Abstract
The evolutionally conserved DNA damage response (DDR) and cell cycle checkpoints preserve genome integrity. Central to these genome surveillance pathways is a protein kinase, Chk1. DNA damage induces activation of Chk1, which then transduces the checkpoint signal and facilitates cell cycle arrest and DNA damage repair. Significant progress has been made recently towards our understanding of Chk1 regulation and its implications in cancer etiology and therapy. Specifically, a model that involves both spatiotemporal and conformational changes of proteins has been proposed for Chk1 activation. Further, emerging evidence suggests that Chk1 does not appear to be a tumor suppressor; instead, it promotes tumor growth and may contribute to anticancer therapy resistance. Recent data from our laboratory suggest that activating, but not inhibiting, Chk1 in the absence of chemotherapy might represent an innovative approach to suppress tumor growth. These findings suggest unique regulation of Chk1 in cell biology and cancer etiology, pointing to novel strategies for targeting Chk1 in cancer therapy.
Keywords: Chk1, cell cycle checkpoints, cancer, cancer therapy
Introduction
The act of DDR and cell cycle checkpoints requires the activation of four protein kinases that form the canonical ATR-Chk1 and ATM-Chk2 pathways. While the ATM-Chk2 pathway primarily responds to DNA double-strand breaks (DSB), the ATR-Chk1 pathway recognizes a broad spectrum of DNA abnormalities ranging from UV light, to DNA replication inhibition, to virus infection, to inter-strand DNA crosslinking, and to DSB end resection 1–9.
Chk1 was initially identified by David Beach’s group in 1993 as a Ser/Thr protein kinase that controls the G2/M phase transition in response to DNA damage in fission yeast 10. Shortly after that, Antony Carr’s group reported the identification of the same gene, named Rad27, in budding yeast 11. In 1997, Chk1 orthologs from fruit fly (drosophila ‘grapes’), human and mouse (CHK1) were identified 12–15. In this review, we will summarize how Chk1 is regulated with a particular focus on human Chk1. Further, we will discuss the role of Chk1 in cancer and therapy.
Phosphorylation and Activation of Chk1
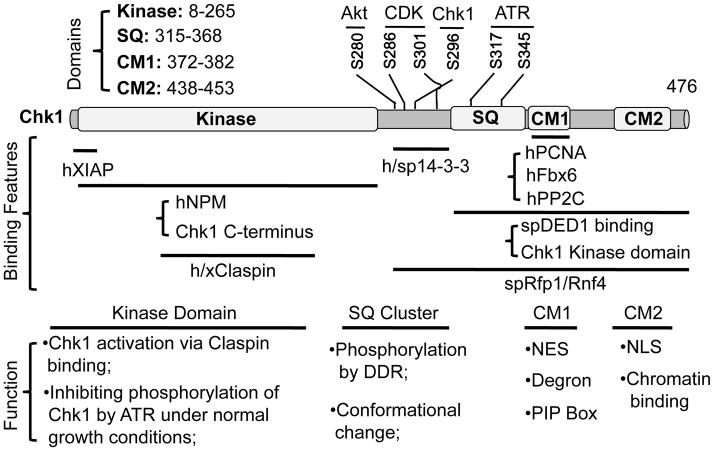
In 1996, Rene Bernards and Nancy Walworth firstly showed that DNA damage induces Chk1 phosphorylation in pombe 16. Similar observations were reported for human and Xenopus Chk1 in a caffeine-dependent manner 13, 17. Caffeine inhibits the activity of ATR and ATM, but not Chk1 18, 19, indicating that Chk1 might be a target of ATR or ATM, which preferentially phosphorylates proteins at Ser/Thr followed by Gln. Human Chk1 contains four Ser/Gln (SQ) residues (317/345/357/366). Phosphorylation at Ser-317 and 345 (or Ser-344 in Xenopus), and to a much lesser extent, at Ser-366, by ATR was demonstrated experimentally, but phosphorylation at Ser-357 has not been detected (Figure 1) 20–22. Importantly, such ATR-dependent phosphorylation of Chk1 is conserved from yeast to human 23, 24.
Figure 1. Human Chk1 domain structure, interacting proteins and function of each domain.
SQ, Ser/Gln cluster; CM, conserved motif; NES, nuclear export signal; NLS, nuclear localization signal; h, human; sp, S pombe; x, Xenopus; PIP, PCNA-interacting protein. Location of each domain is based on human Chk1. Lines illustrate regions of Chk1 that interact with other proteins. Phosphorylation sites are indicated.
Phosphorylation of Chk1 requires the generation of structures containing single-stranded DNA (ssDNA) adjacent to dsDNA, in which ssDNA is coated with replication protein A (RPA) complex. A major route of ssDNA generation is likely through the uncoupling between the active helicase and stalled DNA polymerase at replication forks during S phase 25. This is consistent with observations that activation of Chk1 often requires the presence or activity of proteins involved in DNA replication. The ssDNA-RPA structure then functions as a platform to attract ATR and its regulatory unit ATRIP, as well as a number of regulatory factors including Rad17, TopBP1, the 9-1-1 complex, Tim/Tipin and Claspin (or scMrc1) to damage sites. Formation of this multi-subunit complex stimulates the activity of ATR, which then phosphorylates Chk1 at Ser-317 and Ser-345, leading to the activation of Chk1 and eventually the entire checkpoint pathway 25. Many other proteins, such as BRCA1, MCPH1 and p300/CBP, seem to participate in inducing maximal phosphorylation of Chk1 during DDR 26–30.
Phosphorylation of Chk1 by ATR is important for the DDR and checkpoints. This is illustrated by studies showing that mutating Ser-345 or Ser-317 to Ala led to checkpoint defects, as well as increased sensitivity to replicative stress 31. Interestingly, these two sites exhibit different roles in checkpoint function. Phosphorylation at Ser-317 is required for phosphorylation at Ser-345 22, 32–34. Yet, phosphorylation at Ser-317 alone is not sufficient for inducing maximal phosphorylation at Ser-345 34. The distal C-terminus of Chk1 is required for maximal phosphorylation at Ser-345 likely through providing an optimal conformation 34, 35. Consistent with these observations, mutation of Ser-317 to Ala in somatic cells only abrogated the G2/M phase checkpoint, whereas mutation of Ser-345 to Ala resulted in loss of both checkpoints and cell viability 33. These results led to the idea that Ser-317 phosphorylation triggers the checkpoints, whereas phosphorylation at Ser-345 is the final determinant for maximal checkpoint activation 32, 33.
Function of Chk1
Activated Chk1 in turn phosphorylates a number of downstream effectors to trigger a pleiotropic cellular response including transcription regulation, energy consumption alteration, cell-cycle arrest or delay, DNA repair, or cell death if the damage is too severe to repair. Here we summarize the key checkpoint functions of Chk1.
S phase DNA replication
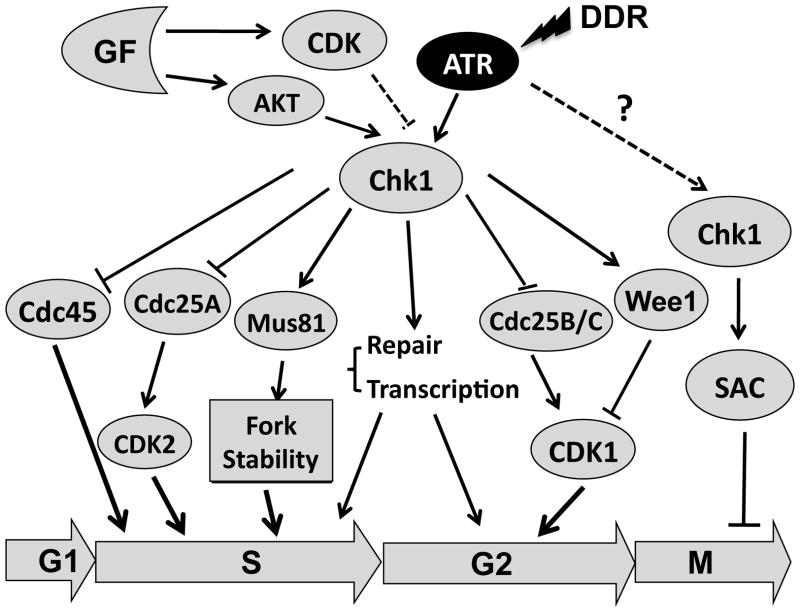
Chk1 responds largely to genotoxic stresses in S phase (Figure 2), in which a key target is the dual-specificity phosphatase, Cdc25A. Cdc25A undergoes Chk1-dependent phosphorylation and proteasomal degradation 36–40. As a result, the activity of Cdk2/cyclin E or Cdk2/cyclin A complex is reduced, leading to the slowing or stalling of DNA replication. Chk1 may also induce chromatin release of Cdc45, an important factor for DNA replication 41. In addition, Chk1 monitors the DNA replication during unperturbed S phase 42. Consistently, Chk1 forms complexes with a number of proteins involved in the DNA replication machinery, including PCNA, Pol alpha and Tim/Tipin 43–45.
