Abstract
Immune-related genes are often characterized by adaptive protein evolution. Selection on immune genes can be particularly strong when hosts encounter novel parasites, for instance, after the colonization of a new habitat or upon the exploitation of vacant ecological niches in an adaptive radiation. We examined a set of new candidate immune genes in East African cichlid fishes. More specifically, we studied the signatures of selection in five paralogs of the human immunodeficiency virus type I enhancer-binding protein (Hivep) gene family, tested their involvement in the immune defense, and related our results to explosive speciation and adaptive radiation events in cichlids. We found signatures of long-term positive selection in four Hivep paralogs and lineage-specific positive selection in Hivep3b in two radiating cichlid lineages. Exposure of the cichlid Astatotilapia burtoni to a vaccination with Vibrio anguillarum bacteria resulted in a positive correlation between immune response parameters and expression levels of three Hivep loci. This work provides the first evidence for a role of Hivep paralogs in teleost immune defense and links the signatures of positive selection to host–pathogen interactions within an adaptive radiation.
Keywords: positive selection, immune response, adaptive radiation, molecular evolution
The interplay between hosts and their parasites (i.e., macroparasites, bacteria, and viruses) represents one of the strongest biological interactions (Haldane 1949). Pathogens impose strong selection pressures on their hosts and have the potential to rapidly change the genotypic composition of host populations, which may ultimately alter the structure of entire ecosystems (Thompson 1988; Ebert and Hamilton 1996; Keesing et al. 2010). To counteract the permanently evolving pathogen virulence, hosts evolve resistance through diverse immune response mechanisms (Hamilton 1980). These include the discrimination between self and nonself, facilitating the recognition of pathogen-derived epitopes (Altizer et al. 2003; Boots et al. 2008). Invertebrates and vertebrates share the architecture of the innate immune system, i.e., a conserved immediate defense mechanism including Toll-like receptors, lysozymes, and cellular defenses (Janeway et al. 2001). Immune memory, however, is the hallmark of the adaptive immune system of vertebrates (Cooper and Alder 2006; Flajnik and Kasahara 2010). The unique somatic diversification of receptors of the immunoglobulin family during ontogeny [i.e., V(D)J recombination] mediates a dramatic increase in the number of foreign pathogen epitopes that the adaptive immune system can remember and attack (Janeway et al. 1996; Zhu et al. 2012).
Because of the constant interplay between host–parasite adaptation and counteradaption, immune genes are commonly characterized by signatures of positive selection through elevated rates of adaptive protein evolution (Hughes and Nei 1988; Hughes and Nei 1989; Jansa et al. 2003; Schlenke and Begun 2003; Sawyer et al. 2004; Nielsen et al. 2005; Jiggins and Kim 2007; Sackton et al. 2007; Tschirren et al. 2011). Selection on immune system diversification can be particularly strong when hosts encounter novel pathogens that induce primary immune challenges. This is the case after the colonization of a new habitat or upon the exploitation of vacant ecological niches (Scharsack et al. 2007; Matthews et al. 2010; Jones et al. 2012). For instance, it has been shown that migratory birds that encounter two or more different parasite faunas have larger immune defense organs (e.g., bursa and spleen) than closely related resident birds (Møller and Erritzøe 1998). Freshwater sticklebacks differ in their immune competence potential depending on their ecotype (e.g., lower parasite diversity in rivers than in lakes) (Scharsack et al. 2007), which is furthermore supported by a correlation between major histocompatibility complex (MHC) genotype and foraging habitat in benthic and limnetic ecomorphs (Matthews et al. 2010). Given the recognized evolutionary importance of the immune system (Janeway et al. 2001; Rodríguez et al. 2012) and the range of available functional and theoretical knowledge, the next step would be to assess to which degree immune genes contribute to, or even trigger, macroevolutionary events such as divergence, rapid speciation, and adaptive radiation.
East African cichlid fishes are a classic example of adaptive radiation (Schluter 2000). Because of their great numbers of closely related endemic species and their high levels of phenotypic and ecological diversity, cichlids are an important model system to study the genetic basis of diversification, adaption, and speciation (Kornfield and Smith 2000; Kocher 2004; Seehausen 2006; Salzburger 2009; Santos and Salzburger 2012). Previous studies of cichlid adaptive radiations have mainly focused on the understanding of ecologically important traits (and their genetic basis), such as the feeding apparatus (Terai et al. 2002b; Albertson et al. 2005; Fraser et al. 2008; Fraser et al. 2009; Muschick et al. 2011; Muschick et al. 2012), as well as on sexually selected traits such as coloration and pigmentation (Terai et al. 2002a; Terai et al. 2003; Salzburger et al. 2007; Roberts et al. 2009). Fewer studies have addressed the evolution of the immune system or, more generally, physiology in relation to diversification and rapid speciation in cichlids (Blais et al. 2007; Gerrard and Meyer 2007; Dijkstra et al. 2011). Dijkstra et al. (2011), for example, showed that divergence in coloration is accompanied by differentiation in immune function in Lake Victoria cichlids, and divergence in alleles of the MHC has previously been proposed as trigger of speciation in Lake Malawian cichlids through MHC-mediated mate choice (Blais et al. 2007). Several genes related to the immune system, including MHC loci, have been found to show signs of positive selection in East African cichlids (Gerrard and Meyer 2007), suggesting a role for immune genes during cichlid adaptive radiations.
In this study, we focused on the function and molecular characterization of a novel family of immune genes in (cichlid) fishes, which have been implicated to have immunological and developmental functions in mammals and insects (Seeler et al. 1994; Wu et al. 1996; Torres-Vazquez et al. 2001). In a previous study that focused on a candidate gene family for neural crest-derived structures in cichlids (i.e., the endothelin family of ligands and receptors), we detected strong signatures of positive selection in a gene adjacent to one of the focal loci, the zinc finger protein human immunodeficiency virus type I enhancer-binding protein 1 (Hivep1) (Diepeveen and Salzburger 2011). Hivep1 is a transcription factor with functions in a variety of biological and developmental processes, e.g., HIV-1 gene expression (Maekawa et al. 1989; Muchardt et al. 1992; Seeler et al. 1994), in the Decapentaplegic signaling pathway important for cell fate specification during embryogenesis (Grieder et al. 1995; Dai et al. 2000; Marty et al. 2000; Torres-Vazquez et al. 2001), in V(D)J recombination of immunoglobulins (Wu et al. 1993; Wu et al. 1996), and in MHC enhancer binding (Baldwin et al. 1990; William et al. 1995). Although a single copy of this gene is found in Drosophila, mammals are typically characterized by three copies (Hicar et al. 2001; Dürr et al. 2004). Teleost fish, however, possess up to five duplicates (see Braasch et al. 2009), which is in accordance with the 3R hypothesis of a teleost-specific genome duplication event after the 2R duplications in the vertebrate linage (Sidow 1996; Taylor et al. 2003; Meyer and Van De Peer 2005; Volff 2005).
The goal of the current study was threefold. First, we characterized the signatures of selection (i.e., dN/dS ratios) in the five Hivep paralogs in 40 East African cichlid fish species to determine whether adaptive protein evolution is commonly observed in the Hivep gene family. To this end, we performed phylogenetic analyses of the Hivep loci and estimated dN/dS ratios on both codon sites and in individual cichlid lineages. Second, we examined the role of the Hivep paralogs in the immune defense in the cichlid Astatotilapia burtoni. We evaluated the functional connection between Hivep expression levels and several cellular immune parameters after an experimental vaccination with Vibrio anguillarum bacteria. This fish pathogen was chosen to simulate a novel immune challenge, as the host was expected to be immunologically naïve against these Vibrio bacteria. Finally, we examined putative pleiotropic developmental functions through analyses of cis-regulatory regions to obtain insights into other functions of the Hivep paralogs in teleosts that could be linked to the observed signatures of adaptive protein evolution and related our findings to the explosive speciation events in East African cichlid fishes.
Materials and Methods
Sampling, DNA and RNA extraction, and housing conditions
Samples for the DNA analyses were collected during two expeditions to Lake Tanganyika in 2007 and 2008 using a standard operating procedure described by Muschick et al. (2012). In total, 40 different cichlid species from 14 different lineages, including all major cichlid lineages in East Africa (so-called tribes) (Muschick et al. 2012) were examined (Supporting Information, Table S1). RNA for the gene expression assays was extracted from gill, brain, and liver tissue of adult A. burtoni (laboratory strain, both sexes; see Experimental Vaccination section). DNA and RNA extractions were performed as described elsewhere (Diepeveen and Salzburger 2011), with one exception: the tissue homogenization during the RNA extraction was performed on a BeadBeater (FastPrep-24; MP). Animals being part of the experimental vaccination study were kept under standard conditions (12 hr light, 12 hr dark, 25°) in the animal facilities at the Zoological Institute in Basel before transportation to the Helmholtz Centre for Ocean Research Kiel, where they were kept under the following conditions: 14 hr light; 10 hr dark; and 25° for ≥38 hr before the start of the experimental vaccination.
