Abstract
Fluorescent imaging of RNA in living cells is a technically challenging problem in cell biology. One strategy for genetically encoding fluorescent RNAs is to express them as fusions with ‘RNA mimics of GFP’. These are short aptamer tags that exhibit fluorescence upon binding otherwise nonfluorescent fluorophores that resemble those found in GFP. We find that the brightest of these aptamers, Spinach, often exhibits reduced fluorescence after it is fused to RNAs of interest. We show that a combination of thermal instability and a propensity for misfolding account for the low fluorescence of various Spinach-RNA fusions. Using systematic mutagenesis, we identified nucleotides that account for the poor folding of Spinach, and generated Spinach2, which exhibits markedly improved thermal stability and folding in cells. Furthermore, we show that Spinach2 largely retains its fluorescence when fused to various RNAs. Using Spinach2, we detail the cellular dynamics of the CGG trinucleotide-repeat containing “toxic RNA” associated with Fragile-X tremor/ataxia syndrome, and show that these RNAs form nuclear foci with unexpected morphological plasticity that is regulated by the cell cycle and by small molecules. Together, these data demonstrate that Spinach2 exhibits improved versatility for fluorescently labeling RNAs in living cells.
Introduction
RNA localization is dynamically regulated in cells1,2. A major goal has been to develop genetically encoded systems analogous to green fluorescent protein (GFP) that enable imaging of tagged RNAs in living cells (see Supplementary Note 1). We developed Spinach, a 98-nt RNA aptamer that binds 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), a small molecule mimic of the GFP fluorophore3. Spinach and DFHBI are essentially nonfluorescent when separate, but interact to form a brightly fluorescent complex. Cells can be engineered to express RNAs fused to Spinach which can be imaged in live cells. We labeled the 5S RNA with Spinach in mammalian cells and observed changes in localization under stress conditions3.
Detection of 5S-Spinach in mammalian cells requires 1 sec exposure times despite its high expression level3. In contrast, imaging abundant GFP-tagged proteins in mammalian cells typically requires 10 – 100 msec exposure times under these imaging conditions. Additionally, the low brightness of 5S-Spinach in cells contrasts with the high brightness of Spinach measured in vitro3.
Here we show that Spinach exhibits thermal instability and poor folding, which reduces its brightness. Moreover, Spinach fluorescence is reduced when it is fused to target RNAs. Using systematic mutagenesis guided by brightness, thermostability, and a novel assay to measure folding, we identified mutations that confer thermostability and substantially increase the fraction of properly folded aptamer. The resulting RNA, Spinach2, is a “superfolder” variant of Spinach, which exhibits reduced context-dependence and is markedly brighter than Spinach in living cells. Using Spinach2, we explored the localization and dynamics of toxic CGG-repeat-containing RNAs. Imaging of these RNAs using short exposure times reveals that these RNAs exhibit dynamic localizations, which can be readily altered by cell division and small molecules. These data show that the enhanced folding and thermal stability of Spinach2 make it a versatile tool for imaging RNA in living cells.
Results
Low fluorescence of Spinach-tagged RNAs
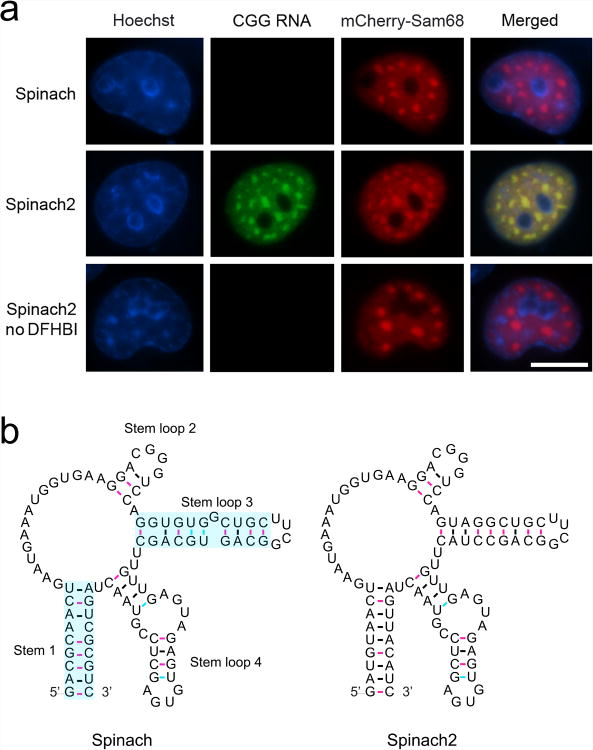
We sought to use Spinach to label “toxic RNA” localization. To do this, we expressed an RNA containing 60 CGG repeats that was previously shown to form intranuclear foci that resemble those seen in Fragile-X tremor/ataxia syndrome (FXTAS) patients4 with a 3′-Spinach tag. However, expression of the CGG60-Spinach construct did not result in readily detectable nuclear foci in COS-7 cells in the presence of DFHBI (Fig. 1). Although fluorescence was not detectable, Spinach-tagged RNA formed nuclear foci as measured by FISH (Supplementary Fig. 1a).
Figure 1. CGG-Spinach2 containing RNAs can be imaged in living cells.
(a) COS-7 expressing (CGG)60-Spinach (top row) or (CGG)60-Spinach2 (middle row) and mCherry-hSam68 in the presence of DFHBI. Cells expressing (CGG)60-Spinach2 and mCherry-hSam68 in the absence of DFHBI are also shown (bottom row). Nuclei were stained using Hoechst. Scale bar, 10 μm.
(b) Secondary structure of Spinach and Spinach2. Spinach is predicted to form one stem and three stem-loops. Stems 1 and stem-loop 3 (shaded cyan) were mutated to generate Spinach2.
We asked whether the Spinach tag was unstable or degraded from the CGG-repeat RNA. To test this, FISH was carried out with a probe against Spinach. This experiment confirmed that Spinach is present in these foci (Supplementary Fig. 1a, bottom row). In addition, we tested whether Spinach-tagged CGG-repeat RNA is destabilized using quantitative RT-PCR (qRT-PCR). However, tagged and untagged versions of (CGG)60-repeat RNA were equally stable (Supplementary Fig. 1b). The observation that the Spinach-tagged CGG-repeat RNA was abundant in foci but not fluorescent indicates that Spinach is not fluorescent in the context of the CGG-repeat RNA, and requires modifications to enhance its fluorescence in cells.
Folding and thermostability of Spinach
To understand the lower-than-expected fluorescence of Spinach in mammalian cells, we considered several factors that could affect its brightness. These include low DFHBI cell permeability, low intrinsic brightness, and poor folding in cells. DFHBI permeability is unlikely to account for the low fluorescence since permeability of DFHBI matches that of Hoechst in mammalian cells, with maximal fluorescence achieved in approximately 30 min (Supplementary Fig. 2). Additionally, in vitro measurements of Spinach-DFHBI fluorescence show that its overall brightness is 80% of GFP and 53% of eGFP3, which is bright enough for imaging. We therefore considered the possibility that Spinach misfolds in cells, reducing the number of Spinach-tagged RNAs that can bind and activate the fluorescence of DFHBI.