Figure 2. Cell cycle checkpoint function of Chk1.<.
br>Chk1 regulates DDR and cell cycle checkpoints during the S, G2 and M phases of the cell cycle. Growth factors (GF) also regulate Chk1. Dash lines represent as-yet confirmed signaling. SAC, spindle assembly checkpoint. See text for detail.
A large body of evidence suggests that Chk1 regulates at least three aspects of DNA replication in S phase: (1) controlling late origin firing; (2) controlling the elongation process; and (3) maintaining stalled replication fork stability 46–53. Accordingly, inhibition of Chk1 often leads to increased origin firing 51, 54. However, the overall DNA replication velocity is inhibited in Chk1-deficient cells 50, 51, 55, 56. As a result, cells may be stuck in the S phase with a 2N DNA content 33, 55. Subsequently, cells with incompletely duplicated DNA enter mitosis prematurely due to the lack of the Chk1-controlled G2/M checkpoint, undergoing a cell death process termed ‘mitotic catastrophe’. The role of Chk1 in stabilizing stalled replication forks is less well understood. Chk1-inhibited cells exhibited Mus81/Eme1-dependent cleavage of stalled replication forks and the generation of DSBs 55, indicating that the DNA endonuclease, Mus81/Eme1, is involved in fork stability regulated by Chk1.
G2/M phase transition
In response to DNA damage at G2 phase, Chk1 phosphorylates and activates the Wee1 kinase, leading to phosphorylation of Cdk1 at the inhibitory Tyr-15 residue 57, 58 (Figure 2). In addition, Chk1 phosphorylates the phosphatase, Cdc25C, at Ser-216, whose activity is required for de-phosphorylation of Cdk1 at Tyr-15 and its subsequent activation 13, 17, 59. This phosphorylation of Cdc25C also creates a docking site for the 14-3-3 (or pombe Rad24) family proteins 14. Association with 14-3-3 leads to nuclear export of Cdc25C, preventing it from activating Cdk1/cyclin B1 in the nucleus 60. However, nuclear export of Cdc25C may be a secondary effect of Cdc25C phosphorylation 61.
While Chk1 is critical for holding cells at the G2 phase in response to DNA damage, it has to be kept inactive for normal G2/M phase transition or after the damage is repaired. This can be firstly achieved through Plk1-dependent phosphorylation followed by proteasomal degradation of Claspin, the key mediator for Chk1 phosphorylation 62, 63. Secondly, when cells are ready for mitotic entry, Chk1 cannot be phosphorylated and activated by DNA damage. This appears to be due to the lack of recruitment of important mediator proteins required for Chk1 phosphorylation, including RPA, ATR and CtIP, to DNA damage sites 64. Interestingly, this event is controlled by PKB/AKT 64, although exactly how PKB/AKT inhibits the recruitment of these factors to DNA damage sites remains unknown. Thirdly, the rapid increase in Cdk1/Cyclin B activity at late G2 phase leads to phosphorylation of Chk1 at Ser-280/301, which somehow limits phosphorylation of Chk1 by ATR, preventing its activation by DNA damage 65. Together, these findings suggest that Chk1 activity is finely tuned to ensure a proper and timely progression of the G2/M transition.
M phase
The idea of M phase DDR is less clear than those in G1, S and G2 phases. Similarly, whether Chk1 is involved in M phase DDR is unclear, as this is a stage where ssDNA/dsDNA structures, the key element for Chk1 activation, can hardly be generated. Even though immunostaining results showed that Ser-345-phosphorylated Chk1 is detected both in normal and damaged M phase cells 33, 66, a potential caveat is that these phospho-antibodies recognize more than just phosphorylated Chk1 in cells.
On the other hand, increasing evidence clearly points out a role of Chk1 in normal M phase progression. When cells enter M phase, Chk1 undergoes Cdk-dependent phosphorylated (Ser-286/301) 65. This phosphorylation seems to induce the nuclear export of Chk1 to relieve its inhibitory effect on cyclin B/Cdk1 in the nucleus, leading to mitotic progression 67. Inhibition or depletion of Chk1 leads to various mitotic abnormalities, such as chromosome misalignment on the spindle and kinetochore defects 66, 68, 69. Potential targets of Chk1 include the spindle assembly checkpoint proteins, Aurora A kinase 70, Plk1 68, and Aurora B kinase 69. In this regard, co-depletion of the spindle assembly checkpoint proteins rescued the M phase abnormalities caused by Chk1 depletion 68. In Drosophila, Chk1 (grapes) regulates chromosome condensation 71, 72, indicating conserved roles of Chk1 in M phase progression.
In mammals, a small portion of Chk1 was reported to localize to centrosomes to block premature activation of the cyclin B/Cdk1 complex, preventing abnormal M phase entry73. However, recent research results showed that the anti-Chk1 antibodies used for the staining cross-reacted with another centrosomal protein 74, suggesting that the centrosomal localization of Chk1 warrants further investigation. Nevertheless, these findings clearly point out critical roles of Chk1 in M phase progression.
Other functions of Chk1
While the major function of Chk1 is to coordinate the DDR and cell cycle checkpoint response, it also regulates a number of other cellular functions, including DNA damage repair, gene transcription, embryo development and somatic cell viability 20, 75–77. Other protein kinases, including PKB/AKT and the MAPKAPK kinase, p90/RSK, can phosphorylate Chk1 at sites different from ATR 78, 79. Further, Chk1 also regulates cellular response to HIV virus infection, low or high level of oxygen exposure, protein misfolding stress, or heat shock 8, 9, 80–83.
Mechanisms Controlling Function and Expression of Chk1
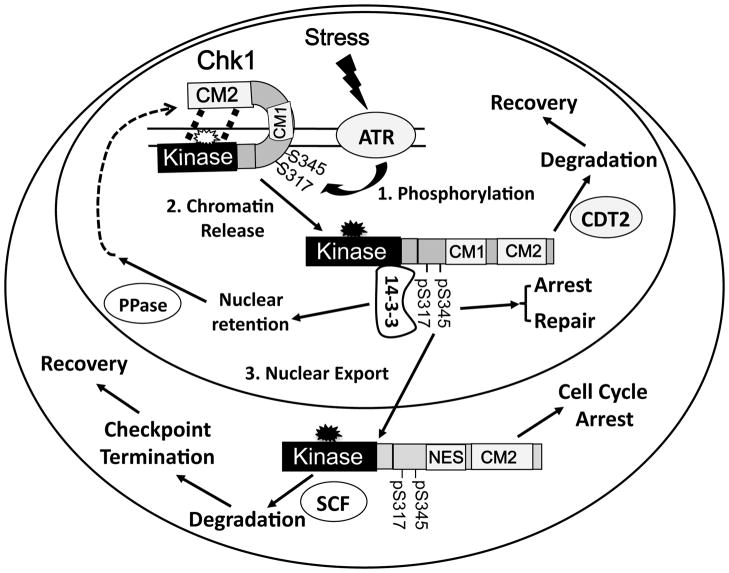
In addition to mechanisms regulating upstream events governing the phosphorylation and activation of Chk1, recent studies from our laboratory and others illustrated a number of novel mechanisms that regulate both activation and expression of Chk1. These involve phosphorylation-coupled protein conformational change and cellular re-distribution, as well as proteasome-dependent degradation of Chk1 (Figure 3).
Figure 3. Spatiotemporal and conformational regulation of Chk1.
The intra-molecular interaction between the N-terminal kinase domain and the C-terminal domain requires structural studies to confirm. Thus, it is shown as dash lines here. For simplicity, proteins and protein complexes involved in the signaling pathway, such as RPA, TopBP1, Tim/Tipin, are omitted in this model. The catalytic site is illustrated in open and solid stars for ‘closed’ and ‘open’ conformation, respectively. The inner and outer circles represent the nucleus and the cytoplasm, respectively. The CM1 motif bears non-canonical NES activity, facilitating nuclear export of Chk1.
