Abstract
Pesticide resistance poses a major challenge for the control of vector-borne human diseases and agricultural crop protection. Although a number of studies have defined how mutations in specific target proteins can lead to insecticide resistance, much less is known about the mechanisms by which constitutive overexpression of detoxifying enzymes contribute to metabolic pesticide resistance. Here we show that the Nrf2/Keap1 pathway is constitutively active in two laboratory-selected DDT-resistant strains of Drosophila, 91R and RDDTR, leading to the overexpression of multiple detoxifying genes. Disruption of the Drosophila Nrf2 ortholog, CncC, or overexpression of Keap1, is sufficient to block this transcriptional response. In addition, a CncC-responsive reporter is highly active in both DDT-resistant strains and this response is dependent on the presence of an intact CncC binding site in the promoter. Microarray analysis revealed that ~20% of the genes differentially expressed in the 91R strain are known CncC target genes. Finally, we show that CncC is partially active in these strains, consistent with the fitness cost associated with constitutive activation of the pathway. This study demonstrates that the Nrf2/Keap1 pathway contributes to the widespread overexpression of detoxification genes in insecticide-resistant strains and raises the possibility that inhibitors of this pathway could provide effective synergists for insect population control.
Keywords: insecticide resistance, gene regulation, transcriptional control, Nrf2
1. INTRODUCTION
Insects pose a constant threat to agricultural crop production and transmit a number of vector-borne human diseases, including malaria, filaria and dengue. Although insecticides provide an effective approach for vector control, their widespread use over the last few decades has led to the development of pesticide resistance in a number of insect species (Heckel, 2012). Cross-resistance to different classes of insecticides has further complicated efforts to control insect populations. As a result, the development of effective pest control strategies has become a major focus for current research and has led to widespread efforts to understand the molecular mechanisms that underlie insect pesticide resistance.
Resistance to insecticides arises through several mechanisms, two of which have been most extensively studied, target site resistance and metabolic resistance (Ffrench-Constant et al., 2004; Perry et al., 2011). Target site resistance can result from mutations that decrease the binding affinity of insecticides toward the molecules with which they interact, and are most commonly encountered in ion channels and enzymes that play critical roles in synaptic transmission. For example, mutations in the para sodium channel result in resistance to DDT and pyrethroids while mutations in GABA gated chloride channels result in resistance to dieldrin (Ffrench-Constant et al., 1991; Ffrench-Constant et al., 1993; 2000). These target proteins are essential for survival and therefore only a few conserved point mutations can be tolerated that decrease their insecticide sensitivity while maintaining normal protein function. Similarly, cross-resistance is limited to compounds that act at the same active site in the target protein.
Metabolic resistance, on the other hand, arises from an increase in the overall metabolic capacity of organisms to detoxify pesticides and other xenobiotics. Insects employ an extensive array of enzymes, including cytochrome P450 monooxygenases (P450s), glutathione S-transferases (GSTs) and carboxylesterases, which detoxify a wide range of endogenous and exogenous toxic compounds (Li et al., 2007). These Phase I and Phase II enzymes can be transcriptionally activated in a constitutive manner due to mutations in either cis-acting elements or trans-acting factors, conferring pesticide resistance. Metabolic resistance can also arise due to mutations that increase the catalytic activity of these detoxification enzymes. In contrast to the genes involved in target site resistance, many genes associated with metabolic resistance are not vital for survival and thus tend to be more tolerant of genomic changes that alter enzyme function and/or expression. Furthermore, due to the broader spectrum of substrate specificity, cross-resistance to different classes of insecticides is more common in metabolism-based resistance.
Drosophila melanogaster has been used extensively as a model system to understand the molecular mechanisms underlying insecticide resistance. Detailed studies of target site resistance have led to the identification of mutations in several key genes including those that encode the sodium channel, GABA gated chloride channel, acetylcholinesterase, and n-acetylcholine receptor (Perry et al., 2011). Similarly, metabolic insecticide resistance has been identified in a number of field-isolated and laboratory-selected strains of Drosophila (Li et al., 2007). Overexpression of a single P450 gene Cyp6g1 is associated with resistance to DDT and imidacloprid in field-derived Drosophila strains (Daborn et al., 2002) and ectopic overexpression of this enzyme in transgenic animals is sufficient to confer resistance to DDT, dicyclanil, and nitenpyram (Daborn et al., 2007). Similarly, Cyp12a4 overexpression provides resistance to Lufenuron (Bogwitz et al., 2005), while overexpression of Tribolium castaneum Cyp6BQ9 in the nervous system of Drosophila confers resistance to dieldrin (Zhu et al., 2010). In contrast to target site resistance, however, the molecular mechanisms underlying many forms of metabolic resistance remain unknown. This is primarily due to our limited understanding of how the genes that encode xenobiotic detoxifying enzymes are regulated in insects.
In an effort to better understand the mechanisms of metabolic resistance, several resistant strains of Drosophila have been developed in the laboratory. Two such strains are currently available, 91R and RDDTR, which were established by recurrent selection of wild caught flies on increasing concentrations of DDT over several generations. The 91R strain was established in Minnesota, USA (Merrell and Underhill, 1956; Dapkus and Merrell, 1977), while the RDDTR strain was developed from the Raleigh strain in France (Cùany et al., 1990). Despite their distinct origins, both of these strains exhibit a high degree of resistance to a common spectrum of insecticides. Previous genetic analysis of the 91R strain has shown that it overexpresses Cyp6g1 due to the insertion of an Accord transposable element in its 5’ UTR (Daborn et al., 2002). A number of other putative detoxifying genes are also overexpressed in the 91R strain (Pedra et al., 2004; Qiu et al. 2013). Attempts to identify the molecular mechanisms underlying this coordinate up-regulation suggested that it is due to a trans-acting factor or factors located on the third chromosome (Maitra et al., 2000). However, the identity of this factor and the mechanism by which it regulates detoxification gene expression remain unknown. In comparison to the 91R strain, less is known about RDDTR. This strain has been reported to overexpress Cyp6a2, which also carries point mutations that increase its catalytic activity (Amichot et al., 2004). However, it is not known if the overexpression of Cyp6a2 in this strain arises from cis or trans-regulatory mutations. Further, it remains to be determined if other detoxification genes are overexpressed in the RDDTR genetic background.