Loci, PCR amplification, and sequencing
Previously, five nuclear Hivep paralogs (i.e., Hivep1, Hivep2a, Hivep2b, Hivep3a, and Hivep3b) were identified in teleost fishes (Braasch et al. 2009). Ensemble (versions 61 and 67) sequences from the following species were downloaded: zebrafish (Danio rerio); cod (Gadus morhua); medaka (Oryzias latipes); spotted green pufferfish (Tetraodon nigroviridis); fugu (Takifugu rubripes); tilapia (Oreochromis niloticus); and stickleback (Gasterosteus aculeatus) (Table S2). For two loci, we performed in-house tblastx searches on the server of the Zoological Institute (University of Basel) to determine Hivep protein sequences in the preliminary cichlid genomes of A. burtoni, Neolamprologus brichardi, and Pundamilia nyererei (BROAD Institute, unpublished data). These teleost and cichlid sequences were aligned with Codon Code Aligner 3.7.1 (CodonCode Corporation) to determine exon–intron structure and to design cichlid-specific primers (Table S3).
Subsequent PCR and sequencing reactions were performed as described elsewhere (Diepeveen and Salzburger 2011). PCR products were visualized with GelRed (Biotium) on a 1.5% agarose gel. Sequences were aligned and visually inspected using Codon Code Aligner 3.7.1 (CodonCode Corporation) and exon/intron boundaries were determined based on homology with the obtained other teleost sequences. Total sequenced regions (TSR), protein-coding regions, and concatenated (TSRs of all five loci) data sets were constructed. All generated cichlid Hivep sequences have been deposited into GenBank (GenBank KF049218–KF049416) (Table S1).
Phylogenetic analyses and tests for selection pressure
Phylogenetic analyses and tests for selection pressure were performed as described elsewhere (Diepeveen and Salzburger 2011; Diepeveen et al. 2013). In short, models of nucleotide substitution were chosen based on likelihood ratio tests (LRTs) conducted in jModeltest 0.1.1 (Guindon and Gascuel 2003; Posada 2008) and used in maximum likelihood searches in PAUP* (Swofford 2002) and Bayesian Inference in MrBayes 3.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) for each individual paralog and for the concatenated dataset. Bootstrap analyses with 100 replicates were performed in PAUP* and MrBayes was run for 10,000,000 generations. Tylochromis polylepis and/or Oreochromis tanganicae were used as the outgroup in these analyses (Salzburger et al. 2002). The consensus tree based on the concatenated dataset was used as a common input tree in the subsequent analyses.
Both site and branch-site models, as implemented in Codeml, Phylogenetic Analysis by Maximum Likelihood (PAML) 4.2 (Yang 1997; Yang 2007), were used to test for selection pressure. The nonsynonymous/synonymous substitution rate ratio, ω or dN/dS, the proportion of sites assigned to an ω category, the p0,1,2, and the p and q parameters of the β distribution were estimated for all five datasets under different models. LRTs of the following model comparisons were performed to detect sites under positive selection: M1a (nearly neutral) with M2a (positive selection); M7 (β) with M8 (β and ωs ≥1); and M8a (β and ωs = 1) with M8. The comparison between M0 (one ratio) and M3 (discrete) was used as a test of variable ω among sites. Posterior probabilities for site classes were calculated with the Bayes empirical Bayes (BEB) (Yang et al. 2005). Next, LRTs between the null model (ωs = 1) and the alternative model (ωs ≥1) were performed to determine if focal, or foreground, lineages evolved under non-neutral evolution. These foreground branches were chosen based on the results from the phylogenetic and PAML analyses.
Subsequent sliding window analyses (window size = 20) were performed with TreeSAAP (selection on amino acid properties) 3.2 (Woolley et al. 2003) for the four loci for which positively selected sites were observed with the PAML analyses (Hivep1, Hivep2b, Hivep3a, and Hivep3b). Amino acids were categorized based on 31 physicochemical properties to identify regions of positive selection. Selection on amino acids was subsequently screened for positive destabilizing selection by means of categorizing the substitutions into eight categories (categories 1–8) based on the magnitude of radicality (i.e., 1 is the most conservative amino acid substitutions and 8 is the most radical). The three highest categories (6–8; P ≤ 0.001) were used as indicative of radical amino acid substitutions. Next, these substitutions were analyzed with the program SIFT (sorting intolerant from tolerant) (Ng and Henikoff 2003) to screen for possible effect on protein function.
Analyzing cis-regulatory regions
The five Hivep sequences from A. burtoni were compared with the obtained teleost sequences of O. niloticus, O. latipes, T rubripes, T. nigroviridis, and D. rerio with mVISTA (Mayor et al. 2000; Frazer et al. 2004). Sequences were globally aligned with Shuffle-LAGAN (Brudno et al. 2003) and the minimum sequence similarity was set to 50%. Intragenic conserved noncoding elements were predicted and analyzed with rVISTA (Loots et al. 2002) to identify potential transcription factor binding sites.
Experimental vaccination and immune response measurements
To examine the expression patterns of the Hivep paralogs after an experimental vaccination, we exposed adult cichlid fish of the species A. burtoni to Vibrio bacteria following Roth et al. (2011). V. anguillarum was physically isolated from the stomach of freshly caught broad-nosed pipefish (Syngnathus typhle) (Roth et al. 2012). Strain confirmation was performed via sequencing of the 16S rRNA, recA, and pyrH loci (GenBank reference numbers provided in Roth et al. 2012). On day 1 of the experiment, fish of both sexes were randomly assigned to either the experimental treatment (12 individuals) or the control treatment (11 individuals) and injected intraperitoneally with either 50 µl heat-killed (60 min at 65°) V. anguillarum (phylotype S6M4; 106 cells/ml dissolved in phosphate-buffered saline (PBS), i.e., experimental treatment) or 50 µl PBS (i.e., control treatment), respectively, according to the methods of Roth et al. (2012). Fish were tagged subcutaneously with visible implant elastomer tags (Northwest Marine Technology) according to treatment and kept in a single aquarium system. After ∼21 hr of exposure, fish were killed with MS222 and weight and standard length were noted as in Birrer et al. (2012). Blood was collected from the caudal vessel in heparinized capillaries (Na-heparinized; Brand GMBH + Co. KG), followed by extraction of the head kidneys and spleen, which were forced through a 40-µm nylon sieve to prepare cell suspensions for subsequent cellular immune measurements. All steps were performed on ice. Cells were washed twice with RPMI medium (10 min, 600 rpm, 4°) and resuspended in a final volume of 450 µl.
The number of lymphocytes and monocytes (as proxies for immune response in the form of inflammation and/or stress to the treatment) were measured in blood, head kidneys, and spleen tissues by means of flow cytometry (FACSCalibur; Becton Dickinson) with pre-assessed cichlid-specific settings for each tissue type. The proportions of monocytes, lymphocytes, and the lymphocyte/monocyte ratio were calculated. Furthermore, the activity of the relative number of lymphocytes in the G2-M and synthesis (S) phases of the proliferation cycle compared to the relative number of lymphocytes in the G0-1 phase was measured by killing cells in ethanol and subsequent labeling of the DNA with propidium iodide (Sigma Aldrich) as described by Roth et al. (2011). Lymphocytes were identified by their characteristic FSC/SSC pattern (i.e., cell volume and inner complexity). Proliferating cells in the G2-M phase were distinguished from G0-1 and S phase cells by a more intense red fluorescence because of their higher DNA content. To test whether the obtained data were normally distributed, D’Agnostino and Pearson omnibus normality tests as implemented in graphPad Prism version 5.0a for Mac OS X (http://www.graphpad.com) were conducted. Outliers with values outside 2 SDs from the mean were removed (i.e., up to three individuals per treatment group and tissue type).
The experiment was performed according to current national legislation of the Ministerium für Landwirtschaft, Umwelt und ländliche Räume des Landes Schleswig-Holstein (project entitled “Effects of global change on the immunological interaction of pipefish and cichlids with their natural bacteria communities”). One fish from the control treatment died during the experiment.