Conformational change of Chk1
In the absence of DNA damage, Chk1 appears to adopt a ‘closed’ conformation through an intra-molecular interaction between the N-terminus and the C-terminus 84, 85. This ‘closed’ conformation probably physically blocks the active site of the kinase domain of Chk1, as well as stabilizes the protein in the absence of DNA damage 85 (Figure 3). Yeast Chk1 contains a pseudosubstrate motif at the C-terminus (D469) that facilitates such intramolecular interaction 86. However, this residue is not conserved in Chk1 from other species and does not regulate the auto-inhibitory effect of Chk1 86. In response to DNA damage, Chk1 is phosphorylated by ATR on chromatin 56, 87. This phosphorylation disrupts the intra-molecular interaction in an unidentified manner, leading to the exposure of the catalytic domain of Chk1 (Figure 3). Since the catalytic site of Chk1 always adopts an active conformation 88, this ‘open’ conformation then unleashes the catalytic activity to turn on the checkpoint through phosphorylating downstream effector proteins. Interestingly, such auto-inhibitory regulation seems to be quite common for protein kinases, such as Src and MLCK, etc 89, 90.
Our recent studies revealed a new layer of function of this ‘closed’ conformation, which is to prevent Chk1 from being phosphorylated at ATR sites in the absence of DNA damage 34. Since Chk1 phosphorylation functions as a trigger to release the intra-molecular restraint, preventing Chk1 from being phosphorylated by ATR is important to maintain Chk1 in the ‘closed’ inactive conformation under normal growth conditions. These mutual inhibitory effects between the N-terminal kinase domain and the C-terminal regulatory domain of Chk1 significantly increase the threshold of accidental checkpoint activation in the absence of DNA damage, which could be achieved either through exposing the catalytic domain or through phosphorylating the ATR sites of Chk1. Keeping Chk1 inactive under normal growth condition is critical for cell viability, as constitutive activation of Chk1 leads to cell cycle arrest and eventually cell death 34.
Spatiotemporal regulation of Chk1
Since Chk1 functions as the ‘messenger’ to deliver the DNA damage alarm, it is conceivable that Chk1 does not form stable DNA damage-induced foci. This is consistent with observations that phospho-Chk1 is expressed throughout the nucleus after DNA damage 91. This phosphorylation not only induces the protein conformational change, as discussed above, but also triggers a rapid release of Chk1 from damaged chromosomal sites into the soluble nucleoplasm, and later on into the cytoplasm 44, 56, 75, 87. Phosphorylated Chk1 undergoes SCFFbx6- and Cul4ACDT2- dependent degradation in the cytoplasm and nucleus, respectively 56, 85, 92, 93. Interestingly, compared to the rapid checkpoint activation, degradation of Chk1 occurs at a relatively slow rate 56, leading to around 4 hr difference between Chk1 phosphorylation and the onset of detectable Chk1 protein reduction. This may be partially determined by the intensity of DNA damage and the time required for the mobilization of phospho-Chk1 from chromatin to the nucleoplasm and eventually to the cytoplasm, where the E3 ligases are located 85. Retention of Ser-345 phospho-Chk1 in the nucleus might be facilitated through its association with 14-3-3 proteins 94–96. This delayed degradation of Chk1 has significant biological implications. First, it allows active Chk1 molecules enough time to turn on the checkpoint. Second, degrading Chk1 after checkpoint activation would fine-tune the cellular checkpoint level, contributing to the ultimate checkpoint termination, cell cycle resumption and cell survival 97. While numerous studies have reported degradation of mammalian Chk1 (see review 97), yeast Chk1 does not seem to undergo degradation. One possible explanation is that the putative degron for Chk1 degradation, the CM1 motif, is highly conserved in mammals, but not in yeast. This difference suggests that mammalian Chk1 may have acquired additional regulatory steps during evolution, reminiscent of the fact that Chk1 is essential in mammals, but not in yeast 98.
The nuclear-cytoplasm shuttling of Chk1 is Crm1-dependent 95, 99. Chk1 does not contain a canonical NES (a Leu zipper, LxxxLxxLxL). Our recent studies suggest that the CM1 motif functions as a non-canonical NES (LxxxMxxFxxxL) that regulates nuclear export of Chk1. In contrast, the CM2 motif and flanking region contains canonical nuclear localization signals (NLS) and are responsible for nuclear localization of human Chk1, as for Xenopus Chk1 84, 99. Probably the most interesting observation of our studies is that even though the cytoplasmic pool of Chk1 is important for checkpoint function, it is the nuclear pool of Chk1 that supports cell viability 99.
Transcriptional and post-translational control of Chk1 expression
Chk1 isoforms
Mammalian CHK1 mainly transcribes as isoform 1, which encodes the full-length Chk1 protein. However, short Chk1 isoforms exist due to alternative splicing or translation start sites or protein cleavage 100–104. A question remaining unanswered is whether and how Chk1 isoforms affect cellular checkpoint function. Given the fact that the N- and C-termini interact each other, it is tempting to speculate that Chk1 isoforms or short fragments might interfere with the function of endogenous Chk1 through competitively interacting with endogenous Chk1 molecules, although overexpressing the N-terminal kinase domain of Chk1 did not activate checkpoints in yeast 35.
The full-length transcript of human CHK1
Expression of mammalian Chk1 isoform 1 (both mRNA and protein) peaks at S and G2 phases of the cell division cycle 105, in line with its major roles in regulating DDR and checkpoints in these cell cycle phases 34, 64. The CHK1 gene appears to be a target of the E2F family of transcription factors 106, which controls the rise of Chk1 at the G1/S transition. Therefore, stresses that can activate p53-dependent G1/S arrest often lead to reduced Chk1 expression through inhibiting E2F-dependent transcription of CHK1 107. However, the reduction in the protein level of Chk1 under both normal cell cycle transition and genotoxic stress conditions is mainly controlled by proteasome-dependent degradation 97. The de-ubiquitination enzyme, USP1, on the other hand, is involved in de-ubiquitination and stabilization of Chk1 108. Interestingly, nutrient restriction (e.g., glucose deprivation or hypoxia), cytokine treatment, heat shock or histone deacetyltransferase inhibition, or altering the expression of Chk1-interacting proteins also triggered proteasome-dependent degradation of Chk1 26, 56, 109–116, leading to up to 90% decrease in the level of Chk1.
Why are Chk1 protein levels affected by such a wide range of stresses and physiological changes? This is probably because the expression level of Chk1 is critical for its function, especially for cell viability maintenance and tissue development. Even a 50% reduction in Chk1, caused by gene disruption or chemical inhibition 56, 68, 117–119, significantly increased spontaneous cell death and development defects. Loss of one copy of CHK1 leads to anemia and erythropoiesis defects and sudden death in mice 120. This suggests that Chk1 haplo-insufficiency compromises cell viability. How exactly a reduction in the level of Chk1 leads to such defects is currently unknown. It is tempting to speculate that the overall reduction in Chk1 kinase activity due to decreased Chk1 protein levels is responsible for such defects. In line with this hypothesis, a hypomorphic R156Q mutation in the kinase domain of mouse Chk1 that reduces its catalytic activity but not protein levels, failed to support normal cell proliferation and also leads to anemia and erythropoiesis abnormalities (personal communication, Yolanda Sanchez). Together, these data indicate that Chk1 protein expression is tightly controlled both during normal growth conditions and under stressful situations.
Known Chk1 Targets
A number of studies had intended to identify Chk1 substrates using either in vitro peptide screening or in vivo cell labeling 121–123. An overall consensus motif for Chk1 substrate phosphorylation is R/K-R/K-d/e-t-S/T-X-r/k-r, in which upper and lower case letters represent preferred and non-preferred residues, respectively 123. Chk1 substrates that have been confirmed in cell cultures are summarized in Table 1.
Table 1.
