Abstract
A review of the published and unpublished literature on bacterial resistance in human and animals was performed. Sixty-eight articles/reports from the Democratic Republic of Congo (DRC), Mozambique, Tanzania and Zambia were reviewed. The majority of these articles were from Tanzania. There is an increasing trend in the incidence of antibiotic resistance; of major concern is the increase in multidrug- resistant Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Vibrio cholera, non-typhoid Salmonella and other pathogens responsible for nosocomial infections. The increase in methicillin- resistant Staphylococcus aureus and extended-spectrum beta-lactamase (ESBL) producers in the countries under review confirms the spread of these clones worldwide. Clinical microbiology services in these countries need to be strengthened in order to allow a coordinated surveillance for antimicrobial resistance and provide data for local treatment guidelines and for national policies to control antimicrobial resistance. While the present study does not provide conclusive evidence to associate the increasing trend in antibiotic resistance in humans with the use of antibiotics in animals, either as feed additives or veterinary prescription, we strongly recommend a one-health approach of systematic surveillance across the public and animal health sectors, as well as the adherence to the FAO (Food and Agriculture Organization)-OIE (World Organization of animal Health) –WHO(World Health Organization) recommendations for non-human antimicrobial usage.
Introduction
The introduction of penicillin, in the early 1940s, was perceived as marking the end of infectious diseases [1]. However, the emergence of resistant strains was reported just a few years after its use. Since then, resistant clones to various classes of antibiotics have been found to spread worldwide [2]. In some areas, more than 90% resistance has been reported to commonly used antibiotics such as penicillin, ampicillin, co-trimoxazole and gentamicin [3]. The overuse of antibiotics in human and animals has contributed to the emergence of resistant clones [4,5]. It is a fact that the availability of antimicrobials and their proper use have reduced morbidity and mortality due to infectious disease [http://www.cdc.gov/drugresistance/index.html]. In developed countries, the use of antibiotics is strictly controlled, which is not the case in developing countries. The treatment of bacterial infections in Africa is largely empirical and in most instances, there are no laboratory results to guide therapy. Moreover, there are no data on common bacterial isolates and their susceptibility patterns from larger surveillance studies aimed at developing tools for therapeutic guidance. Developing countries bear 95% of the global infectious diseases burden and rely on empirical antimicrobial treatment to counteract these diseases [6]. This has resulted in many infectious diseases, once easily curable, to become untreatable [7-9].
The burden of antimicrobial resistance (AMR) is rapidly growing across antibiotic classes. The emergence of methicillin- resistant Staphylococcus aureus (MRSA) clones spreading among animals and human has made AMR an issue of public health importance [10]. Recently, the emergence of NDM-1 has made infection due to multi-resistant gram negative bacteria untreatable, especially in developing countries where there is no alternative treatment available [11]. Unfortunately, coordinated surveillance of the clones involved is lacking, especially in developing countries in Africa.
This review was undertaken to summarize the patterns and trends of resistance to commonly used antibiotics among common bacterial isolates from humans and animals from 1990s to 2012 in Democratic Republic of Congo (DRC), Mozambique, Tanzania and Zambia. The data from this review will be used to provide recommendations on priority research area that could address the development and spread of antibiotic resistance in humans and animals.
Methods
In this review, literature from four different countries is appraised and reported. Though these countries present differences in terms of local agriculture, economy, state of health care services, political situation, etc., they all have no clear policy on antibiotic use hence at risk of increased antibiotic resistance. Also, these countries have all introduced the one-health concept under the South African Centre of Infectious Diseases (SACIDS) aiming at providing data for evidence based management of infectious diseases.
Study design and search strategy
A systematic literature review was conducted for original articles on bacterial isolates, resistance patterns in humans and animals from DRC, Mozambique, Tanzania and Zambia, published from 1990 to the end of July 2012. The study focused on all bacterial pathogens with the exception of Mycobacterium tuberculosis. A systematic search of online databases including PubMed/Medline, Embase, Popline, Global Health, Google and Web of Knowledge was undertaken. We used the search terms “bacterial isolate resistance patterns”, “antibiotic use”, “antimicrobial resistance”, “microbial resistance”, “susceptibility”, “resistance surveillance” in “human or animal use” combined with the name of the different countries of interest. References of all articles were searched to identify further articles. Articles were reviewed and publications using original data on resistance in animals and humans were included. The studies’ design, setting, demographic data and microbiological methods were appraised. All studies involved human were approved by ethics committee and informed consent obtained. We also consulted the WHO, OIE and FAO websites for relevant publications. New links displayed beside the abstracts were followed and retrieved.
Results
Overview of study design and microbiological susceptibility methods
A total of 68 articles published between 1990 and 2012 describing bacterial pathogens and/or antibiotic resistance in humans and animals were retrieved; all articles were included in this review. Of 68 articles 40 (59%) reported data on the susceptibility pattern; of these 88% used disc diffusion method (Additional file 1: Table S1). The CLSI guideline was used in 60% of articles, 3 articles used French Society for Microbiology and 1 article used European Committee of Antimicrobial and Susceptibility Testing (EUCAST) and 12 articles did not state the guideline used. Thirteen (33%) articles did not report the use of control strains. The majority of the studies were short-term clinical studies and retrospective clinical studies while 4 were surveillance studies (3 from Tanzania and 1 from Mozambique). The distribution of articles by country is seen in Table 1. No articles describing antibiotic resistance in animals were found from DRC and Mozambique. Most of the human studies from DRC, Mozambique, Tanzania and Zambia were carried out in urban settings in tertiary hospitals, with only one study from Tanzania involving primary health facilities, while 3/11 (27%) of human studies from Mozambique involved children from a rural hospital.
Table 1.
Articles distribution per country and source of samples
| Country | Human | Animal |
|---|---|---|
|
Tanzania |
33 |
3 |
|
Zambia |
6 |
5 |
|
DRC |
9 |
1 |
|
Mozambique |
11 |
0 |
| Total | 59 | 9 |
Escherichia coli and Klebsiella pneumoniae causing Urinary tract infections (UTI) and blood stream infections
Escherichia coli and Klebsiella pneumoniae have been identified as the most common causes of Urinary Tract Infection (UTI) in Tanzania; contributed 47% of all UTI. Ten studies from Tanzania reported on UTI and the majority of them (9/10) were short clinical studies involving pregnant women and children [12-21]. No data regarding these species on UTI were found from DRC, Mozambique and Zambia. Resistance rates in different studies are seen in Table 2; Klebsiella spp showed higher resistance rates than Escherichia coli. Among Escherichia coli mean resistance ranges from 14% for ciprofloxacin to 85% for ampicillin while among Klebsiella spp mean resistance ranges from 20% for nitrofurantoin to 85% for ampicillin. There is an increased trend of these isolates to become resistant to commonly used antibiotics. Among Escherichia coli isolates from urine, ampicillin resistance has been found to increase from 17% in 1995–1996 [20] to more than 93% in 2009 [12,19]. In addition, an increasing resistance trend to gentamicin among Escherichia coli isolated from urine specimens has been observed; the rate of gentamicin resistance in Tanzania was found to range from 7% at the Muhimbili National hospital (MNH) in 2003 to more than 44% in the same hospital in 2011 [15,20].
