Abstract
Background
Acetabularia acetabulum is a giant unicellular green alga whose size and complex life cycle make it an attractive model for understanding morphogenesis and subcellular compartmentalization. The life cycle of this marine unicell is composed of several developmental phases. Juvenile and adult phases are temporally sequential but physiologically and morphologically distinct. To identify genes specific to juvenile and adult phases, we created two subtracted cDNA libraries, one adult-specific and one juvenile-specific, and analyzed 941 randomly chosen ESTs from them.
Results
Clustering analysis suggests virtually no overlap between the two libraries. Preliminary expression data also suggests that we were successful at isolating transcripts differentially expressed between the two developmental phases and that many transcripts are specific to one phase or the other. Comparison of our EST sequences against publicly available sequence databases indicates that ESTs from the adult and the juvenile libraries partition into different functional classes. Three conserved sequence elements were common to several of the ESTs and were also found within the genomic sequence of the carbonic anhydrase1 gene from A. acetabulum. To date, these conserved elements are specific to A. acetabulum.
Conclusions
Our data provide strong evidence that adult and juvenile phases in A. acetabulum vary significantly in gene expression. We discuss their possible roles in cell growth and morphogenesis as well as in phase change. We also discuss the potential role of the conserved elements found within the EST sequences in post-transcriptional regulation, particularly mRNA localization and/or stability.
Background
High-throughput sequencing of partial cDNAs, or expressed sequence tags (ESTs), provides relatively fast and cost-effective access to the gene expression profile of an organism [1,2]. EST libraries provide access to the population of genes transcribed, making analyses of ESTs informative in determining which genes are expressed at specific developmental ages, in specific tissues, or under specific environmental conditions.
EST analyses are especially useful when studying organisms for which little sequence data exists and for which sequencing of the genome is either not planned, or not easily feasible due to genome size. To date, there is little genomic data available for the Chlorophytes (green algae), a group far more diverse and evolutionarily divergent than all land plants combined. From this group, only Chlamydomonas reinhardtii has been the object of an extensive EST project [3,4]. Genomic information from this project proved critical to elucidating the function, biosynthesis, and regulation of the photosynthetic apparatus [4].
Acetabularia acetabulum (Fig. 1), also known as the "Mermaid's Wineglass", is a giant unicellular green alga whose size and complex life cycle make it an attractive model system for understanding morphogenesis and subcellular localization [5]. Reaching 3 cm in height at maturity, this unicell contains just a single diploid nucleus for most of its life cycle. It undergoes a complex morphogenetic program, most of which takes place at the apex [6], centimeters away from the nucleus. Classic experiments on A. acetabulum [7,8] provided the first compelling evidence for the role of the nucleus in morphogenesis and for the existence of "products of the nucleus", later presumed to be mRNAs [9].
Figure 1.

Juvenile, adult and reproductive morphologies of Acetabularia acetabulum. This giant alga has a complex life cycle and undergoes distinct developmental phases. From a spherical microscopic zygote, it initiates polarized growth elongating primarily at the tip (or apex) and periodically forming whorls of branched hairs. The reproductive phase starts as the unicell initiates a terminal apical whorl or "cap". When mature, the cap will house gametangia in which gametes form. The thallus and the diploid nucleus are drawn to scale. The number and complexity of the whorls of hairs was reduced for the sake of clarity.
The life cycle of A. acetabulum is composed of several developmental phases (Fig. 1). Like multicellular land plants, juvenile and adult phases of A. acetabulum are temporally sequential, but morphologically distinct [10]. Juvenile phase comprises the first centimeter of growth while adult phase comprises the remaining 2 to 3 cm [10]. Juvenile whorls of hairs are stacked closer to each other along the stalk, and the branching pattern of the hairs within each whorl is simpler than in adults [10]. Physiologically, these two phases differ as well. For example, juveniles grow well in crowded conditions and poorly at low population densities, while adults grow well only at low population densities. Similar to land plants, the transition between phases is associated with a change in the reproductive competence of the apex [11,12]. In A. acetabulum, adult apices are competent to produce a terminal reproductive whorl, the cap, while juvenile apices are not (J Messmer and DF Mandoli, unpublished). At the molecular level however, the difference is gene expression patterns between adult and juvenile phases are virtually unknown.
To reveal differences in gene expression between adult and juvenile phases, we constructed two subtracted EST libraries from A. acetabulum. These libraries were designed to contain transcripts specific to one phase or the other, presumably enriched in transcripts involved in morphogenesis or phase change. We randomly sequenced and analyzed 941 ESTs from these two libraries. Our analyses of these sequences indicate that juvenile and adult phases differ significantly in their gene expression patterns. We also identified 3 consensus sequences, shared mainly by adult ESTs, that have identity with introns and the 3'UTR from carbonic anhydrase genes we previously cloned [13]. We discuss the potential role of these conserved elements in mRNA post-transcriptional regulation, particularly mRNA localization and/or stability.
Results
General characterization of the ESTs
Suppressive subtractive hybridization, or SSH [14], results in the isolation and amplification of mRNAs present in one population (the tester population) and absent in the other (the driver population). Using SSH, we created two subtracted libraries, one putatively enriched in juvenile-specific transcripts and one putatively enriched in adult-specific transcripts (Additional file 1). From now on, we will refer to these libraries as the "juvenile library" and the "adult library", respectively.
To test the differential expression of the ESTs, 96 clones from each library were randomly chosen and spotted in the same pattern onto two nylon membranes. Each replicate membrane was hybridized with one of two probes, created either from adult or juvenile mRNA samples (Fig. 2). Out of the 96 randomly-chosen, putative juvenile clones, 53 were only expressed in juveniles, 13 were expressed at a higher level in juveniles than in adults, 5 were expressed at similar levels in adult and juveniles and 25 did not generate any signal with either probe (Fig 2., top panels). Out of the 96 randomly-chosen, putative adult clones, 44 were expressed only in adults, 14 were expressed at a higher level in adults than in juveniles, 10 were expressed at similar levels in adults and juveniles, 5 were expressed at a higher level or only in juveniles and 23 did not generate any signal with either probe (Fig 2., bottom panels). In addition, differential expression of three clones was confirmed by virtual northern blots (data not shown). Virtual Northern blots differ from Northern blots in that phase-specific cDNA is blotted on the nylon membranes instead of mRNA [15]. These data provide solid preliminary evidence that the SSH was successful at isolating many transcripts differentially expressed in these two phases.