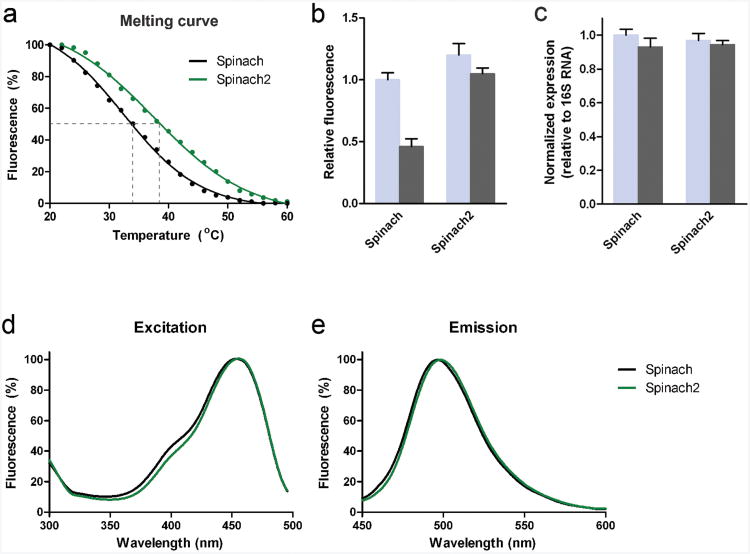
We first asked if Spinach can fold in cells at 37°C. We determined the melting temperature (Tm) of Spinach by monitoring the fluorescence of the RNA-DFHBI complex in vitro between 20°C and 60°C. These experiments showed that Spinach has a Tm of 34 ± 0.6°C (Fig. 3a, Supplementary Table 1), indicating that a substantial fraction of Spinach molecules may be unfolded when imaging at 37°C.
Figure 3. Properties of Spinach and Spinach2 in vitro and in bacteria.
(a) Spinach2 has increased thermostability relative to Spinach. The fluorescence of Spinach (black) and Spinach2 (green) was measured in the presence of DFHBI from 20 to 60 °C. Shown are representative data (dots) along with the best fit curve from fitting with the Boltzmann sigmoidal equation (line).
(b) Excitation spectra of Spinach and Spinach2. Fluorescence excitation was measured from 300-500 nm with emission recorded at 510 ± 10 nm.
(c) Emission spectra of Spinach and Spinach2. Fluorescence was excited with 420 ± 10 nmlight and emission was recorded from 450-600 nm. In all cases, spectra are normalized to maximal signal.
(d) Normalized fluorescence signal from E. coli expressing either Spinach or Spinach2. All values shown are normalized to Spinach signal at 25 °C. Values plotted are mean and s.e.m. values of three independent experiments.
(e) Normalized Spinach and Spinach2 expression in E. coli. Total RNA from samples used in fluorescence measurements were then subjected to reverse transcription followed by qRT-PCR. Spinach and Spinach2 signal were normalized to 16S RNA. Data represent mean and s.e.m. values for three independent replicates.
Mutational analysis of Spinach
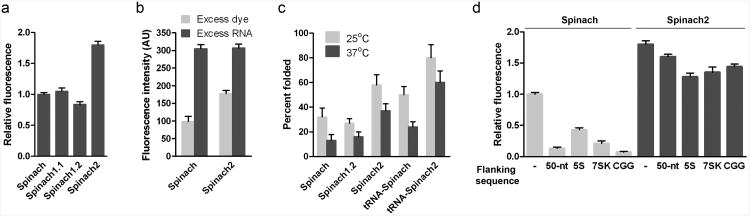
We next sought to identify mutations that could increase the thermostability of Spinach by correcting bulges and mismatches in the predicted structure of Spinach (Fig. 1b). These results led to the generation of Spinach1.1 and Spinach1.2, which have perfect complementarity in stem 1 and stem 1 and stem loop 3, respectively (see Online Methods and Supplementary Fig. 3). Spinach1.1 showed slightly enhanced thermostability, with a Tm of 35 ± 0.5°C and was as bright as Spinach (Fig. 2a, Supplementary Table 1). Spinach1.2 displayed higher thermostability relative to Spinach and Spinach1.1, with Tm value of 38 ± 0.3°C (Supplementary Table 1). However, the observed brightness of Spinach1.2 was 16% lower than Spinach (Fig. 2a). Taken together, these data indicate that mutations in stem 1 and stem loop 3 enhance thermostability, but do not improve brightness.
Figure 2. Spinach2 is brighter than Spinach due to improved folding.
(a) Normalized brightness values of Spinach and derivatives. The fluorescence signal from 1 μM RNA and 10 μM DFHBI was measured at 25°C and normalized to Spinach fluorescence. Data represent mean and s.e.m. values for three independent replicates.
(b) Results of folding assay. Data represent mean and s.e.m. values for three independent replicates.
(c) Percent folded values of Spinach and derivatives at 25°C and 37°C. Data represent mean and s.e.m. values for three independent replicates.
(d) Normalized brightness values of Spinach and derivatives. The fluorescence signal from 1 μM RNA and 10 μM DFHBI was measured at 25°C and normalized to Spinach fluorescence. Data represent mean and s.e.m. values for three independent replicates.
Development of Spinach2
We next sought to understand the basis for the reduced brightness of Spinach1.2. We considered that these mutations either (1) reduce the extinction coefficient or quantum yield of Spinach-DFHBI; or (2) increase the misfolded fraction of Spinach that is unable to bind DFHBI. To determine if these mutations increase the percent of Spinach that is misfolded, we developed an assay to measure the fraction of Spinach that is properly folded (see Online Methods).
Using this assay with buffers that mimic ion concentrations normally found in the cytoplasm, we found that 32 ± 4.2% and 13 ± 2.8% of Spinach is folded at 25 and 37°C, respectively. Spinach1.2 was largely misfolded, with 27 ± 2.1 and 16 ± 2.3% folded at 25 and 37°C, respectively (Fig. 2b,c). These data indicate that Spinach folds poorly, and that the increased thermostability of Spinach1.2 did not correspond to a higher folded fraction of Spinach at 25°C.
We next carried out systematic mutagenesis to identify Spinach mutants that exhibited the enhanced thermostability of Spinach1.2, but also exhibited improved folding (see Online Methods). Using this approach, we identified six positions in Spinach that maintained or enhanced brightness at 25°C and maintained Spinach1.2 thermostability. These mutations were tested alone and in combination (Supplementary Fig. 4, Supplementary Table 2). The winner from this screen contained all six mutations and was 1.8- and 2.8-fold brighter than Spinach in vitro at 25 and 37°C, respectively, and has Tm of 38 ± 0.4°C (Fig. 3a, Supplementary Table 1). We named this mutant Spinach2 (Fig. 1b).
Characterization of Spinach2 fluorescence properties
We next asked whether Spinach2 exhibits improved folding relative to Spinach using the assay described above. These experiments showed that a substantially higher fraction of Spinach2 is folded compared to Spinach, with 58 ± 4.8% and 37 ± 3.3% folded at 25 and 37°C, respectively (Fig. 2c). Thus, the mutations in Spinach2 result in markedly enhanced folding.
The mutations in Spinach2 could affect its ability to activate the fluorescence of DFHBI. To test this, we calculated its extinction coefficient and quantum yield of Spinach2. In these experiments, we used excess RNA and 0.1 μM DFHBI, so that we could compare the properties of 0.1 μM Spinach-DFHBI and 0.1 μM Spinach2-DFHBI, regardless of any difference in the percent of each RNA that is folded. We found that both Spinach and Spinach2 have nearly identical photophysical properties (Supplementary Table 1). Moreover the excitation and emission spectra, as well as the KD for DFHBI binding are nearly identical (Fig. 3b,c, Supplementary Table 1). These data suggest that the enhanced brightness seen with Spinach2 reflects an increase in the folding efficiency of this RNA.