Table 2.
Antimicrobial resistance rates of Escherichia coli and Klebsiella pneumoniae from Urine Tanzania (ref: [12-15,19-21])
| Antibiotic(N*) | Resistance rates in different studies%Escherichia coli(range) | Mean resistance n/N* (%) |
|---|---|---|
| Ampicillin (518) |
92, 96, 53, 98, 100, 69, 80 (53–100) |
438/518 (85) |
| Augmentin (501) |
53, 70, 88, 85, 37 (37–88) |
337/501 (67) |
| Gentamicin (518) |
7, 38, 6, 23, 32, 44 (6–44) |
130/518 (25) |
| Co-trimoxazole (339) |
80, 65, 95,97, 50 (50–97) |
262/339 (77) |
| Ciprofloxacin (501) |
8, 30, 12, 9, 13 (8–30) |
72/501 (14) |
| Ceftriaxone (339) |
51, 29, 14, 27, 19 (14–51) |
95/339 (28) |
| Ceftazidime (306) |
50, 11, 14 (11–50) |
77/306 (25) |
| Nitrofurantoin (289) |
23, 6, 13, 31(6–31) |
53/289 (18) |
| |
Resistance rates in different studies Klebsiella pneumoniae % (range) |
|
| Ampicillin (193) |
56, 100, 98 (56–100) |
163/193(85) |
| Augmentin (193) |
11, 78, 86 (11–86) |
112/193(58) |
| Gentamicin (193) |
11, 28, 38 (11–38) |
50/193 (26) |
| Co-trimoxazole (193) |
56, 83, 95 (95–56) |
150/193 (78) |
| Ciprofloxacin (193) |
19, 19, 44 (19–44) |
53/193 (27) |
| Ceftriaxone (193) |
33, 66, 46 (33–66) |
93/193 (48) |
| Ceftazidime (52) |
52 |
27/52 (52) |
| Nitrofurantoin (193) | 18, 21, 22 (18–22) | 37/193 (20) |
N*=Total number of isolates tested.
Escherichia coli and Klebsiella pneumoniae also have been found to cause blood stream infections, especially in neonates [22-26]. Escherichia coli was the most common isolate in blood stream community- acquired infections while Klebsiella spp was the commonest isolate in health care associated infection [22-24]. Mean resistance rates of Escherichia coli from the blood were 2%, 15%, 28%, 37%, 38%, 55%, 57%, 76% and 95% for meropenem, ciprofloxacin, ceftazidime, gentamicin, ceftriaxone, augmentin, tetracycline, co-trimoxazole and ampicillin respectively (Table 3). With exception for meropenem, and ciprofloxacin Klebsiella spp had higher resistance rates than Escherichia coli. Escherichia coli and Klebsiella pneumoniae strains from the blood displayed higher rates of resistance than those from urine specimens against common antibiotics such as ampicillin, co-trimoxazole and gentamicin [22,24,25]. In this review resistance to third generation cephalosporins was observed in more than 20% of hospital-acquired Klebsiella spp and Escherichia coli.
Table 3.
Antimicrobial resistance rates of Escherichia coli and Klebsiella pneumoniae from the blood, Mozambique and Tanzania (ref: [22-26])
| Antibiotic | Resistance rate in different studies%Escherichia coli(range) | Mean resistance n/N* (%) |
|---|---|---|
| Ampicillin |
84, 96 , 85, 100, 96 (84–100) |
227/240 (95) |
| Augmentin |
40, 25, 69, 86 (25–86) |
47/86 (55) |
| Tetracycline |
55, 59 (55–59) |
28/49(57) |
| Gentamicin |
13, 29, 46, 68, 28 (13–68) |
87/234 (37) |
| Co-trimoxazole |
72, 87, 54, 77, 90 (54–90) |
173/227 (76) |
| Ciprofloxacin |
40, 8, 8, 4 (4–40) |
13/86 (15) |
| Ceftriaxone |
12, 54, 50 (12–54) |
23/59(38) |
| Ceftazidime |
0.0, 12, 50, 50 (0–50) |
24/86 (28) |
| Meropenem |
0.0, 0.0, 4 (0–4) |
1/59 (2) |
| |
Resistance rate in different studies % Klebsiella pneumoniae(range) |
|
| Ampicillin |
91, 100, 100, 100, 100 (91–100) |
202/206 (98) |
| Augmentin |
32, 47, 38, 84 (32–84) |
97/194 ( 50) |
| Tetracycline |
67, 62 (62–67) |
91/141(65) |
| Gentamicin |
40, 47, 47, 67, 18 (18–67) |
91/206 (44) |
| Co-trimoxazole |
63, 63, 94, 79, 70 (64–94) |
151/206 (74) |
| Ciprofloxacin |
0.00, 0.00, 0.00, 8 (0–8) |
4/194 (2) |
| Ceftriaxone |
50 |
25/50 (50) |
| Ceftazidime |
6, 21, 15, 49 (6–49) |
42/184 (22) |
| Meropenem | 0.00, 0.00, 2 (0–2) | 1/160 (1) |
N*=Total number of isolates tested.
One study in Tanzania reported data from surveillance study [24]. In that study a total of 1936 and 1771 of E.coli and Klebsiella pneumoniae were isolated in 2 year period from 1st January 1998 to 31st December 1999. Majority of these isolates were from urine specimens, others were from pus, blood and other routine specimens. Escherichia coli were 80%, 28%, 5%, 77%, 8%, 76%, 32% and 13% resistant to ampicillin, amoxicillin/clavulanic acid, ceftazidime, tetracycline, gentamicin, co-trimoxazole, nitrofurantoin and quinolones while the rates of resistance to similar antibiotics for Klebsiella pneumoniae were 85%, 32%, 6%,66%, 14%, 69%, 53% and 6% respectively.
Extended –spectrum beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae
Data on ESBL producing E. coli and Klebsiella pneumoniae are limited in all the countries under review. Of 302 E. coli tested for ESBL production in these countries 88/302 (29%) were found to be ESBL producers while of 283 Klebsiella pneumoniae tested 116/282 (41%) were ESBL producers [3,19,27-30]. Different ESBL alleles have been reported including CTX-M-15 Escherichia coli ST 131 which has been reported worldwide [28]. Other reported alleles include TEM-63, SHV-2a and SHV-12 [29].
