Abstract
The transcription factor CREB is an important regulator of many adaptive processes in neurons, including sleep, cellular homeostasis, and memory formation. The Drosophila dCREB2 family includes multiple protein isoforms generated from a single gene. Overexpression of an activator or blocker isoform has been shown to enhance or block memory formation, but the molecular mechanisms underlying these phenomena remain unclear. In the present study, we generate isoform-specific antibodies and new transgenic flies to track and manipulate the activity of different dCREB2 isoforms during memory formation. We find that nuclear accumulation of a dCREB2 activator-related species, p35+, is dynamically regulated during memory formation. Furthermore, various dCREB2 genetic manipulations that enhance or block memory formation correspondingly increase or decrease p35+ levels in the nucleus. Finally, we show that overexpression of S6K can enhance memory formation and increase p35+ nuclear abundance. Taken together, these results suggest that regulation of dCREB2 localization may be a key molecular convergence point in the coordinated host of events that lead to memory formation.
Introduction
The cAMP Response Element Binding Protein (CREB) is a critical transcriptional regulator of numerous adaptive brain functions, including memory formation (Frank et al., 1994; Carlezon et al., 2005; Alberini 2009). Mammals have three genes in the CREB/CREM/ATF-1 superfamily, and numerous molecular mechanisms are used to produce different isoforms off of these genes. Drosophila has a single bZIP-encoding gene (dCREB2) that contains the well-conserved kinase inducible domain (KID or P-Box). However, this single gene produces multiple protein isoforms. Two of these isoforms, activator and blocker, have been shown to enhance or suppress memory, respectively, when overexpressed (Yin et al., 1994; Yin et al., 1995a; Tubon et al. 2013). While these isoforms have been shown to interact in vitro and in transfected cells (Yin et al., 1995b), the specific endogenous isoforms and molecular mechanisms involved during memory formation remain unclear.
Mammalian CREB is phosphorylated in response to a variety of cellular stimuli, and phosphorylation is a key regulatory step in the activation of the protein (Kornhauser et al., 2002). In the baseline state, phosphorylation is very low, and external stimuli activate signaling pathways (eg. CaM kinase, PKA, MAPK) that phosphorylate serine 133 (Parker et al., 1996). The dCREB2 equivalent of serine 133 (serine 231), however, is highly phosphorylated at baseline (Horiuchi et al., 2004). Although there is evidence for regulatory phosphorylation at other sites of dCREB2 (Horiuchi et al., 2004), it is likely that the rate-limiting step in the activation of dCREB2 resides elsewhere. The dCREB2 gene is more similar in its overall properties to the mammalian CREM gene than it is to CREB (Yin et al., 1995b). CREM makes a large variety of protein isoforms, many of which display subcellular localization outside of the nucleus (Lalli et al., 1996). It is likely that CREM blocker isoforms limit nuclear entry of the activators, or facilitate their nuclear export (Fenaroli et al., 2004).
In addition to post-translational modifications on CREB itself, the activity of co-activators such as CREB Binding Protein (CBP) and Transducers of Regulated CREB activity (TORC or CRTC) influence mammalian CREB activity (Kwok et al., 1994; McManus & Hendzel, 2001; Altarejos & Montminy, 2011). Hippocampal CBP knock-out mice retain activity-dependent changes in CREB phosphorylation yet display deficits in CREB-dependent transcription and long-term memory (Barrett et al., 2011). TORCs are dynamically shuttled into the nucleus in an activity-dependent manner (Bittinger et al., 2004; Ch’ng et al, 2012) and perturbing TORC activity in the rodent hippocampus affects long-term memory (Sekeres et al., 2012). These co-activators directly interact and influence CREB activity, and are important in relaying stimulus information into the nucleus. The Drosophila homologs dCBP and dTORC have been shown to participate in different types of memory formation (Hirano et al., 2013). However, it is unclear if additional regulatory mechanisms exist to control dCREB2 activity.
To understand the regulation of dCREB2-dependent transcription during memory formation, we created new transgenic flies to manipulate dCREB2 activity. New antibodies were raised against predicted dCREB2 isoforms and employed to examine the subcellular localization of dCREB2 isoforms in the Drosophila brain. Unlike mammalian CREB, but similar to mammalian CREM, the majority of the dCREB2 protein exists outside the nucleus. Behavioral training that produces long-term memory increases the levels of an activator-related protein species in the nucleus. This behavior-dependent increase in the nuclear activator protein is necessary for memory formation, and cellular signaling involving the S6K kinase can influence distribution of this protein. Our findings suggest that dynamic regulation of dCREB2 nuclear entry, influenced by different molecular pathways, is a necessary convergence point for the control of dCREB2 transcription during memory formation.
2. Materials and Methods
2.1. Animals
Fly stocks were maintained at 21°C on standard food supplemented with yeast paste. Experimental animals were collected 1–3 days post eclosion, housed at 100 flies per vial, and entrained to a 12:12 light:dark schedule for 3–5 days before behavioral or molecular experiments.
2.2. Drosophila Genetics
Wild-type flies used in expression experiments after memory formation were the w iso(CJ1) strain, known colloquially as 2U. UAS-S6K and hs-Gal4 flies were crossed together, and the doubly transgenic progeny were used for inducible S6K experiments. dCREB2 manipulations utilized the previously generated transgenic fly lines: hs-807 (activator), hs-17–2 (blocker), and hs-581 (mutant blocker with two point mutations in the dimerization domain). New transgenic flies used the heat shock promoter to drive expression of: blocker with exons four and/or six of the dCREB2 gene, blocker with the nuclear localization signal mutated (936), and the bZIP only domain of dCREB2 (568). Flies receiving heat shock induction of transgenes were placed at 37°C for 30 minutes and allowed to recover at 21°C for the specified amount of time before molecular or behavioral assays.
2.3. Generation of new DNA constructs and flies
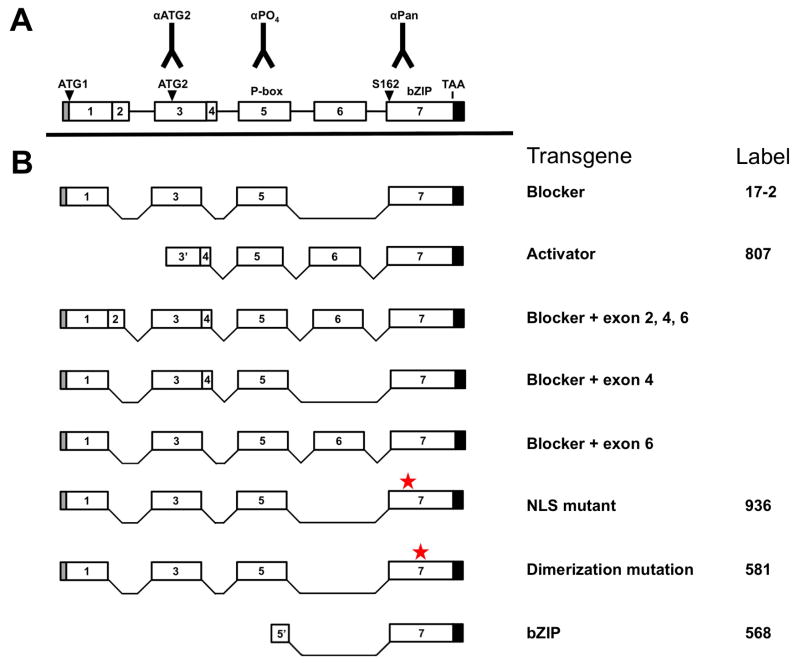
All of the open reading frames (ORFs) shown in Figure 1 were used to make DNA constructs and transgenic flies. PCR amplification was used to add restriction sites onto the ends of the ORFs, and these were subcloned into the CaSpeR-hs vector, oriented and sequenced. Oligonucleotide-mediated, site-directed mutagenesis was used to create the two amino acid substitutions in the nuclear localization signal (RRKKK changed to RAKAK) of the blocker isoform, creating 936. The 568 construct and transgenic flies make a protein that begins downstream of S231 and contains a truncated exon 5, and all of exon 7. Therefore, it codes for a protein that essentially contains only the bZIP domain.