Gene expression assays and analyses
Gill, brain, and liver tissues of the 22 experimental animals were extracted and directly stored in RNA later (Invitrogen). RNA extraction and reverse-transcriptase were conducted as described elsewhere (Diepeveen and Salzburger 2011). Subsequent gene expression analyses were performed by means of quantitative PCR on a BioMark HD system at the Genetic Diversity Centre of the ETH Zurich, following the manufacturer’s protocol. Levels of gene expression were measured in 48.48 dynamic array integrated fluidic circuits with EvaGreen DNA binding dye. Primers were designed and tested for the five focal Hivep loci, two housekeeping loci [i.e., elongation factor 1 (EF1) and ribosomal protein SA 3 (RpSA3)] (Colombo et al. 2013) and four control loci with demonstrated immune-related functions [i.e., allograft inflammatory factor 1 (AIF1), anti-inflammatory response (Watano et al. 2001); coagulation factor II receptor-like 1 (F2RL1), inflammation and immunity (Rothmeier and Ruf 2012); interleukin 10 (IL10), immunosuppression (Jankovic and Trinchieri 2007); Toll-like receptor 5 (TLR5), pathogen recognition (Janeway and Medzhitov 2002; Akira et al. 2006)] (Table S3). Data were visualized, amplification plots were checked, and outliers were removed with the Fluidigm Real-Time PCR analysis software version 3.1 (Fluidigm). Further comparative analyses were performed with the qBasePLUS2 software package (Biogazelle). EF1 and RpSA3 were used as reference targets for the multiple reference gene normalization approach as implemented in qBasePLUS2 software (Biogazelle). Three different positive controls were included in this study; RNA was extracted from whole nonexperimental A. burtoni juveniles and two mixes composed of nine samples (gill, liver, and brain tissues of three randomly chosen individuals) each for the control group and the experimental group separately. Variation between PCR replicates and deviation of normalization factors were checked and outliers with values outside 2 SDs from the mean were removed. Data were controlled for inter-run variation.
Unpaired t tests were performed for the control immune genes expression levels between the control and experimental treatment groups with the qBasePLUS2 software (Biogazelle). To test for a correlation between the expression of Hivep paralogs [i.e., quality-controlled and normalized relative quantities (CNRQ, here just RQ)] and the immune response measurements, Pearson correlations were calculated in GraphPad Prism in the control and experimental treatment groups.
Results
Phylogenetic analyses of cichlid Hivep sequences
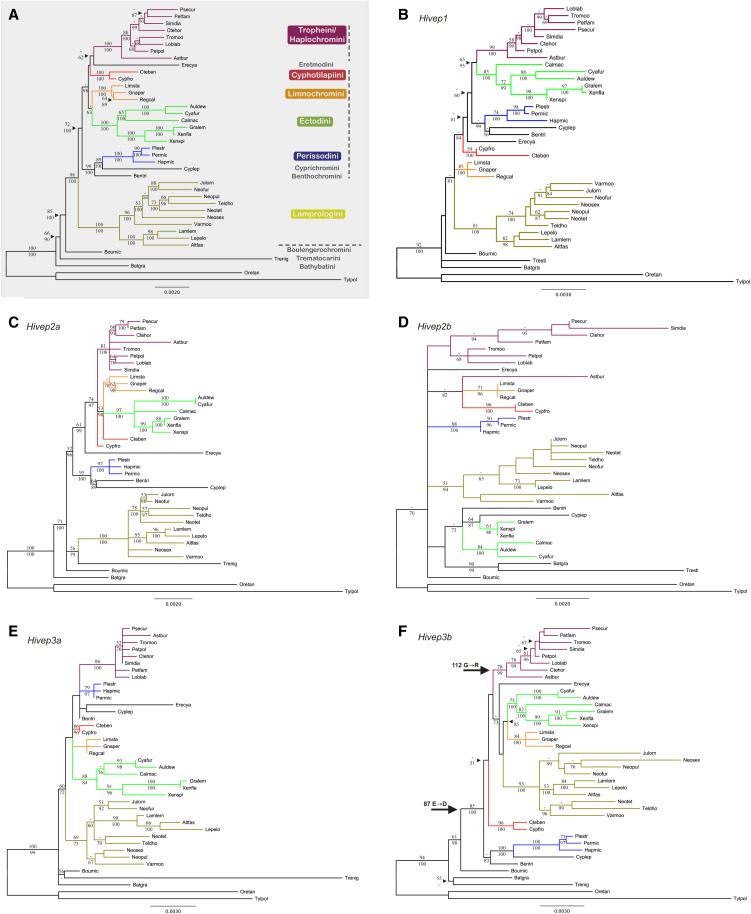
To examine the molecular evolutionary history of the Hivep paralogs, we performed maximum likelihood and Bayesian Inference phylogenetic analyses based on the total sequenced region per locus, and the concatenated dataset including all loci. The phylogenetic topologies of the obtained partial cichlid Hivep gene sequences and the concatenated dataset of 13.5 kb are displayed in Figure 1, A–F. Generally, the observed topologies of the gene trees, and the concatenated tree in particular, correspond with the available species trees (Salzburger et al. 2002; Takahashi 2003; Salzburger and Meyer 2004; Clabaut et al. 2005; Salzburger et al. 2005; Muschick et al. 2012), with T. polylepis, O. tanganicae, B. graueri, B. microlepis, and T. nigrifrons as most basal species, followed by the Lamprologini, the Eretmodini, and the species belonging to the "C-lineage" (Salzburger et al. 2002; Clabaut et al. 2005; Day et al. 2008) (Figure 1). As previously observed (Diepeveen and Salzburger 2011), E. cyanostictus was found at a different position within the C-lineage, whereas this species has been commonly resolved outside the C-lineage in previous studies (Salzburger et al. 2002; Clabaut et al. 2005; Day et al. 2008). Also, the relationships between the individual lineages of the C-lineage altered between the individual gene trees. Long branches were observed for T. polylepis, the lamprologines, individual lamprologine species, different branches within the ectodines in several gene trees, and for the haplochromines in Hivep2b and T. nigrifrons in the Hivep3a gene tree.
Figure 1.
Phylogenetic hypotheses based on maximum likelihood for the concatenated dataset and the individual Hivep loci consisting of 40 taxa. (A) Concatenated dataset (13,543 base pairs (bp); best-fitting model of nucleotide substitution: HKY+I+G). Lineages are recovered with maximum support values, whereas relationships within and between lineages are supported with relative high values. The horizontal dotted line separates the five most basal species from the derived lineages: the lamprologines, the eretmodines, and the species belonging to the C-lineage, with the latter marked by the vertical dotted line. (B) Hivep1 (3440 bp; TPM1uf+I+G) well-resolved with all major lineages recovered with high support values. (C) Hivep2a (3143 bp; TIM2+G). The lamprologines plus the five most basal species are found basal of the C-lineage plus Eretmodini. All major lineages are monophyletic, except the Cyphotilapiini. (D) Hivep2b (1517 bp; TrN+I+G). Mostly unresolved tree with a basal polytomy, excluding the two outgroup species from all other species. Polytomous relationships were further found for the haplochromine and ectodine lineages. (E) Hivep3a (2142 bp; TPM1uf+G). The lamprologines plus the five most basal species are found basal of the C-lineage plus Eretmodini. (F) Hivep3b (3301 bp; HKY+I+G). The lamprologines are positioned within the C-lineage. Black arrows represent the two branches for which ω > 1 was found in the branch-site analyses and their lineage-specific amino acid substitutions. Bootstrap values (PAUP*) and Bayesian posterior probabilities (MrBayes) >50% are shown, respectively, above and below the branches. Cichlid lineage names and a color key for the six cichlid lineages with more than one species included in this study are provided in the gray box in (A). Abbreviations of species names consist of the first three characters of the genus name followed by the first three characters of the species name (Table S1 shows full species names). Branch lengths of T. polylepis were shortened by 50% in all phylogenies and for T. nigrifrons in (E).
Selection pressure on sites and branches
To investigate signatures of selection pressure in the Hivep paralogs, we used site and branch-site models (Yang 1997, 2007) to obtain, e.g., nonsynonymous/synonymous substitution rate ratios (dN/dS). The maximum likelihood parameter estimations for ω, p0,1,2 and p, and q under different evolutionary models can be found in Table 1 for all five Hivep loci. Estimations of ω under the one ratio model (M0) suggest that the Hivep genes evolved under purifying selection, with ω ranging from 0.093 (Hivep2a) to 0.303 (Hivep1). A small proportion of sites, 1.4% (Hivep2a) to 11.8% (Hivep3b), was estimated to have evolved neutrally (ω = 1) under the neutral model (M1a). By using models that allow ω to vary among sites (M2a, M3, and M8), up to 4.3% of sites were detected to have evolved with ω > 1 in Hivep1, Hivep2b, Hivep3a, and Hivep3b, with more than 89.4% of sites evolving under purifying selection.