Blood stream infection due to Salmonella spp
There is increase trend of Salmonella spp causing blood stream infection in developing countries especially among HIV/AIDS patients. In these countries most of Salmonella spp from the blood were found to be resistant to ampicillin, co-trimoxazole and chloramphenicol (Table 4) while being sensitive to ciprofloxacin [26,31-37]. An increased resistance trend of Salmonella spp to ciprofloxacin in DRC from 0.0% in 2009 to 15.4% in 2012 was noted [32,33].
Table 4.
Antimicrobial resistance rates of Salmonella spp from the blood, Zambia, DRC, Mozambique and Tanzania ref: [26,32,33,37]
| Antibiotic | Resistance rates in different studies%(range) | Mean resistance n/N* (%) |
|---|---|---|
| Ampicillin |
100, 65, 74, 69 (65–100) |
477/620(77) |
| Co-trimoxazole |
100, 58, 66, 38 (38–100) |
404/620 (65) |
| Chloramphenicol |
100, 40, 55, 85 (40–100) |
434/620 (70) |
| Gentamicin |
16, 23 (16–23) |
78/400(20) |
| Ciprofloxacin | 0, 8, 15 (0–15) | 32/225 (14) |
Salmonella spp, Shigella spp, Vibrio cholera, Campylobacter spp and diarrheagenic E.coli; and their susceptibility patterns
Diarrheal diseases account for approximately 25% of all deaths in <5 year-old children in developing countries [38,39] and common pathogens (Campylobacter spp, Salmonella spp, Shigella spp and Diarrheagenic E. coli) causing diarrhea in human have been found in animals. Except for erythromycin resistance rates to ampicillin, co-trimoxazole, chloramphenicol and tetracycline are higher among Shigella spp than Salmonella spp in these countries (Table 5) [40-45]. Shigella spp have been found to be common in DRC and it was noted that more than 50% of Shigella spp from that country were multi-drug resistant strains. This is a 4-fold increase compared to a study which was done in 1964 which demonstrated resistance to two antibiotics to be 10% [42,45].
Table 5.
Antimicrobial resistance rates of Salmonella spp, Shigella spp and diarrheagenic E. coli from the stool, Zambia, DRC, Mozambique and Tanzania
| Antibiotic | Resistance rate in different studies%Salmonellaspp(range):ref:[[41,42]] | Mean resistance n/N* (%) |
|---|---|---|
| Ampicillin |
25 |
10/40(25) |
| Co-trimoxazole |
78, 18 (18–78) |
29/61(48) |
| Chloramphenicol |
15 |
6/40 (15) |
| Tetracycline |
94, 15 (15–94) |
33/61(55) |
| Cefotaxime |
14 |
3/21(14) |
| Erythromycin |
86 |
18/21(86) |
| |
Resistance rate in different studies % Shigella spp (range): ref: [43-45] |
|
| Ampicillin |
97, 56 (56–97) |
130/171 (77) |
| Co-trimoxazole |
97, 84 (84–97) |
155/171 (91) |
| Chloramphenicol |
94, 52 (52–94) |
125/171(73) |
| Tetracycline |
98, 66 (66–98) |
140/171 (82) |
| Erythromycin |
19 |
12/62 (19) |
| |
Resistance rate in different studies % diarrheagenic E. coli(range): ref [51,52,54] |
|
| Ampicillin |
72, 100 (72–100) |
89/104 (86) |
| Co-trimoxazole |
71, 60, 58 (58–71) |
74/118 (63) |
| Chloramphenicol |
45, 100 (45–100) |
75/104 (73) |
| Tetracycline |
48, 100 (48–100) |
77.104 (74) |
| Erythromycin |
83 |
12/14 (83) |
| Ciprofloxacin |
1 |
1/94 (1) |
| Nitrofurantoin | 23 | 3/14 (23) |
N*=Total number of isolates tested.
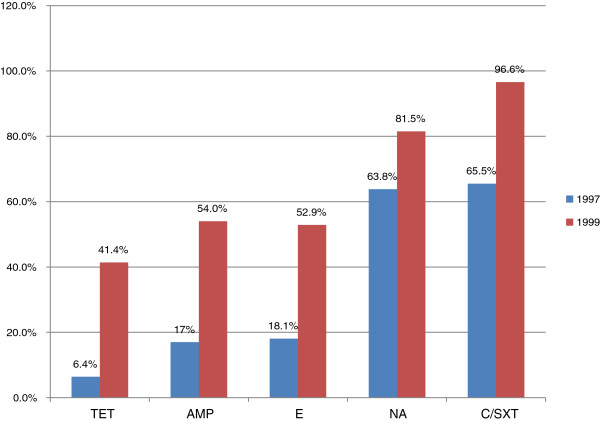
Five studies [46-50] reported on Vibrio cholera with four of them reporting on the susceptibility pattern of Vibrio cholera among isolates obtained during outbreaks [47-50]. In Zambia [47], Vibrio cholera 01 strains isolated from first two outbreaks (1990–1991), showed a low level resistance (2-3%) to tetracycline. The use of tetracycline caused the resistance to increase to 95% in the subsequent outbreaks in 1992. Similar observations were reported in Tanzania [48], whereby it was noted that there was an increase in resistance patterns against V. cholera between outbreaks occurred in 1997 and later outbreaks in1999 (Figure 1). In Mozambique; of 175 rectal swabs or stool samples from patients with diarrhea, 58 strains of V. cholerae 01 were isolated between January 7 and March 8, 2004. The antimicrobial susceptibility of 15 isolates showed that all strains were sensitive to tetracycline, ampicillin, furazolidine, erythromycin and ciprofloxacin but resistant to co-trimoxazole [49].
Figure 1.
Pattern of resistance of Vibrio cholerae isolates from Tanzania to tetracycline (TET), erythromycin (E), nalidixic acid (NA) and chloramphenicol / co-trimoxazole in 1997 and 1999.
Five studies [51-55] investigated the presence diarrheagenic E. coli strains in these countries with 3 of studies [51,52,54] describing susceptibility pattern to various antibiotics. The mean resistance rates of diarrheagenic E. coli isolated in these countries for ciprofloxacin, nitrofurantoin, co-trimoxazole, chloramphenicol, tetracycline, erythromycin and ampicillin are seen in Table 5.
Campylobacter spp is one the commonest causes of diarrhea in children but only four short-term studies could be found on the subject from Tanzania and Mozambique [44,56-58]. The prevalence of campylobacter infection in human in Tanzania was found to range from 2.6% to 9.3% [56-58]. All these human studies from Tanzania did not test for the susceptibility pattern of the isolates. The other study [59] in the same area investigated the presence of Campylobacter spp in domestic ducks; found that 80% of ducks had thermophilic campylobacter infection. Of 50 isolates tested for susceptibility pattern 48%, 74% and 82% were resistant to cefuroxime, tetracycline and amoxycillin respectively while 35% of the isolates were resistant to erythromycin. The resistance rates to norfloxacin and ciprofloxacin were 10% and 16% respectively (Table 6). In Mozambique of 529 samples from children 9 (1.7%) had campylobacter infection, all nine Campylobacter spp strains isolated from children with diarrhea were susceptible to erythromycin while 1 (11) was resistant to ciprofloxacin [44].