Figure 2.
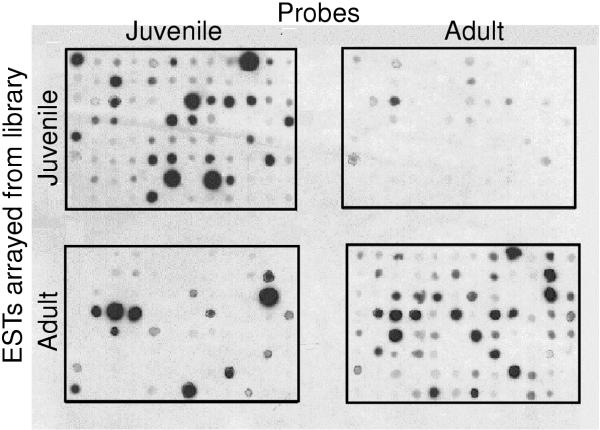
Dot blot analysis of the level of expression randomly chosen ESTs. 96 clones randomly chosen from the juvenile library and 96 clones randomly chosen from the adult library were spotted onto nylon membranes and the membranes were probed with either a "juvenile" probe (created from mRNA isolated from juveniles) or an "adult" probe (created from mRNA isolated from adults).
In total, 604 and 601 ESTs were sequenced from the adult and juvenile libraries respectively. Sequences containing no insert or unreliable data (as evidenced by the sequence trace) were excluded, leaving 478 ESTs from the adult library and 463 ESTs from the juvenile library for further analysis. Sequences were cleaned in silico of contaminating fragments (vector and primer sequences; see Materials and Methods). For 87% (411) of the adult clones and 83% (392) of the juvenile clones, this single-pass sequencing provided the complete sequence of the insert, i.e. vector sequence bordered both ends of the insert. ESTs ranged from 68 bps to 855 bps in length. On average, juvenile clones were longer than adult clones, averaging 474 bps and 408 bps respectively.
Due to the way the libraries were created (Additional file 2), some ESTs in the final library contained either a polyA or polyT tract [16]. These tracts originated from the polyA tail of the corresponding original mRNAs, indicating that these ESTs probably contained untranslated regions. Because the ESTs were not cloned directionally, sequences containing polyA or polyT tracts were obtained according to which strand was sequenced. The occurrence of these tracts was significantly higher in the juvenile than in the adult library (40 vs 15% respectively).
Redundancy and overlap between the two libraries
All ESTs were aligned and partitioned into clusters (see Material and Methods for details). All ESTs that were not part of a cluster remained singletons (279 adult and 233 juvenile ESTs). A consensus sequence was derived from clusters containing two or more ESTs. The juvenile ESTs formed 77 clusters and the adult ESTs formed 84 clusters. Clusters contained up to 14 ESTs (Fig. 3). This probably over-estimates the true number of clusters, as non-overlapping ESTs would be placed into two or more separate clusters or remain singletons even if they originated from the same initial mRNA. In addition, it is possible that sequences that only differed because of sequencing errors or regions of poor sequence quality were not clustered together.
Figure 3.

Number of ESTs in each cluster class. A cluster class included all the clusters that contain the same number of ESTs. Most ESTs remained singletons (class 1). The largest cluster contains 14 ESTs (class 14). INSET: Redundancy of and overlap between the two libraries. ''Adult clusters'' contained only adult ESTs, ''juvenile clusters'' contained only juvenile ESTs and ''mixed clusters'' contained at least one adult and one juvenile EST. The majority of the ESTs remained singletons.
Clusters containing ESTs from both libraries were labeled as "mixed clusters". Only 2 such mixed clusters were found, representing a mere 0.3% of the total number of clusters (Fig. 3). Thus, the overlap between the two libraries is minimal, providing additional evidence that SSH probably successfully isolated ESTs specific to each developmental phase.
Gene functions of the ESTs
All ESTs or cluster sequences were analyzed for homology using BLASTN, TBLASTX and InterPro (see Materials and Methods). Hits with E values that were <1.00E-06 for BLASTN and TBLASTX searches were considered significant. In total, ESTs representing only 162 clusters or ESTs produced significant BLAST hits, 144 of which were associated with a putative gene function (Table 1). 45% of these putative functions were independently confirmed by InterPro searches. Interestingly, the only two mixed clusters were both associated with the large subunit ribosomal RNA (rrnL) gene, the only chloroplast encoded gene found in our analyses.
Table 1.
Number of clusters or singletons that produced significant BLAST hits in different database searches.
| Significant hit against | # clusters or singletons |
| All three databases | 108 |
| Genbank1 and Arabidopsis2 | 19 |
| Genbank1and Chlamydomonas3 | 9 |
| Arabidopsis2 and Chlamydomonas3 | 2 |
| Genbank1 only | 26 |
| Arabidopsis2 only | 1 |
| Chlamydomonas3 only | 13 |
| No significant hit | 497 |
| Total | 675 |
All singletons and clusters were also analyzed for homologies using BLASTX against the Arabidopsis thaliana protein database [17] and using TBLASTX against the current draft of the Chlamydomonas reinhardtii genome [18]. For both searches, hits with E values < 10E-06 were considered significant. In general, the same sequences produced significant hits against each of the different databases (Table 1). This independently confirmed the results of the first searches and the low percentage of coding sequences within our ESTs. In total, 178 ESTs (only 26%) produced significant hits in at least one of the BLAST searches (Table 1).
The 178 ESTs or clusters that produced significant hits were sorted into functional categories according to the classification scheme developed for plants [19] (Fig. 4). The largest functional class from both libraries contained genes associated with photosynthesis ("energy" in Fig. 4). In general, a higher percentage of juvenile ESTs have functions related to transcription, and protein synthesis, transport and storage while a higher percentage of adult ESTs have functions related to cell structure (Fig. 4 and Additional file 3).
Figure 4.