Figure 1. Schematic of dCREB2 gene, transgenic animals, and new antibodies.
(A) Schematic of the dCREB2 gene showing all seven exons (numbered 1–7), peptide locations for newly generated antibodies, and the various functional regions of the proteins (such as the phosphorylation box [P-Box or KID], the bZIP domain, the site of the S162 mutation, and the two translational start sites [ATG1 and ATG2]). (B) Schematic of different transgenic flies used in this report. The blocker backbone is comprised of exons 1, 3, 5, and 7. Two transgenic lines (936 and 581) involve mutations in the blocker backbone, with red stars indicating the approximate sites of the mutations. Three other lines contain additional exons (2, 4 and/or 6) in addition to the blocker backbone. Exons 3′ through 7, beginning at ATG2, constitute the Activator. The bZIP transgene (568) encodes a small part of exon 5′ and all of exon 7 (which contains the bZIP domain).
2.4. Behavioral Training
Flies were trained in the olfactory avoidance-training paradigm developed by Tully and Quinn and modified to allow for automated training sessions. A single-cycle of training consists of 90 sec exposure to ambient air; 60 sec, 70V pulses of electric shock (the unconditioned stimulus) lasting 1.5 sec and administered every 5 sec (12 total) accompanied by simultaneous exposure to 1 odor (the conditioned stimulus condition, CS+); 45 seconds of ambient air exposure to clear the first odor; 60 seconds of exposure to the second odor, with no shock (the CS− condition), and 45 sec of ambient air to clear the second odor. Testing was done by placing flies in a choice point for 2 minutes and allowing them to choose between the CS+ and CS− stimuli. Spaced training consists of 10 single cycles separated by 15-min rest intervals. We used 3-octanol and 4-methylcyclohexanol as odors. The performance index=[the number of flies making the correct choice] − [the number of flies making the incorrect choice]/total number of flies, multiplied by 100. To avoid odor-avoidance biases, we calculate the performance index of each single N by taking an average performance of 2 groups of flies, 1 trained with 3-octanol as CS+, the other with 4-methylcyclohexanol.
2.5. dCREB2 antibodies
The antibodies used in this study and their epitopes are shown in Figure 1. The αPan, αPO4, and αATG2 antibodies are all polyclonal antibodies raised in rabbits. The αPan antibody was raised against a sequence of amino acids in the basic region of dCREB2, and affinity purified using the antigenic peptide linked to a column. The αPO4-specific antibody was raised against a peptide that contains the S231 residue in a phosphorylated state. Serum was passed over a peptide column containing the unphosphorylated peptide, and the flow through fraction was bound and eluted from the phospho-peptide column. The αATG2 antibody has been described previously (Tubon et al. 2013). In brief, it was raised against a peptide that is just C-terminal to the ATG2 codon. A second peptide that contains residues N- and C-terminal to ATG2 was used to “subtract” antibodies from whole sera raised against the first peptide. This procedure should eliminate most antibodies whose epitopes contain amino acids upstream of ATG2. This second peptide was attached to an affinity column, and the flow through from this column was bound to an affinity column made from the first peptide. The bound fraction was eluted and designated αATG2. The monoclonal antibody has been described previously (Belvin et al., 1999). In Figure 5B, the commercially available (Santa Cruz), mammalian CREB antibody (αC21) was used. Its validity in flies has been previously demonstrated (Tubon et al. 2013).
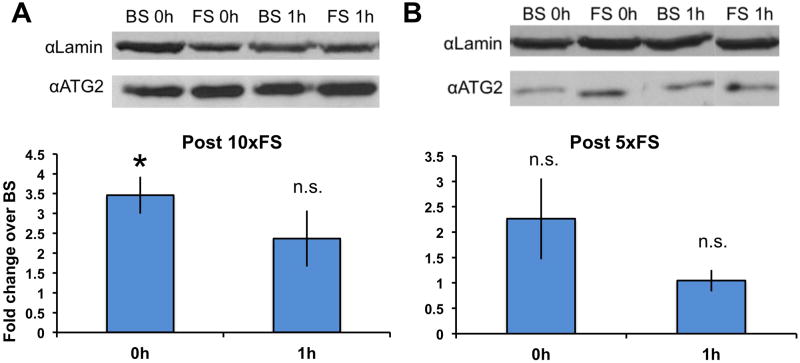
Figure 5. Memory formation increases nuclear abundance of a 35+ kD isoform in wild-type flies.
Western analysis of head extracts from behaviorally trained flies probed with αATG2. Groups of flies were trained with forward-paired, spaced (FS) or backwards-paired spaced (BS) training. Representative images and quantification of nuclear proteins is shown for flies that received 10× (A) or 5× (B) Spaced training trials. Quantification shows p35+ bands normalized to lamin and displays fold change of FS over BS Training. The data are presented as experimental means, and the error bars represent s.e.m. Comparisons are made using a two sample ta-test. Significant differences between groups are indicated as *p<0.05. 10xFS Training produces a significant 3.5-fold increase over BS training immediately after training (n=3, p<0.05), with a 2-fold increase that shows a trend towards significance (n=4, p=0.07) one hour after training. 5xFS Training results in a trend towards significance immediately after training (n=4, p=0.1), with no difference detected at one hour after training (n=4, p=0.4).
2.6. Tissue Preparation
2.6.1. Tissue preparation for Western blot
Flies were flash-frozen in liquid nitrogen and heads were separated using sieves on dry ice. Heads were homogenized on ice for 30 seconds in a homogenization buffer containing 15 mM Hepes, pH 7.5, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail tablets (Roche cOmplete ULTRA Tablets, Mini, EDTA-free, EASYpack 05 892 791 001). Debris was spun down at 200g and the supernatant was used for experiments. Protein concentrations were determined used a Bradford assay and concentrations were normalized across samples in an experiment. Samples were mixed with 2× protein loading buffer and boiled for 5 minutes before loaded into an SDS-PAGE gel or being stored at −80°C for later use.