Chk1 substrates, including the phosphorylation site and potential checkpoint functions
| Substrate | P. Site | Substrate Regulation | Function | Ref. | |
|---|---|---|---|---|---|
| DDR & Checkpoints | Cdc25A | S76/123 | Degradation | S phase arrest | 37 |
| T507(X504) | 14-3-3 binding | S & G2 arrest | 127, 128 | ||
| Cdc25B | S230/563 | Reduced activity | G2/M arrest | 129 | |
| Cdc25C | S216 | Cytoplasm location | G2/M arrest | 13, 14, 60 | |
| p53 | S20 | Stabilization | G1/S or G2/M arrest | 124, 125 | |
| 14-3-3γ | S367 | P53 stabilization | G1/S arrest by UV | 143 | |
| Nek11 | S273 | Phosphorylates Cdc25A | Cdc25A degradation | 130 | |
| spPds1 | multiple | Stabilization | Inhibiting anaphase | 131, 132 | |
| XWee1 | S549 | 14-3-3 binding, Cdk1pY15 | G2/M arrest | 133 | |
| Tlk | S695 | Chromatin assembly | S checkpoints | 135 | |
| KAP1 | S473 | Heterochromatin binding | Unclear | 123, 134 | |
| Chk1 | S296 | Autophosphorylation | Checkpoint marker | 146, 147 | |
| Aurora B | S331 | Aurora B activation | Kinetochore attachment | 69, 149 | |
|
| |||||
| Repair | Rad51 | T309 | Unclear | Homologous repair | 77 |
| BLM | S646 | Stabilization | Damage repair | 138, 139 | |
| FANCE | T346/S374 | Degradation | Damage repair | 144 | |
| FANCD2 | S331 | Mono-ubiquitination | Damage repair | 145 | |
| Metnase | S495 | DNA binding | Damage repair | 140 | |
|
| |||||
| Gene Expression & Cell Death | p73 | S47 | Trans-activation | Cell death | 136 |
| H3 | T11 | GCN5 recruitment | Transcription | 75 | |
| p53 | S366/T387 | Reduced Acetylation | Transcription repression | 126 | |
| p65 | T505 | Unclear | Transcription repression | 141 | |
| p50 | S329 | Inhibition of DNA binding | Transcription repression | 142 | |
| ING1b | S126 | Stabilization | CycB1 transcription | 148 | |
| p57 | S19 | Unclear | Preventing differentiation | 151 | |
| p21 | T140/S141 | Unclear | Preventing differentiation | 151 | |
Chk1-interacting Proteins
Chk1 exerts its function often through interacting with other proteins. Numerous proteins have been reported to interact with Chk1 (Figure 1 & Table 2), although not all of them have been reported to have biological significance. Some of these interacting proteins are also Chk1 substrates.
Table 2.
Chk1 interacting proteins
| Interacting Proteins | Method | Requirement | Function | Ref. |
|---|---|---|---|---|
| Claspin | CO-IP | Phosphorylation by CK1γ1 | Checkpoints | 152, 153 |
| XIAP | CO-IP | Metaphase | Unknown | 154 |
| BAD | CO-IP | Unknown | Unknown | 155 |
| Timeless | CO-IP | G1/S arrest by UV | G1/S arrest by UV | 43 |
| PCNA | MS/CO-IP | Constitutive | Checkpoints | 44 |
| Polα | CO-IP | Constitutive | Checkpoints | 45 |
| Wip1 | CO-IP | Constitutive | Checkpoint Off | 157 |
| MCPH1 | CO-IP | Constitutive | Unclear | 158 |
| DNMT1 | CO-IP | Constitutive | Chk1 cell localization | 159 |
| Fbx6 | CO-IP | Phosphorylation | Chk1 degradation | 85 |
| Cul4A/DDB1 | MS/CO-IP | Phosphorylation | Chk1 degradation | 56, 85, 93 |
| NPM | CO-IP | Constitutive | NPM Chromatin loading | 160 |
| MutSα | CO-IP | Constitutive | Chromatin loading | 161 |
| SpRfp1/Rnf4 | Y2H | No CO-IP to confirm | Damage repair | 162 |
| Fem1b | Y2H | Constitutive | Checkpoints | 163 |
Chk1 in Cancer
Human CHK1 is located in 11q22–23, a region that not only contains the highly mutated gene ATM, but also has frequent deletions and loss of heterozygosity in human tumors 15. Given its critical roles in DDR and cell cycle checkpoints, Chk1 was initially thought to function as a tumor suppressor, and numerous efforts were made to look for CHK1 mutations in human tumors. However, so far no homozygous loss-of-function mutation of CHK1 has been detected in a wide range of human tumors 164–169. These results suggest that a homozygous mutation that affects CHK1 function is not able to support tumor clone expansion. Thus, those tumor clones might have died out before being detected. Since the expression level is as important as Chk1’s catalytic function, an extension of this speculation is that mutations identified so far do not compromise Chk1’s expression below a critical level.
Based on these observations, we hypothesize that Chk1 is unlikely a canonical tumor suppressor. Mice studies supported such a notion. Conditional knockout studies in mouse thymus and mammary gland indicate that Chk1 depletion does not increase spontaneous tumor incidence 68, 69, 117, 118. In fact, loss of CHK1 reduces tumorigenicity in mice driven by TP53-null mutation or carcinogen exposure 170, 171. This is similar to the suppression of mouse skin tumorigenesis when the catalytic activity of ATR is ablated in a XPC-/- background 172. These findings suggest that Chk1 (or the ATR-Chk1 axis) is a weak tumor suppressor at best. Instead, increasing evidence suggests that the ATR-Chk1 axis, or at least Chk1, may actually promote tumor growth.
Roles of Chk1 in Tumor Etiology and Therapy Resistance
Chk1 has been found to be overexpressed in a variety of human tumors, including breast, colon, liver, gastric, nasopharyngeal carcinoma, etc. 150, 173–178. Remarkably, its expression often positively correlates with tumor grade and disease recurrence 150, 176, 179. A transgenic mouse line carrying an extra copy of CHK1 facilitates cell transformation probably due to the enhanced ability of those cells to deal with replicative stress 180. These observations are well in line with the idea that Chk1 promotes tumor growth.
Further, Chk1 may also contribute to therapy resistance. Enhanced activation of Chk1 led to resistance of cancer cells, including cancer stem cells from brain glioblastoma, prostate and lung NSCLC, to chemotherapy or radiotherapy, as well as to other anticancer therapies, for instance, HDAC inhibitors 85, 181–186. Conversely, inhibiting Chk1 by RNAi or small molecules reversed such therapy resistance. Further, elevated levels of Ser-345 phosphorylated Chk1 proteins correlated with increased radio-resistance in metastatic brain and lung cancer patients 187. In addition, cancer cells may acquire chemotherapy resistance through increased expression of Chk1 188. Together with the fact that Chk1 is essential for the maintenance of cell viability, these findings suggest a tumor-promoting model of Chk1. In this model, tumor cells that have increased Chk1 expression possess survival advantages over their neighbors, because the more Chk1 protein cells have, the better able they are to handle the DNA damage stress caused either by the harsh tumor microenvironment (e.g., replicative stress 180) or by chemotherapy/radiotherapy. Eventually, these Chk1-proficient cells will grow out and dominate within the tumors, fueling the generation of more malignant, drug-resistant clones to give rise to tumor recurrence and disease relapse in the clinic. This model provides strong support to target Chk1 in human cancer therapy.
Targeting Chk1 in Human Diseases
Chemotherapy and radiotherapy kill proliferating cancer cells through generating massive DNA lesions. In the meantime, they activate the Chk1-dependent DDR and cell cycle checkpoints to facilitate cell survival. A conventional idea is that when Chk1 is inhibited, cancer cells lose their ability to respond to and repair DNA damage, enhancing the cell killing effect of chemotherapy or radiotherapy. Therefore, combining Chk1 inhibition with chemotherapy or radiotherapy provides the so-called ‘synthetic lethality’ effect in cancer therapy (Figure 4). Consistent with this idea, a number of siRNA screening studies identified Chk1 as a target, which when depleted by siRNA, led to the most significantly enhanced cell killing effect among kinases by chemotherapy or radiation therapy in ovarian, triple negative breast and brain cancers 189–191. Furthermore, since a large portion of tumors lost the p53-dependent G1 phase checkpoint, they rely much more heavily on the Chk1-dependent S and G2/M checkpoints for survival. Therefore, Chk1 inhibition should be especially effective against p53-deficient cancer cells compared to p53-proficient cancer cells 192. Recent work using a humanized mouse model of triple negative breast cancer confirmed such an idea 193.
Figure 4. Strategies for targeting Chk1 in cancer therapy.
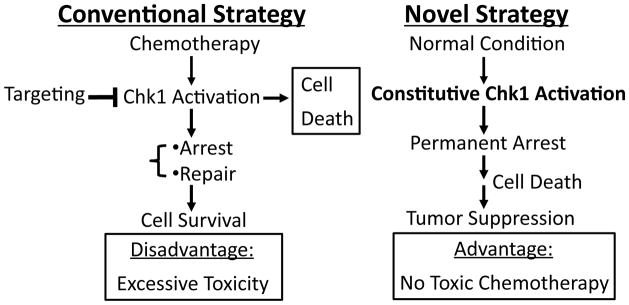
Under conventional idea, a Chk1 inhibitor is combined with chemotherapy to enhance the therapy effect. In the novel approach derived from our latest research results, constitutively activating Chk1 under normal growth condition is sufficient to induce permanent cell cycle arrest and cell death. The new approach should significantly reduce the toxicity of chemotherapy since it does not require concurrent use of chemotherapy.