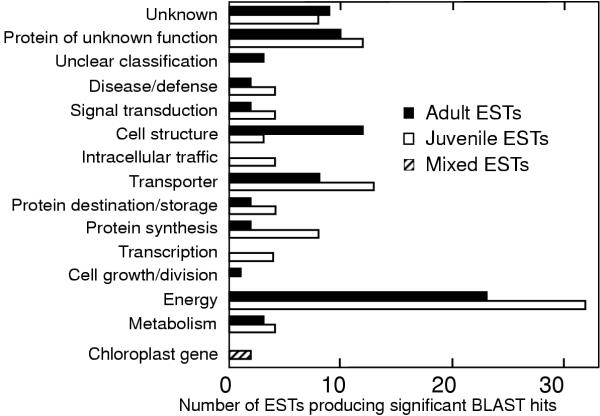
Classification of the ESTs according to their putative function. Those juvenile and adult ESTs whose function could be predicted based on searches of public databases were classified according to those putative functions. Only two ESTs were found in both the adult and the juvenile libraries. These ESTs are labeled "mixed".
Phylogenetic analysis of the ESTs
To assess if the putative functions of the ESTs also occurred in land plants or in other green algae, we identified (for each of the 178 ESTs that generated significant BLAST hits) the sequence giving the lowest E value ("best match") in the BLAST searches and the organism to which this sequence belongs. As expected, most of these "best-match" sequences belong either to Chlamydomonas reinhardtii or the Streptophyta (land plants). This is not surprising, as the number of sequences available for most green algal lineages remains extremely limited. Specifically, A. acetabulum belongs to the class Ulvophyceae for which very little sequence information is available [20].
Conserved sequences within the ESTs
Some ESTs showed similarity to each other over short regions. These ESTs clustered into three groups of 5, 9 and 2 sequences respectively (Fig. 5). The length of the common stretch of sequence varies between 30–70 bps for group 1, 45–90 bps for group 3 and 170 to 250 bps for group 2 (Fig. 5). Within each group, these ESTs showed no similarity to each other outside of these regions but within these regions, the level of identity was high. Most of these ESTs belonged to the adult library. None of these 16 ESTs produced any relevant BLAST hit, making it difficult to predict whether or not they contain coding sequences. Among the sequences sharing the second consensus sequence (Fig. 5c), all but two of the ESTs ended with a polyA or polyT tract, indicating that they probably contain 3' UTRs. None of the ESTs sharing the first or third consensus sequences (Fig. 5b or 5d) ended with a polyA or polyT tract. These 3 conserved elements may be specific to A. acetabulum, because they were not found in any other sequence in Genbank (nucleotide database) except for the carbonic anhydrase 1 and 2 (CA1 and CA2) genes from A. acetabulum [13]. All three of these conserved sequences fell in the non-coding regions of the two CA genes, either in introns or the 3'UTRs (Fig. 5a).
Figure 5.

Conserved sequences among independent ESTs and their position relative to the A. acetabulum carbonic anhydrase (AaCA1) genomic sequence. a: Structure of the AaCA1 genomic sequence. Positions in the intron and 3'UTR where sequence was omitted are indicated by slanted, heavy double lines. The white boxes represent AaCA1 exons, the hatched box represents the 3' UTRs. The black boxes represent three regions of strong homology between different EST sequences and the AaCA1 gene and the AaCA2 mRNA. b, c and d: sequence alignments of the AaCA1 gene and different ESTs (singletons or clusters) over these three conserved regions. The name of the EST or gene from which each sequence originates is indicated in front of the sequence. The regions of the AaCA1 and AaCA2 genes shown in the alignment are indicated in parentheses. * indicates a consensus in at least 70% of the sequences aligned. - indicates a gap in the alignment. The presence of a polyA/T tract at the end of an EST is indicated by "(A/T)" in front of the EST name.
Discussion
Adult and juvenile phases in A. acetabulum differ significantly in gene expression
The expression analysis (Fig. 2) and the fact that there is virtually no overlap between the two libraries suggest that the subtraction succeeded in isolating differentially expressed transcripts.
The 941 ESTs were organized into 675 independent clusters or singletons. Although this number is probably an overestimate – non-overlapping ESTs originating from the same transcript may partition to different clusters or remain singletons – these ESTs only represent a portion of all the ESTs present in the libraries. These data provide strong evidence that the adult and juvenile phases in A. acetabulum differ significantly in gene expression and that a large number of genes are probably phase-specific.
Physiological differences between the two developmental phases
The functions of the transcripts expressed at the different phases partition differently into functional classes (Fig. 4). Given that the libraries were created such that only ESTs specific to one developmental phase would be isolated, the distribution of gene functions among the ESTs is not expected to reflect that of a typical photosynthetic cell, but merely the functions that are specific to one developmental phase or the other.
Juveniles seem to devote much of their unique gene expression to transcription, protein synthesis, transport and storage, consistent with the general idea that juveniles are fast growing, more dedicated to growth than morphogenesis. During juvenile phase, the unicells increase about 10-fold in height (from <1 mm to 1 cm) in 2 weeks and are not competent to make a cap. On the other hand, adults increase in height about 3-fold, another 3 centimeters. Adult cells are much more complex than juvenile cells, with numerous whorls of hair that are highly branched. There is thus an increased requirement for cell wall synthesis during the adult phase. Adults also are competent to execute complex cap morphogenesis, and are preparing for nuclear divisions, nuclei transport and gametogenesis [21]. There is not a large increase in cytoplasmic volume between adult and juveniles because most of the volume within an adult is occupied by a central vacuole (Ngo et al., submitted). This more complex adult development is consistent with a lower percentage of transcripts dedicated to protein synthesis and a higher percentage of transcripts involved in cell structure (e.g. cytoskeletal proteins, enzymes involved in cell wall synthesis or maintenance, histones).