2.6.2. Subcellular fractionation by differential centrifugation
As with regular tissue preparation, flies were flash frozen and heads were separated on dry ice. Samples were homogenized on ice 3 times, for 30 seconds, in a homogenization buffer containing 15 mM Hepes, pH 7.5, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail tablets (Roche cOmplete ULTRA Tablets, Mini, EDTA-free, EASYpack 05 892 791 001). Three debris spins were performed at 200g for five minutes and supernatant was collected for the subsequent spin. Nuclei were collected three times by centrifuging samples at 1000g for five minutes and combining pellets. Nuclear pellets were washed once with homogenization buffer and re-spun for purer nuclear samples. Cytoplasmic protein consisted of the supernatant following the third nuclear spin. Protein concentrations were determined using a Bradford assay and normalized across samples in an experiment. Samples were mixed with a 2× loading buffer and boiled before loading into an SDS-PAGE gel or storing at −80°C for future use.
2.7. Western blotting
Samples were resolved with 10% SDS-PAGE and transferred to a Whatman Protran nitrocellulose membrane using electrophoresis. Membranes were rinsed with TBST and blocked for 30 minutes at room temperature with milk (5% milk in 1xTBST). Membranes were incubated overnight at 4°C with primary antibodies in milk. The following noncommercial CREB antibodies were used: αPan-CREB (1:5000), αATG2 (1:1000), and αPO4-CREB (1:1000). A Lamin antibody (Drosophila Studies Hybridoma Bank, ADL67.10) was used at 1:5000. The mammalian CREB antibody, αC21, was used in Figure 5B (1:1000). Following incubation in primary antibody, membranes were rinsed in TBST 4×15 minutes, blocked in milk, and incubated in secondary antibody (Jackson ImmunoResearch laboratories, Peroxidase AffiniPure Goat Anti-Rabbit IgG 111-035-003 or Goat Anti-Mouse 111-030-003) for 1 hour at room temperature. After secondary antibody incubation, membranes were rinsed in TBST 4×15 minutes then incubated for 4 minutes in Pierce Western Blotting Substrate ECL reagent (Prod #32106). Membranes were exposed to film (Denville Scientific Premium Radiography Film, Cat #E3018) for time periods ranging from 10 seconds to 2 hours prior to developing the film.
Images of film were scanned and digitally saved using HP Scanner, version 2.4.4 (3), and analyzed using ImageJ software. For post-memory formation quantification, dCREB2 bands were quantified and normalized to Lamin bands, of the same sample, from the linear range. Lamin-normalized values from Forward Spaced (FS) trained animals were then normalized to samples from Backward Spaced animals in the same experiment so that values could be compared across experiments.
2.8. Statistical analysis
Data are presented as mean ± SEM. All statistical tests are performed using R (R Development Core Team, 2012) software. For all assessments, the alpha value was set to 0.05. To analyze Western blots, the signal from each experimental band was normalized to the signal from a loading control. Samples from Forward-Spaced and Backward-Spaced animals were then normalized to samples from Backward-Spaced animals so that values can be compared across experiments. The differences of the means of these normalized values were then assessed using two sample t tests. For all behavioral experiments, two-sample t-tests were performed to provide between-subjects comparisons of heat-shock and non heat-shock treated animals of the same genotype.
3. Results
3.1 Validation of the new dCREB2 antibodies
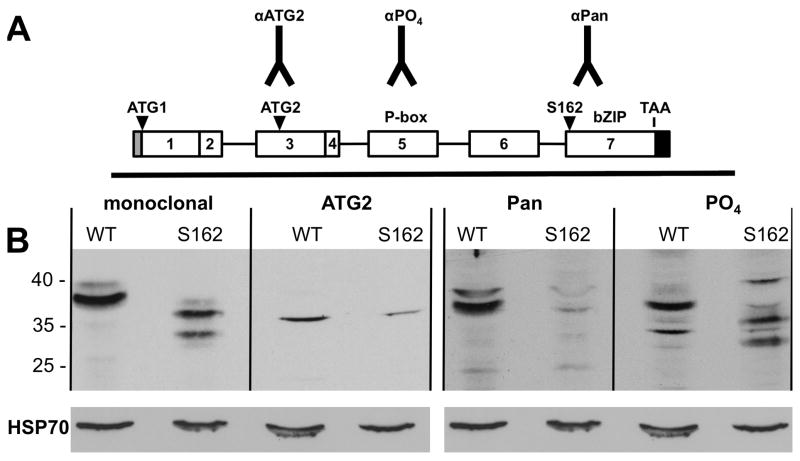
We utilized the S162 mutant fly, which contains a glutamine-to-stop codon single base substitution in the dCREB2 gene (Hendricks et al. 2001) to validate our new antibodies. Figure 1 shows the location of the S162 mutation on the schematic map of the dCREB2 locus. The S162 mutation is located late in the dCREB2 open reading frame, truncating about 60 amino acids from any protein isoform that contains the C-terminal bZIP domain. In Figure 2, we use wild type and S162 head extracts to evaluate the specificity of our new antibodies. These comparisons are semi-quantitative, since we use HSP70 immunoreactivity to demonstrate equal loading in each sample. The monoclonal antibody detects a set of truncated protein bands very similar to what was previously reported, except the current gel does not resolve the bands into doublets (Belvin et al. 1999). The αPO4 antibody detects a very similar set of bands, and this pattern is consistent with the location of the stop codon, deleting amino acids from the C-terminal end. Therefore, these antibodies show specificity for dCREB2-encoded proteins. The αPan antibody does not recognize any protein bands in the S162 extract with high affinity. This is the expected result since the antigenic peptide used to raise this antibody is “downstream” of the stop codon in S162 and should be missing from all of its truncated proteins. The αATG2 antibody weakly recognizes an S162 band that seems slightly slower in its mobility than the wild type band. In summary, the monoclonal, αPan and αPO4 antibodies are clearly dCREB2-specific. While the αATG2 antibody shows altered immunoreactivity between wild type and S162 mutant flies, the evidence for dCREB2-specificity of the αATG2 antibody is less strong. However, further comparisons (discussed below) make it very likely that αATG2 is also dCREB2-specific.
Figure 2. dCREB2 antibodies recognize altered protein isoforms in S162 mutant flies.
(A) Schematic of the dCREB2 gene showing peptide locations for newly generated antibodies and functional regions of the gene, including the site of the S162 mutation. (B) Western analysis of extracts from wild type (2U) or S162 mutant flies using a previously generated monoclonal antibody (left panel), the αATG2 antibody (left middle panel), the αPan antibody (right middle panel), and the αPO4 antibody (right panel). Blots were also probed with the HSP70 to indicate relative protein levels in each lane.
