiation through iNOS up-regulation. Proc Natl Acad Sci U S A. 2012;109:14007–14012. doi: 10.1073/pnas.1211509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei J, et al. p53 Family: Role of Protein Isoforms in Human Cancer. J Nucleic Acids. 2012;2012:687359. doi: 10.1155/2012/687359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bacolod MD, Barany F. Gene dysregulations driven by somatic copy number aberrations-biological and clinical implications in colon tumors: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2010;12:552–561. doi: 10.2353/jmoldx.2010.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. e2073. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albuquerque C, et al. Colorectal cancers choosing sides. Biochim Biophys Acta. 2011;1816:219–231. doi: 10.1016/j.bbcan.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 63.Sharma S, et al. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crea F, et al. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83:184–193. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 65.Bronevetsky Y, Ansel KM. Regulation of miRNA biogenesis and turnover in the immune system. Immunol Rev. 2013;253:304–316. doi: 10.1111/imr.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fabbri M. TLRs as miRNA receptors. Cancer Res. 2012;72:6333–6337. doi: 10.1158/0008-5472.CAN-12-3229. [DOI] [PubMed] [Google Scholar]
- 68.Kong YW, et al. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 69.Turchinovich A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu CW, et al. MicroRNA-18a Attenuates DNA Damage Repair through Suppressing the Expression of Ataxia Telangiectasia Mutated in Colorectal Cancer. PLoS One. 2013;8:e57036. doi: 10.1371/journal.pone.0057036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stracker TH, et al. The ATM signaling network in development and disease. Front Genet. 2013;4:37. doi: 10.3389/fgene.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sperka T, et al. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- 73.Migheli F, et al. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS One. 2013;8:e52501. doi: 10.1371/journal.pone.0052501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan XY, et al. Association between RUNX3 promoter methylation and gastric cancer: a meta-analysis. BMC Gastroenterol. 2011;11:92. doi: 10.1186/1471-230X-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nishio M, et al. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res. 2010;30:2673–2682. [PubMed] [Google Scholar]
- 77.Lee BB, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 78.Mirchev MB, et al. DNA methylation in patients with colorectal cancer--correlation with some clinical and morphological features and with local tumour invasion. Folia Med (Plovdiv) 2010;52:22–30. doi: 10.2478/v10153-010-0043-9. [DOI] [PubMed] [Google Scholar]
- 79.Drini M, et al. Investigating the potential role of genetic and epigenetic variation of DNA methyltransferase genes in hyperplastic polyposis syndrome. PLoS One. 2011;6:e16831. doi: 10.1371/journal.pone.0016831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 81.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 82.Ito K, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14:226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 83.Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29:2605–2615. doi: 10.1038/onc.2010.88. [DOI] [PubMed] [Google Scholar]
- 84.Ku JL, et al. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23:6736–6742. doi: 10.1038/sj.onc.1207731. [DOI] [PubMed] [Google Scholar]
- 85.Kim JH, et al. Subsets of microsatellite-unstable colorectal cancers exhibit discordance between the CpG island methylator phenotype and MLH1 methylation status. Mod Pathol. 2013 doi: 10.1038/modpathol.2012.241. [DOI] [PubMed] [Google Scholar]
- 86.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 87.Feinberg AP, et al. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 88.Baker SJ, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 89.Tulchin N, et al. c-myc protein distribution. Neoplastic tissues of the human colon. Am J Pathol. 1992;140:719–729. [PMC free article] [PubMed] [Google Scholar]
- 90.Tulchin N, et al. C-myc protein distribution - mammary adenocarcinomas of mtv/myc transgenic mice. Int J Oncol. 1995;7:5–9. [PubMed] [Google Scholar]
- 91.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 93.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 94.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 95.Grimm C, et al. DNA-methylome analysis of mouse intestinal adenoma identifies a tumour-specific signature that is partly conserved in human colon cancer. PLoS Genet. 2013;9:e1003250. doi: 10.1371/journal.pgen.1003250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kan PY, et al. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 98.Hassan AH, et al. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 99.Akhavan-Niaki H, Samadani AA. DNA Methylation and Cancer Development: Molecular Mechanism. Cell Biochem Biophys. 2013 doi: 10.1007/s12013-013-9555-2. [DOI] [PubMed] [Google Scholar]
- 100.Wilson AJ, et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res. 2010;70:609–620. doi: 10.1158/0008-5472.CAN-09-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee YM. Control of RUNX3 by histone methyltransferases. J Cell Biochem. 2011;112:394–400. doi: 10.1002/jcb.22969. [DOI] [PubMed] [Google Scholar]
- 102.Icardi L, et al. The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev. 2012;23:283–291. doi: 10.1016/j.cytogfr.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 103.Fahrner JA, et al. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- 104.Park SY, et al. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410–416. doi: 10.1002/path.2596. [DOI] [PubMed] [Google Scholar]
- 105.Ding SZ, et al. Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis. PLoS One. 2010;5:e9875. doi: 10.1371/journal.pone.0009875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kellermayer R, et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–1460. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rius M, Lyko F. Epigenetic cancer therapy: rationales, targets and drugs. Oncogene. 2012;31:4257–4265. doi: 10.1038/onc.2011.601. [DOI] [PubMed] [Google Scholar]
- 108.Licciardi PV, et al. Epigenome targeting by probiotic metabolites. Gut Pathog. 2010;2:24. doi: 10.1186/1757-4749-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ritchie ML, Romanuk TN. A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS One. 2012;7:e34938. doi: 10.1371/journal.pone.0034938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mohamadzadeh M, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khazaie K, et al. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A. 2012;109:10462–10467. doi: 10.1073/pnas.1207230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saber R, et al. Lipoteichoic acid-deficient Lactobacillus acidophilus regulates downstream signals. Immunotherapy. 2011;3:337–347. doi: 10.2217/imt.10.119. [DOI] [PubMed] [Google Scholar]
- 113.Lightfoot YL, et al. Targeting aberrant colon cancer-specific DNA methylation with lipoteichoic acid-deficient Lactobacillus acidophilus. Gut Microbes. 2013;4:84–88. doi: 10.4161/gmic.22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hedin C, et al. Evidence for the use of probiotics and prebiotics in inflammatory bowel disease: a review of clinical trials. Proc Nutr Soc. 2007;66:307–315. doi: 10.1017/S0029665107005563. [DOI] [PubMed] [Google Scholar]
- 115.Marteau PR, et al. Protection from gastrointestinal diseases with the use of probiotics. Am J Clin Nutr. 2001;73:430S–436S. doi: 10.1093/ajcn/73.2.430s. [DOI] [PubMed] [Google Scholar]
- 116.Ghadimi D, et al. Epigenetic imprinting by commensal probiotics inhibits the IL-23/IL-17 axis in an in vitro model of the intestinal mucosal immune system. J Leukoc Biol. 2012;92:895–911. doi: 10.1189/jlb.0611286. [DOI] [PubMed] [Google Scholar]