A surprisingly high number of ESTs from both libraries are associated with photosynthesis (Additional file 3, class 2). For example, five ESTs were putative homologs of the rbcS protein. It is possible that these ESTs truly come from different transcripts. It is also possible that they originate from the same transcript but have not been clustered together because they do not overlap or because of regions of poor sequence quality. To address this question, we aligned these sequences to the A. acetabulum rbcS mRNA sequence present in Genbank (Fig. 6). We observed in each EST or cluster, a region of high sequence identity to the Aa-rbcS sequence (identity varied between 84 and 89% at the nucleotide level). Two of the clusters (cn115 and cn116) were identical except for sequence ambiguities, which were frequent enough for these two sequences not to be clustered together. With the exception of cn116 and cn115, the other sequences were different enough from each other and from the Aa-rbcS sequence to conclude that they did not actually originate from the same transcript. In support of this hypothesis, regions outside of these fragments of very high sequence identity could not be aligned (<50% identity). At the amino acid level, the sequence identity between these regions (white boxes in Fig. 6) and the Aa-rbcS sequence was almost complete, varying between 94 and 96%. A closer look at the sequences confirmed that most of the nucleotide differences present in the ORFs occurred at the third position of a codon, often resulting in the conservation of the amino acid sequence. Finally, one of the clusters (cn115) ends with a polyA tract, indicating the end of the 3'UTR, while the transcript that generated cluster 146 seems to possess a much longer 3'UTR that contains no polyA tract. Taken together, these results suggest that there may be at least 3 different loci coding for the rbcS protein in Acetabularia (cn169, J538 and Aa-rbcS, Fig 6). Confirmation of these surprising results with different methods and detailed expression analysis of the different loci will be of great interest. A similar analysis of the chlorophyll a/b binding proteins might also determine whether different proteins are expressed during different phases and whether they are functionally distinct and/or differentially regulated. The LHC (light-harvesting complex) binding proteins form a very large family that has been best characterized in land plants [22]. Analysis of the 22 ESTs with identity to LHC binding proteins from land plants will improve our currently poor knowledge of this gene family in algae.
Figure 6.
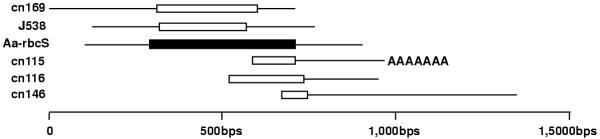
Alignment of the putative rbcS clusters and ESTs with the A. acetabulum rbcS sequence from Genbank. The 5 ESTs or clusters that showed homology to the rbcS gene were aligned to the A. acetabulum rbcS mRNA sequence (Aa-rbcS) published in Genbank (sequence id: X51815). The black box represents the ORF of the Aa-rbcS sequence. The white boxes represent, for each EST or cluster, regions of high identity with the Aa-rbcS sequence. The percentage of identity in these regions varied between 84% and 89%. The rest of the sequences (black line) showed little or no identity (<50%) to the Aa-rbcS sequence or to each other. At the amino acid level, the conserved regions (white boxes) were 94 to 96% identical to each other or to the Aa-rbcS sequence.
Putative gene functions of particular interest
Immuno-cytochemistry has been used to visualize tubulin and actin proteins in A. acetabulum during development [23,24]. Actin microfilaments were found in thalli of all ages forming continuous, parallel bundles along the entire stalk. Microtubules, conversely, could not be detected in the alga prior to meiosis. Microtubules were detected during reproduction, surrounding haploid nuclei as they are transported up into the cap. Consistent with these results, our data suggest that juveniles express actin transcripts (but not tubulin) while adults express both alpha and beta tubulin (but not actin) transcripts, presumably in preparation for reproduction.
Two of our ESTs were putative expansin homologues. Expansins promote cellulose walls extension in land plants. Typical of the "mannan weeds", the wall of the diplophase of A. acetabulum is predominantly a para-crystalline mannan framework (Dunn et al., submitted). Only gametangia are enclosed in a cellulosic wall, itself surrounded by the mannan wall of the cap. Consistent with this, our results indicate that the expansin gene was expressed during adult but not juvenile phase. So far, expansins have only been found in land plants [25] where expansin acts within the cell wall and is activated by an acidic pH [26]. If these transcripts code for expansin proteins that play a role in loosening walls in A. acetabulum, it would be interesting to see if their mechanism of action is similar to that in land plants, and whether their substrate is also a cellulose wall.
A. acetabulum, for most of its life cycle, contains only one nucleus, which is located in the rhizoid. This nucleus undergoes replication at the end of adult phase, during reproduction [27]. At this juncture, there is a tremendous need for nucleotides and histones to make the millions of haploid nuclei needed for gametogenesis [21]. Consistent with this, histone mRNAs were found only in the adult library (Additional file 3, class 9). It would be interesting to look more deeply into when these transcripts are expressed and how this organism is able to produce histone proteins in such high quantities in such a short period of time.
Finally, one of the ESTs has homology to an argonaute protein (Additional file 3, class 11, E value to Oryza sativa argonaute of 5e-15). Argonaute proteins are highly conserved and play a major role in RNA interference in animals (a.k.a. quelling in fungi or post-transcriptional gene silencing in plants [28]). These processes are involved in the silencing of specific genes via double stranded RNA [29] and their importance in post-transcriptional regulation is just starting to be deciphered. Argonaute proteins have been found in land plants, ciliates, animals and fungi but, to the best of our knowledge, this EST is the first identified algal sequence of an argonaute protein.
Why most of the ESTs do not correspond to any previously described sequences
We can think of three reasons why only 28.6% of the ESTs, a particularly low number, were assigned a putative homolog based on BLAST and InterPro searches. First, these are subtracted libraries, created with the objective of identifying rare, phase-specific transcripts or transcripts involved in morphogenesis, apical growth, or phase change. Hence, these ESTs should include fewer housekeeping transcripts, abundant transcripts, or transcripts common to both phases or to other organisms.
Second, the ESTs were generated by a reverse transcriptase using a poly-T primer that often does not generate full-length cDNAs. Our libraries therefore tend to be enriched in 3' ends of the transcripts, which contain non-coding sequences and which would not be recognized in homology searches. The high percentage of ESTs containing a polyA or polyT stretch supports this hypothesis.
Finally, A. acetabulum belongs to the order Dasycladales, in the green algal class Ulvophyceae, for which very little sequence data is currently available. Before the addition of our ESTs, only 73 DNA sequences from A. acetabulum were available in Genbank, representing just 37 different genes. Although complete genomes of several land plants and green algae are now at least partially available, it is plausible that most of the A. acetabulum sequences are too divergent from those of other algae or land plants to be recognized as orthologs when entered in BLAST searches [30]. To test this hypothesis, we raised the cut-off value for the BLASTN and BLASTX searches against the Genbank databases from 10E-06 to 10E-03. Most additional hits obtained originated from algal or land plant sequence as opposed to a random distribution of the organisms represented in Genbank. This supports the hypothesis that these ESTs are probably homologous to these algal or plant sequences but too divergent for the homologies to be trusted.