3.2. dCREB2 antibodies detect endogenous and overexpressed dCREB2 proteins
To complement our analysis with mutant S162 proteins, we used our new antibodies to assess whether they detect overexpressed dCREB2 proteins. The lower part of Figure 1 shows a series of DNA constructs used to make transgenic flies, and their structure, description and nomenclature is described. All of the constructs were placed under the control of the heat-shock promoter, and transgenic flies (with the corresponding names) were generated. Some of these transgenic flies (17-2, 581 [known as A2-2] and 807) have been published previously (Yin et al., 1994, Yin et al., 1995a, Yin et al., 1995b; Tubon et al., 2013), while the remaining ones are new. The simplest functional categorization of these constructs is between activators (807), blockers (17-2, 568), and mutant blockers (936 and 581). The 936 construct and flies contain two amino acid substitutions (RRKKK to RAKAK) in the conserved nuclear localization signal (NLS) of all CREB proteins, including dCREB2 (Waeber and Habener, 1991). These mutations have been shown to block nuclear entry when the protein is expressed in mammalian cells. The previously described 581 (A2-2) construct and flies contains two substitutions in the leucine zipper, which prevents it from forming homo- or heterodimeric species (Dwarki et al., 1990). The relative location of the NLS and leucine zipper mutations are indicated with red stars.
Head extracts from induced and non-induced flies were used to test the newly generated dCREB2 antibodies, and the results are shown in Figure 3. The uninduced samples show what endogenous protein bands are detected, while the induced samples reveal detectability of overexpressed proteins. The αPan-antibody, which was raised using residues in the basic region, recognizes a 38/40 kD doublet that has been previously described. This doublet is also detectable in most of the extracts probed with the αPO4 antibody. All antibodies detected induction of blocker- or modified blocker-containing transgenes, but the details differed. For some transgenes (blocker, 581 and 936), both bands in the doublet increased after transgene induction (columns 4–6). For others, only the upper (blocker + exon4) or lower (blocker + exon6) band increased (columns 1 and 2). When exon 2 is included in addition to exons 4 and 6, the induced protein migrates above the doublet band (column 3). The one-to-one correspondence between these coding exons and their resultant bands remains somewhat unclear. Induction of the 568 transgene produces a 10 kD protein which is only detectable using the pan-antibody (column 7). The most complex pattern of bands occurs when the 807 transgene is induced, and multiple antibodies detect a suite of bands that correspond to 15, 24, 28 and 35+ kD (column 8). Whether some of these bands result from differences in coding capacity, antibody affinity for specific proteins, post-translational modifications and antibody affinity, or even compensatory synthesis off of the endogenous dCREB2 gene, is not clear. However, the simple generalizations are that blocker- and mutant blocker-containing transgenes induce proteins at or near the 38/40 kD doublet bands, that 568 inducibly makes a very small 10 kD protein, and that the 807 transgene produces a suite of bands that migrate between 15–35+ kD. Behavioral experiments are consistent with the 24, 28, 32 and 35+ kD bands corresponding to differentially modified activator proteins (see Tubon et al., 2013). In summary, all of our antibodies detect endogenous and overexpressed proteins, and many of these bands are common to more than one antibody. However, there are unique bands that are only detected with a particular antibody.
Figure 3. dCREB2 antibodies recognize different protein isoforms after transgene induction.
Each of the columns (1–8) of the Figure (with its description) corresponds to a specific transgenic fly that encodes a different protein isoform. These flies were heat-shock induced (+), or not (−), and total head extracts were made and analyzed using western blots probed with different antibodies, which are shown on the left (in rows). The apparent mobilities of different protein species are shown relative to the mobility of molecular weight markers. The induction of various blocker-derived transgenes (columns 1–6) show increases in species that migrate around 38/40 kDa, and these are best detected using the αPO4 and αPan antibodies. Induction of the 568 transgene produces a p10 species that is only detected by the αPan antibody (column 7). Induction of the activator (807) transgene shows increases in multiple protein species, differentially detected by various antibodies, which migrate from 15 to 35 kD (column 8).
3.3. dCREB2 isoforms differ in subcellular localization
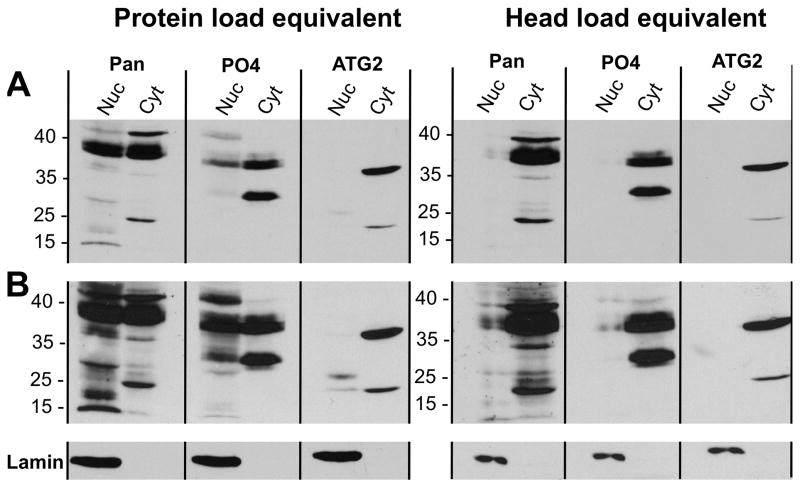
Differential centrifugation was used to fractionate wild type head extracts into crude, nuclear- and cytoplasmic-enriched samples. The new antibodies were then employed in western blot analyses to examine the subcellular localization of different proteins, and the data are shown in Figure 4. In the left panels, head extracts were loaded onto gels normalized across samples for total protein content. The images in the right panels were from gels loaded with the same number of head-equivalents in each lane. Figure 4A (the top panels) show the short exposure of the western blot, while Figure 4B shows the images from a longer exposure of the same blots. Lamin immunoreactivity was used to demonstrate the validity of our nuclear fractions.
Figure 4. The endogenous dCREB2 protein isoforms are preferentially localized in the cytoplasm.
Western analysis of fractionated protein from wild type, head nuclear (Nuc) and cytoplasmic (Cyt) extracts. Samples were probed using the new antibodies. The relative mobilities of protein markers are indicated. (A) Short exposure of samples loaded with either protein (left) or head (right) equivalents across lanes. (B) Long exposure of the same blots.
As expected, each antibody displayed preferential affinity to different protein isoforms within the samples. The αPan antibody shows strongest affinity to the 38/40 kD species and a 25 kD species in the cytoplasm, and numerous bands in the nucleus. While the αPO4 antibody also strongly detects the 38/40 kD bands, it recognizes an additional 30 kD band in the cytoplasm. In contrast, the αATG2 antibody most strongly detects both a 35+ and 25 kD species in the cytoplasm. Next, samples were loaded based on head equivalents to visualize relative amounts of protein in each sub-compartment. Unlike reports in the mammalian literature, the majority of all dCREB2 protein isoforms, regardless of molecular weight, reside in the cytoplasm compared to the nucleus. This distribution is clearly seen when equal head equivalents of extract are loaded and analyzed using all of the antibodies. Nuclear protein can be visualized when the nuclear fraction is overloaded relative to the cytoplasmic one (based on cellular or head equivalents). The relative enrichment in the cytoplasm is true for blocker-related protein isoforms (p38-p40 kD), as well as the cluster of activator-related species that range from p24-p35+ kD.