Table 6.
Antimicrobial resistance rates of Escherichia coli from animals and Campylobacter spp in Mozambique, Tanzania and Zambia (ref: [51,59,74,77])
| Antibiotic | Resistance rate in different studies: Escherichia coli%(range) | Mean resistance n/N* (%) |
|---|---|---|
| Ampicillin |
80, 90, 88 (80–90) |
356/414 (86) |
| Augmentin |
82, 74, 83 (74–83) |
276/346 (79) |
| Gentamicin |
63, 67 (63–67) |
191/294(65) |
| Co-trimoxazole |
36, 71, 91 (36–91) |
282/427 (66) |
| Tetracycline |
4, 96 (4–96) |
154/307 (50) |
| |
Resistance rates Campylobacter jejuni %(range) |
|
| Amoxicillin |
82 |
41/50 (82) |
| Tetracycline |
74, 22 (22–74) |
28/59 (48) |
| Erythromycin |
35, 0.0 (0–35) |
18/59 (30) |
| Cefuroxime |
48 |
24/50 (48) |
| Norfloxacin |
10 |
5/50 (10) |
| Ciprofloxacin | 16, 11 (11–16) | 9/59 (15) |
N*=Total number of isolates tested.
Methicillin-resistant Staphylococcus aureus (MRSA) and Health care-associated infections (HCAIs)
The WHO estimates that health care-associated infections (HCAIs) in developing countries are found in 15.5% of admissions, which is 3 times higher than the rate in developed countries [60,61]. One form of infection, the surgical site infection, is particularly common in developing countries due to the economic constraints of hospitals and lack of hygiene, infection prevention strategies and staff [29,62-66]. Staphylococcus aureus has been found to be the predominant pathogen causing SSI in these countries with 13% of the bacteria exhibiting MRSA phenotype (Table 7). Interestingly, the prevalence of MRSA in Tanzania has been increasing, at the Muhimbili National Hospital, the prevalence was 0.4% in 1999 [66], 2% in 2004 [24], and 23.3% in 2010 [25]. At the Bugando Medical Center (BMC), the prevalence of MRSA was 16.3% [67] in 2009 and 18.8% in 2011 [61].
Table 7.
Antimicrobial resistance rates of Staphylococcus aureus , Zambia, DRC, Mozambique and Tanzania (ref: [26,62,68-72])
| Antibiotic | Resistance rate in different studies%(range) | Mean resistance n/N* (%) |
|---|---|---|
| Penicillin |
90, 90, 85 (85–90) |
291/329 (88) |
| Erythromycin |
35, 5, 7 (5–35) |
51/327 (16) |
| Clindamycin |
22 |
10/46 (22) |
| Co-trimoxazole |
31, 73 (31–73) |
145/278 (52) |
| Chloramphenicol |
37, 7 (7–37) |
63/287 (22) |
| Gentamicin |
5 |
9/182 (5) |
| Tetracycline |
48, 28 (28–48) |
56/147 (38) |
| MRSA | 31, 16, 2, 27, 2, 8 (2–31) | 317/2472 (13) |
N*=Total number of isolates tested.
Nine studies [26,62,63,67-72] reported the resistance patterns of Staphylococcus aureus with only two studies providing molecular insights of MRSA. The mean resistance rates ranges from 5% to 88% to various antibiotics as seen in Table 7. Regarding spa types, sixteen different spa types (t012, t021, t122, t186, t279, t701, t1855, t1877, t224, t084, t304, t934, t1247, t2864, t7722 and t7723) were observed in one study in Zambia [69], while in Tanzania; the new MRSA clone ST1797/t7231 has been isolated thus emphasizing the diversity of MRSA clones in Africa [72]. In Mozambique; of 24 MRSA isolates 6(25%), 3(12.5%) and 1(4.2%) were also resistant to tetracycline, erythromycin and co-trimoxazole and only 1 strain was resistant to all antibiotics tested [26].
Antibiotic resistance in isolates from animals
Eight reviewed studies [51,59,73-78] investigated the magnitude of bacterial diseases in animals with only 4 studies [51,59,77,78] reporting resistance profile to various antibiotics. As for Escherichia coli from human; Escherichia coli from animals exhibited high resistance rates ranging from 50% for tetracycline to 86% for ampicillin (Table 7). Salmonella enteritidis was found to contaminate 3.8% and 4.7% of eggs and chicken carcasses respectively. All Salmonella enteritidis isolates were found to be sensitive to gentamicin, ampicillin, tetracycline, co-trimoxazole, amoxicillin, furazolidine and chloramphenicol.
Discussion
In the developing countries under review, there are limited data from large surveillance studies on antimicrobial resistance. In addition few short clinical studies document the susceptibility pattern of common pathogens from human and animals. This may partly be due to the lack of microbiological facilities in many health facilities in developing countries [79]. Our findings emphasize the need for coordinated efforts to improve the diagnosis of infectious diseases in developing countries coupled with surveillance of antimicrobial resistance in these countries. Together with the increased effort by WHO to control malaria transmission, other potential causes of fever should be taken into consideration; and appropriate antibiotic treatment will reduce morbidity and mortality resulting from other causes of fever.
Despite few studies blood stream and urinary tract infections have been found to be common in these countries as demonstrated in this review and it should be noted that the endemicity of HIV has changed their epidemiology in Africa. Apart from Salmonella spp; multi-drug Escherichia coli and Klebsiella pneumoniae were found to be common causes of blood stream infections and UTI. Increased trend of these isolates to become resistant to ampicillin, gentamicin and third generations’ cephalosporins was noted; this could be due to overuse of these drugs in the community and hospitals, in all countries reviewed no clear antibiotic policy was found.
ESBL has been found to be a threat, especially as a cause of nosocomial infections. Prevalence as high as 50% have been observed among Klebsiella pneumoniae from inpatients in these countries. The occurrence of the Escherichia coli clone ST 131 in Tanzania confirms that resistant clones can spread from one part of the world to another [28]. Low mean resistance rates to meropenem were observed in these countries; this could be explained by the fact that this drug is expensive and not available in the market. There is an urgent need of antibiotic policy in these countries because in countries where carbapenems have been misused, such as India or Pakistan, outbreaks of carbapenems resistant Escherichia coli and Klebsiella pneumoniae have been experienced [11]. Recently, the emergence of NDM-1 plasmid mediated carbapenems resistance has been noted, spreading from India to Europe, USA and Africa. Joint efforts are needed to control the spread of NDM-1.