Recently we have demonstrated that the evolutionarily conserved Nrf2/Keap1 pathway plays a central role in regulating the coordinate transcriptional response to xenobiotic compounds in Drosophila (Misra et al., 2011). CncC is the Drosophila ortholog of Nrf2, which is a CNC-bZIP transcription factor. Under normal physiological conditions, it is retained in the cytoplasm by the actin-associated protein Keap1, which also functions as an adapter for the Cullin-3-based ubiquitination machinery, facilitating CncC proteasomal degradation (Kensler et al., 2007; Sykiotis and Bohmann, 2008). Electrophiles and reactive oxygen species disrupt the interaction between Keap1 and CncC, causing CncC stabilization and subsequent nuclear translocation. In the nucleus, CncC forms a heterodimer with Maf-s, binds to antioxidant response elements/electrophile response elements in target promoters, and up-regulates their transcription. Activation of this pathway is necessary and sufficient for xenobiotic induced transcription of a wide range of detoxification genes in Drosophila (Misra et al., 2011). Moreover, ectopic activation of the pathway provides resistance against the insecticide malathion. We thus conclude that exposure of insects to xenobiotic compounds induces a defensive response mediated by the CncC/Keap1 pathway that provides drug resistance and promotes survival.
Interestingly this pathway also provides a possible mechanism to explain the widespread expression of detoxification genes in the 91R and RDDTR DDT-resistant strains. We show here that the CncC/Keap1 pathway is constitutively active in both the 91R and RDDTR strains, leading to broad ectopic expression of detoxification genes. Overexpression of Keap1 or disruption of CncC function is sufficient to block this transcriptional response. Moreover, we show that the mutation(s) that cause constitutive activation of this pathway are located on the third chromosome. These studies demonstrate that the CncC/Keap1 pathway contributes to the overexpression of detoxification genes in the 91R and RDDTR strains, and suggest that efforts to inhibit this response may improve pesticide efficacy.
2. MATERIALS AND METHODS
2.1. Drosophila stocks
Canton S, tub-Gal80ts, and w;Act5-Gal4 were obtained from the Bloomington Drosophila Stock Center. The w;UAS-Keap1 and y, w;UAS-CncC-RNAi lines were kindly provided by D. Bohmann. The 91C and 91R strains were obtained from R. Ganguly and the RDDTR strain was obtained from M. Amichot. The WT-lacZ, Δ15-lacZ, 5XWT-lacZ and 5XMUT-lacZ lines have been described previously (Misra et al., 2011). To generate the flies carrying the third chromosomes from the 91R and RDDTR strains, in combination with various reporters, the 91R and RDDTR flies were crossed to the balancer line w1118; T(2,3) apXa/SM5; TM3, Sb1 to replace the X chromosomes. Subsequently, the second chromosomes were replaced with those carrying the appropriate reporter transgenes. Flies were raised on standard cornmeal/molasses/agar food at 20–25°C and matured to five days of age before all studies. Stocks with tub-Gal80ts were reared at 18°C. Progeny were allowed to emerge and then shifted to the restrictive temperature of 29°C for 3–6 days.
2.2. Northern blot hybridizations
Total RNA was isolated from 5–7 day old control and DDT-resistant flies using Tripure (Roche), following the manufacturer’s instructions. Equal amounts of RNA were fractionated by formaldehyde agarose gel electrophoresis and analyzed by northern blot hybridization, as described (Misra et al., 2011). Probes were generated by PCR, purified using Qiaquick gel extraction columns (Qiagen), and labeled with a Prime-It II kit (Stratagene). The PCR primers used to generate each probe are as published (Misra et al., 2011).
2.3. Histochemical detection of β-galactosidase in adult tissues
Mature adult animals were dissected in PBS. Tissues were fixed in 1.5% formaldehyde in PBS for 20 minutes and stained with 0.2% X-gal (Roche) for 15–30 minutes at 37°C.
2.4. Microarray experiments
RNA was isolated from mature 91R or 91C male flies. All samples were prepared in four replicates to facilitate subsequent statistical analysis. Total RNA was extracted with TriPure (Roche) followed by purification with RNAeasy columns (QIAGEN). Probe labeling, hybridization to two-color Agilent Drosophila 44K arrays and scanning, were performed by the University of Utah Microarray Core Facility. The data were Lowess normalized using R, and the fold changes in gene expression and t-statistics were determined using GeneSifter (VizX Labs, Seattle, WA). p-values were calculated using the Benjamimi and Hochberg correction for false-discovery rate. Comparison between microarray datasets was performed using Genevenn and the p-value for significance of overlap between gene sets was calculated by hypergeometric probability. Microarray data from this study can be accessed online at NCBI GEO (accession number: GSE48952).
3. RESULTS
3.1. The 91R and RDDTR insecticide resistant strains coordinately overexpress CncC target genes
The 91R strain has been previously shown to overexpress a number of detoxifying genes, some of which are inducible by phenobarbital (PB) and regulated by the CncC/Keap1 pathway (Pedra et al., 2004; Misra et al., 2011; Qiu et al. 2013). This raises the interesting possibility that constitutive activation of the CncC/Keap1 pathway could contribute to the overexpression of detoxifying genes in this strain. Similarly, the RDDTR strain is known to overexpress Cyp6a2, which is regulated by CncC. We therefore examined the transcription of several known CncC-regulated detoxification genes in these two strains by northern blot hybridization (Fig. 1A). As a control we used the 91C strain for the 91R strain and the wild-type laboratory strain Canton S as a control for RDDTR. The 91C strain was established from the same flies from which the 91R strain was developed, except that they were not exposed to DDT (Dapkus and Merrell, 1977). Interestingly, both 91R and RDDTR flies overexpress Cyp6a2, Cyp6a8, Jheh1, GstD2 and CG6188, all of which are known to be regulated by PB as well as the CncC/Keap1 pathway (Misra et al., 2011). These results raise the possibility that the CncC/Keap1 pathway is active in both the 91R and RDDTR resistant strains.