Table 1. Site model parameter estimates for the five Hivep paralogs.
| Parameter Estimates Using Different Models | |||||||
|---|---|---|---|---|---|---|---|
| Locus | M0 (One Ratio) | M1a (Neutral)a | M2a (Selection)a | M3 (Discrete)a | M7 (β)a | M8 (β and ω)a | M8a (β and ωs = 1)a |
| Hivep1 | ω = 0.303 | p0 = 0.896, ω0 = 0.000 | p0 = 0.899, ω0 = 0.012 | p0 = 0.997, ω0 = 0.106 | p = 0.005, q = 0.046 | p = 0.013, q = 0.091 | p = 0.005, q = 4.470 |
| p1 = 0.104, ω1 = 1.000 | p1 = 0.099, ω1 = 1.000 | p1 = 0.003, ω1 = 18.416 | p0 = 0.998 | p0 = 0.896 | |||
| p2 = 0.002, ω2 = 21.464 | p2 = 0.000, ω2 = 62.335 | p1 = 0.002, ω = 21.496 | p1 = 0.104, ω = 1.000 | ||||
| Hivep2a | ω = 0.093 | p0 = 0.986, ω0 = 0.080 | p0 = 0.986, ω0 = 0.080 | p0 = 0.543, ω0 = 0.000 | p = 0.554, q = 5.217 | p = 0.554, q = 5.221 | p = 0.672, q = 6.412 |
| p1 = 0.014, ω1 = 1.000 | p1 = 0.001, ω1 = 1.000 | p1 = 0.270, ω1 = 0.203 | p0 = 1.000 | p0 = 0.999 | |||
| p2 = 0.012, ω2 = 1.000 | p2 = 0.186, ω2 = 0.203 | p1 = 0.000, ω = 1.00 | p1 = 0.001, ω = 1.000 | ||||
| Hivep2b | ω = 0.120 | p0 = 0.942, ω0 = 0.000 | p0 = 0.941, ω0 = 0.000 | p0 = 0.957, ω0 = 0.000 | p = 0.005, q = 0.049 | p = 0.010, q = 0.160 | p = 0.005, q = 2.777 |
| p1 = 0.058, ω1 = 1.000 | p1 = 0.059, ω1 = 1.000 | p1 = 0.043, ω1 = 1.439 | p0 = 1.000 | p0 = 0.942 | |||
| p2 = 0.000, ω2 = 38.425 | p2 = 0.000, ω2 = 38.861 | p1 = 0.000, ω = 38.446 | p1 = 0.058, ω = 1.000 | ||||
| Hivep3a | ω = 0.251 | p0 = 0.919, ω0 = 0.000 | p0 = 0.909, ω0 = 0.000 | p0 = 0.984, ω0 = 0.040 | p = 0.005, q = 0.046 | p = 0.005, q = 0.049 | p = 0.005, q = 0.047 |
| p1 = 0.081, ω1 = 1.000 | p1 = 0.090, ω1 = 1.000 | p1 = 0.016, ω1 = 4.264 | p0 = 0.999 | p0 = 1.000 | |||
| p2 = 0.001, ω2 = 31.219 | p2 = 0.000, ω2 = 47.201 | p1 = 0.001, ω = 31.868 | p1 = 0.000, ω = 1.000 | ||||
| Hivep3b | ω = 0.277 | p0 = 0.882, ω0 = 0.000 | p0 = 0.894, ω0 = 0.014 | p0 = 0.000, ω0 = 0.000 | p = 0.005, q = 0.046 | p = 0.023, q = 0.170 | p = 0.005, q = 2.855 |
| p1 = 0.118, ω1 = 1.000 | p1 = 0.102, ω1 = 1.000 | p1 = 0.992, ω1 = 0.090 | p0 = 0.996 | p0 = 0.882 | |||
| p2 = 0.004, ω2 = 13.855 | p2 = 0.008, ω2 = 10.193 | p1 = 0.004, ω = 13.814 | p1 = 0.118, ω = 1.000 | ||||
The maximum likelihood parameter estimations for ω, p0,1,2 and p and q under different evolutionary models for all five Hivep loci. Estimations of ω under the one ratio model (M0) suggest that the Hivep genes evolved under purifying selection. A small proportion of sites was estimated to have evolved neutrally (ω = 1) under the M1a model in all loci. By using the M2a, M3, and M8 models, a small proportion of sites were detected to have evolved under ω > 1 in Hivep1, Hivep2b, Hivep3a, and Hivep3b. Hivep, human immunodeficiency virus type I enhancer-binding protein.
p0-2 are the proportions of sites assigned to the ω category or to a beta distribution with p and q as parameters; ω ratios >1 and their corresponding proportions are depicted in bold.
Likelihood ratio tests of several model comparisons (Table 2) were performed to detect positively selected amino acids. This approach resulted in the rejection of the null models in the M1a vs. M2a, M7 vs. M8, and M8a vs. M8 comparisons for all loci except Hivep2a. Positively selected sites were detected with the BEB for Hivep1 (16 sites), Hivep2b (2 sites), Hivep3a (18 sites), and Hivep3b (13 sites).
Table 2. LRT statistics of three site model comparisons and positively selected sites.
| Locus | Test | LRT (2∆l) | P | Selected Sites (BEB)* |
|---|---|---|---|---|
| Hivep1 | M1a vs. M2a | 210.047 | <0.001 | 37S, 49Q, 61V, 81N, 106T, 114R, 130T, 248A, 292Q, 472Q, 530V, 546L, 558T, 582P, 587H, 656N |
| M7 vs. M8 | 210.084 | <0.001 | 37S, 49Q, 61V, 81N, 106T, 114R, 130T, 248A, 292Q, 472Q, 530V, 546L, 558T, 582P, 587H, 656N | |
| M8a vs. M8 | 210.024 | <0.001 | See M7 vs. M8 comparison | |
| Hivep2a | M1a vs. M2a | 0.000 | 1.000 | 0 |
| M7 vs. M8 | 0.000 | 1.000 | 0 | |
| M8a vs. M8 | 0.007 | 0.932 | 0 | |
| Hivep2b | M1a vs. M2a | 35.537 | <0.001 | 143P |
| M7 vs. M8 | 37.555 | <0.001 | 143P, 330Q | |
| M8a vs. M8 | 34.250 | <0.001 | See M7 vs. M8 comparison | |
| Hivep3a | M1a vs. M2a | 304.843 | <0.001 | 62S, 63A, 64A, 82S, 141A, 200S, 265I, 329Q, 343P, 371D, 436V, 445A, 484T, 565E, 573T, 703P |
| M7 vs. M8 | 307.001 | <0.001 | 62S, 63A, 64A, 82S, 141A, 168N, 200S, 265I, 329Q, 343P, 371D, 436V, 437K, 445A, 484T, 565E, 573T, 703P | |
| M8a vs. M8 | 307.003 | <0.001 | See M7 vs. M8 comparison | |
| Hivep3b | M1a vs. M2a | 111.745 | <0.001 | 87E, 112G, 218H, 286T, 326S, 335I, 352G, 392G, 399P, 401P, 403R, 447I, 511K |
| M7 vs. M8 | 113.003 | <0.001 | 87E, 112G, 218H, 286T, 326S, 335I, 352G, 392G, 399P, 401P, 403R, 447I, 511K | |
| M8a vs. M8 | 111.749 | <0.001 | See M7 vs. M8 comparison |
LRTs resulted in the rejection of the null models in the M1a vs. M2a, M7 vs. M8, and M8a vs. M8 comparisons for all loci except Hivep2a. Positively selected sites were detected for Hivep1, Hivep2b, Hivep3a, and Hivep3b. LRT, likelihood ratio test; BEB, Bayes empirical Bayes; Hivep, human immunodeficiency virus type I enhancer-binding protein.
P = 0.01 (bold) and P = 0.05 (italic).
LRTs of the branch-site analyses were performed to test whether focal lineages evolved under non-neutral evolution. Significant LRTs were only observed for Hivep3b (Table 3), indicating that although the ω ratios do vary among sites for three of the other four Hivep loci, they do not seem to vary significantly among lineages. For Hivep3b, the following two branches were observed with ω > 1: the derived lineages (excluding the five basal species; P < 0.001) and the haplochromines (P = 0.031) (Table 3 and Figure 1).