Numerous attempts have been made by various pharmaceutical companies to identify specific Chk1 inhibitors to enhance the effect of chemotherapy 194–196. However, a major issue with this conventional strategy is the off-target effects and toxicity associated with Chk1 inhibitors, chemotherapy or a combination of them. So far, no therapy has reached the bedside even though theoretically a highly selective Chk1 inhibitor would synergize with chemotherapy and have a therapeutic window in combination with a DNA damaging agent (refer reviews 194, 195, 197). In addition, loss of CHK1 caused developmental defects of normal blood system 120. Therefore, alternative strategies need to be developed to target Chk1 in cancer therapy.
Recently we discovered that disrupting the ‘closed’ conformation of Chk1 (e.g., by mutating one of two absolutely conserved residues G448 or L449 at the CM2 motif) leads to activation of Chk1 in the absence of DNA damage 34. Remarkably, expression of this constitutively active Chk1 mutant completely blocked cancer cell proliferation and eventually led to cell death under normal growth conditions 34. This is probably because the mutant sounds a significantly high ‘false’ alarm signal as if the cell had massive DNA damage beyond its capability to repair. These results lead to the novel concept that too much activation of Chk1 is detrimental to cell survival in the absence of DNA damage. This in turn indicates that artificially activating, but not inhibiting, Chk1 in the absence of DNA damage could be developed into an innovative strategy in cancer therapy (Figure 4). This novel strategy does not involve the use of toxic chemotherapeutic drugs; therefore, it has the potential to significantly reduce the side effect of anticancer therapy compared with conventional strategies.
Conclusion
Significant progress has been made during the last decade in understanding Chk1 regulation and its potential as a cancer therapy target. Meanwhile, many new questions have been raised. To name a few important ones: (1) How exactly does Chk1 support cell viability? Is this related solely to its checkpoint function or an as yet to be unidentified role of Chk1? (2) What are specific targets of Chk1 in the nucleus versus in the cytoplasm? Given the fact that cytoplasmic Chk1 does not support cell viability, it is important to answer this question in the near future. (3) How exactly does phosphorylation at Ser-317/345 by ATR lead to Chk1 activation? A crystal structure of the full-length Chk1 protein is clearly the key for answering this critical question. (4)How can one develop a more specific targeting strategy towards Chk1 in cancer therapy? We propose that artificially activating, but not inhibiting, Chk1 under normal growth conditions might represent a novel idea in suppressing tumor growth. In conclusion, exciting results regarding Chk1 regulation and targeting are expected to continuously emerge in the near future.
Acknowledgments
We apologize to colleagues whose work was not cited due to space limitations. We thank Paul MacDonald for critical reading of the manuscript. Y.W.Z was supported by the NCI Howard Temin Career Development Award (R00CA126173), and is currently supported by NCI R01 (CA163214) and a pilot grant from American Cancer Society (IRG-91-022-15). T.H. is supported by NCI CA82683 and CA80100. T.H. is a Frank and Else Schilling Cancer Society Professor, and the Renato Dulbecco Chair in Cancer Research.
Footnotes
Conflict of Interest: We declare no potential conflict of interest
References
- 1.Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol Cell. 2009;35:704–15. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–50. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 5.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22:8552–61. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–6. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 7.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–89. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 8.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem. 2003;278:25879–86. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman ES, Sherman MP, Blackett JL, Neidleman JA, Kreis C, Mundt P, Williams SA, Warmerdam M, Kahn J, Hecht FM, Grant RM, de Noronha CM, et al. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J Virol. 2006;80:10407–18. doi: 10.1128/JVI.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–71. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 11.al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–60. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–26. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 14.Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–5. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 15.Flaggs G, Plug AW, Dunks KM, Mundt KE, Ford JC, Quiggle MR, Taylor EM, Westphal CH, Ashley T, Hoekstra MF, Carr AM. Atm-dependent interactions of a mammalian chk1 homolog with meiotic chromosomes. Curr Biol. 1997;7:977–86. doi: 10.1016/s0960-9822(06)00417-9. [DOI] [PubMed] [Google Scholar]
- 16.Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–6. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 17.Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–69. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–82. [PubMed] [Google Scholar]
- 19.Moser BA, Brondello JM, Baber-Furnari B, Russell P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol Cell Biol. 2000;20:4288–94. doi: 10.1128/mcb.20.12.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–59. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–39. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT. Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene. 2009;28:2314–23. doi: 10.1038/onc.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–56. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Girona A, Tanaka K, Chen XB, Baber BA, McGowan CH, Russell P. Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc Natl Acad Sci U S A. 2001;98:11289–94. doi: 10.1073/pnas.191557598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Lee J, Stern DF. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J Biol Chem. 2004;279:34091–4. doi: 10.1074/jbc.C400139200. [DOI] [PubMed] [Google Scholar]
- 27.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet. 2002;30:285–9. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 28.Stauffer D, Chang B, Huang J, Dunn A, Thayer M. p300/CREB-binding protein interacts with ATR and is required for the DNA replication checkpoint. J Biol Chem. 2007;282:9678–87. doi: 10.1074/jbc.M609261200. [DOI] [PubMed] [Google Scholar]
- 29.Yoo HY, Jeong SY, Dunphy WG. Site-specific phosphorylation of a checkpoint mediator protein controls its responses to different DNA structures. Genes Dev. 2006;20:772–83. doi: 10.1101/gad.1398806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato K, Sundaramoorthy E, Rajendra E, Hattori H, Jeyasekharan AD, Ayoub N, Schiess R, Aebersold R, Nishikawa H, Sedukhina AS, Wada H, Ohta T, et al. A DNA-damage selective role for BRCA1 E3 ligase in claspin ubiquitylation, CHK1 activation, and DNA repair. Curr Biol. 2012;22:1659–66. doi: 10.1016/j.cub.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 31.Capasso H, Palermo C, Wan S, Rao H, John UP, O’Connell MJ, Walworth NC. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 2002;115:4555–64. doi: 10.1242/jcs.00133. [DOI] [PubMed] [Google Scholar]
- 32.Niida H, Katsuno Y, Banerjee B, Hande MP, Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol Cell Biol. 2007;27:2572–81. doi: 10.1128/MCB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci U S A. 2008;105:20752–7. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Han X, Zhang Y. Autoregulatory mechanisms of phosphorylation of checkpoint kinase 1. Cancer Res. 2012;72:3786–94. doi: 10.1158/0008-5472.CAN-12-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosoy A, O’Connell MJ. Regulation of Chk1 by its C-terminal domain. Mol Biol Cell. 2008;19:4546–53. doi: 10.1091/mbc.E08-04-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–7. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 37.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–58. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 38.Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 2002;21:3694–703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci U S A. 2002;99:14795–800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–9. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 41.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem. 2006;281:30631–44. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 42.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scorah J, Dong MQ, Yates JR, 3rd, Scott M, Gillespie D, McGowan CH. A conserved proliferating cell nuclear antigen-interacting protein sequence in Chk1 is required for checkpoint function. J Biol Chem. 2008;283:17250–9. doi: 10.1074/jbc.M800369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taricani L, Shanahan F, Parry D. Replication stress activates DNA polymerase alpha-associated Chk1. Cell Cycle. 2009;8:482–9. doi: 10.4161/cc.8.3.7661. [DOI] [PubMed] [Google Scholar]