In this review few studies were found to address enteric pathogens, it was noted that Shigella spp were more resistance than Salmonella spp; an increase trend of multi--drug resistant Shigella spp in DRC was noted. Similarly most of diarrheagenic Escherichia coli were resistant to commonly used antibiotics (ampicillin, co-trimoxazole, tetracycline and erythromycin). This could be due to self prescription of these antibiotics for the treatment of diarrhea episodes as evidenced by increased resistance of Vibrio cholerae strains between outbreaks in Zambia and Tanzania. In all countries under review, many patients buy antibiotics from private pharmacies and drug shops for self-medication before seeking medical professional care [80-82]. In addition Campylobacter spp were found to be resistant to ciprofloxacin, cefuroxime and erythromycin. This situation needs to be further investigated with standardize microbiological method so that the real magnitude can be established or confirmed.
In addition, MRSA appears to be an emerging problem; the problem might be underestimated because not all laboratories in these countries are performing MRSA identification. However Staphylococcus aureus has been the major cause of SSI, as documented in few studies from these countries. The increase trend of MRSA as noted in Tanzania; necessitate the coordinated surveillance to determine the evolution of these strains in Africa. MRSA isolates have been isolated from animals and we need, therefore, to compare the genotypes between animals and humans, as evidenced by the diversity of MRSA genotypes in Tanzania and Zambia [68-70].
Few laboratories are routinely conducting testing for ESBL and MRSA detection. This observation stresses the need for governmental and non- governmental organizations to provide sustainable support to improve laboratory capacity in developing countries. This should go hand in hand with the establishment of a quality assurance system to ensure quality microbiological results from all laboratories. As noted in Tanzania, there is an increased resistance trend to ceftriaxone and ceftazidime among isolates causing nosocomial infections. Improving diagnostic facilities and research capacity to determine the evolution of ESBL and MRSA clones in Africa is no longer an option but is mandatory, especially when the treatment of ESBL producing isolates and MRSA becomes too expensive for countries like DRC, Mozambique, Tanzania and Zambia.
Unauthorized use of antibiotics seems to be common, both in medical and veterinary settings. While we have found no evidence linking antimicrobial resistance in human cases with the use of similar antibiotics in animals, there is a need for a coordinated one health based surveillance of the antimicrobial resistance in humans and animals. It appears also relevant to follow the recommendations of the Joint FAO-OIE-WHO on non-human antimicrobial usage.
Conclusions
Relevant information on bacterial diseases is limited in DRC, Mozambique, Tanzania and Zambia. Moreover, there are no national policies guiding surveillance. Lack of detailed microbiological method was noted in various studies with no detailed information regarding quality assurance; but despite this limitation an increased trend of resistance to commonly used antibiotics such as ampicillin, co-trimoxazole, gentamicin, erythromycin, tetracycline and third generation cephalosporins was noted. This might be exacerbated by behavioral factors, incentive motivate prescribing, and the dispensing and purchasing of antibiotics when they are inappropriate for treating a specific condition. Other contributing factors include the drugs’ quality or the use of partial doses, and patients’ demand for symptomatic eradications. A systematic national surveillance system is urgently needed to provide data on the levels of bacterial diseases and drug resistance in common pathogens from hospitals and communities. In addition, a comparative molecular epidemiology study to compare human and animals isolates is urgently needed to shade a light of the transmission of bacterial pathogens, especially at the community level. The rising incidence of MRSA and ESBL infections points to the need to include the heightened the regimes for hospital facility cleanliness as part of the policy for the use of antibiotics and antibiotic stewardship in order to minimize the risk of hospital-acquired infections with MRSA and ESBL producing isolates.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SEM, MM and MR participated in the literature search; SEM prepared the first draft of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Showing Methodology used for susceptibility testing, population sampled and specimens collected in various studies.
Contributor Information
Stephen E Mshana, Email: mshana72@yahoo.com.
Mecky Matee, Email: mmateemi@yahoo.com.
Mark Rweyemamu, Email: mark.rweyemamu@sacids.org.
Acknowledgements
This study was supported mainly by a grant from the Rockefeller Foundation (2011-DSN-307) to SACIDS at Sokoine University and secondarily by the grant from the Wellcome Trust (WT087546MA) also to SACIDS.
The authors acknowledge the support provided by their respective universities and the encouragements to undertake this one health based review.
References
- Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;146:837. [PubMed] [Google Scholar]
- Manges AR, Johnson JR, Foxman B, O'Bryan TT, Fullerton KE, Riley LW. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N Engl J Med. 2001;345:1007–1013. doi: 10.1056/NEJMoa011265. [DOI] [PubMed] [Google Scholar]
- Mshana SE, Kamugisha E, Mirambo M, Chakraborty T, Lyamuya EF. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza Tanzania. BMC Res Notes. 2009;2:49. doi: 10.1186/1756-0500-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MD, Wilkins J. Antibiotic resistance in bacteria isolated from poultry. Rural Industries Research and Development Corporation Publication No 1/105 Report of RIRDC Project No USA-9A. 2001. In.
- Aarestrup FM, Wegener HC. The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1999;1(8):639–644. doi: 10.1016/S1286-4579(99)80064-1. [DOI] [PubMed] [Google Scholar]
- Hart CA, Kariuki S. Antimicrobial resistance in developing countries. BMJ. 1998;317(7159):647–650. doi: 10.1136/bmj.317.7159.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum PC, Bhamjee A, Scragg JN, Hallett AF, Bowen AJ, Cooper RC. Streptococcus pneumoniae resistant to penicillin and chloramphenicol. Lancet. 1977;2(8046):995–997. doi: 10.1016/s0140-6736(77)92892-6. [DOI] [PubMed] [Google Scholar]
- WHO. Emergence of multi-drug resistant Neisseria gonorrhoeae - Threat of global rise in untreatable sexually transmitted infections. WHO/RHR. 2011;11:14. [Google Scholar]
- Vlieghe E, Phoba M, Tamfun J, Jacobs J. Antibiotic resistance among bacterial pathogens in Central Africa: a review of the published literature between 1955 and 2008. Inter J Antimicro Agents. 2009;34(4):295–303. doi: 10.1016/j.ijantimicag.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Ferreira JP, Anderson KL, Correa MT, Lyman R, Ruffin F, Reller LB, Fowler VG Jr. Transmission of MRSA between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS One. 2011;6(11):e26978. doi: 10.1371/journal.pone.0026978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicro Agents Chemother. 2011;55(2):934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festo E, Kidenya BR, Hokororo A, Mshana SE. Predictors of urinary tract infection among febrile children attending at Bugando Medical Centre, Northwestern Tanzania. Arch Clin Microbiol. 2011;2(5):2. [Google Scholar]