Table 3. Parameter estimations and LRTs for the null and alternative hypotheses of the branch-site model for two different cichlid lineages for Hivep3b.
| Clade | Model | Site Class | 0 | 1 | 2a | 2b | LRT (P) |
|---|---|---|---|---|---|---|---|
| DL | Model A (Null) | Proportion | 0.766 | 0.123 | 0.096 | 0.015 | |
| Background ω | 0.000 | 1.000 | 0.000 | 1.000 | |||
| Foreground ω | 0.000 | 1.000 | 1.000 | 1.000 | |||
| Model A (Alternative) | Proportion | 0.897 | 0.087 | 0.014 | 0.001 | 18.509 (<0.001) | |
| Background ω | 0.070 | 1.000 | 0.070 | 1.000 | |||
| Foreground ω | 0.070 | 1.000 | 7.974 | 7.974 | |||
| HC | Model A (Null) | Proportion | 0.725 | 0.167 | 0.088 | 0.020 | |
| Background ω | 0.000 | 1.000 | 0.000 | 1.000 | |||
| Foreground ω | 0.000 | 1.000 | 1.000 | 1.000 | |||
| Model A (Alternative) | Proportion | 0.806 | 0.185 | 0.008 | 0.002 | 4.628 (0.031) | |
| Background ω | 0.000 | 1.000 | 0.000 | 1.000 | |||
| Foreground ω | 0.000 | 1.000 | 24.33 | 24.33 |
LRTs of the branch-site analyses indicate that Hivep3b evolved under non-neutral evolution (ω > 1) in the following two focal lineages: the derived lineages (excluding the five basal species) and the haplochromines. LRT, likelihood ratio test; Hivep, human immunodeficiency virus type I enhancer-binding protein; DL, derived lineage; HC, Haplochromini.
Sliding windows and amino acid substitution characteristics
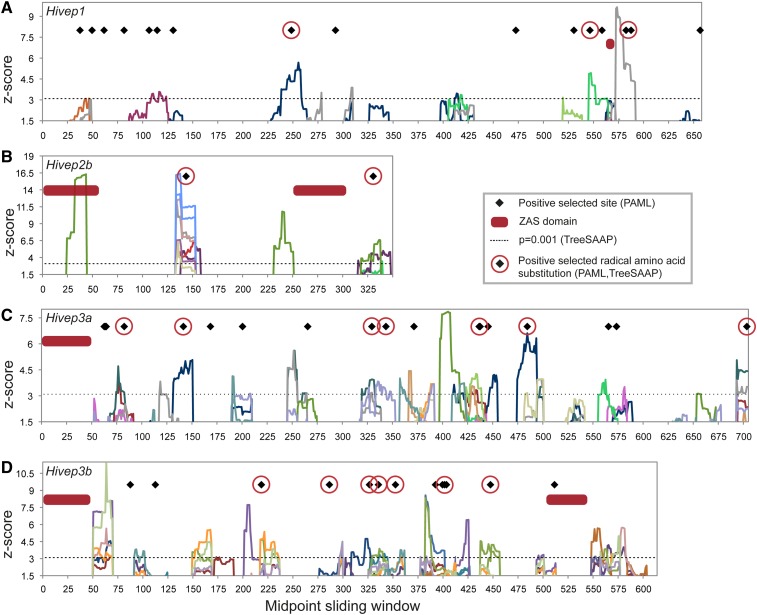
To visualize regions with elevated dN/dS values and to connect such regions with the physiochemical properties of the respective amino acid substitutions, we performed sliding window analyses. The sliding window plots of Hivep1, Hivep2b, Hivep3a, and Hivep3b are depicted in Figure 2, A–D. Regions of positive selection (z-score ≥ 3.09 corresponding with P ≤ 0.001) were observed for all four loci, with highest z-scores for Hivep2b, and the most numerous regions with a z-score ≥ 3.09 observed for Hivep3a. Interestingly, not all of these retrieved regions of positive selection correspond with the obtained positively selected sites as identified by the PAML analyses and vice versa. Relative few regions of positive selection are observed in the ZAS domains that contain the zinc fingers that bind specific DNA motifs. Notable exceptions are the ZAS-N domain of Hivep2b and the ZAS-C domain of Hivep3b (Figure 2); the latter is furthermore characterized by a positively selected site identified by the PAML analyses. Most commonly observed positively selected amino acid properties among the four paralogs affect the alpha-helical tendencies, the compressibility, the equilibrium constant (ionization of COOH), and the surrounding hydrophobicity. The SIFT analyses of the observed substitutions to screen for possible effect on protein function showed that all substitutions are tolerant and thus have no predicted damaging effect on function (data not shown).
Figure 2.
Sliding window plots and radical amino acid properties for four Hivep paralogs. (A) For Hivep1, multiple sliding windows were observed with z-score ≥ 3.09. For several positively selected sites identified with PAML, no radical substitutions were found and vice versa. (B) In Hivep2b, four regions of positive selection are observed of which two correspond with positively selected sites identified by the PAML analyses. (C) The observed regions of positive selection for Hivep3a are characterized by many different physicochemical properties. (D) Many radical amino acid substitutions were found for Hivep3b that correspond with most of the positively selected sites identified by PAML analyses. Each physicochemical amino acid property is individually color coded (see Figure S2 for details). Black diamonds (♦) represent positively selected amino acid sites obtained by the PAML analyses and red circles around them represent positively selected radical nonsynonymous substitutions (category 6-8). The dotted line at z-score = 3.09 represents P = 0.001, whereas z-score = 1.64 represents P = 0.05. Red rectangles represent the following ZAS domains: Hivep1 ZAS-C; Hivep2b ZAS-N, ZAS-C; Hivep3a ZAS-N; and Hivep3b ZAS-N, ZAS-C.
Analyzing cis-regulatory regions
We investigated noncoding regions within the Hivep paralogs for potential cis-regulatory elements to determine possible binding sites for transcription factors, indicative of putative functional involvement in signaling pathways. Vistaplots of Hivep1, Hivep2a, Hivep2b, and Hivep3b are depicted in Figure S1, A–D. Because of a limited number of retrieved teleost sequences for Hivep3a, the Vistaplot was not informative for this locus and was therefore excluded from further analyses. For all four analyzed loci, conserved noncoding elements (CNEs) were observed in A. burtoni. Interestingly, a similar pattern of two CNEs surrounding a single exon was observed in all loci (arrows in Figure S1). Although this pattern seems common among teleost fish for Hivep1 and Hivep2a, for Hivep2b and Hivep3b this pattern seems to be restricted to cichlid fishes (O. niloticus is the reference sequence in these analyses). A third cichlid-specific CNE was observed in a subsequent intron in both Hivep1 and Hivep2a, whereas for Hivep3b two more cichlid-specific CNEs were identified.
Because the particular pattern of two CNEs surrounding an exon was observed in all four analyzed loci, the subsequent search for potential transcription factor binding sites was mainly focused on these regions to determine any overlap in possible function of these regions. The analyses resulted in similar hits among Hivep paralogs and suggested a possible association between the Hivep paralogs and different types of signaling pathways involved in, e.g., sex determination [androgen receptor (AR); pre-B-cell leukemia transcription factor 1 (PBX1); sex-determining region Y protein (SRY)], immune system [B-cell lymphoma 6 protein (BCL6); H2.0-like homeobox protein (HB24); signal transducer and activator of transcription1,3,5a (STAT1,3,5a)], developmental patterning [homeobox protein Hox-A3 (Hoxa3); homeobox protein MSX-1 (Msx1)], and several members of the paired box protein Pax (PAX) and bone morphogenetic protein (BMP) pathways.