3.4. Memory formation increases nuclear abundance of the p35+ isoform
To determine what molecular changes occur to dCREB2 proteins in response to memory formation, animals were trained in a classical conditioning paradigm in which flies learn the association between an odor and a foot shock. Since the most robust form of this conditioning involves 10 (10x) spaced repetitions (trials) of the odor-shock pairing, an experimental and control version of this protocol was used. Some flies were exposed to Forward Spaced training (FS), where the presentation of odor and shock overlapped during each individual trial. Repetitive training using this type of pairing produces robust long-term memory (LTM) that lasts up to a week. Other flies were exposed to the same number of training trials, but ones where the odor and shock presentations did not overlap in time (Backward Spaced [BS] training). This type of training does not produce associative learning or memory of the stimuli.
Heads were collected immediately (0h) or one hour (1h) after 10xFS or 10xBS training, extracts prepared and fractionated using differential centrifugation. Samples were separated using SDS-PAGE and visualized using western blots probed with the αATG2 antibody to detect changes in activator nuclear abundance. Since dCREB2 functions in the nucleus, we chose to use the αATG2 antibody to measure changes in the nuclear abundance of the ATG2-initiated activator. This antibody has low background and preferentially recognizes the ATG2-initiated protein. Lamin, a nuclear marker, was used to validate the crude subcellular fractionation, and the results are presented in Figure 5.
After quantifying dCREB2 and Lamin bands within the linear range and normalizing dCREB2 to Lamin, we found that a 35+ kD species (p35+) has higher nuclear expression after FS training compared to BS training. To compare across experiments, the quantified FS values were normalized to BS values and the data were represented as the fold increase in p35+ when FS is compared to BS training. There is a significant 3-fold increase in nuclear expression of the p35+ immediately after 10xFS training (n=3, p<0.05), and a 2-fold increase that trended towards significance one hour after 10xFS training (n=4, p=0.07) (Figure 5A).
To assess whether weaker conditioning can also induce this increase in nuclear expression of the p35+, wild-type flies were trained with a 5xSpaced paradigm. 5xFS training can also induce LTM, but memory levels are lower and retention is shorter. Again, heads were collected immediately and one hour after 5xFS or 5xBS training. Immediately after 5xFS training we observed a trend toward an increase in nuclear expression of p35+ (n=4, p=0.1), but one hour after training any memory formation-induced increase was undetectable (n=4, p=0.4) (Figure 5B). The general specificity and validity of these changes were verified using identical samples probed with the αPO4 antibody (data not shown).
3.5. Transgenic dCREB2 manipulations affect nuclear abundance of the p35+ isoform
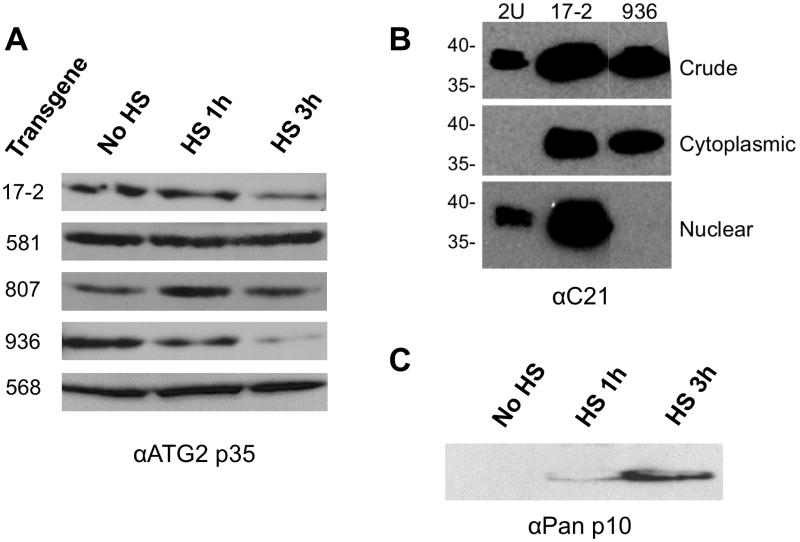
Previously we showed that induction of the blocker-encoding transgene interfered with long-term memory formation, and that induced overexpression of the activator transgene could enhance it (Yin et al. 1994, Yin et al. 1995a). In Figure 5 we demonstrate a correlation between long-term memory formation and an increase in p35+ nuclear abundance. Like all members of this protein family, dCREB2 isoforms physically interact through their leucine zipper domains. Using our new transgenic flies, we wanted to determine mechanisms that contribute to these causal and correlative findings. Induction of the activator transgene (807) increases levels of nuclear p35+, as detected by the αATG2 antibody, at one- and three-hours post-induction (Figure 6A). This result is expected, since transgenic overexpression should increase the amounts of an activator-related protein isoform in the nucleus. Conversely, induction of blocker protein (17-2) decreases nuclear levels of endogenous p35+ three hours after induction. Additionally, induction of a blocker protein that contains a mutation preventing dimerization (581) does not affect steady-state nuclear levels of p35+ at any time point (Figure 6A). Importantly, induction of a blocker protein containing a mutation in the nuclear localization signal (936) reduces the amount of p35+ in the nucleus at one and three hours after transgene induction. Finally, overexpression of the bZIP domain of dCREB2 (568) does not affect levels of nuclear p35+.
Figure 6. Induction of dCREB2-expressing transgenes can affect nuclear abundance of the 35+ kD activator isoform.
(A) Western analysis of total head proteins after transgene inductions. Various transgenes differ in their effects on αATG2-detected nuclear p35+. Induction of blocker- (17-2 and 936) and activator- (807) encoding transgenes decrease and increase nuclear p35+ abundance respectively, while induction of 581 and 568 do not affect its levels. (B) The C21 antibody was used to visualize protein localization of the blocker (17-2) and NLS mutant blocker (936) transgenic protein. The induction of 17-2 produces blocker protein that distributes to both the nuclear and cytoplasmic compartments, while the 936 transgene produces a mutant blocker protein that is cytoplasmic. (C) 568 induction produces a 10 kDa protein that is detected in the nucleus using the αPan antibody.
To determine the subcellular localization of the transgenic proteins, samples from induced flies were fractionated using differential centrifugation and analyzed using western blots (see Figures 6B–6C). The amount of total protein that was loaded for each compartment was adjusted to minimize detection of endogenous protein isoforms, thus allowing clear visualization of induced, overexpressed transgenic protein. However, the total amount of protein loaded on the gel for all genotypes within a given compartment was equivalent. As a control, wild type (2U) flies were induced even though they do not contain a heat-shock inducible transgene. For wild type flies (2U), the majority of the protein in the 38 kD doublet actually resides in the cytoplasm (see Figure 4A, head load equivalent). There is no signal in the wild type cytoplasmic lane in Figure 6B because the protein amount that was loaded was less than the amount that can be detected. When the 17-2 (blocker) transgene is induced, there is a large increase in the amounts of p38–40 in all compartments relative to the induced (but non-transgenic) wild type flies. This result shows that the induced blocker protein accumulates in both the cytoplasm and nucleus. In contrast, the 936 transgenic protein only accumulates in the cytoplasm. Interestingly, this cytoplasmic localization actually decreases the amount of nuclear, endogenous p38–p40, since those species decrease to an undetectable level. The 568 transgenic flies induce a protein that migrates with a mobility of approximately 10 kD (Figure 6C), which is found in mostly in the nuclear compartment.