- Msaki BP, Mshana SE, Hokororo A, Mazigo HD, Morona D. Prevalence and predictors of urinary tract infection and severe malaria among febrile children attending Makongoro health centre in Mwanza city North-Western Tanzania. Arch Public Health. 2012;70(1):4. doi: 10.1186/0778-7367-70-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masinde A, Gumodoka B, Kilonzo A, Mshana SE. Prevalence of urinary tract infection among pregnant women at Bugando Medical Centre, Mwanza Tanzania. Tanzan J Health Res. 2009;11(3):154–159. doi: 10.4314/thrb.v11i3.47704. [DOI] [PubMed] [Google Scholar]
- Lyamuya EF, Moyo SJ, Komba EV, Haule M. Prevalence, antimicrobial resistance and associated risk factors for bacteriuria in diabetic women in Dar es Salaam Tanzania. Afr J Microbiol Res. 2011;5(6):683–689. [Google Scholar]
- Isaack H, Mbise R, Hirji K. Nosocomial bacterial infections among children with severe protein energy malnutrition. East Afr Med J. 1992;69(8):433–436. [PubMed] [Google Scholar]
- Moyo SJ, Aboud S, Kasubi M, Maselle SY. Bacterial isolates and drug susceptibility patterns of urinary tract infection among pregnant women at Muhimbili National Hospital in Tanzania. Tanzan J Health Res. 2010;12(4):233–236. doi: 10.4314/thrb.v12i4.52997. [DOI] [PubMed] [Google Scholar]
- Mshana S, Kamugisha E, Mirambo M, Kataraihya J. The role of uncentrifuged urine microscopy in the diagnosis of urinary tract infection. Tanzan Med J. 2011;25(1):17–18. [Google Scholar]
- Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY. Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC Res Notes. 2010;3(1):348. doi: 10.1186/1756-0500-3-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoy G, Justin-Temu M, Mndolwa M. Antibiotic sensitivity of bacterial pathogens in urinary tract infections at Muhimbili National Hospital, Dar es Salaam, Tanzania. East Cent Afr J Pharma Sci. 2007;9(3):67–70. [Google Scholar]
- Blomberg B, Olsen BE, Hinderaker SG, Langeland N, Gasheka P, Jureen R, Kvåle G, Midtvedt T. Antimicrobial resistance in urinary bacterial isolates from pregnant women in rural Tanzania: implications for public health. Scand J Infect Dis. 2005;37(4):262–268. doi: 10.1080/00365540410021045. [DOI] [PubMed] [Google Scholar]
- Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DS, Urassa WK, Fataki M, Msangi V, Tellevik MG, Maselle SY. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J Clin Micro. 2005;43(2):745–749. doi: 10.1128/JCM.43.2.745-749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr. 2010;10:39. doi: 10.1186/1471-2431-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg B, Mwakagile DS, Urassa WK, Maselle SY, Mashurano M, Digranes A, Harthug S, Langeland N. Surveillance of antimicrobial resistance at a tertiary hospital in Tanzania. BMC Public Health. 2004;4(1):45. doi: 10.1186/1471-2458-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo S, Aboud S, Kasubi M, Maselle S. Bacteria isolated from bloodstream infections at a tertiary hospital in Dar es Salaam, Tanzania: antimicrobial resistance of isolates. SAMJ: South Afri Med J. 2010;100(12):835–838. doi: 10.7196/samj.4186. [DOI] [PubMed] [Google Scholar]
- Mandomando I, Sigaúque B, Morais L, Espasa M, Vallès X, Sacarlal J, Macete E, Aide P, Quintò L, Nhampossa T. Antimicrobial drug resistance trends of bacteremia isolates in a rural hospital in southern Mozambique. Am J Trop Med Hyg. 2010;83(1):152. doi: 10.4269/ajtmh.2010.09-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg B, Manji KP, Urassa WK, Tamim BS, Mwakagile DS, Jureen R, Msangi V, Tellevik MG, Holberg-Petersen M, Harthug S. Antimicrobial resistance predicts death in Tanzanian children with bloodstream infections: a prospective cohort study. BMC Infect Dis. 2007;7(1):43. doi: 10.1186/1471-2334-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect. 2011;17(8):1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- Ndugulile F, Jureen R, Harthug S, Urassa W, Langeland N. Extended Spectrum β-Lactamases among Gram-negative bacteria of nosocomial origin from an Intensive Care Unit of a tertiary health facility in Tanzania. BMC Inf Dis. 2005;5(1):86. doi: 10.1186/1471-2334-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boeck H, Lunguya O, Muyembe J-J, Glupczynski Y, Jacobs J. Presence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in waste waters, Kinshasa, the Democratic Republic of the Congo. Eur J Clin Microbiol Infect Dis. 2012;31(11):3085–3088. doi: 10.1007/s10096-012-1669-8. [DOI] [PubMed] [Google Scholar]
- Kariuki S, Cheesbrough J, Mavridis A, Hart C. Typing of Salmonella enterica serotype paratyphi C isolates from various countries by plasmid profiles and pulsed-field gel electrophoresis. J Clin Micro. 1999;37(6):2058–2060. doi: 10.1128/jcm.37.6.2058-2060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyembe-Tamfum J, Veyi J, Kaswa M, Lunguya O, Verhaegen J, Boelaert M. An outbreak of peritonitis caused by multidrug-resistant< i> Salmonella Typhi</i> in Kinshasa, Democratic Republic of Congo. Travel Med Infect Dis. 2009;7(1):40–43. doi: 10.1016/j.tmaid.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Lunguya O, Lejon V, Phoba M-F, Bertrand S, Vanhoof R, Verhaegen J, Smith AM, Keddy KH, Muyembe-Tamfum J-J, Jacobs J. Salmonella typhi in the democratic republic of the congo: fluoroquinolone decreased susceptibility on the rise. PLoS Negl Trop Dis. 2012;6(11):e1921. doi: 10.1371/journal.pntd.0001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoba M-F, Lunguya O, Mayimon DV, di Mputu PL, Bertrand S, Vanhoof R, Verhaegen J, Van Geet C, Muyembe J-J, Jacobs J. Multidrug-resistant Salmonella enterica, Democratic Republic of the Congo. Emerg Infect Dis. 2012;18(10):1692. doi: 10.3201/eid1810.120525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz J, Herrera-Leon S, Mandomando I, Macete E, Puyol L, Echeita A, Alonso PL. Detection of Salmonella enterica serotype Typhimurium DT104 in Mozambique. Am J Trop Med Hyg. 2008;79(6):918–920. [PubMed] [Google Scholar]
- Sigaúque B, Roca A, Mandomando I, Morais L, Quintó L, Sacarlal J, Macete E, Nhamposa T, Machevo S, Aide P. Community-acquired bacteremia among children admitted to a rural hospital in Mozambique. Pediatr Infec Dis J. 2009;28(2):108–113. doi: 10.1097/INF.0b013e318187a87d. [DOI] [PubMed] [Google Scholar]
- Meremo A, Mshana SE, Kidenya BR, Kabangila R, Peck R, Kataraihya JB. High prevalence of non-typhoid salmonella bacteraemia among febrile HIV adult patients admitted at a tertiary Hospital North-Western Tanzania. Int Arch Med. 2012;5(1):28. doi: 10.1186/1755-7682-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon J, Vargas M, Schellenberg D, Urassa H, Casals C, Kahigwa E, Aponte J, Mshinda H, Vila J. Diarrhea in children under 5 years of age from Ifakara, Tanzania: a case–control study. J Clin Micro. 2000;38(12):4459–4462. doi: 10.1128/jcm.38.12.4459-4462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack RB, Rahman M, Yunus M, Khan EH. Antimicrobial resistance in organisms causing diarrheal disease. Clin Infect Dis. 1997;24(1):S102–S105. doi: 10.1093/clinids/24.supplement_1.s102. [DOI] [PubMed] [Google Scholar]
- Mwansa J, Mutela K, Zulu I, Amadi B, Kelly P. Antimicrobial sensitivity in enterobacteria from AIDS patients Zambia. Emerging Infec Dis. 2002;8(1):92. doi: 10.3201/eid0801.010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa A, Foster S, Ochieng D, Grimm D, Mwenya D. Resistant acute respiratory infections and diarrheal diseases in Zambia: A situation and needs assessment. APUA. 2010.