Experimental vaccination and immune response measurements
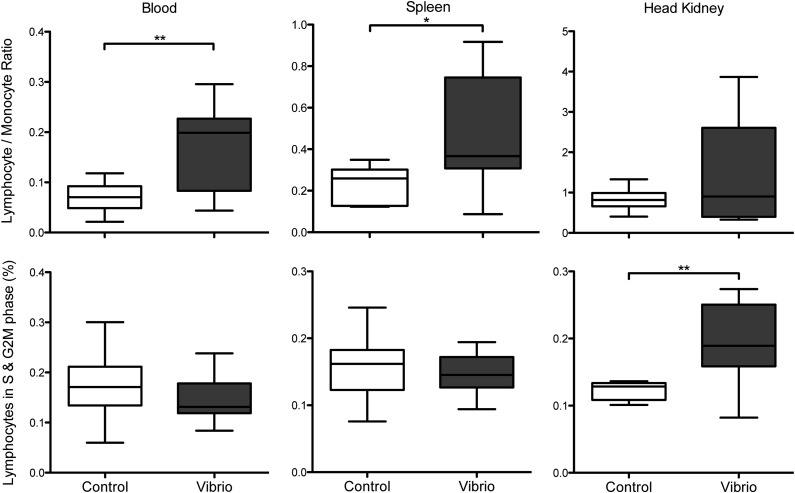
We performed experimental vaccinations to test whether the Hivep paralogs are involved in an induced immune response. The experimental vaccination was realized by exposure to heat-killed V. anguillarum for ∼21 hr, following the methods of Birrer et al. (2012). Several immune response measurements were performed to determine induction of immune defense dynamics. The lymphocyte/monocyte ratio and the relative number of lymphocytes in the G2-M and S phases of the proliferation cycle were measured in blood, spleen, and head kidney (Figure 3). Data were normally distributed. The experimental treatment resulted in an elevated lymphocyte/monocyte ratio in blood (P = 0.008; unpaired t test) and spleen (P = 0.018), indicative of a higher proportion of cells from the adaptive immune system (i.e., immune response). A higher proportion of lymphocytes in the S and G2-M phases was found in the head kidney of the experimental group (P = 0.005), indicative of induced lymphocyte proliferation.
Figure 3.
Immune response measurements after the experimental vaccination in A. burtoni adults. Lymphocyte/monocyte ratios (top) and proportions of cells in the S and G2-M phases (bottom) measured in blood (left), spleen (center), and head kidney (right) for the control treatment (white boxplot) and experimental treatment (Vibrio; gray boxplot). *P < 0.05; **P < 0.01. Depicted are the median, lower and upper quartiles (box), and the minimum and maximum observed values (error bars).
Gene expression assays
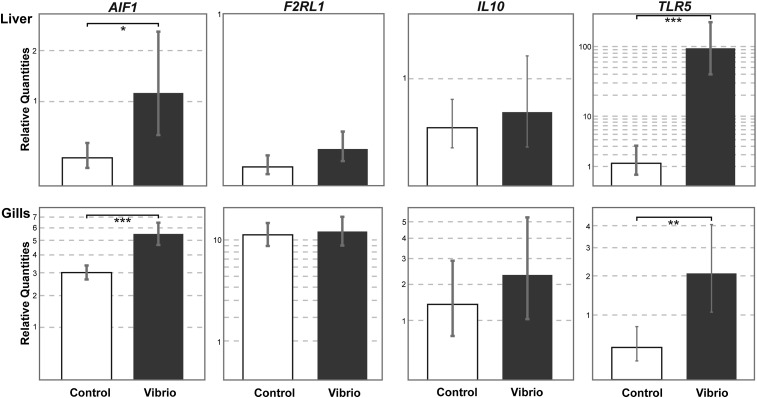
We measured the expression levels of four control immune loci, AIF1, F2RL1, IL10, and TLR5 in liver and gill tissues. These relative expression levels are depicted in Figure 4. For AIF1 and TLR5, we found significantly higher levels of relative expression in liver (P = 0.014 and P < 0.001; unpaired t test) and gills (P < 0.001 and P = 0.006) in the experimental treatment group.
Figure 4.
Gene expression assays for the control immune genes in A. burtoni adults. The relative gene expression levels (relative quantities) of AIF1, F2RL1, IL10, and TLR5 measured in liver (top) and gills (bottom) for the control treatment (white bars) and experimental treatment (Vibrio, blue bars). *P < 0.05; **P < 0.01; ***P < 0.001. Depicted are the mean and the 95% CI (error bars).
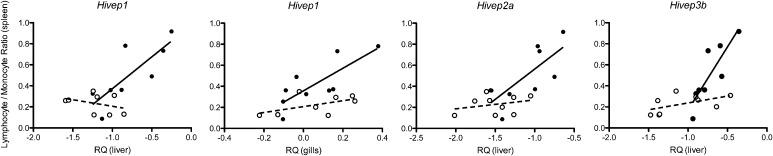
To analyze the effect of Vibrio exposure on the expression levels of the Hivep paralogs in detail, we assessed their expression levels in relation to an immune response parameter (i.e., lymphocyte/monocyte ratio) per treatment group (i.e., control and experimental). Four correlations were significant between the lymphocyte/monocyte ratio of the spleen and the relative expression of Hivep1 (liver; Pearson r = 0.798, P = 0.018), Hivep1 (gills; Pearson r = 0.794, P = 0.011), Hivep2a (Pearson r = 0.745, P = 0.021), and Hivep3b (Pearson r = 0.852, P = 0.007) (Figure 5). In these cases, the expression level of the Hivep paralogs thus correlates positively with the level of the immune response parameter.
Figure 5.
Correlations between immune response measurements and Hivep gene expression levels in A. burtoni adults. The relative gene expression levels [relative quantities (RQ)] of Hivep1, Hivep2a, and Hivep3b measured in gills and/or liver correlated with the observed lymphocyte/monocyte ratios measured in spleen for the control treatment (open circles and dashed fitted trend) and experimental treatment (closed circles and black fitted trend lines). Significant correlations were only observed for the experimental group: Hivep1 (liver; Pearson r = 0.798, P = 0.018); Hivep1 (gills; Pearson r = 0.794, P = 0.011); Hivep2a (Pearson r = 0.745, P = 0.021); and Hivep3b (Pearson r = 0.852, P = 0.007).
Discussion
In this study, we examined the molecular evolutionary history of the Hivep gene family members in relation to their presumed immune-related function in a renowned model system for evolutionary biology, the East African cichlid fishes. We performed comparative phylogenetic analyses and detailed screens of dN/dS ratios, analyzed putative cis-regulatory regions within the loci, and, in particular, investigated the expression levels of the Hivep paralogs after an experimental vaccination with V. anguillarum. We show, for the first time to our knowledge, that the Hivep paralogs play a putative role in the response to vaccination in fish, and that they are characterized by signatures of long-term positive selection. Our findings regarding the Hivep paralogs indicate that they are important candidate genes for immune-related functions in teleost fish and suggest broader implications in relation to speciation events, such as the adaptive radiations in East African cichlid fishes.
Exposure to V. anguillarum causes an immune response in A. burtoni
To test whether the exposure to a vaccination with heat-killed V. anguillarum resulted in an upregulation of the cellular fish immune response, we measured lymphocyte/monocyte ratios, the proportions of proliferating lymphocytes, and the expression levels of four control immune genes with demonstrated functions in the inflammatory response and immunity (Watano et al. 2001; Janeway and Medzhitov 2002; Akira et al. 2006; Jankovic and Trinchieri 2007; Rothmeier and Ruf 2012) (see Materials and Methods section). Consistent with an elevated immune response upon Vibrio vaccination, we found an increased lymphocyte production in the head kidney, the organ where clonal lymphocyte production takes place (Rombout et al. 2005; Abdel-Aziz et al. 2010). We also found a higher proportion of lymphocytes both in blood and spleen, indicating lymphocyte migration toward peripheral organs. Although lymphocytes are transported via blood, the spleen is the major lymphoid tissue associated with the clearance of blood-borne antigens (Press and Evensen 1999; Whyte 2007). Finally, the significant upregulation of both AIF1 and TLR5 in the Vibrio-exposed group indicates activation of the immune system (Watano et al. 2001; Janeway and Medzhitov 2002; Akira et al. 2006). However, we did not find a significant upregulation for two other immune genes with demonstrated functions in the immune response, F2RL1 and IL10 (Déry et al. 1998; Savan et al. 2003; Rothmeier and Ruf 2012). These were possibly missed by the choice of our measuring time point (21 hr after vaccination), as suggested by Savan et al. (2003), who found elevated IL10 expression in carp liver tissue after LPS stimulation only within the first 6 hr of incubation.
Hivep expression levels correlate with cellular immune response parameters in an East African cichlid fish
Although several functions of the Hivep paralogs have been demonstrated in the fruitfly, Xenopus frog, human, and mouse (Wu 2002), the Hivep paralogs have, so far, not been examined in teleost fishes. We tested, for the first time, whether there is a correlation between immune response parameters and the expression level of the Hivep paralogs in fish as an indicator of putative function(s) in the immune response.
Although our study does not determine the exact function of the Hivep paralogs within the immune response, the positive correlations between the lymphocyte/monocyte ratio and the expression levels of three Hivep paralogs indicate that the expression of —at least—Hivep1, Hivep2a, and Hivep3b is upregulated upon the experimental vaccination. This implies that Hivep paralogs play a role during the immune response of fish. These results provide, to our knowledge, the first indication of an immunological function of the Hivep paralogs in teleost fish, which is congruent with preliminary findings in pipefish (O. Roth, personal communication). The Hivep gene family thus offers a potential novel family of immune genes for teleost fish that, when their functions are characterized in more detail, can be used in future immunological screens.