3.6. dCREB2 manipulations that interfere with nuclear accumulation of p35+ also interfere with memory formation
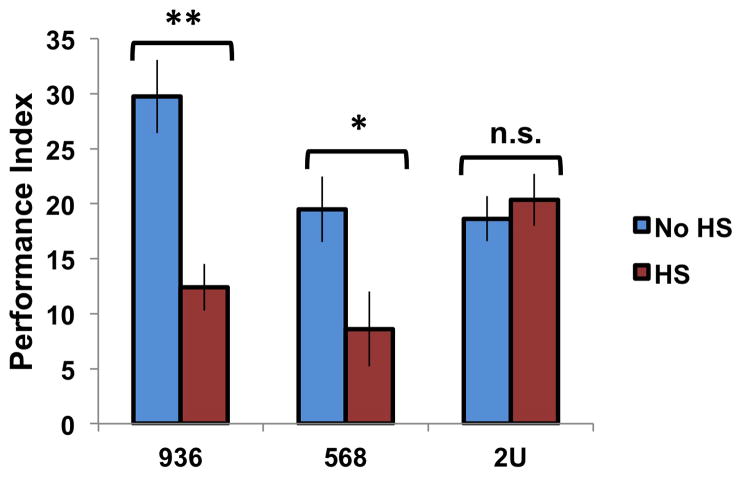
To determine the functional consequences of inducing the new transgenic lines, we assayed the effects of 936 and 568 induction on memory formation. Groups of flies from each genotype, as well as wild-type (2U) flies, were subjected to a 30-minute heat shock prior to 10xFS training and tested for 24-hour memory. Heat-shock induction of both dCREB2 transgenes resulted in a significant decrease in memory (n=10–12, p<0.0005 and p<0.005), while heat-shock induction itself did not affect the performance of wild-type flies (n=14, p=0.67) (Figure 7).
Figure 7. Transgenic dCREB2 manipulations impair long-term memory.
Three different groups of flies were exposed to heat-shock induction (red histograms) or not (blue histograms), behaviorally-trained, and tested for 24-hour memory. Heat-shock induction of the dCREB2 blocker with a mutation in the NLS (936) (n=12, p<0.0005) and induction of the bZIP domain of dCREB2 (568) (n=10, p<0.005) both impair long-term memory, while heat-shock does not impair memory in wild type (2U) flies (n=14, p=0.67). Data are presented as means, and the error bars are s.e.m. Comparisons were made using a two sample t-test. Significant differences between groups are indicated as *p<0.01 or **p<0.0005.
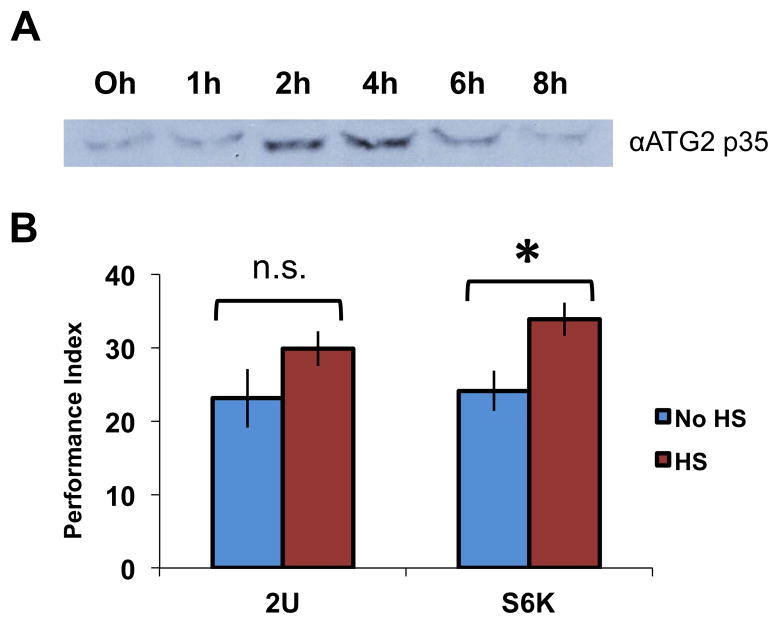
3.7 Overexpression of S6K increases nuclear abundance of p35 and enhances memory
After finding genetic manipulations that decrease p35+ nuclear levels and also decrease memory formation, we next asked if there might be a way to increase the amounts of its nuclear accumulation. We induced, or not, S6K flies that contained a heat-shock driven transgene, and collected fly heads at various time points. The 35+ kD dCREB2 isoform shows increased nuclear expression from 2–4 hours after S6K transgene induction (Figure 8A). Next, S6K induction was tested for effects on memory formation. Three hours after heat-shock induction of S6K, or a control treatment with no S6K induction, flies were subjected to 5× Spaced training in the olfactory associative paradigm. When tested 24 hours later, flies that received S6K overexpression exhibited significantly better memory than flies without S6K overexpression (n=8, p<0.01), while heat-shock treatment of wild-type flies did not affect memory (n=8, p=0.17) (Figure 8B). When the same samples were probed with the αPO4 antibody, an identical increase was detected in the nuclear compartment (data not shown). Thus both behavioral training and S6K induction increase nuclear p35+ detected using the αATG2 or αPO4 antibodies.
Figure 8. S6K induction induces nuclear translocation of dCREB2 activator and enhances memory.
(A) Heat-shock induction of S6K increases nuclear abundance of the 35+ kD dCREB2 species in a time window two to four hours after induction, as detected by the αATG2 antibody. (B) Wild type (2U) or S6K transgenic flies were exposed to heat-shock (red histograms) or not (blue histograms) three hours prior to 5 cycles of spaced training. Induction of S6K enhances 24-hour memory (n=8, p<0.01), while heat-shock has no affect on memory formation in wild type flies (n=8, p=0.17). The data are presented as means, and the error bars are s.e.m. Comparisons were made using a two sample t-test. Significant differences between groups are indicated as *p<0.01.
Conclusion/Discussion
To investigate the biological roles of specific dCREB2 protein isoforms, we raised new antibodies and made the transgenic flies depicted in Figure 1. Surprisingly, our antibodies showed that the majority of dCREB2 protein, regardless of structure and apparent mobility on protein gels, is localized in the cytoplasmic compartment. This finding is true for both the blocker and activator isoforms. The blocker primarily consists of the “backbone” structure (exons 1, 3, 5 and 7 in Figure 1), which migrates as a doublet species around 38–40 kD. When the alternative exons (2, 4 and/or 6) are spliced into this backbone, the apparent mobility of the resulting proteins does not change dramatically (see Figure 3). The activator-related molecular species (p24, p28, p30, p35+, p60; see Figures 2 and 3 and Tubon et al. 2013) also are enriched in the cytoplasm, and this is most clearly demonstrated when nuclear and cytoplasmic samples are loaded on a per head-equivalent basis. All of the dCREB2 protein isoforms shown in Figure 1 contain the amino acid residues RRKKK in their basic regions, and this sequence has been shown to be necessary and sufficient to localize mammalian CREB proteins into the nucleus (Waeber and Habener, 1991). Since the mammalian CREB protein is mostly found in the nucleus, the cytoplasmic enrichment for dCREB2 protein is unexpected. This localization is maintained even when transgenes are induced and different dCREB2 protein isoforms are overexpressed (H. Zhou, data not shown). These data suggest that a significant fraction of dCREB2-encoded proteins are tethered in the cytoplasmic compartment through an unknown mechanism.