Fig. 1.
CncC target genes are coordinately up-regulated in the RDDTR and 91R strains. RNA was isolated from RDDTR and 91R flies along with the corresponding control strains, Canton S (CS) and 91C, and analyzed by northern blot hybridization to detect the transcription of CncC-regulated genes, as shown. Hybridization to detect rp49 mRNA was used as a control for loading and transfer.
3.2. The CncC/Keap1 pathway is constitutively active in the 91R and RDDTR strains
If the CncC/Keap1 pathway is active in the 91R and RDDTR strains, then a CncC-responsive reporter gene should be expressed in these genetic backgrounds and this expression should be dependent on CncC binding to the promoter. To test these possibilities, we used transgenic flies that carry a lacZ reporter gene fused to either a wild-type 313 bp Cyp6a2 promoter fragment (WT-lacZ) or the same promoter fragment that carries a 15 bp deletion disrupting the CncC binding site (Δ15-lacZ) (Misra et al., 2011). Transgenic animals carrying these constructs were crossed to the 91R and RDDTR strains and examined for lacZ transcription (Fig. 2A,B). As expected, both the WT-lacZ and Δ15-lacZ reporters are not expressed in the control genotypes. In contrast, the WT-lacZ reporter is expressed in parallel with the endogenous Cyp6a8 gene in both the 91R and RDDTR genetic backgrounds, while the Δ15-lacZ reporter remains silent in these strains (Fig. 2A,B). These results support the proposal that the CncC/Keap1 pathway is constitutively active in both of these DDT-resistant lines. In addition, activation of the reporters in animals that carry only one chromosomal homolog from each of the resistant strains suggests that the pathway is activated in a dominant manner. To test this possibility we examined the transcription of Cyp6a8 in animals that are homozygous or heterozygous for the 91R or RDDTR chromosomes (Fig. 2C,D) and observed that, although the heterozygous animals overexpress Cyp6a8 compared to the wild-type controls, the expression level is lower than that in the homozygous animals, indicating that the effects of 91R and RDDTR on the CncC/Keap1 pathway are semidominant in nature. We then examined how these strains interact with each other with respect to their effect on the CncC/Keap1 pathway. To address this, the 91R and RDDTR strains were mated with either Canton S wild-type flies or with each other and their heterozygous progeny were analyzed for transcription of Cyp6a8 (Fig. 2E). As expected from our earlier results, 91R and RDDTR activate the CncC/Keap1 pathway in a semidominant manner (Fig. 2E, lanes 4,5). In addition, combining chromosomes from the 91R and RDDTR strains have an additive effect on Cyp6a8 expression, suggesting that these two DDT-resistant strains activate the pathway by either the same or parallel mechanisms (Fig. 2E, lane 6).
Fig. 2.
The effects of 91R and RDDTR on the CncC/Keap1 pathway are semidominant and additive. (A) Transgenic flies carrying a lacZ reporter gene fused to either a wild-type 313 bp Cyp6a2 promoter fragment (WT-lacZ) or the same fragment carrying the 15 bp deletion that disrupts the CncC binding site (Δ15-lacZ) were mated with either 91R or control 91C strain, after which RNA was extracted from the resulting heterozygous progeny and analyzed by northern blot hybridization for the transcription of lacZ and Cyp6a8. (B) Flies carrying either the WT-lacZ or Δ15-lacZ transgene were mated with either RRDTR or control CS strain, after which RNA was extracted from the resulting heterozygous progeny and analyzed by northern blot hybridization for transcription of lacZ and Cyp6a8. (C) The 91R strain was mated with CS flies and RNA was extracted from either CS, 91R or their heterozygous progeny and analyzed by northern blot hybridization for transcription of Cyp6a8. (D) The RDDTR strain was mated with CS flies and RNA was extracted from either CS, RDDTR or their heterozygous progeny and analyzed by northern blot hybridization for transcription of Cyp6a8. (E) The 91R and RRDTR strains were mated with either CS flies or with each other and RNA was extracted from either CS, 91R, RDDTR or their heterozygous progeny and analyzed by northern blot hybridization for transcription of Cyp6a8. Hybridization to detect rp49 mRNA was used as a control for loading and transfer in all panels.
3.3. The third chromosome of the 91R and RDDTR strains is sufficient for CncC activation
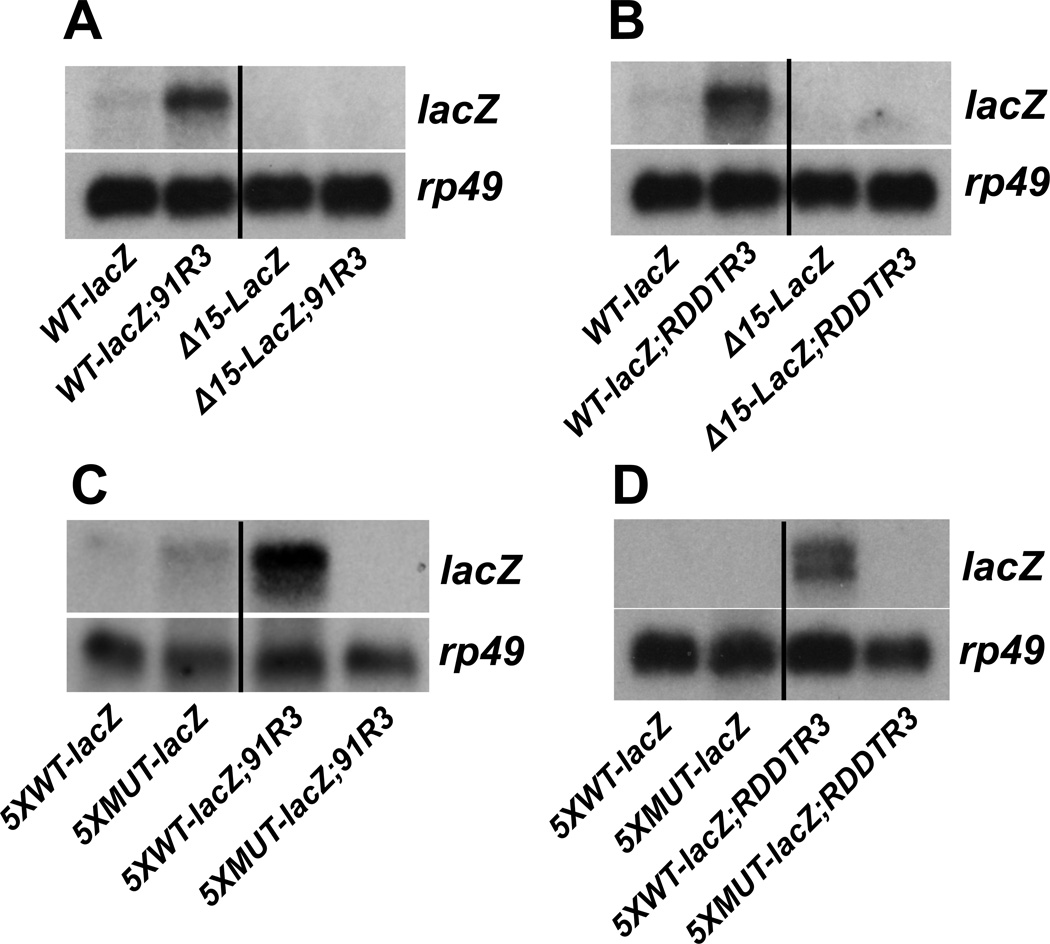