- 46.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 47.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–23. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–7. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 49.Zachos G, Rainey MD, Gillespie DA. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22:713–23. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–26. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:3184–9. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–5. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6:2760–7. doi: 10.4161/cc.6.22.4932. [DOI] [PubMed] [Google Scholar]
- 54.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–64. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forment JV, Blasius M, Guerini I, Jackson SP. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One. 2011;6:e23517. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–18. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 57.O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–54. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–11. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 59.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–7. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–5. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 61.Lopez-Girona A, Kanoh J, Russell P. Nuclear exclusion of Cdc25 is not required for the DNA damage checkpoint in fission yeast. Curr Biol. 2001;11:50–4. doi: 10.1016/s0960-9822(00)00026-9. [DOI] [PubMed] [Google Scholar]
- 62.Mailand N, Bekker-Jensen S, Bartek J, Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell. 2006;23:307–18. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 63.Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–29. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 64.Xu N, Hegarat N, Black EJ, Scott MT, Hochegger H, Gillespie DA. Akt/PKB suppresses DNA damage processing and checkpoint activation in late G2. J Cell Biol. 2010;190:297–305. doi: 10.1083/jcb.201003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiromizu T, Goto H, Tomono Y, Bartek J, Totsukawa G, Inoko A, Nakanishi M, Matsumura F, Inagaki M. Regulation of mitotic function of Chk1 through phosphorylation at novel sites by cyclin-dependent kinase 1 (Cdk1) Genes Cells. 2006;11:477–85. doi: 10.1111/j.1365-2443.2006.00955.x. [DOI] [PubMed] [Google Scholar]
- 66.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci U S A. 2009;106:5159–64. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Enomoto M, Goto H, Tomono Y, Kasahara K, Tsujimura K, Kiyono T, Inagaki M. Novel positive feedback loop between Cdk1 and Chk1 in the nucleus during G2/M transition. J Biol Chem. 2009;284:34223–30. doi: 10.1074/jbc.C109.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Natl Acad Sci U S A. 2006;103:11964–9. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, Gillespie DA. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–60. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krystyniak A, Garcia-Echeverria C, Prigent C, Ferrari S. Inhibition of Aurora A in response to DNA damage. Oncogene. 2006;25:338–48. doi: 10.1038/sj.onc.1209056. [DOI] [PubMed] [Google Scholar]
- 71.Yu KR, Saint RB, Sullivan W. The Grapes checkpoint coordinates nuclear envelope breakdown and chromosome condensation. Nat Cell Biol. 2000;2:609–15. doi: 10.1038/35023555. [DOI] [PubMed] [Google Scholar]
- 72.Royou A, Macias H, Sullivan W. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr Biol. 2005;15:334–9. doi: 10.1016/j.cub.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 73.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–91. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 74.Matsuyama M, Goto H, Kasahara K, Kawakami Y, Nakanishi M, Kiyono T, Goshima N, Inagaki M. Nuclear Chk1 prevents premature mitotic entry. J Cell Sci. 2011;124:2113–9. doi: 10.1242/jcs.086488. [DOI] [PubMed] [Google Scholar]
- 75.Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–32. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 76.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14:1439–47. [PMC free article] [PubMed] [Google Scholar]
- 77.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 78.Shtivelman E, Sussman J, Stokoe D. A role for PI 3-kinase and PKB activity in the G2/M phase of the cell cycle. Curr Biol. 2002;12:919–24. doi: 10.1016/s0960-9822(02)00843-6. [DOI] [PubMed] [Google Scholar]
- 79.Li P, Goto H, Kasahara K, Matsuyama M, Wang Z, Yatabe Y, Kiyono T, Inagaki M. P90 RSK arranges Chk1 in the nucleus for monitoring of genomic integrity during cell proliferation. Mol Biol Cell. 2012;23:1582–92. doi: 10.1091/mbc.E11-10-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–43. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Das KC, Dashnamoorthy R. Hyperoxia activates the ATR-Chk1 pathway and phosphorylates p53 at multiple sites. Am J Physiol Lung Cell Mol Physiol. 2004;286:L87–97. doi: 10.1152/ajplung.00203.2002. [DOI] [PubMed] [Google Scholar]
- 82.Furusawa Y, Iizumi T, Fujiwara Y, Zhao QL, Tabuchi Y, Nomura T, Kondo T. Inhibition of checkpoint kinase 1 abrogates G2/M checkpoint activation and promotes apoptosis under heat stress. Apoptosis. 2012;17:102–12. doi: 10.1007/s10495-011-0660-7. [DOI] [PubMed] [Google Scholar]
- 83.Malzer E, Daly ML, Moloney A, Sendall TJ, Thomas SE, Ryder E, Ryoo HD, Crowther DC, Lomas DA, Marciniak SJ. Impaired tissue growth is mediated by checkpoint kinase 1 (CHK1) in the integrated stress response. J Cell Sci. 2010;123:2892–900. doi: 10.1242/jcs.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katsuragi Y, Sagata N. Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell. 2004;15:1680–9. doi: 10.1091/mbc.E03-12-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, Manning G, Abraham RT, Hunter T. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–53. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palermo C, Hope JC, Freyer GA, Rao H, Walworth NC. Importance of a C-terminal conserved region of Chk1 for checkpoint function. PLoS One. 2008;3:e1427. doi: 10.1371/journal.pone.0001427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16:150–9. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 88.Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, Tempczyk-Russell A, Nguyen B, Myers P, Lundgren K, Kan CC, O’Connor PM. The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell. 2000;100:681–92. doi: 10.1016/s0092-8674(00)80704-7. [DOI] [PubMed] [Google Scholar]
- 89.Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–27. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 90.Gallagher PJ, Herring BP, Trafny A, Sowadski J, Stull JT. A molecular mechanism for autoinhibition of myosin light chain kinases. J Biol Chem. 1993;268:26578–82. [PMC free article] [PubMed] [Google Scholar]
- 91.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huh J, Piwnica-Worms H. CRL4CDT2 targets CHK1 for PCNA-independent destruction. Mol Cell Biol. 2012 doi: 10.1128/MCB.00847-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leung-Pineda V, Huh J, Piwnica-Worms H. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–7. doi: 10.1158/0008-5472.CAN-08-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen L, Liu TH, Walworth NC. Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev. 1999;13:675–85. doi: 10.1101/gad.13.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y. Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J Biol Chem. 2003;278:25207–17. doi: 10.1074/jbc.M300070200. [DOI] [PubMed] [Google Scholar]
- 96.Dunaway S, Liu HY, Walworth NC. Interaction of 14-3-3 protein with Chk1 affects localization and checkpoint function. J Cell Sci. 2005;118:39–50. doi: 10.1242/jcs.01570. [DOI] [PubMed] [Google Scholar]
- 97.Merry C, Fu K, Wang J, Yeh IJ, Zhang Y. Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle. 2010;9:279–83. doi: 10.4161/cc.9.2.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–54. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang J, Han X, Feng X, Wang Z, Zhang Y. Coupling cellular localization and function of checkpoint kinase 1 (chk1) in checkpoints and cell viability. J Biol Chem. 2012;287:25501–9. doi: 10.1074/jbc.M112.350397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haruki N, Saito H, Tatematsu Y, Konishi H, Harano T, Masuda A, Osada H, Fujii Y, Takahashi T. Histological type-selective, tumor-predominant expression of a novel CHK1 isoform and infrequent in vivo somatic CHK2 mutation in small cell lung cancer. Cancer Res. 2000;60:4689–92. [PubMed] [Google Scholar]
- 101.Shann YJ, Hsu MT. Cloning and characterization of liver-specific isoform of Chk1 gene from rat. J Biol Chem. 2001;276:48863–70. doi: 10.1074/jbc.M108253200. [DOI] [PubMed] [Google Scholar]
- 102.Pabla N, Bhatt K, Dong Z. Checkpoint kinase 1 (Chk1)-short is a splice variant and endogenous inhibitor of Chk1 that regulates cell cycle and DNA damage checkpoints. Proc Natl Acad Sci U S A. 2012;109:197–202. doi: 10.1073/pnas.1104767109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Okita N, Kudo Y, Tanuma S. Checkpoint kinase 1 is cleaved in a caspase-dependent pathway during genotoxic stress-induced apoptosis. Biol Pharm Bull. 2007;30:359–62. doi: 10.1248/bpb.30.359. [DOI] [PubMed] [Google Scholar]
- 104.Matsuura K, Wakasugi M, Yamashita K, Matsunaga T. Cleavage-mediated activation of Chk1 during apoptosis. J Biol Chem. 2008;283:25485–91. doi: 10.1074/jbc.M803111200. [DOI] [PubMed] [Google Scholar]
- 105.Kaneko YS, Watanabe N, Morisaki H, Akita H, Fujimoto A, Tominaga K, Terasawa M, Tachibana A, Ikeda K, Nakanishi M. Cell-cycle-dependent and ATM-independent expression of human Chk1 kinase. Oncogene. 1999;18:3673–81. doi: 10.1038/sj.onc.1202706. [DOI] [PubMed] [Google Scholar]