Spinach2 retains fluorescence in diverse contexts
RNA folding can be affected by flanking sequences, which can potentially form interactions with the RNA aptamer. To test whether sequence context affects Spinach and Spinach2 folding, we monitored the fluorescence of Spinach and Spinach2 inserted into different RNAs. First, both Spinach and Spinach2 were synthesized with an additional 50 nt of RNA on both the 5′ and 3′ sides. We then compared the fluorescence of identical concentrations (0.1 μM) of flanked Spinach to Spinach alone. Flanked Spinach was only 20% as bright as Spinach (Fig. 2d). We next asked if Spinach2 is affected by these flanking sequences. Flanked Spinach2 was 90% as bright as Spinach2 alone, and 10-fold brighter than flanked Spinach (Fig. 2d), indicating that Spinach2 is relatively insensitive to flanking sequence.
Spinach fluorescence in vivo is improved by inserting Spinach into the tRNALys3 sequence3,5, which acts as a folding scaffold6. In the case of Spinach, the folding is increased from 32 ± 4.2 to 50 ± 3.9% at 25°C and 13 ± 2.8 to 24 ± 2.4% at 37°C by the presence of the tRNA (Fig. 2c). In the case of Spinach2, the folding is increased from 58 ± 4.8 to 80 ± 6.1% at 25°C and 37 ± 3.3 to 60 ± 5.4% at 37°C. For this reason, we used tRNALys3-Spinach and tRNALys3-Spinach2 in all subsequent tagged constructs and imaging experiments. We next compared the folding of Spinach or Spinach2 fused to 5S. 5S-Spinach2 was 3-fold brighter than 5S-Spinach (Fig. 2d), indicating that Spinach2 folds better than Spinach when fused to this RNA, even when Spinach is present in the context of the tRNALys3 scaffold. 5S-Spinach2 folding was only ∼30% lower than Spinach2 (Fig. 2d). We also examined the folding of Spinach and Spinach2 fused to the 5′ end of the nuclear 7SK RNA7. In this case, Spinach2-7SK was 6-fold brighter than Spinach-7SK in vitro (Fig. 2d). Moreover, Spinach2-7SK folding was only ∼25% lower than Spinach2 (Fig. 2d).
Lastly, we examined Spinach2 folding in the context of CGG-repeat-containing RNA. Spinach and Spinach2 were appended to the 3′ end of (CGG)60 RNA. The in vitro synthesized (CGG)60-Spinach was nearly nonfluorescent, while (CGG)60-Spinach2 was 80% as bright as Spinach2 alone (Fig. 2d). Together, these data show that Spinach2 retains substantial fluorescence when tagged to diverse RNAs.
Spinach2 exhibits increased fluorescence in E. coli
We next asked whether the increased stability of Spinach2 in vitro would correspond to a brighter signal in E. coli. Spinach2 was 1.4-fold brighter at 25°C and 2.1-fold brighter at 37°C than Spinach (Fig. 3d). Aptamer abundance was normalized to 16S RNA and found to be essentially identical for all samples (Fig. 3e).
Imaging 5S-Spinach2 and Spinach2-7SK
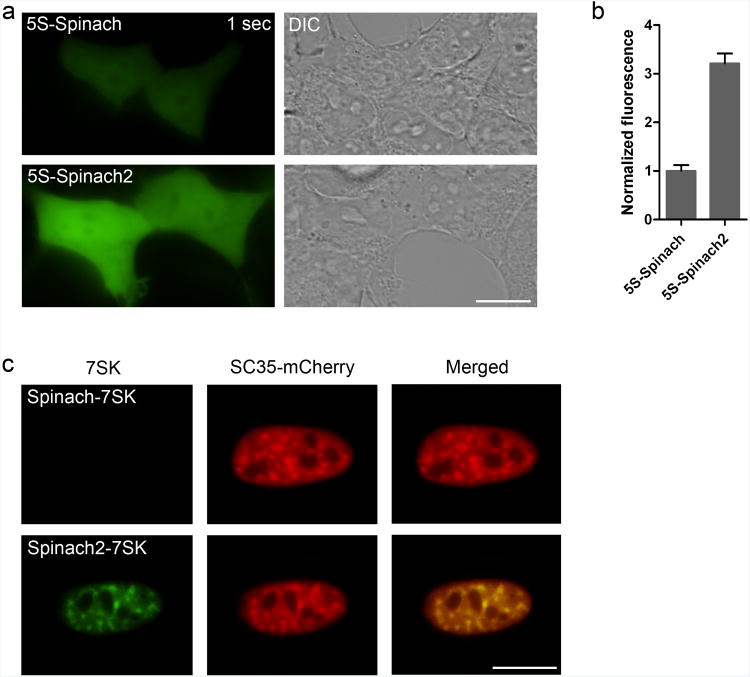
We next asked if 5S-Spinach2 is brighter in mammalian cells. HEK293T cells expressing 5S-Spinach or 5S-Spinach2 exhibited the expected diffuse nuclear and cytoplasmic distribution (Fig. 4a)3. When quantified, 5S-Spinach2 showed 3.2-fold higher signal relative to 5S-Spinach (Fig. 4b).
Figure 4. 5S-Spinach2 is brighter than 5S-Spinach in mammalian cells.
(a) HEK293T cells were transiently transfected to express either 5S-Spinach or 5S-Spinach2 under the control of the 5S promoter. Cells were incubated with 20 μM DFHBI and imaged with a 1 sec (left column) exposure time. Green fluorescence and DIC (right column) images are shown. Scale bar, 10 μm.
(b) The brightness for cells labeled with either 5S-Spinach or 5S-Spinach2 were determined and normalized for area. 5S-Spinach signal was normalized to one. 5S-Spinach2 was 3.2-fold brighter than Spinach. Data shown represent mean and s.e.m. values for 20 cells per condition.
(c) HeLa cells were transiently transfected to express either Spinach-7SK or Spinach2-7SKunder the control of the CMV promoter. Cells were cotransfected with SC35-mCherry, which labels nuclear speckles. Cells were incubated with 20 μM DFHBI and imaged for 200 ms. Green and red fluorescence images are shown along with overlaid images. Scale bar, 10 μm.
We also compared the brightness of Spinach-7SK and Spinach2-7SK in HeLa cells. 7SK localizes to nuclear speckles8. Expression of Spinach-7SK showed no detectable signal, but expression of Spinach2-7SK labeled intranuclear foci that colocalize with SC35, a known protein component of nuclear speckles8,9, tagged with mCherry (Fig. 4c). These data demonstrate improved RNA imaging in live cells using Spinach2.
Imaging (CGG)60 RNA with Spinach2
Many “toxic RNAs” contain extensive trinucleotide repeats10. FMR1 transcripts containing 55-200 CGG repeats11-13 lead to neurodegeneration associated with FXTAS14. These transcripts aggregate to form intranuclear foci14 that cause cytotoxicity by sequestering proteins and affecting splicing and microRNA biogenesis4,15,16, although the translation of these RNAs may also contribute to disease phenotypes17.
Little is known about the dynamics of CGG-repeat-containing RNA localization in the nucleus. Because these RNA complexes are highly G/C rich, it has been proposed that they form highly stable hairpins that may be difficult to disrupt4,15,18. Previous studies have shown that the splicing factor Sam68 dynamically associates with CGG-repeat nuclear foci4,19. However, these studies do not address whether the RNAs themselves are dynamic or immobile in nuclei.