Previous genetic analysis suggests that the overexpression of Cyp6a2 and Cyp6a8 genes in the 91R strain is due to a factor or factors located on the third chromosome (Maitra et al., 2000). Similar studies in two different strains have indicated that factors located near cytological locations 86D and 90D on the third chromosome cause overexpression of P450s (Houpt et al., 1988; Waters and Nix, 1988). If this is true, then the third chromosome alone from 91R and RDDTR should be sufficient to activate the CncC-responsive reporter gene. To test this possibility, we created animals that carry either the WT-lacZ or the Δ15-lacZ transgenes on the second chromosome (as homozygotes) in combination with a homozygous third chromosome from either 91R or RDDTR, and examined the transcription of lacZ mRNA by northern blot hybridization (Fig. 3A,B). Both the reporter transgenes and DDT-resistant strains were outcrossed to a w1118 background to eliminate any contribution from the X chromosome. As expected, both the WT-lacZ and the Δ15-lacZ reporters are not expressed when they are combined with wild-type third chromosomes (Fig. 3A,B). In contrast, the WT-lacZ reporter is expressed when it is in combination with only the third chromosome from either the 91R or the RDDTR strain, while the Δ15-lacZ reporter remains silent in these backgrounds. We also tested if the third chromosome from the 91R and RDDTR strains is sufficient to activate gene expression from an isolated CncC binding site upstream from a reporter construct. For this purpose, we used transgenic animals that carry a lacZ reporter gene fused to either five tandem copies of a wild-type CncC binding site in the Cyp6a2 promoter (5XWT-lacZ) or a mutant version of this promoter fragment lacking the Nrf2/Maf-s binding site (5XMUT-lacZ) (Misra et al., 2011) on their second chromosome (as homozygotes) in combination with homozygous third chromosomes from either 91R or RDDTR, and examined lacZ transcription (Fig. 3C,D). As expected, both the 5XWT-lacZ and 5XMUT-lacZ reporters are not expressed when they are combined with wild-type third chromosomes. In contrast, the 5XWT-lacZ reporter is expressed when it is in combination with the third chromosome from either the 91R or RDDTR strain, while the 5XMUT-lacZ reporter remains unexpressed in these backgrounds. Taken together, these experiments indicate that the mutation or mutations that cause constitutive activation of the CncC/Keap1 pathway is located on the third chromosome in both of these strains.
Fig. 3.
Mutation(s) that activate the CncC/Keap1 pathway are located on the third chromosomes of the 91R and RDDTR strains. (A) RNA was extracted from flies carrying the WT-lacZ or Δ15-lacZ transgenes on the second chromosome along with the third chromosomes from the 91R strain, and analyzed by northern blot hybridization to detect the transcription of lacZ. (B) RNA was extracted from flies carrying the WT-lacZ or Δ15-lacZ transgenes on the second chromosome along with the third chromosomes from the RDDTR strain, and analyzed by northern blot hybridization to detect the transcription of lacZ. (C) RNA was extracted from flies carrying a lacZ reporter gene fused to either five tandem copies of a 25 bp sequence that encompasses the Nrf2/Maf binding site in the Cyp6a2 promoter (5XWT-lacZ) or a mutant version of this promoter fragment lacking the Nrf2/Maf binding site (5XMUT-lacZ) on the second chromosome, along with the third chromosomes from the 91R strain, and analyzed by northern blot hybridization to detect the transcription of lacZ. (D) RNA was extracted from flies carrying either the 5XWT-lacZ or 5XMUT-lacZ transgenes on the second chromosome, along with the third chromosomes from the RDDTR strain, and analyzed by northern blot hybridization to detect the transcription of lacZ. Hybridization to detect rp49 mRNA was used as a control for loading and transfer.
3.4. CncC is activated in the appropriate tissues of 91R and RDDTR animals
Spatial expression studies of P450 genes have revealed that the midgut and Malpighian tubules are predominant sites for Phase I detoxification (Chung et al., 2009). To determine if the activated CncC/Keap1 pathway functions in a similar tissue-specific manner in the 91R and RDDTR strains, we examined β-galactosidase activity in tissues dissected from animals that carry either the WT-lacZ or the Δ15-lacZ reporter on the second chromosome along with the third chromosomes from either wild-type, 91R or RDDTR strains (Fig. 4). As a control, we used a heat-inducible hs-CncC transgene to ectopically activate the CncC pathway in animals that carry either the WT-lacZ or Δ15-lacZ reporter (Misra et al., 2011). As expected, β-galactosidase activity was clearly detectable in heat-treated hs-CncC animals carrying the WT-lacZ transgene (Fig. 4D), and no expression was evident in non-heat-treated hs-CncC; WT-lacZ animals (Fig. 4A) or in heat-treated animals carrying the Δ15-lacZ reporter (Fig. 4G). Similarly, no β-galactosidase activity was detected when either the WT-lacZ or Δ15-lacZ reporter were carried in wild-type flies (Fig. 4B,C). In the presence of either the 91R or RDDTR third chromosomes, however, the WT-lacZ transgene was abundantly expressed in both the midgut and Malpighian tubules (Fig. 4E,H), while no β-galactosidase was expressed from the Δ15-lacZ reporter in either genetic background (Fig. 4F, I). Interestingly, these patterns are similar to those described for many P450 genes (Chung et al., 2009), suggesting that CncC/Keap1 pathway activation in the 91R and RDDTR strains recapitulates the wild-type expression patterns of detoxification gene expression.