Do adult and juvenile transcripts differ in structure? Insights into post-transcriptional regulation
Curiously, 40% of the juvenile clones but only 15% of the adult clones end with a polyA or polyT tract. If these tracts correspond to the mRNA polyA tail, then these ESTs contain some or all of the 3' untranslated regions (3' UTR) of the transcript from which they originated. We have diagrammed hypotheses explaining the differential occurrence of these tracts in adult versus juvenile clones (Fig. 7). The first explanation presumes an artifact of the techniques used to create the libraries. ESTs result from the amplification of cDNA fragments that have been digested by RsaI, each RsaI fragment having an equal chance of being amplified and cloned. If the adult cDNAs were more completely digested than the juvenile cDNAs, then the adult cDNAs would have generated a higher number of ESTs, a lower proportion of which would contain polyA tracts (Fig. 7a). A second hypothesis presumes differential mRNA length: if adult cDNAs were, on average, longer than juvenile cDNAs, each adult cDNA would produce more ESTs, yielding a lower proportion of ESTs containing the polyA tract. Adult cDNAs could be longer if on average they have longer coding sequences (Fig. 7b) or longer 3'UTRs (Fig. 7c). If they have longer 3'UTRs, the proportion of coding sequences as well as the proportion of ESTs with polyA tracts will be higher in juvenile ESTs than in adult ESTs, consistent with our findings.
Figure 7.
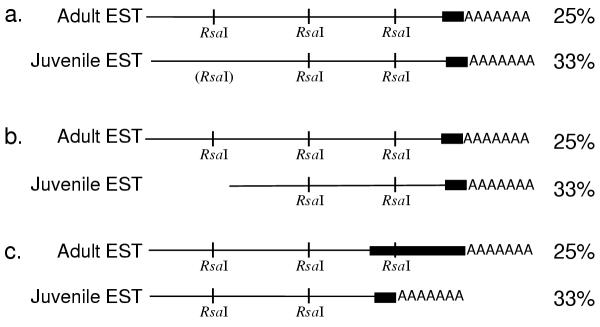
Hypothetical explanations for the difference in frequency of poly A/T tracts within the two libraries. The black boxes represent the 3' UTR of hypothetical transcripts. On the right are the calculated percentages of ESTs containing a polyA or polyT tract that would result from the creation of ESTs from the hypothetical mRNA shown. a: Differential digestion of the initial cDNAs. ESTs resulted from the amplification of cDNA fragments that have been digested by RsaI, each RsaI fragment having an equal chance of being amplified and cloned. If the adult cDNAs were more completely digested than the juvenile cDNAs, the adult cDNAs generated a higher number of ESTs (4 instead of 3 in this case), a lower proportion of which would contain polyA tracts. b and c: Differential mRNA length in vivo. Adult cDNAs were, on average, longer than juvenile cDNAs, so each adult cDNA produced more ESTs (4 instead of 3), yielding a lower proportion of ESTs containing the polyA tract. Adult cDNAs could be longer because they have, on average, longer coding sequences (b) or longer 3'UTRs (c).
Why would 3' UTRs be longer in adult transcripts than in juvenile transcripts? In adult A. acetabulum, growth and morphogenesis occurs almost exclusively at the stalk apex, centimeters away from the unique nucleus located in the rhizoid. Therefore, aspects of post-transcriptional regulation, such as mRNA stability and mRNA localization, are probably very important to the regulation of gene expression in these unicells. Indeed, more than half of the transcripts (9/16) studied to date in A. acetabulum are localized to one end or the other of the unicell, most often to its apex [31-33]. To achieve this localization, each transcript must contain cis-acting elements within its sequence, also called 'zipcodes' [34]. In yeast and animal cells, 'zipcodes' are part of the 3' UTR of the localized transcripts [35]. Also, considering the rate at which mRNA molecules move along cytoskeletal elements along the stalk of A. acetabulum [36,37], to reach the apex, any mRNA must be at least three days old, classifying them among the "ultra-stable" mRNA species [38]. In plants, the cis-acting elements responsible for stability of an mRNA molecule are also located in its 3' UTR [38]. Transcripts 3' UTR might therefore play an important role in the regulation of gene expression in this species, especially in adults.
To achieve these stability and localization patterns, adult mRNAs probably contain several post-transcriptional regulatory elements within their 3' UTRs, potentially explaining why these would be longer. What are these regulatory elements? The fact that three conserved elements (Fig. 5) were found within several unrelated ESTs, most of which originate from the adult library is promising. These 3 elements also appear to be specific to A. acetabulum and are located in a non-coding region of the carbonic anhydrase gene, whose transcript is apically localized [13]. Five of the ESTs containing the second conserved element also contain a polyA tract, suggesting that these ESTs may code for 3' UTRs. The first and second conserved elements fall within introns of AaCA1 (Fig. 5). It is possible that these sequence elements of AaCA1 are part of alternatively spliced introns and sometimes contained in the mature mRNAs produced from this gene. Future research will focus on elucidating the function of these conserved elements and their spatial expression during development.
Conclusion
These results presented here provide strong evidence supporting the hypothesis that adult and juvenile phases in A. acetabulum differ significantly in gene expression patterns and that a large number of genes are phase-specific. Our next goal is to identify among these genes those that might be involved in morphogenesis or phase change. The ESTs from the two phases also partition into different functional classes, underlining further the physiological differences between the two phases. Finally, we identified conserved elements within the EST sequences. While the functional significance of these conserved elements remains to be elucidated, it is tempting to suggest that these sequences might be involved in the post-transcriptional regulation of these transcripts, possibly in sub-cellular localization and/or stability.
Methods
Culture of A. acetabulum
Unicells were grown in artificial seawater until they reached the desired developmental age. Axenic cultures were obtained by decontaminating mature caps and then using the axenic gametangia they housed for mating [21]. Zygotes were grown in sterile artificial seawater, Ace27, which is identical to Ace25 [39] except that the KCl prestock was purified over a chelex-100 column and it contains urea hydrogen peroxide at a final concentration of 10-15 M. Cultures were grown under cool white fluorescent lights at a photon flux density of 170 μmol m-2 s-1 on a 14 h light/10 h dark photoperiod, at 21°C ± 2°C and repeatedly diluted to suit their developmental age [21].
mRNA extraction
Juveniles were harvested by filtration and adults were harvested using sterile dental tools. The unicells were dried briefly on a Kimwipe, and weighed on aluminum foil. Packets of algae of the same age were flash-frozen in liquid nitrogen. 7.15 g of juveniles (approximately 18,000 unicells) and 18.2 g of adults (approximately 4,000 unicells) were ground to a fine powder under liquid nitrogen. The powder was transferred to Oakridge tubes containing extraction buffer (0.1 to 0.2 g of ground unicells/ml extraction buffer). RNA was extracted according to Chang et al. [40].