Other functional implications
The experimental vaccination did not lead to upregulated expression levels of all five Hivep paralogs. We found no correlation between the expression levels of Hivep2b and Hivep3a and the immune response measurements. These paralogs either have immunological functions beyond the scope of our experimental design or are not involved in the immune response in teleost fishes. Previously, it had been shown that Hivep genes are involved in functions other than the immune system in insects and vertebrates, e.g., in murine osteoclastogenesis (Liu et al. 2011), in BMP/Dpp signaling (Grieder et al. 1995; Dai et al. 2000; Marty et al. 2000; Torres-Vazquez et al. 2001; Yao 2006; Saita et al. 2007; Yin et al. 2010), and in the development of the nervous system (Campbell and Levitt 2003; Takagi et al. 2006). Interestingly, several of the potential transcription factor binding sites identified within the observed CNEs correspond with these known functions of Hivep paralogs. For instance, we found multiple hits for components of the BMP signaling pathway, as well as several other developmental patterning loci, suggesting a putative role of the Hivep paralogs in developmental patterning and bone formation in cichlid fishes. Hivep paralogs have been found to play a role in the specification of Drosophila wing and halter discs (Torres-Vazquez et al. 2001), multiple dpp-dependent patterning events of both Drosophila ectoderm and mesoderm (Arora et al. 1995), and male tail patterning in Caenorhabditis elegans (Liang et al. 2007). The roles of Hivep in the BMP pathway, possibly through alternative splicing (Hicar et al. 2001; Hong and Wu 2005; Yin et al. 2010), together with several indications of functions in appendage specification and patterning make them candidate genes for fin patterning and anal fin egg-spot formation, a sexually selected trait involved in courtship and spawning behavior and intrasexual communication of haplochromine cichlid species (Wickler 1962; Hert 1989; Salzburger et al. 2005; Salzburger 2009; Theis et al. 2012). Future detailed expression and functional analyses should elucidate whether the Hivep paralogs are involved the development of egg spots in haplochromine cichlid fishes.
Implications of positive selection on Hivep paralogs, immune genes, and speciation events
Positive selection, or adaptive sequence evolution, is the hallmark of evolutionary change and molecular adaptation. By comparing the nonsynonymous substitution rate (dN) to the synonymous substitution rate (dS) of protein coding genes, the selection regime (i.e., neutral, purifying or positive) per amino acid can be inferred (Yang and Bielawski 2000). This method is widely used and has led to the identification of many cases of positive selection (Yang and Bielawski 2000). Genes involved in (evading) defensive systems or immunity are typically found with signatures of positive selection (Endo et al. 1996; Yang and Bielawski 2000; Schlenke and Begun 2003; Nielsen 2005; Nielsen et al. 2005; Biswas and Akey 2006; Yang 2006; Jiggins and Kim 2007; Montoya-Burgos 2011). As discussed, several functions within the immune response have been described for the Hivep paralogs in other species, and our detailed inferences of the dN/dS ratios provide evidence for positive selection acting on four out of five Hivep paralogs. Interestingly, signs of positive selection have been found before in vertebrate Hivep paralogs. Hivep2 has been found with a signature of positive selection in Tetraodon (Montoya-Burgos 2011) and the cow lineage (Toll-Riera et al. 2011), whereas Hivep3 showed signs of positive selection in the human lineage (Vamathevan et al. 2008). At least for the human Hivep paralog, it has been suggested that the immune function is the cause for the signature of positive selection. Together, these results indicate that it is likely that the immune-related functions of the Hivep paralogs are the cause for the elevated dN/dS ratios observed across vertebrate lineages, including the 14 cichlid lineages examined here.
Positively selected genes are typically only loosely connected to reproductive isolation in Drosophila (Wu and Ting 2004). This is in contrast to the vertebrate MHC loci, known for their signatures of positive selection (Hughes and Nei 1988; Hughes and Nei 1989; Yang and Bielawski 2000; Montoya-Burgos 2011), which have been proposed as pleiotropic speciation genes (Eizaguirre et al. 2009). Because these genes are involved in adaptation to novel habitats in response to different pathogenic communities and assortative mating via mate choice, their pleiotropic effects are hypothesized to induce and accelerate speciation (Eizaguirre et al. 2009). Our work shows that several of the Hivep paralogs also have putative pleiotropic roles in immune defense and an important sexually selected trait—the anal fin egg-spot in East African haplochromine cichlids—subject to mate choice (Hert 1989; Couldridge 2002; but see Theis et al. 2012). Mate choice for the most attractive male anal fin could thus select a certain Hivep genotype and thereby facilitate adaptation to pathogenic environments at the same time. Similar to the MHC loci, the Hivep paralogs might have played important roles during the explosive speciation events of cichlid fishes and therefore are exciting new putative speciation genes.
Hivep3b: Selective patterns in haplochromines and other derived cichlid lineages
That we found evidence of lineage-specific positive selection acting on Hivep3b indicates that this locus underwent adaptive protein evolution in both the derived cichlid lineages, including the lamprologines, eretmodines, and the C-lineage, and the most species-rich cichlid lineage, the haplochromines. Adaptive protein evolution underlies the adaptive evolution of traits and is thus ultimately responsible for species divergence and evolutionary innovation (Yang 2006). Interestingly, the elevated dN/dS ratios were observed in lineages that are characterized by explosive speciation and diversification events (Salzburger et al. 2005; Day et al. 2008), which can be seen as further support for the hypothesis that the pleiotropic functions of the Hivep paralogs —Hivep3b specifically— can be linked to speciation events. During such events genes could have been recruited to perform altered functions to generate novel or modified traits, which ultimately may have played a role in the divergence between species. A lineage-specific amino acid substitution in Hivep3b was observed for all the haplochromines (position 112 G → R) and the derived lineages (position 87 E → D), as well as several substitutions in a subset of the species belonging to these lineages. Functional analyses are now needed to test whether these substitutions have a fitness advantage for these species and, above all, their function in these cichlid lineages.
Supplementary Material
Acknowledgments
We thank past and current members of the Salzburger group for their contribution to sampling during fieldwork; Brigitte Aeschbach and Nicolas Boileau for assistance during laboratory work; and Verena Klein, Anne Beemelmanns, and Susanne Landis for help with the fish dissections and the experimental vaccinations. We also thank Jörn Scharsack for support in establishing the immune parameter measurements. The Inflammation Center at the University of Kiel kindly provided access to the flow cytometer facilities to perform immune measurements. Gene expression data analyzed in this article were generated in the Genetic Diversity Centre of ETH Zurich. We express our gratitude to Louis Du Pasquier, Christian Michel, Christophe Eizaguirre, Joost Raeymaekers, two anonymous reviewers, and the Editor for their constructive comments and suggestions. We also thank the BROAD Institute for sharing unpublished cichlid genome sequence data with the community. This work was supported by the European Research Council (starting grant ‘‘INTERGENADAPT’’ to W.S.), the Swiss National Science Foundation (grant 3100A0_122458 to W.S.), the Freiwillige Akademische Gesellschaft Basel (dissertation support grant to E.T.D.), the Volkswagenstiftung (grant to O.R.), and the German Research Foundation (grant to O.R. and W.S.).