Thus, we tested whether Spinach2 could be used to image CGG-repeat RNA. Although (CGG)60-Spinach was not detected (Fig. 1a, top row), expression of (CGG)60-Spinach2 resulted in bright intranuclear foci that were readily detected using wide-field microscopy with 50-100 msec exposure times (Fig. 1a, middle row). These foci colocalized with mCherry-hSam68, a marker of CGG-containing nuclear foci4 (Fig. 1a). The foci were highly heterogeneous in appearance (Supplementary Fig. 4b). Thus (CGG)60-Spinach2 can be used to study the dynamics of toxic RNA aggregates.
Live-cell imaging of CGG-repeat RNA aggregates
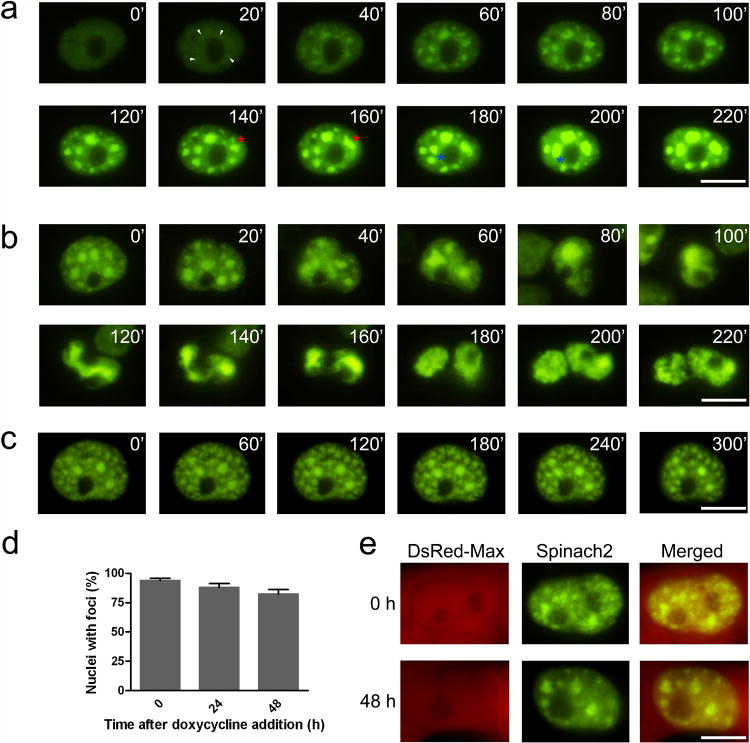
We next monitored the formation of (CGG)60-Spinach2 foci in transiently transfected COS-7 cells. Spinach2 fluorescence was detectable as early as 3 h post-transfection. (CGG)60-Spinach2 signal was initially diffusely nucleoplasmic, with foci formation evident within 1 h (Fig. 5a, Supplementary Movie 1). Foci number, size, and brightness increased over the course of the experiment. These data indicate that CGG-repeat RNA aggregates rapidly following expression.
Figure 5. Imaging RNA foci in COS-7 cells.
(a) Foci formation in COS-7 cells after transient transfection with a CGG-Spinach2 vector. At 2 h post-transfection, cells were incubated with imaging medium containing DFHBI and imaged every 20 min for 6 h. In these images, time zero indicates the first frame that displayed fluorescence above background. Small foci formed de novo (white arrowheads) are highlighted. Merging foci are also highlighted foci (red and blue asterisks). Scale bar, 10 μm.
(b) RNA foci are partitioned and divided during cell division. Scale bar, 10 μm.
(c) (CGG)60-Spinach2 signal persists in foci after transcriptional silencing. Cells containing CGG aggregates were treated with 1 μg/mL actinomycin D to inhibit transcription and monitored for changes in foci. Images obtained every hour for 5 h are shown. Scale bar, 10 μm.
(d) (CGG)60-Spinach2 signal persists in foci for over 48 h after transcriptional silencing. A TET-Off expression system was used in COS-7 cells to test foci stability in the absence of new transcription over longer time periods. Data shown represent the mean and s.e.m. values for three independent replicates in which 100 DsRed-positive cells were counted for each treatment.
(e) Representative nuclei for 0 and 48 h after doxycycline addition. Scale bar, 10 μm.
We next asked if the aggregated CGG-repeat RNA is highly stable in cells. To examine the stability of (CGG)60-Spinach2 RNA, we measured fluorescence after treatment of cells with actinomycin D, a potent transcription inhibitor. Here, we observed that Spinach2 signal was stable and remained unchanged for up to 8 h (Fig. 5c), at which point actinomycin D-mediated cytotoxicity was observed.
To test the stability of (CGG)60-Spinach2 foci over longer time periods, we controlled (CGG)60-Spinach2 transcription using the TET-Off system20 (see Online Methods). Immediately following transcription inhibition, 94 ± 1.7% of transfected cells contained foci. These foci were long-lived, as 88 ± 5.6 and 82 ± 6.5% of cells retained foci after 24 and 48 h, respectively (Fig. 5d,e). These results were supported by qRT-PCR results, which demonstrate that (CGG)60 and (CGG)60-Spinach2 RNA are highly stable. The stability of these RNAs is most likely due to its incorporation into nuclear foci as (CGG)30 RNA, which do not form foci4, are markedly less stable (Supplementary Fig. 1b).
CGG-repeat RNA undergo rearrangements during cell division
Since these RNAs are relatively resistant to degradation and form thermodynamically stable duplexes18,21, we sought to test whether they form static foci. To test this idea, we monitored foci morphology in transfected cells. Time-lapse imaging revealed that foci were mobile and can merge to form larger foci (Fig. 5a, Supplementary Movie 1).
This dynamic nature was also apparent in dividing cells (Fig. 5b). Prior to cell division, typical cells contain multiple foci. During division, the foci coalesce to form a large single aggregate that then extends into a long linear structure. This long aggregate is divided between daughter cells. The RNA then appears to become diffusely nucleoplasmic before reaggregating into foci. These results suggest that CGG-repeat RNA foci aggregate and disaggregate during the cell cycle.
A small molecule can disrupt RNA aggregates
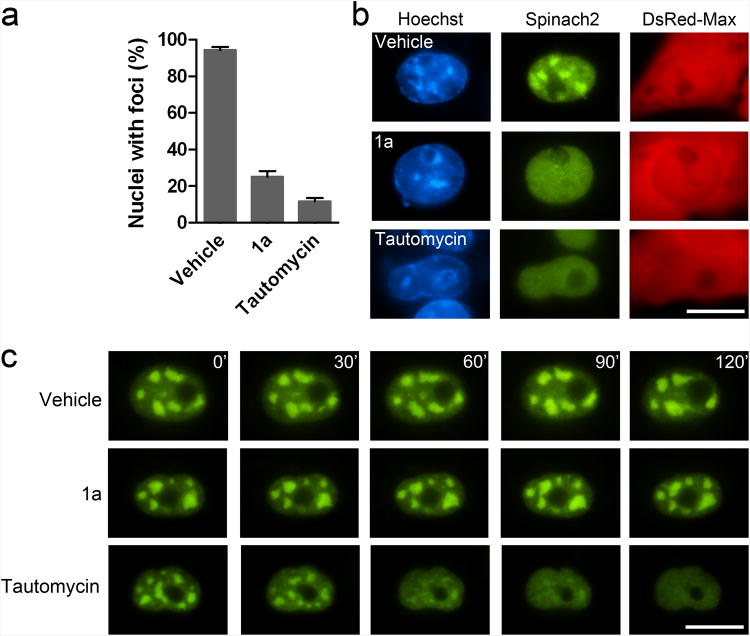
We next asked if small molecules can induce disaggregation of CGG-repeat RNA foci. No molecules have been shown to disrupt existing aggregates, although two drugs prevent the formation of CGG-repeat RNA foci in transfected cells. These are tautomycin4 and 1a, a small molecule that binds CGG-repeat RNA and disrupts its binding to a CGG-binding protein, DGCR84, 22. We confirmed that both drugs prevent (CGG)60-Spinach2 foci formation (Fig. 6a,b).