Suppressive Subtractive hybridization (SSH)
cDNA synthesis and SSH were performed according to the manufacturer's recommendations using the PCR cDNA Synthesis Kit (Clontech Laboratories, Inc.) and the PCR-Select cDNA Subtraction Kit (Clontech Laboratories, Inc.) respectively. A summary of the steps involved in SSH and a more detailed figure of the formation of the ESTs from mRNA can be found in Additional file 1 and 2.
Cloning of the ESTs to make the libraries
DNA was precipitated using a standard ethanol precipitation protocol [41]. In order to add 3' A-overhangs to the PCR products for subsequent cloning, the DNA was resuspended into 25 μl of PCR reaction cocktail (2.5 μl of 10X buffer, 1.5 μl MgCl2, 2 μl 10 mM dNTPs, 18.875 μl water and 0.125 μl Taq polymerase (Promega)) and incubated at 72°C for 8–10 minutes. The DNA was precipitated again [41] and resuspended in TE to the starting volume of the DNA amplification reaction. Following the manufacturer's recommendations, each library was cloned into 2 different cloning vectors using the AdvanTAge™ PCR Cloning Kit (Clontech Laboratories, Inc., now a discontinued product) and the TOPO™-TA Cloning Kit (Invitrogen).
Dot blot and virtual Northern blot analysis of the libraries
The quality of subtraction was controlled as recommended by the PCR-Select protocol provided by Clontech. PCR-amplified inserts of 96 randomly picked clones from both libraries were duplica-spotted onto nylon membranes and hybridized with the radioactively labeled subtraction mix from both subtractions. In addition, differential expression of cDNA inserts of three clones was confirmed by virtual northern blots using SMART cDNA synthesis (Clontech) [15]. The clones used in these dot blots and virtual northern blots were not sequenced and are not part of the following sequence analysis.
EST sequencing
Colonies were randomly picked from each library using sterile toothpicks. Plasmid DNA from each colony was isolated and eluted with 2 × 40 μl of elution buffer (Plasmid Miniprep Kit, Qiagen).
DNA sequencing was carried out at the Plant-Microbe Genomics Facility, Ohio State University. The sequencing reactions were prepared by mixing 400 ng of plasmid DNA and 4 pmol of primer (M13F (5'-GTAAAACGACGGCCAG-3') or M13R (5'-CAGGAAACAGCTATGAC-3') with water for a total volume of 10 μl. Next, 2 μl of BigDyeTerminator mixture, version 2 (Applied Biosystems), 4 μl BetterBuffer (The Gel Company) and 4 μl water were added. The cycling parameters were those recommended by the manufacturer except that the reactions were run for 35 cycles instead of 25. The reactions were cleaned up with Millipore Multiscreen/Sephadex columns, according to the manufacturers recommendations (Millipore Technical Note TN053). The resulting 20 μl of clean sequencing reaction product (in water) was placed in an Applied Biosystems 3700 DNA Analyzer for separation and analysis.
Sequence analysis
Sequence preparation
Each clone was sequenced once using the M13 forward primer. If the sequence was of poor quality, the clone was sequenced again using the M13 reverse primer. Using Sequencher (Gene Codes Inc.), each nucleotide sequence was cleaned in silico of contaminating vector or primer sequence individually by aligning the EST sequence to that of the vector and those of the primers used in the creation of the libraries (nested PCR primer 1 (5'-TCGAGCGGCCGCCCGGGCAGGT-3') and nested PCR primer 2 (5'-AGCGTGGTCGCGGCCGAGGT-3'). These steps insured that the remaining sequence was devoid of contaminating DNA fragments that could potentially generate erroneous hits in BLAST searches [16]. A high proportion of the sequences also contained polyA or polyT tracts. These DNA fragments were also removed in silico from the corresponding sequences before performing homology searches.
Homology searches
Each EST was queried as follows:
- BLASTN searches [42] (database searched: nr (non-redundant nucleotide sequences)) [43].
- TBLASTX searches [42] (database searched: nr (non-redundant nucleotide sequences), genetic code: 6 (used by ciliates and Dasycladales [44]), defaults were used for the rest of the parameters) [43].
- InterPro searches [45] queries protein motifs databases from the European Bioinformatics Institute [46]. InterPro hits were mapped to the Gene Ontology [47].
- BLASTX searches against the Arabidopsis thaliana database [17].
- TBLASTX searches against the June 2003 draft of the Chlamydomonas reinhardtii genome [18].
Clustering of the ESTs
Clustering and alignments of the ESTs were performed using StackPack software (Electric Genetics, Cape Town, South Africa) [48,49]. Clustering was performed in two phases. The first phase used the 'd2' algorithm, which is part of the 'd2' cluster [50,51]. The second phase used Phrap [52].
The analysis was run with the following parameters:
d2_cluster: word_size = 6, similarity_cutoff = 0.96, minimum_sequence_size = 50, window_size = 150 and reverse_comparison = 1.
Phrap: old_ace = 1, vector_bound = 0, trim_score = 20, forcelevel = 0, penalty = -2, gap_init = -4, gap_ext = -3, ins_gap_ext = -3, del_gap_ext = -3, maxgap = 30, flags = -retain_duplicates.
Organization of the data
Bioinformatics scripts and the database systems used to store and query sequence/annotation data were provided by the Specialized Plant Resources in Informatics and Genomics (SPRIG) project http://bioinformatics.org/sprig, in particular, the SPRIG generic EST database and support script [53].
Availability of the sequences
The EST sequences analyzed in this study have been submitted to dbEST division of Genbank under accession numbers: CF 258288 to CF259228.
List of abbreviations
3' UTR: 3' untranslated region
BLAST: Basic local alignment search tool
SSH: Suppressive subtractive hybridization
AaCA1: Carbonic anhydrase 1 from Acetabularia acetabulum
EST: Expressed Sequence Tag
RTase: Reverse transcriptase.