- 106.Carrassa L, Broggini M, Vikhanskaya F, Damia G. Characterization of the 5’flanking region of the human Chk1 gene: identification of E2F1 functional sites. Cell Cycle. 2003;2:604–9. [PubMed] [Google Scholar]
- 107.Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–76. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guervilly JH, Renaud E, Takata M, Rosselli F. USP1 deubiquitinase maintains phosphorylated CHK1 by limiting its DDB1-dependent degradation. Hum Mol Genet. 2011;20:2171–81. doi: 10.1093/hmg/ddr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brazelle W, Kreahling JM, Gemmer J, Ma Y, Cress WD, Haura E, Altiok S. Histone deacetylase inhibitors downregulate checkpoint kinase 1 expression to induce cell death in non-small cell lung cancer cells. PLoS One. 2010;5:e14335. doi: 10.1371/journal.pone.0014335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim AJ, Kim HJ, Jee HJ, Song N, Kim M, Bae YS, Chung JH, Yun J. Glucose deprivation is associated with Chk1 degradation through the ubiquitin-proteasome pathway and effective checkpoint response to replication blocks. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamcr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arlander SJ, Eapen AK, Vroman BT, McDonald RJ, Toft DO, Karnitz LM. Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress. J Biol Chem. 2003;278:52572–7. doi: 10.1074/jbc.M309054200. [DOI] [PubMed] [Google Scholar]
- 112.Kim KS, Choi KJ, Bae S. Interferon-gamma enhances radiation-induced cell death via downregulation of Chk1. Cancer Biol Ther. 2012:13. doi: 10.4161/cbt.20990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc Natl Acad Sci U S A. 2005;102:15105–9. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 116.Li DQ, Ohshiro K, Khan MN, Kumar R. Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. J Biol Chem. 2010;285:19802–12. doi: 10.1074/jbc.M109.085258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 118.Zaugg K, Su YW, Reilly PT, Moolani Y, Cheung CC, Hakem R, Hirao A, Liu Q, Elledge SJ, Mak TW. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc Natl Acad Sci U S A. 2007;104:3805–10. doi: 10.1073/pnas.0611584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Greenow KR, Clarke AR, Jones RH. Chk1 deficiency in the mouse small intestine results in p53-independent crypt death and subsequent intestinal compensation. Oncogene. 2009;28:1443–53. doi: 10.1038/onc.2008.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boles NC, Peddibhotla S, Chen AJ, Goodell MA, Rosen JM. Chk1 haploinsufficiency results in anemia and defective erythropoiesis. PLoS One. 2010;5:e8581. doi: 10.1371/journal.pone.0008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.O’Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277:16102–15. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- 122.Kim MA, Kim HJ, Brown AL, Lee MY, Bae YS, Park JI, Kwak JY, Chung JH, Yun J. Identification of novel substrates for human checkpoint kinase Chk1 and Chk2 through genome-wide screening using a consensus Chk phosphorylation motif. Exp Mol Med. 2007;39:205–12. doi: 10.1038/emm.2007.23. [DOI] [PubMed] [Google Scholar]
- 123.Blasius M, Forment JV, Thakkar N, Wagner SA, Choudhary C, Jackson SP. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 125.Craig AL, Chrystal JA, Fraser JA, Sphyris N, Lin Y, Harrison BJ, Scott MT, Dornreiter I, Hupp TR. The MDM2 ubiquitination signal in the DNA-binding domain of p53 forms a docking site for calcium calmodulin kinase superfamily members. Mol Cell Biol. 2007;27:3542–55. doi: 10.1128/MCB.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ou YH, Chung PH, Sun TP, Shieh SY. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell. 2005;16:1684–95. doi: 10.1091/mbc.E04-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen MS, Ryan CE, Piwnica-Worms H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol. 2003;23:7488–97. doi: 10.1128/MCB.23.21.7488-7497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N. Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J. 2004;23:3386–96. doi: 10.1038/sj.emboj.7600328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schmitt E, Boutros R, Froment C, Monsarrat B, Ducommun B, Dozier C. CHK1 phosphorylates CDC25B during the cell cycle in the absence of DNA damage. J Cell Sci. 2006;119:4269–75. doi: 10.1242/jcs.03200. [DOI] [PubMed] [Google Scholar]
- 130.Melixetian M, Klein DK, Sorensen CS, Helin K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol. 2009;11:1247–53. doi: 10.1038/ncb1969. [DOI] [PubMed] [Google Scholar]
- 131.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–71. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 132.Wang H, Liu D, Wang Y, Qin J, Elledge SJ. Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes Dev. 2001;15:1361–72. doi: 10.1101/gad.893201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee J, Kumagai A, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol Biol Cell. 2001;12:551–63. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu C, Zhang S, Gao X, Xu X, Lv Y, Zhang Y, Zhu Z, Zhang C, Li Q, Wong J, Cui Y, Zhang W, et al. Roles of Kruppel-associated Box (KRAB)-associated Co-repressor KAP1 Ser-473 Phosphorylation in DNA Damage Response. J Biol Chem. 2012;287:18937–52. doi: 10.1074/jbc.M111.313262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Groth A, Lukas J, Nigg EA, Sillje HH, Wernstedt C, Bartek J, Hansen K. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J. 2003;22:1676–87. doi: 10.1093/emboj/cdg151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gonzalez S, Prives C, Cordon-Cardo C. p73alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23:8161–71. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 137.Kristensen LP, Larsen MR, Hojrup P, Issinger OG, Guerra B. Phosphorylation of the regulatory beta-subunit of protein kinase CK2 by checkpoint kinase Chk1: identification of the in vitro CK2beta phosphorylation site. FEBS Lett. 2004;569:217–23. doi: 10.1016/j.febslet.2004.05.069. [DOI] [PubMed] [Google Scholar]
- 138.Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, Halazonetis TD, Harris CC. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J Cell Biol. 2004;166:801–13. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kaur S, Modi P, Srivastava V, Mudgal R, Tikoo S, Arora P, Mohanty D, Sengupta S. Chk1-dependent constitutive phosphorylation of BLM helicase at serine 646 decreases after DNA damage. Mol Cancer Res. 2010;8:1234–47. doi: 10.1158/1541-7786.MCR-10-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hromas R, Williamson EA, Fnu S, Lee YJ, Park SJ, Beck BD, You JS, Laitao A, Nickoloff JA, Lee SH. Chk1 phosphorylation of Metnase enhances DNA repair but inhibits replication fork restart. Oncogene. 2012 doi: 10.1038/onc.2011.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rocha S, Garrett MD, Campbell KJ, Schumm K, Perkins ND. Regulation of NF-kappaB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. EMBO J. 2005;24:1157–69. doi: 10.1038/sj.emboj.7600608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schmitt AM, Crawley CD, Kang S, Raleigh DR, Yu X, Wahlstrom JS, Voce DJ, Darga TE, Weichselbaum RR, Yamini B. p50 (NF-kappaB1) is an effector protein in the cytotoxic response to DNA methylation damage. Mol Cell. 2011;44:785–96. doi: 10.1016/j.molcel.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207–18. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27:3098–108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhi G, Wilson JB, Chen X, Krause DS, Xiao Y, Jones NJ, Kupfer GM. Fanconi anemia complementation group FANCD2 protein serine 331 phosphorylation is important for fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 2009;69:8775–83. doi: 10.1158/0008-5472.CAN-09-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Clarke CA, Clarke PR. DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem J. 2005;388:705–12. doi: 10.1042/BJ20041966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kasahara K, Goto H, Enomoto M, Tomono Y, Kiyono T, Inagaki M. 14-3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010;29:2802–12. doi: 10.1038/emboj.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Garate M, Campos EI, Bush JA, Xiao H, Li G. Phosphorylation of the tumor suppressor p33(ING1b) at Ser-126 influences its protein stability and proliferation of melanoma cells. FASEB J. 2007;21:3705–16. doi: 10.1096/fj.07-8069com. [DOI] [PubMed] [Google Scholar]
- 149.Petsalaki E, Akoumianaki T, Black EJ, Gillespie DA, Zachos G. Phosphorylation at serine 331 is required for Aurora B activation. J Cell Biol. 2011;195:449–66. doi: 10.1083/jcb.201104023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen G, Yang L, Li B, Huang P, Chen D, Liang Y, Zhang R, et al. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J Clin Invest. 2012 doi: 10.1172/JCI61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ullah Z, de Renty C, DePamphilis ML. Checkpoint kinase 1 prevents cell cycle exit linked to terminal cell differentiation. Mol Cell Biol. 2011;31:4129–43. doi: 10.1128/MCB.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–49. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- 153.Meng Z, Capalbo L, Glover DM, Dunphy WG. Role for casein kinase 1 in the phosphorylation of Claspin on critical residues necessary for the activation of Chk1. Mol Biol Cell. 2011;22:2834–47. doi: 10.1091/mbc.E11-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Galvan V, Kurakin AV, Bredesen DE. Interaction of checkpoint kinase 1 and the X-linked inhibitor of apoptosis during mitosis. FEBS Lett. 2004;558:57–62. doi: 10.1016/S0014-5793(03)01488-1. [DOI] [PubMed] [Google Scholar]