Figure 6. The effects of tautomycin and 1a on CGG RNA foci.
(a) Tautomycin and 1a prevent the formation of CGG repeat foci. Transfected cells were treated with vehicle, 20 μM 1a, or 5 μM tautomycin was added to cells. After 24 h, 100 DsRed-positive cells were analyzed for the presence of nuclear foci for each condition. 94 ± 1.4, 25 ± 4.9, and 12 ± 4.2% of nuclei contained foci with vehicle, 1a, and tautomycin, respectively. Data shown are mean and s.e.m. values for three independent experiments.
(b) Representative images of nuclei after 24 h of treatment. DsRed-Max was used as a transfection control.
(c) Preexisting (CGG)60-Spinach2 foci are disaggregated by tautomycin. COS-7 cells expressing (CGG)60-Spinach2 foci were incubated with vehicle, 20 μM 1a, or 5 μM tautomycin and imaged for 2 h after drug treatment. Scale bar 10 μm.
To determine whether 1a can disrupt existing foci, COS-7 cells expressing (CGG)60-Spinach2 were treated with 1a and imaged every 5 min for 2 h. No change in foci was observed under these conditions (Fig 6c). To test whether longer treatments were required for 1a to disrupt foci, cells expressing (CGG)60-Spinach2 were treated with 1a for 48 h. Immediately after addition of 1a, 94 ± 2.8% of examined nuclei contained foci. After 48 h of 1a treatment, 86 ± 3.5% contained foci, indicating that 1a does not substantially disrupt foci, even after long treatments (Supplementary Fig. 5a). Furthermore, a 48 h treatment of 1a did not induce the dissociation of Sam68 from (CGG)60-Spinach2 foci (Supplementary Fig. 5b). These results show that 1a can prevent foci formation, but does not readily disrupt existing foci.
In contrast, we observed that tautomycin induces disaggregation of foci in as little as 1 h (Fig. 6c, Supplementary Movie 2). The (CGG)60-Spinach2 remained as diffuse nucleoplasmic staining in cells (Fig. 6c). Removal of tautomycin after a 2 h treatment was not sufficient to restore foci formation (Supplementary Fig. 6), suggesting that tautomycin induces cellular changes that prevent reaggregation.
To test if the effect of tautomycin on (CGG)60-Spinach2 foci was due to inhibition of its known targets protein phosphatase-1 (PP1) or protein phosphatase-2A (PP2A)23, we treated cells with okadaic acid at a concentration that also inhibits both PP1 and PP2A24. In this case, no foci disruption was observed over 4 h (Supplementary Fig. 7). These results suggest that the disaggregation effect of tautomycin is likely to be due to a different target than PP1 or PP2A.
Discussion
We found that Spinach exhibits poor thermal stability and folding when tagged to other RNAs. To resolve these issues, we developed Spinach2 by targeted mutagenesis of Spinach followed by screening for enhanced brightness and thermostability. Spinach2 has nearly identical photophysical properties to Spinach, yet displays enhanced folding both alone and in the context of flanking RNA. Moreover, Spinach2 exhibits improved folding at both 25°C and 37°C, yielding significant enhancements during imaging. Our results show that improvements in folding and thermostability enable imaging of RNAs that are otherwise not detectable with Spinach.
Although Spinach2 folds more efficiently than Spinach, the improved folding is more apparent when Spinach2 is fused to other RNAs. For example, Spinach2 retains 80% of its fluorescence when fused to the CGG-repeat RNA, while Spinach is essentially nonfluorescent in this context. Thus, the improved performance of Spinach2 in live cells reflects its improved folding when fused to other RNAs. However, it is possible that other flanking sequences will affect Spinach2 fluorescence. Therefore, the fluorescence of a Spinach2-tagged RNA should first be established by in vitro transcription of the Spinach2-tagged RNA and compared with the fluorescence of untagged Spinach2. If the Spinach2-tagged RNA lacks fluorescence in vitro, inserting Spinach2 at other sites may restore fluorescence by providing flanking sequences that are more compatible with Spinach2 folding.
Both (CGG)60-Spinach2 and Spinach2-7SK form RNA-enriched foci within the cell, which make imaging straightforward. However, imaging RNAs present at lower concentrations may require longer imaging times. Since multimerization of fluorescent proteins has been successfully used to enhance the imaging of low abundance proteins25, an analogous strategy could be adapted to label RNAs with Spinach2. Tagging RNAs with multiple Spinach2 sequences may be valuable to enhance the brightness of tagged RNA and aid in imaging lower abundance RNAs.
In order to demonstrate the ability to use Spinach2 in diverse imaging experiments, we imaged and characterized the localizations of CGG repeat-containing RNAs in living cells for the first time. These RNAs were thought to form stable G/C-rich aggregates18,21. Our studies show that the RNA component of these foci is highly dynamic in cells and undergoes considerable morphologic rearrangements, especially during cell division. These results suggest that CGG-repeat RNAs bind to preexisting nuclear structures that are normally partitioned during cell division. This idea is supported by previous studies demonstrating colocalization of CGG-repeat RNAs with various intranuclear markers4.
By imaging (CGG)60-Spinach2 we were able to identify the first compound that can induce disaggregation of toxic RNAs. Previous studies have relied on imaging foci-associated RNA-binding proteins, such as Sam684,16. Direct imaging of toxic RNA provides opportunities to identify small molecules and signaling pathways that affects CGG-repeat RNA localization dynamics in living cells. Assays using (CGG)60-Spinach2 may enable the identification of additional compounds that can disrupt foci and potentially serve as therapeutics for FXTAS.
Online Methods
Spinach2 DNA sequence
5′-GATGTAACTGAATGAAATGGTGAAGGACGGGTCCAGTAGGCTGCTTCGGCAGC CTACTTGTTGAGTAGAGTGTGAGCTCCGTAACTAGTTACATC-3′
Reagents and equipment
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich. Commercially available reagents were used without further purification. Absorbance spectra were recorded with a Thermo Scientific NanoDrop 2000 spectrophotometer with cuvette capability. Fluorescence excitation and emission spectra were measured with a Perkin Elmer LS-55 fluorescence spectrometer.
Preparation and analysis of Spinach and Spinach mutants
RNAs were created by using the appropriate single stranded DNA templates (Integrated DNA Technologies) and PCR amplification using primers which included a 5′ T7 promoter sequence to generate double stranded DNA templates. PCR products were then purified with PCR purification columns (Qiagen) and used as templates for in vitro T7 transcription reactions (Epicentre) as described previously3. RNA is purified using ammonium acetate precipitation, and quantified using both absorbance values and the Riboquant Assay kit (BD Biosciences). Photophysical characterization of Spinach2 was carried out as previously described3.