Authors' contributions
I.H. prepared some of the plasmid DNAs, participated in the bioinformatics analysis of the sequences and the design of the study, analyzed the results and drafted the manuscript. M.W. performed the bioinformatics analysis of the sequences and created the EST database. Z.S-S. created the subtracted libraries. E.G. and J.M.H. coordinated the sequencing and performed the initial analysis. E.G. participated in the design of the study. D.M. conceived of and initiated the project, prepared some of the plasmid DNAs and participated in the design and coordination of the study. All authors read, reviewed and approved the final manuscript.
Supplementary Material
Creation of subtracted libraries. Summary of the steps involved in the creation of subtracted libraries.
cDNA synthesis and RsaI digestion. Detailed description of the steps involved in the formation of double stranded cDNA from mRNA and in the digestion of the cDNA population. This procedure generates a population of ESTs, some of which contain a polyA tract and the size of the ESTs thus depends on the position of the RsaI restriction sites. Non-interrupted lines represent DNA strands and dashed lines represent RNA strands. The gray boxes represent the primers used for these amplification steps. A more detailed description of these steps can be found in Henry and Mandoli [16].
Function and name of the ESTs that produced significant BLAST hits. All ESTs that produced significant hits were classified according to their function and following the classification scheme developed for plants [19].
Acknowledgments
Acknowledgements
We would like to thank Drs Harvey Bradshaw, Rose Ann Cattolico, Richard Olmstead and Elizabeth Van Volkenburgh for useful advice, comments and encouragement. We thank Mr. Richard Ivey for cell culture and RNA extraction, Mr. Eric Blackstone for help with the cloning of the EST libraries and Mr. Matthew Links, Bioinformatics Group (Dept. of Computer Sciences, University of Saskatoon, Canada) for the clustering and alignments of the ESTs. The sequencing of our ESTs was performed at the Plant-Microbe Genomics Facility (Ohio State University, Columbus). This work was supported in part by a Botany Departmental Fellowship, a Fulbright grant-in-aid and a Belgian-American Educational Foundation Fellowship (all to IMH), by NSF IBN #9630618, a University of Washington Friday Harbor Laboratory Apprenticeship course, and a PanWy Foundation grant (all to DFM) and by the Deutsche Forschungsgemeinschaft SFB 57 (to Z.S-S).
Contributor Information
Isabelle M Henry, Email: henryi@u.washington.edu.
Mark D Wilkinson, Email: markw@illuminae.com.
J Marcela Hernandez, Email: hernandez.16@osu.edu.
Zsuzsanna Schwarz-Sommer, Email: schwarzs@mpiz-koeln.mpg.de.
Erich Grotewold, Email: grotewold.1@osu.edu.
Dina F Mandoli, Email: mandoli@u.washington.edu.
References
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST-database for "expressed sequence tags". Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Fillingham JS, Chilcoat ND, Turkewitz AP, Orias E, Reith M, Pearlman RE. Analysis of expressed sequence tags (ESTs) in the ciliated protozoan Tetrahymena thermophila. J Eukaryot Microbiol. 2002;49:99–107. doi: 10.1111/j.1550-7408.2002.tb00350.x. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequenced tags. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- Shrager J, Hauser C, Chang C-W, Harris EH, Davies J, McDermott J, Tamse R, Zhang Z, Grossman AR. Chlamydomonas reinhardtii Genome Project. A Guide to the Generation and Use of the cDNA Information. Plant Physiol. 2003;131:401–408. doi: 10.1104/pp.016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumais J, Serikawa K, Mandoli DF. Acetabularia: a unicellular model for understanding subcellular localization and morphogenesis during development. J Plant Growth Regul. 2000;19:253–364. doi: 10.1007/s003440000035. [DOI] [Google Scholar]
- von Dassow M, Odell GM, Mandoli DF. Relationships between growth, morphology and wall stress in the stalk of Acetabularia acetabulum. Planta. 2001;213:659–666. doi: 10.1007/s004250100538. [DOI] [PubMed] [Google Scholar]
- Hämmerling J. Entwicklung und formbildungsvermogen von Acetabularia mediterranea. Biol Zent Bl. 1932;52:42–61. [Google Scholar]
- Hämmerling J. Uber formbildende Substanzen bei Acetabularia mediterranea, ihre raumliche und zeitliche Verteilung und ihre Herkunft. Wilhelm Roux Arch Entwichklungsmech Org. 1934;131:1–81. doi: 10.1007/BF00649809. [DOI] [PubMed] [Google Scholar]
- Hämmerling J. Nucleo-cytoplasmic interactions in Acetabularia and other cells. Annu Rev Plant Physiol Plant Mol Biol. 1963;14:65–92. [Google Scholar]
- Mandoli DF. Elaboration of body plan and phase change during development of Acetabularia: How is the complex architecture of a giant unicell built? Annu Rev Plant Physiol Plant Mol Biol. 1998;49:173–198. doi: 10.1146/annurev.arplant.49.1.173. [DOI] [PubMed] [Google Scholar]
- Jones CS. An essay on juvenility, phase change, and heteroblasty in seed plants. Int J Plant Sci. 1999;160:S105–S111. doi: 10.1086/314215. [DOI] [PubMed] [Google Scholar]
- Vega SH, Sauer M, Orkwiszewski JA, Poethig RS. The early phase change Gene in Maize. Plant Cell. 2002;14:133–147. doi: 10.1105/tpc.010406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikawa KA, Porterfield DM, Mandoli DF. Asymmetric subcellular mRNA distribution correlates with carbonic anhydrase activity in Acetabularia acetabulum. Plant Physiol. 2001;125:900–911. doi: 10.1104/pp.125.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau Y-F, Campbell AP, Chenchick A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverlov ED, Siebert PD. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Nat Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz. Brucchaus I, Roeder Verification of differential gene transcription using virtual northern blotting. Nucl Acids Res. 1999;27:e3. doi: 10.1093/nar/27.11.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Mandoli DF. Precious cells contain precious information: strategies and pitfalls in expression analysis from a few cells. In: E Grotewold ed, editor. Plant functional genomics: methods and protocols. 236, In press. Totowa, NJ, Humana Press; 2003. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis thaliana protein database http://www.tigr.org/tdb/e2k1/ath1/
- Draft of the Chlamydomonas reinhardtii genome http://genome.jgi-psf.org/chlre1/chlre1.download.html
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, Drost L, Ridley P, Chalwatzis N. Analysis of 1.9 Mb of continuous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Taxonomic classification at NCBI http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/