- Lunguya O Asuni M Mumba D Mabwa L Nsungu M Manienga J Sérotypes et pharmacorésistance des Salmonella et Shigella à Kinshasa (1994–1999) Congo Méd 20054227–233.24105933 [Google Scholar]
- Temu M, Kaatano G, Miyaye N, Buhalata S, Shushu M, Kishamawe C, Changalucha J. Antimicrobial susceptibility of Shigella flexneri and S. dysenteriae isolated from stool specimens of patients with bloody diarrhoea in Mwansa, Tanzania. Tanzan J Health Res. 2008;9(3):186–189. [PubMed] [Google Scholar]
- Mandomando I, Jaintilal D, Pons MJ, Vallès X, Espasa M, Mensa L, Sigaúque B, Sanz S, Sacarlal J, Macete E. Antimicrobial susceptibility and mechanisms of resistance in Shigella and Salmonella isolates from children under five years of age with diarrhea in rural Mozambique. Antimicro Agents Chemother. 2009;53(6):2450–2454. doi: 10.1128/AAC.01282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo J, Niel L, Talarmin A, Dubrous P. [Antibiotic sensitivity to epidemic strains of Vibrio cholerae and Shigella dysenteriae 1 isolated in Rwandan refugee camps in Zaire] Med trop revue du Corps de sante colonial. 1994;55(4):351–353. [PubMed] [Google Scholar]
- Colombo MM, Mastrandrea S, Leite F, Santona A, Uzzau S, Rappelli P, Pisano M, Rubino S, Cappuccinelli P. Tracking of clinical and environmental Vibrio cholerae O1 strains by combined analysis of the presence of the toxin cassette, plasmid content and ERIC PCR. FEMS Immunol Med Micro. 1997;19(1):33–45. doi: 10.1111/j.1574-695X.1997.tb01070.x. [DOI] [PubMed] [Google Scholar]
- Mwansa J, Mwaba J, Lukwesa C, Bhuiyan N, Ansaruzzaman M, Ramamurthy T, Alam M, Balakrish Nair G. Multiply antibiotic-resistant Vibrio cholerae O1 biotype El Tor strains emerge during cholera outbreaks in Zambia. Epidemiol Infect. 2007;135(5):847–853. doi: 10.1017/S0950268806007254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urassa W, Mhando Y, Mhalu F, Mgonja S. Antimicrobial susceptibility pattern of Vibrio cholerae 01 strains during two cholera outbreaks in Dar Es Salaam, Tanzania. East Afr Med J. 2000;77(7):350–353. [PubMed] [Google Scholar]
- Ansaruzzaman M, Bhuiyan N, Nair GB, Sack DA, Lucas M, Deen JL, Ampuero J, Chaignat C-L. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis. 2004;10(11):2057. doi: 10.3201/eid1011.040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Han KH, Choi SY, Lucas ME, Mondlane C, Ansaruzzaman M, Nair GB, Sack DA, von Seidlein L, Clemens JD. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J Med Microb. 2006;55(2):165–170. doi: 10.1099/jmm.0.46287-0. [DOI] [PubMed] [Google Scholar]
- Mubita C, Syakalima M, Chisenga C, Munyeme M, Bwalya M, Chifumpa G, Hang ombe BM, Sinkala P, Simuunza M, Fukushi H. Antibiograms of faecal Escherichia coli and Enterococci species isolated from pastoralist cattle in the interface areas of the Kafue basin in Zambia. Veterinarski arhiv. 2008;78(2):179. [Google Scholar]
- Raji M, Minga U, Machang'u R. Prevalence and characterization of verotocytoxin producing Escherichia coli O157 from diarrhoea patients in Morogoro Tanzania. Tanzan J Health Res. 2008;10(3):151–158. doi: 10.4314/thrb.v10i3.14355. [DOI] [PubMed] [Google Scholar]
- Rappelli P, Folgosa E, Solinas ML, DaCosta JL, Pisanu C, Sidat M, Melo J, Cappuccinelli P, Colombo MM. Pathogenic enteric Escherichia coli in children with and without diarrhea in Maputo Mozambique. FEMS Imm Med Micro. 2005;43(1):67–72. doi: 10.1016/j.femsim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Mandomando IM, Macete EV, Ruiz J, Sanz S, Abacassamo F, Valles X, Sacarlal J, Navia MM, Vila J, Alonso PL. Etiology of diarrhea in children younger than 5 years of age admitted in a rural hospital of southern Mozambique. Am J Trop Med Hyg. 2007;76(3):522–527. [PubMed] [Google Scholar]
- Moyo SJ, Maselle SY, Matee MI, Langeland N, Mylvaganam H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect Dis. 2007;7(1):92. doi: 10.1186/1471-2334-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oketcho R, Nyaruhucha C, Taybali S, Karimuribo E. Influence of enteric bacteria and parasite infection and nutritional status on diarrhoea occurrence in six to 60 month old children admitted at Morogoro Regional Hospital of Tanzania. Tanzan J Health Res. 2012;14(2) doi: 10.4314/thrb.v14i2.3. doi.org/10.4314/thrb.v14i2.3. [DOI] [PubMed] [Google Scholar]
- Mdegela R, Nonga H, Ngowi H, Kazwala R. Prevalence of thermophilic Campylobacter infections in humans, chickens and crows in Morogoro, Tanzania. J Vet Med B. 2006;53(3):116–121. doi: 10.1111/j.1439-0450.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- Vargas M, Gascon J, Casals C, Schellenberg D, Urassa H, Kahigwa E, Ruiz J, Vila J. Etiology of diarrhea in children less than five years of age in Ifakara, Tanzania. Am J Trop Med Hyg. 2004;70(5):536–539. [PubMed] [Google Scholar]
- Nonga HE, Muhairwa A. Prevalence and antibiotic susceptibility of thermophilic Campylobacter isolates from free range domestic duck (Cairina moschata) in Morogoro municipality, Tanzania. Trop Anim Health Prod. 2010;42(2):165–172. doi: 10.1007/s11250-009-9401-0. [DOI] [PubMed] [Google Scholar]