Thermostability measurements
Spinach or Spinach2 (1 μM) was incubated in 20 mM HEPES pH 7.4, 100 mM KCl, 1 mM MgCl2, and 10 μM DFHBI. Fluorescence values were recorded in one degree increments from 20 to 60°C, with a 5 min incubation at each temperature to allow for equilibration. Fluorescence measurements were performed using a Perkin Elmer LS-55 fluorescence spectrometer using the following instrument parameters: excitation wavelength, 460 nm; emission wavelength, 501 nm; slit widths, 10 nm. Curves were fitted using the Boltzmann sigmoidal equation in GraphPad Prism 5 software. Values presented are mean and s.e.m. from three independent measurements.
Folding Assay
Our folding assay involves measuring fluorescence under two conditions, one in which RNA is in excess relative to DFHBI, and one in which the DFHBI is in excess relative to RNA. Since Spinach and DFHBI form a 1:1 stoichiometric complex, the maximum amount of complex that can be formed is determined by the limiting component. In the first condition, the fluorescence was determined by incubation of 0.1 μM DFHBI and 100-fold excess (10 μM) Spinach. This value is used to define the fluorescence of 0.1 μM Spinach-DFHBI complex. We assume that even if nearly all Spinach is misfolded or unfolded, there will be enough properly folded Spinach to stoichiometrically bind 0.1 μM DFHBI. We confirmed this by measuring fluorescence after doubling the RNA to 20 μM, which caused no increase in fluorescence (data not shown). In the second condition, we measure the fluorescence obtained using 10 μM DFHBI and 0.1 μM Spinach. In theory, up to 0.1 μM Spinach-DFHBI can form if all the Spinach is folded. However, if a portion of Spinach is unfolded, the fluorescence will be proportionately less than the fluorescence of 0.1 μM Spinach-DFHBI. Thus, this approach can reveal the fraction of Spinach that is folded under diverse conditions.
Fluorescence was measured for each RNA under the following conditions: (1) 0.1 μM RNA and 10 μM DFHBI and (2) 0.1 μM DFHBI and 10 μM RNA. For each condition, the signal from DFHBI without RNA was subtracted from each signal. Fluorescence was measured in 20 mM HEPES pH 7.4, 100 mM KCl, 1 mM MgCl2 at the designated temperature. Fluorescence measurements were performed using a Perkin Elmer LS-55 fluorescence spectrometer using the following instrument parameters: excitation wavelength, 460 nm; emission wavelength, 501 nm; slit widths, 10 nm. Signal from the first condition (limiting RNA) were divided by the signal from the second condition (limiting dye) to determine the fraction folded.
Generation of Spinach1.1 and Spinach1.2
Spinach is predicted to contain four stems (Fig. 1b). Some of the stems contain bulges and mismatches that likely reduce its thermodynamic stability; however, it is not clear whether these features are also necessary for Spinach-induced DFHBI fluorescence. Our previous work mutating Spinach and designing Spinach-based sensors demonstrated that stem 1 and stem loop 3 can tolerate various mutations and insertions5,7. Therefore, we considered the possibility that the mismatches in stem 1 adversely affect Spinach thermal stability. To test this idea, we generated a mutant of Spinach with perfect complementarity in stem 1. This mutant, called Spinach1.1, was also mutated to convert the last base pair in stem 1 from U-A to C-G, in an attempt to stabilize stem 1 (Supplementary Fig. 4). Spinach1.1 showed slightly enhanced thermostability, with a Tm of 35 ± 0.5°C and was as bright as Spinach (Fig. 2a, Supplementary Table 1).
We next asked if stem loop 3 could be altered to increase Spinach thermostability. We previously found that alterations in stem loop 3 do not substantially reduce Spinach fluorescence7. In Spinach, stem loop 3 contains three mismatches and an internal bulge. We generated Spinach1.2 by retaining the mutations in Spinach1.1 and mutating stem loop 3 to eliminate this bulge and introduce perfect complementarity (Supplementary Fig. 3).
Systematic mutagenesis of Spinach1.2
Because elevated G/C content can lead to stable misfolded structures8,9, we reasoned that decreasing the overall G/C content could promote proper folding. We carried out scanning mutagenesis, mutating every guanidine and cytosine to adenosine or uracil, respectively. In regions where G and C residues were predicted to form a base pair, both residues were mutated to A and U, in order to maintain the complementarity. Each of these 35 mutants was synthesized in vitro, and the fraction folded was measured at 25°C and 42°C. Fluorescence signals that were equal to or greater than Spinach at 25°C indicated equal or greater percent folded. A higher percent signal at 42°C indicated improved thermostability relative to Spinach.
Preparation and in vitro analysis of flanked Spinach and Spinach2 constructs
Spinach and Spinach2 constructs flanked on either side by 50 bps were generated by PCR. For Spinach, the following forward (5′- TAATACGACTCACTATAGGGCGGA CTATGACTTAGTTGCGTTACACCCTTTCTTGACAAAACCTAACTTGACGCAACTGAATGAAA TGGTG-3′) and reverse (5′- AAACAAAAAAAACAAATAAAGCCATGCCAAT CTCATCTTGTTTTCTGCGCGA CGCGACTAGTTACGGAG-3′) primers were used. For Spinach2, the following forward (5′- TAATACGACTCACTATAGGGCGGACTATGACTTA GTTGCGTTACACCCTTTCTTGACAAAACCTAACTTGATGTAACTGAATGAAATG-3′) and reverse (5′-AACAAAAAAAACAAATAAAGCCATGCCAATCTCATCTTGTTTTCT GCGCGATGTAACTAGTTACGGAG-3′) primers were used. The 50 bp sequences were taken from the human β-actin 3′ untranslated region. Both forward primers encode the T7 RNA polymerase promoter for in vitro transcription.
5S-Spinach and 5S-Spinach2 were amplified by PCR from pAV-5S-Spinach and pAV-5S-Spinach2, respectively, using forward (5′-TAATACGACTCACTATAGGGT CTACGGCCATA CCACCCTG-3′) and reverse (5′-TGGCGCCCGAACAGGGAC-3′) primers. (CGG)60-Spinach and (CGG)60-Spinach2 were amplified by PCR from pCDNA-60CGG-Spinach and pCDNA-60CGG-Spinach2, respectively, using forward (5′- TAATA CGACTCACTATAGG -3′) and reverse (5′- GGCAAACAACAGATGGCTGGC AACTAG -3′) primers. PCR products were used as templates for in vitro transcription by Ampliscribe T7 RNA polymerase as previously described3. Fluorescence measurements were recorded for 0.1 μM RNA in the presence of 10 μM DFHBI in buffer composed of 20 mM HEPES pH 7.4, 100 mM KCl, 100 μM MgCl2 as described above.
Cloning Spinach2 for expression in E. coli
Spinach and Spinach2 were PCR amplified with primers containing the EagI restriction sites on both the 5′ and 3′ ends of the Spinach sequence. They were then cloned into a pET28c-based plasmid containing a chimera of the human tRNALys3 scaffold, which we previously used for Spinach and Spinach-based metabolite sensors and has previously been shown to stabilize heterologous expression of RNA aptamers in E. coli6.