- Mandoli DF. What ever happened to Acetabularia? Bringing a once-classic model system into the age of molecular genetics. Int Rev Cytol. 1998;182:1–67. [Google Scholar]
- Jansson S. The light-harvesting chlorophyll a / b-binding proteins. Biochim Biophys Acta. 1994;1184:1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Menzel D. Visualization of Cytoskeletal Changes Through the Life Cycle in Acetabularia. Protoplasma. 1986;132:30–42. [Google Scholar]
- Menzel D. Cell differentiation and cytoskeleton in Acetabularia: Tansley Review Number 77. New Phytol. 1994;128:369–393. doi: 10.1111/j.1469-8137.1994.tb02984.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove D. Loosening of plant cells walls by expansins. Nature Rev. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runft LL, Mandoli DF. Coordination of the cellular events that precede reproductive onset in Acetabularia acetabulum: evidence for a "loop" in development. Development. 1996;122:1187–1194. doi: 10.1242/dev.122.4.1187. [DOI] [PubMed] [Google Scholar]
- Vance V, Vaucheret H. RNA silencing in plants - Defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- Carmell M, Xuan Z, Zhang M, Hannon G. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Cirera S, Wintero AK, Fredholm M. Why do we still find anonymous ESTs? Mamm Genome. 2000;11:689–693. doi: 10.1007/s003350010127. [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Mandoli DF. Aaknox1, a kn1-like homeobox gene in Acetabularia acetabulum, undergoes developmentally regulated subcellular localization. Plant Mol Biol. 1999;41:785–793. doi: 10.1023/A:1006387107071. [DOI] [PubMed] [Google Scholar]
- Vogel H. Untersuchungenn zu spezifischen mRNA-Gradienten in der einzelligen, siphonalen Grunalge Acetabularia acetabulum (L.) Silva. Justus-Liebig-Universitat Giessen.; 1998. [Google Scholar]
- Vogel H, Grieninger GE, Zetsche KH. Differential messenger RNA gradients in the unicellular alga Acetabularia acetabulum. Role of the cytoskeleton. Plant Physiol. 2002;129:1407–1416. doi: 10.1104/pp.010983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Oleynikov Y, Singer RH. The travels of mRNA through all cells large and small. FASEB J. 1999;13:447–454. doi: 10.1096/fasebj.13.3.447. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:355–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Dazy A, Borghi H, Puiseux-Dao S. RNA migration in Acetabularia mediterranea: effects of cytochalasin B, cycloheximide and prolonged dark periods. Plant Sci Lett. 1983;30:285–296. [Google Scholar]
- Kloppstech K, Schweiger HG. Polyadenylated RNA from Acetabularia. Differentiation. 1975;4:115–123. [Google Scholar]
- Gutierrez RA, MacIntosch GC, Green PJ. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci. 1999;4:429–438. doi: 10.1016/S1360-1385(99)01484-3. [DOI] [PubMed] [Google Scholar]
- Hunt BE, Mandoli DF. A new, artificial seawater that facilitates growth of large numbers of cells of Acetabularia acetabulum (Chlorophyta) and reduces the labor inherent in cell culture. J Phycol. 1996;32:483–495. [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning, A Laboratory Manual. Third Edition. Vol. 3. New York, Cold Spring Harbor Laboratory Press; 2001. p. A8.14. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers WE, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- BLAST at NCBI http://www.ncbi.nlm.nih.gov/BLAST/
- Schneider SU, Leible MB, Yang XP. Strong homology between the small subunit of ribulose-1,5-biphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol Gen Genet. 1989;218:445–452. doi: 10.1007/BF00332408. [DOI] [PubMed] [Google Scholar]
- BLAST at NCBI http://www.ncbi.nlm.nih.gov/BLAST/
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MDR, Durbin R, Falquet L, Fleichmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder N, Oinn T, Pagni M, Servant F, Sigrist C, Zdobnov EM. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- InterPro http://www.ebi.ac.uk/interpro/
- Gene Ontology Consortium http://www.geneontology.org/
- The Arabidopsis thaliana protein database http://www.tigr.org/tdb/e2k1/ath1/
- Draft of the Chlamydomonas reinhardtii genome http://genome.jgi-psf.org/chlre1/chlre1.download.html
- Christoffels A, van Gelder A, Greyling G, Miller R, Hide T, Hide W. STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucl Acids Res. 2001;29:234–238. doi: 10.1093/nar/29.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Christoffels A, Gopalafrishnan C, Burke J, Ptitsyn A, Broveak T, Hide W. A comprehensive approach to clustering of expressed human gene sequence: the sequence tag alignment and consensus knowledge base. Genome Research. 1999;9:1143–1155. doi: 10.1101/gr.9.11.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hide W, Burke J, Davison DB. Biological evaluation of d2, an algorithm for high-performance sequence comparison. J Comp Biol. 1994;1:199–215. doi: 10.1089/cmb.1994.1.199. [DOI] [PubMed] [Google Scholar]
- Burke J, Davison D, Hide W. d2_cluster: a validated method for clustering EST and full-length cDNA sequences. Genome Research. 1992;9:1135–1142. doi: 10.1101/gr.9.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Documentation for Phrap and Cross_Match http://www.phrap.org/phrap.docs/phrap.html
- The Open Lab CVS Repository http://bioinformatics.org/cgi-bin/cvsweb.cgi/sprig/database/est/MarksDB/
- The Arabidopsis thaliana protein database http://www.tigr.org/tdb/e2k1/ath1/
- Draft of the Chlamydomonas reinhardtii genome http://genome.jgi-psf.org/chlre1/chlre1.download.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Creation of subtracted libraries. Summary of the steps involved in the creation of subtracted libraries.
cDNA synthesis and RsaI digestion. Detailed description of the steps involved in the formation of double stranded cDNA from mRNA and in the digestion of the cDNA population. This procedure generates a population of ESTs, some of which contain a polyA tract and the size of the ESTs thus depends on the position of the RsaI restriction sites. Non-interrupted lines represent DNA strands and dashed lines represent RNA strands. The gray boxes represent the primers used for these amplification steps. A more detailed description of these steps can be found in Henry and Mandoli [16].
Function and name of the ESTs that produced significant BLAST hits. All ESTs that produced significant hits were classified according to their function and following the classification scheme developed for plants [19].