- Nejad SB, Allegranzi B, Syed SB, Ellis B, Pittet D. Health-care-associated infection in Africa: a systematic review. Bull World Health Organ. 2011;89(10):757–765. doi: 10.2471/BLT.11.088179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawalla B, Mshana SE, Chalya PL, Imirzalioglu C, Mahalu W. Predictors of surgical site infections among patients undergoing major surgery at Bugando Medical Centre in Northwestern Tanzania. BMC Surg. 2011;11:21. doi: 10.1186/1471-2482-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen H, Chugulu S, Kondo S, Lingaas E. Surgical-site infections at Kilimanjaro Christian Medical Center. J Hosp Infect. 2003;55(1):14–20. doi: 10.1016/S0195-6701(03)00225-1. [DOI] [PubMed] [Google Scholar]
- Kayihura V, Osman NB, Bugalho A, Bergström S. Choice of antibiotics for infection prophylaxis in emergency cesarean sections in low‒income countries: A cost‒benefit study in Mozambique. Acta Obstet Gynecol Scand. 2003;82(7):636–641. doi: 10.1034/j.1600-0412.2003.00205.x. [DOI] [PubMed] [Google Scholar]
- Gosling R, Mbatia R, Savage A, Mulligan J-A, Reyburn H. Prevalence of hospital-acquired infections in a tertiary referral hospital in northern Tanzania. Ann Trop Med Parasitol. 2003;97(1):69–73. doi: 10.1179/000349803125002724. [DOI] [PubMed] [Google Scholar]
- Mukwato K, Ngoma C, Maimbolwa M. Compliance with infection prevention guidelines by health care workers at Ronald Ross General Hospital Mufulira District. Med J Zambia. 2008;35(3):110–116. [Google Scholar]
- Urassa W, Haule E, Kagoma C, Langeland N. Antimicrobial susceptibillty of Staphylococcus aureus strains at Muhimbili Medical Centre, Tanzania. East Afr Med J. 1999;76(12):693–695. [PubMed] [Google Scholar]
- Mshana S, Kamugisha E, Mirambo M, Rambau P, Chalya P, Mahalu W, Lyamuya E. Prevalence of clindamycin inducible resistance among methicillin-resistant Staphylococcus aureus at Bugando Medical Centre, Mwanza Tanzania. Tanzan J Health Res. 2009;11(2):59–64. [Google Scholar]
- Kapatamoyo B, Andrews B, Bowa K. Association of HIV with breast abscess and altered microbial susceptibility patterns. Med J Zambia. 2010;37(2):58–63. [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F, Mugisha L, Peck B, Becker K, Gillespie TR, Peters G, Leendertz FH. Drug‒resistant human Staphylococcus Aureus in sanctuary apes pose a threat to endangered wild ape populations. Am J Primatol. 2012;74(12):1071–1075. doi: 10.1002/ajp.22067. [DOI] [PubMed] [Google Scholar]
- Takaisi K, Kandolo DK, Musema MG, Sadiki NH. Resistance aux antibiitques des Enterobacteries et des Staphyloccocques isoles en milieu hospitalier de la ville de Kinshas. Congo Med. 2003;3:1367–1371. [Google Scholar]
- Nyembwe KM, Manienga KJ, Manebwa KJD, Mulumba MP, Verhaeegen J, Myembe TJ. Profil del methillino resistance de Staphylococcus a Kishasa et implication dans I’hygiene hospitaliere. Congo Med. 2004;3:1262–1265. [Google Scholar]
- Moremi N, Mshana SE, Kamugisha E, Kataraihya J, Tappe D, Vogel U, Lyamuya EF, Claus H. Predominance of methicillin resistant Staphylococcus aureus -ST88 and new ST1797 causing wound infection and abscesses. J Infect Dev Ctries. 2012;6(8):620–625. doi: 10.3855/jidc.2093. [DOI] [PubMed] [Google Scholar]
- Ngoma M, Suzuki A, Takashima I, Sato G. Antibiotic resistance of Escherichia coli and Salmonella from apparently healthy slaughtered cattle and pigs, and diseased animals in Zambia. Jpn J Vet Res. 1993;41(1):1–10. [PubMed] [Google Scholar]
- Munang’andu HM, Kabilika SH, Chibomba O, Munyeme M, Muuka GM. Bacteria isolations from broiler and layer chicks in Zambia. J Pathogens. 2012;520564 doi: 10.1155/2012/520564. 2012, doi: 10.1155/2012/520564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang'Ombe B, Sharma R, Tuchili L, Skjerve O. Isolation of bacteria from table eggs in Zambia. Indian J Anim Sci. 1999;69(6):445–447. [Google Scholar]
- Hang'ombe B, Sharma R, Skjerve E, Tuchili L. Occurrence of Salmonella enteritidis in pooled table eggs and market-ready chicken carcasses in Zambia. Avian Dis. 1999;43(3):597–599. doi: 10.2307/1592662. [DOI] [PubMed] [Google Scholar]
- Iqbal M, Shah I, Ali A, Khan M, Jan S. Prevalence and in vitro antibiogram of bacteria associated with omphalitis in chicks. Proteus. 2006;13:5–8. [Google Scholar]
- Pfaller MA, Diseases JSCI. Overview of the epidemiological profile and laboratory detection of extended-spectrum b-lactamases. Clin Infect Dis. 2006;42(Suppl 4):S153–S163. doi: 10.1086/500662. [DOI] [PubMed] [Google Scholar]
- Boogaard J, Semvua H, Boeree M, Aarnoutse R, Kibiki G. Assessment of antibacterial sale by using the Anatomic Therapeutic Chemical classification and defined daily dose methodology in Moshi Municipality, Northern Tanzania. Tanzan J Health Res. 2012;13(3):208–212. [Google Scholar]
- Viberg N, Kalala W, Mujinja P, Tomson G, Lundborg CS. Practical knowledge" and perceptions of antibiotics and antibiotic resistance among drugsellers in Tanzanian private drugstores. BMC Infect Dis. 2010;10:270. doi: 10.1186/1471-2334-10-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfson A, Huebner R, Wasas A, Chola S, Godfrey-Faussett P, Klugman K. Nasopharyngeal carriage of community-acquired, antibiotic-resistant Streptococcus pneumoniae in a Zambian paediatric population. Bull World Health Org. 1997;75(5):453. [PMC free article] [PubMed] [Google Scholar]
- Vila J, Pal T. Update on antibacterial resistance in low-income countries: factors favoring the emergence of resistance. Open Infec Dis J. 2010;4:38–54. doi: 10.2174/1874279301004020038. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Showing Methodology used for susceptibility testing, population sampled and specimens collected in various studies.