Fig. 4.
The mutation(s) located on the third chromosomes of 91R and RDDTR strains activate the CncC/Keap1 pathway in the Malpighian tubules and midgut. Transgenic flies carrying the WT-lacZ CncC reporter along with the heat inducible hsp70-CncC transgene (hs-CncC) were subjected to either no heat shock (−HS)(A) or heat shock (+HS)(D), after which the tissues were dissected and stained for β-galactosidase activity. Flies carrying the Δ15-lacZ reporter gene along with the hsp70-CncC transgene (hs-CncC) were subjected to heat shock (+HS) as a negative control (G), after which the tissues were dissected and stained for β-galactosidase activity. Transgenic flies carrying the WT-lacZ reporter (B, E, H) or the Δ15-lacZ reporter (C, F, I) on the second chromosome along with the third chromosome from either the Canton S (WT) (B, C), 91R (E, F) or RDDTR (H, I) strains were dissected and stained for β-galactosidase activity.
3.5. CncC activation is necessary for detoxification gene expression in the 91R strain
If constitutive activation of the CncC/Keap1 pathway is an underlying cause for overexpression of the detoxifying genes in the 91R and RDDTR strains, then disruption of CncC function should prevent this transcriptional response, as would Keap1 overexpression (which promotes CncC degradation). To test this possibility, we used the GAL4/UAS system to direct RNAi for CncC or to overexpress Keap1. Transgenic flies were established that carry the ubiquitous actin-GAL4 driver along with either UAS-CncC-RNAi or UAS-Keap1 on the second chromosome, in combination with the third chromosomes from the 91R strain. The Tub-Gal80ts construct was included in these lines to control the timing of CncC RNAi or Keap1 overexpression, as these conditions are normally lethal (Sykiotis and Bohmann 2008). Following temperature shifts to activate GAL4, RNA was extracted and examined by northern blot hybridization to detect the expression of Cyp6a2 and Cyp6a8 (Fig. 5A, B). Although both genes are abundantly expressed in the controls that carry either the GAL4 driver or the UAS transgene alone, the expression of these genes is significantly attenuated by either CncC RNAi (Fig. 5A) or Keap1 overexpression (Fig. 5B). These results suggest that the CncC/Keap1 pathway is necessary for the overexpression of detoxification genes in the 91R strain. Similar results were seen in the RDDTR genetic background upon either CncC RNAi or Keap1 overexpression, suggesting that this pathway is required in both strains for detoxification gene overexpression (Fig. 5C).
Fig. 5.
CncC is necessary for Cyp6a2 and Cyp6a8 overexpression in the 91R and RDDTR strains. Flies carrying either the Tub-Gal80ts;Act-GAL4 driver alone, or in combination with either the UAS-CncC-RNAi or UAS-Keap1 transgenes, were combined with the third chromosomes from either the 91R (A,B) or RDDTR (C) strains, as shown. The stocks were shifted to 29°C for five days, after which RNA extracted from these animals was analyzed by northern blot hybridization to detect the transcription of Cyp6a8 and Cyp6a2. Hybridization to detect rp49 mRNA was used as a control for loading and transfer.
3.6. Many CncC-regulated genes are overexpressed in the 91R strain
In order to determine the extent to which CncC activation contributes to the pattern of gene expression in the DDT-resistant strains, we examined the transcriptional profile of 91R adult flies relative to the 91C control line. RNA was isolated from 91C and 91R animals, labeled and hybridized to two color Agilent Drosophila 44K arrays. All experiments were conducted in quadruplicate to facilitate statistical analysis. This study revealed that 1584 genes are differentially expressed 2-fold or higher in the 91R strain relative to the 91C strain, with 762 genes up-regulated and 822 genes down-regulated (Table S1). We compared this list of differentially expressed genes with our previously determined list of genes that alter expression upon ectopic activation of the CncC/Keap1 pathway (Misra et al., 2011) (Fig. 6A). Remarkably, about 20% of the genes differentially expressed in the 91R strain are also regulated by CncC. Similarly, GOstat analysis of the genes that change expression in the 91R strain reveals an enrichment of oxidoreductases, electron carriers and transmembrane transporters in the top GO categories (Fig. 6B), reflecting the predominant gene categories seen upon activation of CncC (Misra et al., 2011).
Fig. 6.
Many genes that change their expression level in the 91R strain relative to 91C strain are regulated by CncC. (A) A Venn diagram is depicted showing the comparison between the genes that change their expression level in the 91R strain compared to the 91C strain with genes that change their expression level upon activation of the CncC pathway (Misra et al., 2011). The p-value for the overlap of the gene sets is shown. (B) Gene ontology (GOstat) analysis of the genes that change expression in the 91R strain relative to 91C. The top GO categories for each gene set are listed in order of significance along with the number of genes affected in that category, the total number of genes in that category (in parentheses), and the statistical significance of the match. (C) Mature 91R or RDDTR flies along with wild-type controls were treated with either no PB (−) or 0.3% PB (+) for two hours, after which RNA was extracted and analyzed by northern blot hybridization to detect the transcription of PB-inducible genes. Hybridization to detect rp49 mRNA was used as a control for loading and transfer.