Since dCREB2 is a transcription factor, its primary function occurs in the nuclear compartment. We previously demonstrated that the majority of dCREB2 protein exists with serine 231 (the fly equivalent to the mammalian serine 133 residue) phosphorylated in the basal state (Horiuchi et al., 2004). This contrasts sharply with the mammalian CREB protein, which exists in the nucleus in a largely unphosphorylated state, awaiting activity-dependent signaling mechanisms that activate it. Taken together, the cytoplasmic localization of dCREB2 protein and the phosphorylation of serine 231 in the basal state suggest strongly that the rate-limiting step in activation of dCREB2 probably involves increasing the nuclear abundance of the activator species. This could at least partially occur through an increase in nuclear entry.
A number of results support the view that p35+ is an endogenous, dCREB2-specific protein product. Different dCREB2-specific antibodies recognize it on western blots. The αATG2, αPan, and αPO4 antibodies recognize an endogenous molecular species that migrates at that mobility (Figures 3–4). p35+ is decreased or missing when head extracts are made from the S162 mutant fly. When blocker-encoding transgenes are induced, they affect the nuclear levels of p35+ (Figure 6). Finally, when the 807 (activator-encoding) transgene is induced, p35+ increases (Figure 3). Although the 807-encoded protein is transgenic and not endogenous, the primary product has an apparent mobility of 24 kD, but when expressed in flies increases the levels of both p28 and p35+ bands.
Three independent lines of evidence support the interpretation that p35+ represents an activator-related species. First, there is a correlation between memory formation and nuclear abundance of p35+. Forward, but not backward, paired training produces long-term memory and results in a greater than 3-fold increase in nuclear p35+ (Figure 5A). Weaker training (5xS instead of 10xS), which produces less robust memory, produces a smaller, non-significant trend towards an increase (Figure 5B). Second, there is a strong correlation between memory formation and a qualitative increase in p35+ in the nucleus. Genetic manipulations of dCREB2 that enhance memory formation (807 induction) increase nuclear abundance of p35+, ones that interfere with long-term memory formation (induction of 17-2 and 936) prevent an increase, while a control manipulation (induction of 581) does not interfere with either memory formation or affect p35+ levels (Figure 5A, Figure 6). Third, manipulating a totally different protein (S6K) results in a parallel set of correlations. Increasing the amount of the S6K protein enhances memory formation and increases p35+ nuclear abundance (Figure 8).
What is the nature of the p35+ isoform? Mammalian CREB has been shown to be SUMOylated and o-glycosylated, and these modifications decrease the apparent mobility of the rsulting proteins on gels (Comerford et al., 2003; Rexach et al., 2012). We have evidence that dCREB2 protein can be o-glycosylated, and one of those species migrates around 35 kD (data not shown). Future experiments will be needed to determine if p35+ o-glycosylated, and how this would affect protein interactions, subcellular localization, or transcriptional activity.
Since a nuclear increase in p35+ correlates so well with memory formation, we believe that p35+ (or its predecessor species p24, p28 and/or p30) enter the nucleus from the cytoplasm, where it is tethered in a heterodimer with other dCREB2 proteins. Our transgenic experiments support this view. The 807 transgene makes p24/p28, overexpression of which increases nuclear abundance of p35+ and can enhance memory formation (Figure 3 and Tubon et al. 2013). The 936 transgene makes a protein that differs in two of the five residues that constitute the nuclear localization signal, which renders the induced protein exclusively cytoplasmic (Figure 6B). Induction of 936 decreases nuclear p35+ levels, and blocks memory formation. Taken together, these data are consistent with the view that 936 blocks memory formation through a cytoplasmic tethering mechanism, where the overexpressed protein binds to, and prevents the nuclear entry of, the endogenous activator species. The induced 17-2 protein is able to form heterodimers with the activator (Yin et al., 1995b), but is less efficient at staying in the cytoplasm (Figure 5B). However, it is still able to interfere with memory formation, since a heterodimeric species (activator:17-2) is less transcriptionally potent (Loriaux et al., 1994). The 17-2 and 581 transgenes differ in two leucine residues in the leucine zipper (Yin et al., 1995b). These amino acid changes are known to disrupt the ability of the mutant (581) protein to form dimers. Therefore, the overexpressed 581 protein cannot form a dimer with the activator, does not prevent a nuclear increase in p35+ after training, and does not affect memory formation. Finally, the overexpressed 568 protein is mostly in the nucleus, and we speculate that it can outcompete activator-containing dimers for binding to CRE sites. Therefore, it blocks memory formation but does not decrease nuclear p35+ abundance. This mechanism of action is identical to the actions of the ICER and S-CREM blockers (Lalli et al., 1996).
Although the data from the transgenic experiments supports the hypothesis that nuclear entry of the activator is rate-limiting for transcription and memory formation, we cannot exclude other possibilities. For example, it is possible that there exists a constant rate of activator import and export from the nucleus, and that memory formation transiently decreases export, thus increasing nuclear abundance. Another possibility is that behavioral training could transiently increase the nuclear stability of the activator protein. More detailed experimentation would be needed to determine the relative contribution of each of these possible mechanisms. However, our transgenic experiments argue that regulating nuclear import of the activator is at least partially involved in controlling the strength and duration of dCREB2-responsive transcription.
We have shown that inducing S6K stimulates nuclear abundance of p35+ and enhances memory formation. These data are the first demonstration that S6K overexpression can enhance long-term memory. Although our correlation between memory formation and an increase in the nuclear abundance of p35+ is clear, we do not know about the underlying molecular mechanisms. Behavioral training could stimulate translation of the activator protein or decrease its turnover. In addition, training might stimulate nuclear entry and/or decrease nuclear export of the activator protein. Similarly, it is not clear if induction of S6K stimulates translation (as this pathway is known to do), affects nuclear entry/export, or modulates protein stability. The kinetics of the increase in dCREB2 protein in both the nuclear and cytoplasmic compartments cannot distinguish among these possibilities. However, taken together with our transgenic experiments, the simplest interpretation is that behavioral training and S6K at least affect nuclear entry of an activator isoform.
One question that arises from our data is why the different antibodies do not recognize a more common pattern of bands. There are nine different dCREB2 protein isoforms reported in the literature (Yin et al., 1995; PubMed; Tubon et al. 2013). Six out of the nine isoforms contain the bZIP region, while the remaining three use alternative splice sites that result in termination of translation prior to the bZIP region. One of these six bZIP-containing isoforms utilizes an early, in-frame ATG codon, and is an activator (Tubon et al., 2013). In addition, we have strong evidence (Figure 4B; data not shown) that internal translation initiation can occur at one or more of the three late, in-frame ATG codons just upstream of the bZIP (near the C-terminus of the protein). Therefore, our current estimate is that there are at least six to nine (6+1, 6+2 or 6+3) distinct coding isoforms made off of this gene that would contain the bZIP region.