- 155.Han EK, Butler C, Zhang H, Severin JM, Qin W, Holzman TF, Gubbins EJ, Simmer RL, Rosenberg S, Giranda VL, Ng SC, Luo Y. Chkl binds and phosphorylates BAD protein. Anticancer Res. 2004;24:3907–10. [PubMed] [Google Scholar]
- 156.Liu HY, Nefsky BS, Walworth NC. The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J Biol Chem. 2002;277:2637–43. doi: 10.1074/jbc.M109016200. [DOI] [PubMed] [Google Scholar]
- 157.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–74. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, Jeggo PA, O’Driscoll M. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat Cell Biol. 2006;8:725–33. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 159.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2’-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen S, Maya-Mendoza A, Zeng K, Tang CW, Sims PF, Loric J, Jackson DA. Interaction with checkpoint kinase 1 modulates the recruitment of nucleophosmin to chromatin. J Proteome Res. 2009;8:4693–704. doi: 10.1021/pr900396d. [DOI] [PubMed] [Google Scholar]
- 161.Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway. J Biol Chem. 2010;285:5974–82. doi: 10.1074/jbc.M109.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kosoy A, Calonge TM, Outwin EA, O’Connell MJ. Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem. 2007;282:20388–94. doi: 10.1074/jbc.M702652200. [DOI] [PubMed] [Google Scholar]
- 163.Sun TP, Shieh SY. Human FEM1B is required for Rad9 recruitment and CHK1 activation in response to replication stress. Oncogene. 2009;28:1971–81. doi: 10.1038/onc.2009.58. [DOI] [PubMed] [Google Scholar]
- 164.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 165.Solyom S, Pylkas K, Winqvist R. Screening for large genomic rearrangements of the BRIP1 and CHK1 genes in Finnish breast cancer families. Fam Cancer. 2010;9:537–40. doi: 10.1007/s10689-010-9360-7. [DOI] [PubMed] [Google Scholar]
- 166.Tort F, Hernandez S, Bea S, Camacho E, Fernandez V, Esteller M, Fraga MF, Burek C, Rosenwald A, Hernandez L, Campo E. Checkpoint kinase 1 (CHK1) protein and mRNA expression is downregulated in aggressive variants of human lymphoid neoplasms. Leukemia. 2005;19:112–7. doi: 10.1038/sj.leu.2403571. [DOI] [PubMed] [Google Scholar]
- 167.Bertoni F, Codegoni AM, Furlan D, Tibiletti MG, Capella C, Broggini M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer. 1999;26:176–80. [PubMed] [Google Scholar]
- 168.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–30. [PubMed] [Google Scholar]
- 169.Kim CJ, Lee JH, Song JW, Cho YG, Kim SY, Nam SW, Yoo NJ, Park WS, Lee JY. Chk1 frameshift mutation in sporadic and hereditary non-polyposis colorectal cancers with microsatellite instability. Eur J Surg Oncol. 2007;33:580–5. doi: 10.1016/j.ejso.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 170.Fishler T, Li YY, Wang RH, Kim HS, Sengupta K, Vassilopoulos A, Lahusen T, Xu X, Lee MH, Liu Q, Elledge SJ, Ried T, et al. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of Chk1 and p53. Oncogene. 2010;29:4007–17. doi: 10.1038/onc.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tho LM, Libertini S, Rampling R, Sansom O, Gillespie DA. Chk1 is essential for chemical carcinogen-induced mouse skin tumorigenesis. Oncogene. 2012;31:1366–75. doi: 10.1038/onc.2011.326. [DOI] [PubMed] [Google Scholar]
- 172.Kawasumi M, Lemos B, Bradner JE, Thibodeau R, Kim YS, Schmidt M, Higgins E, Koo SW, Angle-Zahn A, Chen A, Levine D, Nguyen L, et al. Protection from UV-induced skin carcinogenesis by genetic inhibition of the ataxia telangiectasia and Rad3-related (ATR) kinase. Proc Natl Acad Sci U S A. 2011;108:13716–21. doi: 10.1073/pnas.1111378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–9. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- 174.Madoz-Gurpide J, Canamero M, Sanchez L, Solano J, Alfonso P, Casal JI. A proteomics analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics. 2007;6:2150–64. doi: 10.1074/mcp.M700006-MCP200. [DOI] [PubMed] [Google Scholar]
- 175.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, De Wolf-Peeters C, Christiaens MR, Michiels L, Bouillon R, Verstuyf A. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67:6574–81. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 176.Yao H, Yang Z, Li Y. Expression of checkpoint kinase 1 and polo-like kinase 1 and its clinicopathological significance in benign and malignant lesions of the stomach. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2010;35:1080–4. doi: 10.3969/j.issn.1672-7347.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 177.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C, Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2012 doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]
- 178.Sriuranpong V, Mutirangura A, Gillespie JW, Patel V, Amornphimoltham P, Molinolo AA, Kerekhanjanarong V, Supanakorn S, Supiyaphun P, Rangdaeng S, Voravud N, Gutkind JS. Global gene expression profile of nasopharyngeal carcinoma by laser capture microdissection and complementary DNA microarrays. Clin Cancer Res. 2004;10:4944–58. doi: 10.1158/1078-0432.CCR-03-0757. [DOI] [PubMed] [Google Scholar]
- 179.Lundgren K, Holm K, Nordenskjold B, Borg A, Landberg G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10:R81. doi: 10.1186/bcr2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–61. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Perego P, Gatti L, Righetti SC, Beretta GL, Carenini N, Corna E, Dal Bo L, Tinelli S, Colangelo D, Leone R, Apostoli P, Lombardi L, et al. Development of resistance to a trinuclear platinum complex in ovarian carcinoma cells. Int J Cancer. 2003;105:617–24. doi: 10.1002/ijc.11140. [DOI] [PubMed] [Google Scholar]
- 182.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 183.Cavelier C, Didier C, Prade N, Mansat-De Mas V, Manenti S, Recher C, Demur C, Ducommun B. Constitutive activation of the DNA damage signaling pathway in acute myeloid leukemia with complex karyotype: potential importance for checkpoint targeting therapy. Cancer Res. 2009;69:8652–61. doi: 10.1158/0008-5472.CAN-09-0939. [DOI] [PubMed] [Google Scholar]
- 184.Lee JH, Choy ML, Ngo L, Venta-Perez G, Marks PA. Role of checkpoint kinase 1 (Chk1) in the mechanisms of resistance to histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2011;108:19629–34. doi: 10.1073/pnas.1117544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bartucci M, Svensson S, Romania P, Dattilo R, Patrizii M, Signore M, Navarra S, Lotti F, Biffoni M, Pilozzi E, Duranti E, Martinelli S, et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ. 2012;19:768–78. doi: 10.1038/cdd.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang X, Ma Z, Xiao Z, Liu H, Dou Z, Feng X, Shi H. Chk1 knockdown confers radiosensitization in prostate cancer stem cells. Oncol Rep. 2012;28:2247–54. doi: 10.3892/or.2012.2068. [DOI] [PubMed] [Google Scholar]
- 187.Seol HJ, Yoo HY, Jin J, Joo KM, Kim HS, Yoon SJ, Choi SH, Kim Y, Pyo HR, Lim DH, Kim W, Um HD, et al. The expression of DNA damage checkpoint proteins and prognostic implication in metastatic brain tumors. Oncol Res. 2011;19:381–90. doi: 10.3727/096504011x13123323849654. [DOI] [PubMed] [Google Scholar]
- 188.Roe OD, Szulkin A, Anderssen E, Flatberg A, Sandeck H, Amundsen T, Erlandsen SE, Dobra K, Sundstrom SH. Molecular resistance fingerprint of pemetrexed and platinum in a long-term survivor of mesothelioma. PLoS One. 2012;7:e40521. doi: 10.1371/journal.pone.0040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Arora S, Bisanz KM, Peralta LA, Basu GD, Choudhary A, Tibes R, Azorsa DO. RNAi screening of the kinome identifies modulators of cisplatin response in ovarian cancer cells. Gynecol Oncol. 2010;118:220–7. doi: 10.1016/j.ygyno.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 190.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, Diskin SJ, Attiyeh EF, Sennett R, Norris G, Laudenslager M, Wood AC, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci U S A. 2011;108:3336–41. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bennett CN, Tomlinson CC, Michalowski AM, Chu IM, Luger D, Mittereder LR, Aprelikova O, Shou J, Piwinica-Worms H, Caplen NJ, Hollingshead MG, Green JE. Cross-species genomic and functional analyses identify a combination therapy using a CHK1 inhibitor and a ribonucleotide reductase inhibitor to treat triple-negative breast cancer. Breast Cancer Res. 2012;14:R109. doi: 10.1186/bcr3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20:7453–63. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 193.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A, Aft RL, Ellis MJ, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122:1541–52. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2010;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Carrassa L, Damia G. Unleashing Chk1 in cancer therapy. Cell Cycle. 2011;10:2121–8. doi: 10.4161/cc.10.13.16398. [DOI] [PubMed] [Google Scholar]
- 197.Maugeri-Sacca M, Bartucci M, De Maria R. Checkpoint kinase 1 inhibitors for potentiating systemic anticancer therapy. Cancer Treat Rev. 2012 doi: 10.1016/j.ctrv.2012.10.007. [DOI] [PubMed] [Google Scholar]