There are several reasons why more CncC-regulated genes are not present in the 91R gene list, including the possibility that the CncC/Keap1 pathway is not fully activated in this DDT-resistant line. This possibility is supported by the observation that constitutive activation of this pathway causes embryonic lethality (McGinnis et al., 1998), and no lethality is evident in either the 91R or RDDTR strains. Accordingly, we tested if CncC-regulated genes could be further induced in the 91R and RDDTR strains upon exposure to PB, a compound that is known to cause robust CncC activation. Both 91R and RDDTR animals were fed PB and the expression of Cyp6a2, Cyp6a8 and Jheh1 was examined by northern blot hybridization (Fig. 6C). As described in our previous study (Misra et al., 2011), all three genes are highly overexpressed upon exposure to PB. Interestingly, a similar induction is seen in both the 91R and RDDTR lines. This result indicates that the CncC/Keap1 pathway is not fully active in these two DDT-resistant strains and suggests that this partial response contributes to their viability throughout development.
4. DISCUSSION
The emergence of insecticide resistance has had a major impact on agricultural crop production and the spread of vector-borne human diseases, with a disproportionate effect on developing countries. As a result, considerable effort has been aimed at understanding the mechanisms by which insects acquire pesticide resistance. Here we show that constitutive activation of the CncC/Keap1 pathway is central to the overexpression of detoxifying genes in two insecticide resistant strains of Drosophila, 91R and RDDTR. This is consistent with our previous finding that this pathway plays a key role in the coordinate induction of detoxification gene expression in response to xenobiotic treatment, and ectopic activation of this pathway is sufficient to confer resistance to malathion (Misra et al., 2011). This work provides a molecular mechanism to explain how detoxification gene expression is coordinately up-regulated in insects that have acquired metabolic pesticide resistance.
Genomewide transcriptional profiling revealed that about 20% of the genes that are misexpressed in the 91R strain are regulated by CncC (Fig. 6A). Our identification of many genes that were not reported in a previous microarray study of 91R flies is likely due to our use of the 91C strain as a control (the previous study used Canton S) as well as a different microarray platform and statistical methods (Pedra et al., 2004; Qiu et al. 2013). In addition, our finding that the third chromosomes from either the 91R or RDDTR strains is sufficient to activate the CncC/Keap1 pathway is consistent with a previous mapping study of Cyp6a2 and Cyp6a8 overexpression in 91R flies (Maitra et al., 2000). Mapping experiments in a malathion-resistant strain also showed that the overexpression of two P450s encoded on the second chromosome requires a region located on the right arm of the third chromosome near the striped locus (Houpt et al., 1988). In addition, an increase in dimethyl-nitrosamine demethylase (DMN-d) activity associated with the overexpression of two P450 genes on the second chromosome is dependent on a region located near the Curled locus on the third chromosome (Waters and Nix, 1988). Taken together, these studies suggest that the region between cytological locations 86D and 90D on the right arm of the third chromosome contains one or more factors that might regulate the CncC/Keap1 pathway. Although both Keap1 (89E) and CncC (94E) are located close to this region, no mutation could be detected in their coding regions in either the 91R or RDDTR strain (data not shown), indicating that the pathway is not activated by a sequence change in these two key factors.
Constitutive activation of the CncC/Keap1 pathway causes embryonic lethality in Drosophila (McGinnis et al., 1998). This agrees with the developmental roles of the Nrf2 transcription factor in both Drosophila and C. elegans (Bowerman et al., 1992; McGinnis et al., 1998; Veraksa et al., 2000). Consistent with the high fitness cost associated with activation of the pathway, we observed only partial activation of CncC in the 91R and RDDTR strains (Fig. 6C). From an evolutionary perspective, it is conceivable that such partial activation could provide protection against insecticides such as DDT without compromising the survival of the organism. Partial activation of this pathway could be achieved by restricting this response to certain tissues. Tissue-specific activation of CncC in the major metabolic organs of the fly does not lead to lethality, consistent with this proposal (Misra et al., 2011). Similarly, mutations that activate the pathway only in adults, after development has been completed, could provide protection against chemical toxins. The temporal expression pattern of Cyp6a2 in the RDDTR strain, however, suggests that the CncC/Keap1 pathway is active throughout most of development (Tarès et al., 2007). This raises the possibility that the pathway is either active in a subset of tissues or is ubiquitously active at a low level.
The mechanism(s) by which the CncC/Keap1 pathway is constitutively activated in the 91R and RDDTR strains remains unclear. Indeed, this is a difficult problem to address because there are multiple independent levels at which pathway activation can be achieved. For example, the known effect of reactive oxygen species and electrophiles on Nrf2 means that any mutation that increases ROS levels could potentially activate CncC. Similarly, electrophilic metabolites produced in the body, such as fumarate, can directly modify key cysteine residues in Keap1, disrupting its interaction with Nrf2 (Kinch et al., 2011). Furthermore, the altered expression or activity of a kinase that phosphorylates CncC, or mutations that decrease the efficiency of CncC ubiquitination, could cause pathway activation (Lo and Hannink, 2006; Keum, 2011). To test this latter possibility, we examined if overexpression of USP8, a deubiquitinase located at 93C, could activate CncC, but observed that USP8 overexpression has no effect (data not shown). For efficient ubiquitination, Cullin-3 needs to undergo cycles of neddylation and deneddylation, a process that requires the activity of the COP9 signalosome (CSN) and CAND1 (Wei and Deng, 2003; Duda et al., 2011). Loss of function in any of these components could activate the CncC/Keap1 pathway. Interestingly, the metalloprotease CSN5, which plays a key role in deneddylation of Cullin-3, is down-regulated five-fold in the 91R strain and located at 89B. However, the published microarrays for the csn5 mutants did not reveal any signatures of CncC activation (Oron et al., 2007). In addition, overexpression of proteins that contain the motifs involved in Keap1-CncC interaction can disrupt their association and cause pathway activation (Komatsu et al., 2010). Similarly, sequestration of Keap1 in misfolded protein aggregates can lead to sustained Nrf2 activation (Rajasekaran et al., 2007). Thus, further studies are needed to identify the mutation or mutations that activate CncC in these strains and thereby define the mechanisms by which this is achieved. Indeed, it is possible that a novel pathway may be uncovered that activates this highly regulated system.