There are seven known, identified phosphorylation sites that are conserved between the mammalian CREB and dCREB2 proteins (see Altarejos and Montminy, 2011 and compare to Yin et al., 1995). In addition, mammalian CREB has been shown to be acetylated, SUMOylated, o-glycosylated, and subject to oxidation-reduction responsive modifications. Almost all of these sites that have been identified on the mammalian protein are conserved on the fly molecules. The possible combinations between protein coding capacity and modification state is very large. For reasons that are not clear, each of seven antibodies that we have made, and one mammalian one that we have used extensively, recognizes some common bands, and some unique ones (see Tubon et al., 2013 for a comparison of some of the other antibodies and the bands they recognize). We believe that the combinatorial, [coding+post-translational state] of the protein affects antibody affinity, but the bases for these differences are unclear. However, over the years, our general impression is that certain antibodies recognize certain subsets of bands, and that comparisons need to be made across all of the antibodies to validate specific bands as belonging to dCREB2. Therefore, it is likely that the total number of bands that can be resolved on protein gels is extremely large, and somewhat limited by the availability and relative affinity of each individual antibody.
In this report, we have demonstrated that dCREB2 protein species are enriched in the cytoplasmic compartment. However, at least one species, p35+, increases in nuclear abundance in response to behavioral signals and S6K induction. Moreover, genetic manipulations that block nuclear entry of p35+ also block memory formation. The correlation between behavioral training that produces long-term memory and an increase in p35+ nuclear abundance is consistent with the possibility that nuclear entry is the rate-limiting step for dCREB2-responsive transcription.
Highlights.
Most dCREB2 protein is cytoplasmic.
Behavioral training (in a forward-pairing-dependent manner) increases the nuclear abundance of a a dCREB2-specific protein (p35+) which is probably an activator.
Induction of the S6K gene enhances memory formation, and increases the nuclear abundance of p35+.
Transgenic dCREB2 manipulations that increase nuclear p35+ abundance correlate with memory enhancement.
Transgenic dCREB2 manipulations that decrease nuclear p35+ abundance correlate with memory disruption.
Transgenic dCREB2 manipulations that do not affect nuclear p35+ abundance do not affect memory formation.
Part of the mechanism that regulates dCREB2 activation, and memory formation, probably involves “blocking” isoforms of dCREB2 “tethering” the activator in the cytoplasm.
Acknowledgments
This work was supported through NIH funding to the PI (RO1s NS35575, HL/AR59649, NS063245 and funding from the McKnight Foundation). Additional funding support was provided through the UW-Madison Institute of Aging and the Neuroscience Training Program. Hong Zhou provided important technical assistance and advice. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Robin Fropf, Email: rafrench@wisc.edu.
Thomas C. Tubon, Jr., Email: tubon@madisoncollege.edu.
Jerry C. P. Yin, Email: jcyin@wisc.edu.
References
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiological Reviews. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos YA, Montminy MR. CREB and the CRTC co-activators: sensors for hormonal and metabolic stress. Nature Reviews Molecular Cell Biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36:1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JCP. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–87. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Activation of the cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Current Biology. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Carlezon WA, Duman RS, Nestler EJ. The many faces of CREB. Trends in Neuroscience. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Ch’ng TH, Uzgil B, Lin P, Avliyakulov NK, O’Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan RP, Taylor CT. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A. 2003;100:986–91. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwarki VJ, Montminy M, Verma IM. Both the basic region and the ‘leucine zipper’ domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 1990;9:225–32. doi: 10.1002/j.1460-2075.1990.tb08099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaroli A, Vujanac M, De Cesare D, Zimarino V. A small-scale survey identifies selective and quantitative nucleo-cytoplasmic shuttling of a subset of CREB transcription factors. Exp Cell Res. 2004;299(1):209–226. doi: 10.1016/j.yexcr.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Frank DA, Greenberg ME. CREB: a mediator of long-term memory from mollusks to mammals. Cell. 1994;79:5–8. doi: 10.1016/0092-8674(94)90394-8. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JCP, Sehgal A. A non-cirdadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Masuda T, Naganos S, Matsuno M, Ueno K, Miyashita T, Horiuchi J, Saitoe M. Fasting launches CRTC to facilitate long-term memory formation in Drosophila. Science. 2013;339:443–446. doi: 10.1126/science.1227170. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Jiang W, Zhou H, Wu P, Yin JC. Phosphorylation of conserved casein kinase sites regulates cAMP-reponse element-binding protein DNA binding in Drosophila. Journal of Biological Chemistry. 2004;279:12117–12125. doi: 10.1074/jbc.M212839200. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Cowan CW, Shaywitz AJ, Dolmetsch RE, Griffith EC, Hu LS, Haddad C, Xia Z, Greenberg ME. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proceedings of the National Academy of Sciences. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bächinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lalli E, Lee JS, Lamas M, Tamai K, Zazopoulos E, Nantel F, Penna L, Foulkes NS, Sassone-Corsi P. The nuclear response to cAMP: role of transcription factor CREM. Philos Trans R Soc Lond B Biol Sci. 1996;351:201–9. doi: 10.1098/rstb.1996.0017. [DOI] [PubMed] [Google Scholar]
- Loriaux MM, Brennan RG, Goodman RH. Modulatory function of CREB;CREMα heterodimers depends upon CREMα phosophorylation. Journal of Biological Chemistry. 1994;269:28839–43. [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. CBP, a transcriptional coactivator and acetyltransferase. Biochemistry and Cell Biology. 2001;79:253–266. [PubMed] [Google Scholar]
- Parker D, Ferreri K, Nakajima T, LaMorte VJ, Evans R, Koerber SC, Hoeger C, Montminy MR. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Molecular and Cellular Biology. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing [Computer software manual] Vienna, Austria: 2012. Retrieved from http://www.R-project.org/ [Google Scholar]
- Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, Hsieh-Wilson LC. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nature Chemical Biology. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekeres MJ, Mercaldo V, Richards B, Sargin D, Mahadevan V, Woodin MA, Frankland PW, Josselyn SA. Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising quality. Journal of Neuroscience. 2012;32:17857–68. doi: 10.1523/JNEUROSCI.1419-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubon TC, Jr, Zhang J, Friedman EL, Jin H, Gonzales ED, Zhou H, Drier D, Gerstner JR, Paulson EA, Fropf R, Yin JC. dCREB2-mediated enhancement of memory formation. Journal of Neuroscience. 2013;33(17):7475–87. doi: 10.1523/JNEUROSCI.4387-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waeber G, Habener JF. Nuclear translocation and DNA recognition signals colocalized within the bZIP domain of cyclic adenosine 3′, 5′-monophosphate response element-binding protein CREB. Mol Endocrinol. 1991;5:1431–8. doi: 10.1210/mend-5-10-1431. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995a;81:107–15. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, Quinn WG. A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Molecular Cellular Biology. 1995b;15:5123–5130. doi: 10.1128/mcb.15.9.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]