Whole-cell fluorescence measurements of E. coli
BL21 cells were transformed to harbor either pET28c-tRNA-Spinach or pET28c-tRNA-Spinach2, and grown in Luria broth + 100 μg/mL kanamycin to OD600 0.4 at room temperature. The cells were then induced with addition of 1 mM IPTG for 2 h at room temperature. After induction, cells were normalized for cell density and split into two aliquots. One aliquot per sample was incubated at room temperature, and the other was incubated for 20 min at 37°C. Cells were then measured for total fluorescence using a Tecan SafireII plate reader with 460 ± 10 nm excitation and emission was recorded at 510 ± 10 nm. Data shown represent mean and s.e.m. values for three independent experiments.
qRT-PCR analysis of Spinach and Spinach2 concentration in E. coli
Total RNA samples were collected from E. coli at both 25 and 37 °C using the RNeasy Protect Bacteria Mini Kit (Qiagen). Reverse transcription was carried out on all samples using a reverse primer that bound in the tRNA portion of the tRNA-Spinach transcripts (5′-TGGCGCCCGAACAGGGAC-3′) and a reverse primer against 16S RNA (5′-GTATTACCGCGGCTGCTG-3′) according to the SuperscriptIII reverse transcription kit protocol. qRT-PCR was carried out according to the iQ™ SYBR® Green Supermix (Bio-Rad) protocol with forward (5′-GCCCGGATAGCTCAGTCGGTAG-3′) and reverse (5′-TGGCGCCCGAACAGGGAC-3′) primers against the tRNA portion of either transcript as well as forward (5′-CTCCTACGGGAGGCAGCAG-3′) and reverse (5′-GTATTACCGCGGCTGCTG-3′) primers against 16S RNA. In all cases, Spinach transcript levels were normalized to 16S RNA levels. Data represent mean and s.e.m. values for three independent experiments.
Cloning of 5S-Spinach and Spinach2
pAV-5S-Spinach was generated as previously described3. This construct contains Spinach in the context of the tRNALys scaffold. tRNALys-Spinach was removed from pAV-5S by restriction digest with SalI and XbaI. tRNALys-Spinach2 was amplified from pET28c-tRNA-Spinach2 by PCR using forward (5′-TAGGCGTCGACGCCCGGATAGC TCAGTCGGTAG AGCAG-3′) and reverse (5′- ATATATTCTAGATGGCGCCCGAACA GGGACTTGAACCC-3′) primerse and digesting the resulting PCR products with XbaI and SalI to clone into pAV-5S.
Imaging 5S-Spinach and 5S-Spinach2
Imaging of 5S-Spinach and 5S-Spinach2 was carried out as previously described for 5S-Spinach3. Cells were imaged for either 100 msec or 1 sec. Background signals from cells expressing pAV-5S incubated with DFHBI were also taken at 100 msec and 1 sec and subtracted from the corresponding images using NIS-Elements software.
For brightness quantification, fluorescence signal was measured for 20 background subtracted cells per sample and normalized for total area using NIS-Elements AR 3.2 (Nikon). 5S-Spinach2 signal was normalized to 1.0.
Cloning of Spinach-7SK and Spinach2-7SK
Spinach or Spinach2 in the context of the tRNALys scaffold was amplified by PCR using forward (5′- ATATATGGATCCGCCCGGATAGCTCAGTCGG -3′) and reverse (5′-ATATATAGATCTGGCGCCCGAACAGGGACTTG -3′) primers. The resulting PCR product was digested using BamHI and BglII. This digested PCR producted was then ligated into a version of pLPCXU6PT7SK (Addgene plasmid 27549) that was modified as follows. pLPCXU6PT7SK was used as a template along with forward (5′-ATATATAAGCTTGGATCCATCATCATCGCAGCAAGATCTGGATGTGAGGCGATCTGGC -3′) and reverse (5′- GTCTTGGAAGCTTGACTACCCTACGTTCTCCTAC -3′) primers. This PCR product eliminated the 5′ coat protein binding sequences and encoded BamHI and BglII sites. This PCR product was digested with HindIII and ligated into pLPCXU6PT7SK that was digested with HindIII to generate pLPCXU6PT7SK-fixed. Sequencing was used to verify proper orientation of the insertion. The Spinach and Spinach2 PCR products were then ligated into pLPCXU6PT7SK-fixed at BamHI and BglII to generate pLPC-Spinach-7SK and pLPC-Spinach2-7SK, respectively.
pSC35-mCherry was generated by amplifying mCherry with forward (5′-ATATATGGATCCAATGGTGAGCAAGGGCGAGG -3′) and reverse (5′-TATATATAAGCTTTCACTTGTACAGCTCGTCC -3′) primers and cloning via BamHI and HindIII digestion downstream of SC35 in pcDNA3.1-SC35-cMyc (Addgene plasmid 44721) to generate pcDNA3.1-SC35-mCherry.
Imaging Spinach-7SK and Spinach2-7SK
HeLa cells (ATCC-CRM-CCL-2) were cultured and passaged in DMEM medium supplemented with 50 units of penicillin and 50 μg of streptomycin per mL. For imaging experiments, cells were grown on cells cultured on 24-well glass-bottom dishes and cotransfected with 0.3 μg of pLPC-Spinach-7SK or pLPC-Spinach2-7SK and 0.3 μg of pCDNA3.1-SC35-mCherry using FuGeneHD (Roche) per the manufacturer's instructions in DMEM medium lacking penicillin and streptomycin. Cells were imaged 24 h post-transfection. At 30 min prior to imaging, medium was supplemented with 25 mM HEPES, 5 mM MgSO4, and 20 μM DFHBI. Cells were imaged as described below using FITC and Texas Red filter sets.
Cloning of CGG60-Spinach and Spinach2
Spinach or Spinach2 in the context of the tRNALys scaffold was amplified by PCR using forward (5′-ATATATATCTAGAGCCCGGATAGCTCAGTCGGTAGAGCAG-3′) and reverse (5′-ATATATGGGCCCTGGCGCCCGAACAGGGACTTGAACCC-3′) primers and digesting the resulting PCR products with XbaI and ApaI to clone downstream of the 60 CGG repeats and upstream of the BGH polyadenylation sequence in pCDNA-60CGG to generate pCDNA-60CGG-Spinach and pCDNA-60CGG-Spinach2. For TET-Off experiments, the entire transcript from pCDNA-60CGG-Spinach2 (CGG60-Spinach2-BGH-polyadenylation signal) was excised using NheI and EcoRV and subcloned into pTRE2-Hyg (Clontech) that was cut with NheI and EcoRV.
Transfection of COS-7 cells and live cell imaging
COS-7 cells (ATCC-CRL-1651) were cultured and passaged in DMEM medium supplemented with 50 units of penicillin and 50 μg of streptomycin per mL. For imaging experiments, cells were grown on cells cultured on 24-well glass-bottom dishes and transfected with 0.6 μg of pCDNA-60CGG-Spinach or pCDNA-60CGG-Spinach2 using FuGeneHD (Roche) per the manufacturer's instructions in DMEM medium lacking penicillin and streptomycin. Cells were imaged in CO2-independent medium (Invitrogen) supplemented with L-glutamine. At 30 min-1 h prior to imaging, medium was supplemented with 25 mM HEPES, 5 mM MgSO4, 1 μg/mL Hoechst 33342 (when appropriate), and 20 μM DFHBI or vehicle. Live fluorescence images were acquired in a temperature-controlled chamber at 35-37°C with a CoolSnap HQ2 CCD camera through a 60X oil objective (Plan Apo 1.4 NA) mounted on a Nikon TE2000 epifluorescence microscope and analyzed with the NIS-Elements software. Spinach was imaged with a filter cube typically used for fluorescein/EGFP, with a sputter coated excitation filter 470/40, dichroic mirror 495 (long pass), and emission filter 525/50 (Chroma Technology). DsRed-Max and mCherry were imaged using a filter cube typically used for Texas Red, with a sputter coated excitation filter 560/40 and emission filter 630/75 (Chroma Technology). Background intensity was subtracted from all pixel intensity measurements. Image analyses were completed with NIS-Elements AR 3.2 (Nikon). Drug treatments were carried out as specified in the text. Tautomycin was used at a final concentration of 5 μM in all cases. 1a was used at a final concentration 20 μM in all cases. DMSO was added to a final concentration of 0.1% for vehicle treatments.