Activation of the CncC/Keap1 pathway, however, is unlikely to be the only factor that contributes to insecticide resistance in these strains. Resistance modeling has suggested that selection with sublethal concentrations of insecticides favors the development of polygenic resistance (Schlipalius et al., 2008). Given that these strains were established by recurrent selection on increasing concentrations of DDT, it is likely that they have multiple mutations that confer high resistance. Consistent with this, genetic analysis of the 91R strain identified contributing factors on each major autosome (Dapkus and Merrell, 1977). This strain also carries a transposon insertion that leads to overexpression of Cyp6g1 (Kuruganti et al., 2007), which is known to be sufficient to provide DDT resistance (Daborn et al., 2002). In addition, our microarray experiment suggests that many of the genes that are differentially expressed in the 91R strain do so independently of CncC (Fig. 6A). Moreover, DDT resistance and Cyp6a8 transcript levels do not always correlate in individual isolates from the 91R and RDDTR strains (data not shown). Thus constitutive activation of the CncC/Keap1 pathway likely establishes only one of several components that contribute toward pesticide resistance in these strains.
Finally, misregulation of the xenobiotic detoxification pathway may be a common mechanism underlying resistance to chemical compounds in other organisms. For example, constitutive activation of this pathway has been associated with drug resistance in several types of human cancers, correlating with the overexpression of several Phase II enzymes and multi drug transporters (Singh et al., 2006; Shibata et al., 2008; Shim et al., 2009; Zhang et al., 2010). Similarly, the nematode Nrf2 ortholog SKN-1 regulates a number of detoxification genes in C. elegans (Park et al., 2009). Given that SKN-1 is highly conserved among nematodes (Choe et al., 2012), it is possible that misregulation of this pathway is involved in anthelmintic resistance, which has been reported in a number of parasitic nematodes. If so, then the development of chemical inhibitors of this pathway might be useful for preventing parasitic nematode infections, with major implications for human health and nutrition. Similarly, our results suggest that inhibition of the CncC/Keap1 pathway should sensitize insects to pesticide application. Accordingly, CncC inhibitors might act as effective synergists that could improve our use of pesticides to control insect populations.
Supplementary Material
RNA was extracted from mature adult 91R or 91C flies, labeled, hybridized to two-color Agilent Drosophila 44K arrays, and the data was analyzed using Genesifter following Lowess normalization in R. The columns, in order, show the Fly Base Gene ID, CG number, gene name, cytogenetic location, fold change in expression level, direction of change in expression, p-value, adjusted p-value, biological, cellular and molecular GO function. Genes are sorted by the fold change in expression level between 91C and 91R.
The list of genes that change their expression ≥2-fold in the 91R strain was compared with the list of genes that change their expression ≥2-fold upon ectopic expression of CncC. The columns, in order, show the CG number, Fly Base Gene ID, gene name, fold change in expression level for PB data, direction of change in expression for PB data, p-value for PB data, adjusted p-value for PB data, fold change in expression level for CncC data, direction of change in expression for CncC data, p-value for CncC data, adjusted p-value for CncC data, biological, cellular and molecular GO function. Genes are sorted by the fold change in expression level between 91R and 91C strains.
The Nrf2/Keap1 pathway is constitutively active in DDT-resistant Drosophila
Nrf2/Keap1 activation leads to widespread overexpression of detoxification genes
One or more factors on the third chromosome are sufficient for Nrf2/Keap1 activation
Constitutive activation of Nrf2 may contribute to acquired insecticide resistance
ACKNOWLEDGEMENTS
We thank D. Bohmann, R. Ganguly, M. Amichot, and the Bloomington Stock Center for providing stocks, W. McGinnis for providing the Cnc antibody, and FlyBase for critical information that made these studies possible. We also thank the members of the Thummel lab for their assistance and suggestions during the course of this study. This research was supported by the National Institute of General Medical Sciences (R01GM079197).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
APPENDIX A. SUPPLEMENTARY DATA
Supplementary data related to this article can be found at http://
REFERENCES
- Amichot M, Tarès S, Brun-Barale A, Arthaud L, Bride JM, Berge JB. Point mutations associated with insecticide resistance in the Drosophila cytochrome P450 Cyp6a2 enable DDT metabolism. Eur. J. Biochem. 2004;271:1250–1257. doi: 10.1111/j.1432-1033.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- Bogwitz MR, Chung H, Magoc L, Rigby S, Wong W, O'Keefe M, McKenzie JA, Batterham P, Daborn PJ. Cyp12a4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2005;102:12807–12812. doi: 10.1073/pnas.0503709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;20:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Choe KP, Leung CK, Miyamoto MM. Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance. Drug Metab. Rev. 2012;44:209–223. doi: 10.3109/03602532.2012.684799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Sztal T, Pasricha S, Sridhar M, Batterham P, Daborn PJ. Characterization of Drosophila melanogaster cytochrome P450 genes. Proc. Natl. Acad. Sci. USA. 2009;106:5731–5736. doi: 10.1073/pnas.0812141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cùany A, Pralavorio M, Pauron D, Berge JB, Fournier D, Blais C, Lafont R, Salaun JP, Weissbart D, Larroque C. Characterization of microsomal oxidative activities in a wild-type and in a DDT resistant strain of Drosophila melanogaster. Pest Biochem. Phys. 1990;37:293–302. [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P. A single P450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- Daborn PJ, Lumb C, Boey A, Wong W, Ffrench-Constant RH, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem. Mol. Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Dapkus D, Merrell DJ. Chromosomal analysis of DDT-resistance in a long-term selected population of Drosophila melanogaster. Genetics. 1977;87:685–697. doi: 10.1093/genetics/87.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Mortlock DP, Shaffer CD, MacIntyre RJ, Roush RT. Molecular cloning and transformation of cyclodiene resistance in Drosophila: an invertebrate gamma-aminobutyric acid subtype A receptor locus. Proc. Natl. Acad. Sci. USA. 1991;88:7209–7213. doi: 10.1073/pnas.88.16.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Rocheleau TA, Steichen JC, Chalmers AE. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature. 1993;363:449–451. doi: 10.1038/363449a0. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Anthony N, Aronstein K, Rocheleau T, Stilwell G. Cyclodiene Insecticide Resistance: From Molecular to Population Genetics. Annu. Rev. Entomol. 2000;45:449–466. doi: 10.1146/annurev.ento.45.1.449. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Daborn PJ, Goff GL. The genetics and genomics of insecticide resistance. Trends Genet. 2004;20:163–170. doi: 10.1016/j.tig.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Heckel DG. Insecticide Resistance After Silent Spring. Science. 2012;337:1612–1614. doi: 10.1126/science.1226994. [DOI] [PubMed] [Google Scholar]
- Houpt DR, Pursey JC, Morton RA. Genes controlling malathion resistance in a laboratory-selected population of Drosophila melanogaster. Genome. 1988;30:844–853. doi: 10.1139/g88-136. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharm. Tox. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Keum YS. Regulation of the Keap1/Nrf2 system by chemopreventive sulforaphane: implications of posttranslational modifications. Ann. N. Y. Acad. Sci. 2011;1229:184–189. doi: 10.1111/j.1749-6632.2011.06092.x. [DOI] [PubMed] [Google Scholar]
- Kinch L, Grishin NV, Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell. 2011;20:418–420. doi: 10.1016/j.ccr.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou Y-S, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kuruganti S, Lam V, Zhou X, Bennett G, Pittendrigh BR, Ganguly R. High expression of Cyp6g1, a cytochrome P450 gene, does not necessarily confer DDT resistance in Drosophila melanogaster. Gene. 2007;388:43–53. doi: 10.1016/j.gene.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol. Cell Biol. 2006;26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S, Dombrowski SM, Basu M, Raustol O, Waters LC, Ganguly R. Factors on the third chromosome affect the level of Cyp6a2 and Cyp6a8 expression in Drosophila melanogaster. Gene. 2000;248:147–156. doi: 10.1016/s0378-1119(00)00129-3. [DOI] [PubMed] [Google Scholar]
- McGinnis N, Ragnhildstveit E, Veraksa A, McGinnis W. A cap “n” collar protein isoform contains a selective Hox repressor function. Development. 1998;125:4553–4564. doi: 10.1242/dev.125.22.4553. [DOI] [PubMed] [Google Scholar]
- Merrell DJ, Underhill JC. Selection for DDT Resistance in inbred, laboratory, and wild stocks of Drososphila melanogaster. J. Econ. Entomol. 1956;49:300–306. [Google Scholar]
- Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron E, Tuller T, Li L, Rozovsky N, Yekutieli D, Rencus-Lazar S, Segal D, Chor B, Edgar BA, Chamovitz DA. Genomic analysis of COP9 signalosome function in Drosophila melanogaster reveals a role in temporal regulation of gene expression. Mol. Syst. Biol. 2007;3:108. doi: 10.1038/msb4100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. 2009;8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedra JH, McIntyre LM, Scharf ME, Pittendrigh BR. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proc. Natl. Acad. Sci. USA. 2004;101:7034–7039. doi: 10.1073/pnas.0400580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry T, Batterham P, Daborn PJ. The biology of insecticidal activity and resistance. Insect Biochem. Mol. Biol. 2011;41:411–422. doi: 10.1016/j.ibmb.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Qui X, Sun W, McDonnell CM, Li-Byarlay H, Steele LD, Wu J, Xie J, Muir WM, Pittendrigh BR. Genome-wide analysis of genes associated with moderate and high DDT resistance in Drosophila melanogaster. Pest. Manag. Sci. 2013;69:930–937. doi: 10.1002/ps.3454. [DOI] [PubMed] [Google Scholar]
- Rajasekaran NS, Connell P, Christians ES, Yan L-J, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, Barry WH, Loscalzo J, Odelberg SJ, Benjamin IJ. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlipalius DI, Chen W, Collins PJ, Nguyen T, Reilly PEB, Ebert PR. Gene interactions constrain the course of evolution of phosphine resistance in the lesser grain borer, Rhyzopertha dominica. Heredity. 2008;100:506–516. doi: 10.1038/hdy.2008.4. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- Shim GS, Manandhar S, Shin DH, Kim TH, Kwak MK. Acquisition of doxorubicin resistance in ovarian carcinoma cells accompanies activation of the NRF2 pathway. Free Rad. Biol. Med. 2009;47:1619–1631. doi: 10.1016/j.freeradbiomed.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarès S, Arthaud L, Brun-Barale A, Crochard D, Bride JM, Amichot M. Very high conservation between Cyp6a2 from Drosophila melanogaster and its ortholog Cyp6a26 from D. simulans. Insect Science. 2007;14:15–27. [Google Scholar]
- Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap “n” collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- Waters LC, Nix CE. Regulation of insecticide resistance-related cytochrome P-450 expression in Drosophila melanogaster. Pest Biochem. Phys. 1988;30:214–227. [Google Scholar]
- Wei N, Deng XW. The COP9 signalosome. Ann. Rev. Cell Dev. Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. Loss of Kelch-Like ECH-Associated Protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, Nauen R, Harrison DA, Palli SR. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA. 2010;107:8557–8562. doi: 10.1073/pnas.1000059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA was extracted from mature adult 91R or 91C flies, labeled, hybridized to two-color Agilent Drosophila 44K arrays, and the data was analyzed using Genesifter following Lowess normalization in R. The columns, in order, show the Fly Base Gene ID, CG number, gene name, cytogenetic location, fold change in expression level, direction of change in expression, p-value, adjusted p-value, biological, cellular and molecular GO function. Genes are sorted by the fold change in expression level between 91C and 91R.
The list of genes that change their expression ≥2-fold in the 91R strain was compared with the list of genes that change their expression ≥2-fold upon ectopic expression of CncC. The columns, in order, show the CG number, Fly Base Gene ID, gene name, fold change in expression level for PB data, direction of change in expression for PB data, p-value for PB data, adjusted p-value for PB data, fold change in expression level for CncC data, direction of change in expression for CncC data, p-value for CncC data, adjusted p-value for CncC data, biological, cellular and molecular GO function. Genes are sorted by the fold change in expression level between 91R and 91C strains.