For foci formation experiments, COS-7 cells were transiently transfected with a plasmid expressing (CGG)60-Spinach2. After 2 h, the transfection medium was replaced with imaging medium containing DFHBI. After a 1 h incubation in imaging medium, cells were imaged every 20 min for 6 h.
Analysis of DFHBI cell permeability
COS-7 cells were transfected with pCDNA-60CGG-Spinach2. 24 h post-transfection, cells were medium was supplemented with 25 mM HEPES, 5 mM MgSO4, 1 μg/mL Hoechst 33342, and 20 μM DFHBI. Images were acquired for Hoechst and Spinach2 signal every 5 min for 1 h for 20 cells. All signal were first normalized to area and then normalized to the highest signal for a given nucleus to determine the time for maximal signal to be reached.
FISH of CGG60 RNA
COS-7 cells were grown and transfected as described above on glass coverslips. Cells were fixed and stained as previously described4. CGG repeats were probed using an (CCG)8x-Texas Red DNA oligonucleotide probe (IDT). Spinach was probed using a 3′ Texas Red-labeled DNA oligonucleotide (5′-GCACTGCCGAAGCAGCCACACCTG-3′) (IDT). DAPI was contained in the mounting solution for DNA staining.
qRT-PCR analysis of CGG RNA stability
COS-7 cells were transfected to express either pTet-Off alone or pTet-Off with either pTRE2-Hyg-(CGG)60, pTRE2-Hyg-(CGG)60-Spinach, pTRE2-Hyg-(CGG)60-Spinach2, pTRE2-Hyg-(CGG)30, or pTRE2-Hyg-(CGG)30-Spinach2 in 12 wells each in 24-well plates. At 24 h post-transfection, the transfection media was replaced and doxycycline was added to 1 μg/ml. At 0, 6, 12, and 24 h post-transfection, total RNA from three wells per sample was extracted using TRIzol (Invitrogen) according to manufacturer's protocol.
Reverse transcription was carried out on all samples using a reverse primer that bound downstream of the CGG repeats in all constructs (5′- CTAGAGATATCAGGCTGATCA GC-3′) and a reverse primer against GAPDH mRNA (5′- TCCACCACCCTGTTGCTGTA-3′) according to the SuperscriptIII reverse transcription kit protocol. qRT-PCR was carried out according to the iQ™ SYBR® Green Supermix (Bio-Rad) protocol with forward (5′- GTCAGCTGACGCGTGCTAGCG -3′) and reverse (5′- CTAGAGATATCAGGCTGATCAGC -3′) primers against all CGG transcripts as well as forward (5′- ACCACAGTCCATGCCATCAC -3′) and reverse (5′- TCCACCACCCTGTTG CTGTA -3′) primers against GAPDH mRNA. In all cases, CGG transcript levels were normalized to GAPDH mRNA levels. Data represent mean and s.e.m. values for three independent experiments. We also carried out qRT-PCR of sample RNA compared to in vitro transcribed control RNA to determine the approximate number of CGG repeat-containing RNA in a cell. We obtained roughly 0.2 ng of (CGG)60-Spinach2 RNA from 0.2 × 106 transfected cells. We estimated the molecular weight of polyadenylated (CGG)60-Spinach2 to be roughly 280 kDa. Using these values, we calculated that each transfected cell contained roughly 2000 copies of (CGG)60-Spinach2. On average, each cell contains 10-15 foci, indicating that each aggregate contains roughly 150-200 RNA molecules. It should be noted that foci vary in size in different cells, and foci that are much smaller than the “average” size are readily detectable in cells. Moreover, not we observe some Spinach2 signal in the nucleoplasm that is not in foci. So 150-200 RNA molecules is unlikely to be the limit of detection at 50 ms; however, the precise limit will require more precise quantification methods of these foci that are closer to the limits of detection. Because foci were typically imaged at 50 ms, it is likely that smaller numbers of RNAs would be detectable at longer imaging times such as 500 ms or 1 sec.
Supplementary Material
Acknowledgments
We thank G.S. Filonov, W. Song, N. Svensen, and J. Paige for useful comments and suggestions, and F. Dardel (Université Paris Descartes) for providing plasmids containing the tRNA scaffold sequence. This work was supported by NIH NINDS NS010249 (S.R.J.) and NIH NIHGMS 1R01GM079235 (M.D.D).
Footnotes
Author Contributions: R.L.S., M.D., and S.R.J. conceived and designed the experiments, M.D. provided compounds and assisted in experiments using 1a and CGG foci, R.L.S. performed experiments and analyzed data, and R.L.S., and S.R.J. wrote the manuscript.
Competing Financial Interests: S.R.J. and R.L.S. are authors of a patent application related to technology described in this manuscript.
References
- 1.Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development. 2012;139:3263–3276. doi: 10.1242/dev.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen RP, Niessing D. Assembly of mRNA-protein complexes for directional mRNA transport in eukaryotes--an overview. Curr Protein Pept Sci. 2012;13:284–293. doi: 10.2174/138920312801619493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sellier C, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 7.Gurney T, Eliceiri GL. Intracellular distribution of low molecular weight RNA species in HeLa cells. J Cell Biol. 1980;87:398–403. doi: 10.1083/jcb.87.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasanth KV, et al. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell. 2010;21:4184–4196. doi: 10.1091/mbc.E10-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 10.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu YH, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 12.Dombrowski C, et al. Premutation and intermediate-size FMR1 alleles in 10572 males from the general population: loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Hum Mol Genet. 2002;11:371–378. doi: 10.1093/hmg/11.4.371. [DOI] [PubMed] [Google Scholar]
- 13.Hagerman RJ, et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 14.Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 15.Iwahashi CK, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 16.Sellier C, et al. Sequestration of DROSHA and DGCR8 by Expanded CGG RNA Repeats Alters MicroRNA Processing in Fragile X-Associated Tremor/Ataxia Syndrome. Cell Rep. 2013;3:869–880. doi: 10.1016/j.celrep.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd PK, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–455. doi: 10.1016/j.neuron.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39:7308–7315. doi: 10.1093/nar/gkr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoss O, et al. The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J Biol Chem. 2001;276:8665–8673. doi: 10.1074/jbc.M006851200. [DOI] [PubMed] [Google Scholar]
- 20.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Disney MD, et al. A small molecule that targets r(CGG)(exp) and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem Biol. 2012;7:1711–1718. doi: 10.1021/cb300135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suganuma M, et al. Tautomycin: an inhibitor of protein phosphatases 1 and 2A but not a tumor promoter on mouse skin and in rat glandular stomach. J Cancer Res Clin Oncol. 1995;121:621–627. doi: 10.1007/BF01197780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen P, Klumpp S, Schelling DL. An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. FEBS Lett. 1989;250:596–600. doi: 10.1016/0014-5793(89)80803-8. [DOI] [PubMed] [Google Scholar]
- 25.Genové G, Glick BS, Barth AL. Brighter reporter genes from multimerized fluorescent proteins. Biotechniques. 2005;39(814):816–818. doi: 10.2144/000112056. passim. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.