Footnotes
Literature Cited
- Abdel-Aziz E.-S. H., Abdu S. B. S., Ali T. E.-S., Fouad H. F., 2010. Haemopoiesis in the head kidney of tilapia, Oreochromis niloticus (Teleostei: Cichlidae): A morphological (optical and ultrastructural) study. Fish Physiol. Biochem. 36: 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O., 2006. Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- Albertson R. C., Streelman J. T., Kocher T. D., Yelick P. C., 2005. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. USA 102: 16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altizer S., Harvell C. D., Friedle E., 2003. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 18: 589–596. [Google Scholar]
- Arora K., Dai H., Kazuko S. G., Jamal J., O’Connor M. B., et al. , 1995. The drosophila schnurri gene acts in the Dpp/TGFβ signaling pathway and encodes a transcription factor homologous to the human MBP family. Cell 81: 781–790. [DOI] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, LeClair K. P., Singh H., Sharp P. A., 1990. A large protein containing zinc finger domains binds to related sequence elements in the enhancers of the class I Major Histocompatibility Complex and Kappa Immunoglobulin genes. Mol. Cell. Biol. 10: 1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrer S. C., Reusch T. B. H., Roth O., 2012. Salinity change impairs pipefish immune defence. Fish Shellfish Immunol. 33: 1238–1248. [DOI] [PubMed] [Google Scholar]
- Biswas S., Akey J. M., 2006. Genomic insights into positive selection. Trends Genet. 22: 437–446. [DOI] [PubMed] [Google Scholar]
- Blais J., Rico C., van Oosterhout C., Cable J., Turner G. F., et al. , 2007. MHC adaptive divergence between closely related and sympatric African cichlids. PLoS One 2: e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots M., Best A., Miller M. R., White A., 2008. The role of ecological feedbacks in the evolution of host defence: What does theory tell us? Philos. T. R. Soc. B 364: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I., Volff J. N., Schartl M., 2009. The endothelin system: Evolution of vertebrate-specific ligand-receptor interactions by three rounds of genome duplication. Mol. Biol. Evol. 26: 783–799. [DOI] [PubMed] [Google Scholar]
- Brudno M., Malde S., Poliakov A., Do C. B., Couronne O., et al. , 2003. Glocal alignment: finding rearrangements during alignment. Bioinformatics 19: i54–i62. [DOI] [PubMed] [Google Scholar]
- Campbell D. B., Levitt P., 2003. Regionally restricted expression of the transcription factor c-myc intron 1 binding protein during brain development. J. Comp. Neurol. 467: 581–592. [DOI] [PubMed] [Google Scholar]
- Clabaut C., Salzburger W., Meyer A., 2005. Comparative phylogenetic analyses of the adaptive radiation of Lake Tanganyika cichlid fish: Nuclear sequences are less homoplasious but also less informative than mitochondrial DNA. J. Mol. Evol. 61: 666–681. [DOI] [PubMed] [Google Scholar]
- Colombo M., Diepeveen E. T., Muschick M., Santos E., Indermaur A., et al. , 2013. The ecological and genetic basis of convergent thick-lipped phenotypes in cichlid fishes. Mol. Ecol. 22: 670–684. [DOI] [PubMed] [Google Scholar]
- Cooper M. D., Alder M. N., 2006. The evolution of adaptive immune systems. Cell 124: 815–822. [DOI] [PubMed] [Google Scholar]
- Couldridge V., 2002. Experimental manipulation of male eggspots demonstrates female preference for one large spot in Pseudotropheus lombardoi. J. Fish Biol. 60: 726–730. [Google Scholar]
- Dai H., Hogan C., Gopalakrishnan B., Torres-Vazquez J., Nguyen M., et al. , 2000. The zinc finger protein Schnurri acts as a Smad partner in mediating the transcriptional response to Decapentaplegic. Dev. Biol. 227: 373–387. [DOI] [PubMed] [Google Scholar]
- Day J. J., Cotton J. A., Barraclough T. G., 2008. Tempo and mode of diversification of lake Tanganyika cichlid fishes. PLoS One 3: e1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déry O., Corvera C. U., Steinhoff M., Bunnett N. W., 1998. Proteinase-activated receptors: novel mechanisms of signaling by serine proteases. Am. J. Physiol. 274: C1429–C1452. [DOI] [PubMed] [Google Scholar]
- Diepeveen E. T., Salzburger W., 2011. Molecular characterization of two endothelin pathways in East African cichlid fishes. J. Mol. Evol. 73: 355–368. [DOI] [PubMed] [Google Scholar]
- Diepeveen E. T., Kim F. D., Salzburger W., 2013. Sequence analyses of the distal-less homeobox gene family in East African cichlid fishes reveal signatures of positive selection. BMC Evol. Biol. 13: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra P. D., Wiegertjes G. F., Forlenza M., van der Sluijs I., Hofmann H. A., 2011. The role of physiology in the divergence of two incipient cichlid species. J. Evol. Biol. 24: 2639–2652. [DOI] [PubMed] [Google Scholar]
- Dürr U., Henningfeld K. A., Hollemann T., Knöchel W., Pieler T., 2004. Isolation and characterization of the Xenopus HIVEP gene family. Eur. J. Biochem. 271: 1135–1144. [DOI] [PubMed] [Google Scholar]
- Ebert D., Hamilton W. D., 1996. Sex against virulence: The coevolution of parasitic diseases. Trends Ecol. Evol. 11: 79–82. [DOI] [PubMed] [Google Scholar]
- Eizaguirre C., Lenz T. L., Traulsen A., Milinski M., 2009. Speciation accelerated and stabilized by pleiotropic major histocompatibility complex immunogenes. Ecol. Lett. 12: 5–12. [DOI] [PubMed] [Google Scholar]
- Endo T., Ikeo K., Gojobori T., 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13: 685–690. [DOI] [PubMed] [Google Scholar]
- Flajnik M. F., Kasahara M., 2010. Origin and evolution of the adaptive immune system: Genetic events and selective pressures. Nat. Rev. Genet. 11: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser G. J., Bloomquist R. F., Streelman J. T., 2008. A periodic pattern generator for dental diversity. BMC Biol. 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser G. J., Hulsey C. D., Bloomquist R. F., Uyesugi K., Manley N. R., et al. , 2009. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 7: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Pachter L. S., Poliakov A., Rubin E. M., Dubchak I., 2004. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard D. T., Meyer A., 2007. Positive selection and gene conversion in SPP120, a fertilization-related gene, during the East African cichlid fish radiation. Mol. Biol. Evol. 24: 2286–2297. [DOI] [PubMed] [Google Scholar]
- Grieder N. C., Nellen D., Burke R., Basler K., Affolter M., 1995. Schnurri is required for Drosophila Dpp signaling and encodes a zinc finger protein similar to the mammalian transcription factor PRDII-BF1. Cell 81: 791–800. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum-likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1949. Disease and evolution. Ric. Sci. 19 (Suppl. A): 68–76. [Google Scholar]
- Hamilton W. D., 1980. Sex vs. non-sex vs. parasite. Oikos 35: 282–290. [Google Scholar]
- Hert E., 1989. The function of egg-spots in an African mouth-brooding cichlid fish. Anim. Behav. 37: 726–732. [Google Scholar]
- Hicar M. D., Liu Y., Allen C. E., Wu L.-C., 2001. Structure of the human zinc finger protein HIVEP3: Molecular cloning, expression, exon–intron structure, and comparison with paralogous genes HIVEP1 and HIVEP2. Genomics 71: 89–100. [DOI] [PubMed] [Google Scholar]
- Hong J., Wu L.-C., 2005. Structural characterization of the gene encoding the large zinc finger protein ZAS3: Implication to the origin of multiple promoters in eukaryotic genes. Biochim. Biophys. Acta. 1681: 74–87. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F., 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M., 1988. Pattern of nucleotide substitution at Major Histocompatibility Complex class I loci reveals overdominant selection. Nature 335: 167–170. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M., 1989. Nucleotide substitution at Major Histocompatibility Complex class II loci: Evidence for overdominant selection. Proc. Natl. Acad. Sci. USA 86: 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C. A., Goodnow C. C., Medzhitov R., 1996. Danger–Pathogen on the premises! Immunological tolerance. Curr. Biol. 6: 519–522. [DOI] [PubMed] [Google Scholar]
- Janeway C. A., Medzhitov R., 2002. Innate immune recognition. Annu. Rev. Immunol. 20: 197–216. [DOI] [PubMed] [Google Scholar]
- Janeway C. A., Walport M. J., Travers P., 2001. Immunobiology: The immune system in health and disease, Garland Publisher, New York, USA. [Google Scholar]
- Jankovic D., Trinchieri G., 2007. IL-10 or not IL-10: that is the question. Nat. Immunol. 8: 1281–1283. [DOI] [PubMed] [Google Scholar]
- Jansa S. A., Lundrigan B. L., Tucker P. K., 2003. Tests for positive selection on immune and reproductive genes in closely related species of the murine genus mus. J. Mol. Evol. 56: 294–307. [DOI] [PubMed] [Google Scholar]
- Jiggins F. M., Kim K. W., 2007. A screen for immunity genes evolving under positive selection in Drosophila. J. Evol. Biol. 20: 965–970. [DOI] [PubMed] [Google Scholar]
- Jones F. C., Chan Y. F., Schmutz J., Grimwood J., Brady S. D., et al. , 2012. A Genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr. Biol. 22: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F., Belden L. K., Daszak P., Dobson A., Drew Harvell C., et al. , 2010. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468: 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher T. D., 2004. Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 5: 288–298. [DOI] [PubMed] [Google Scholar]
- Kornfield I., Smith P. F., 2000. African cichlid fishes: model systems for evolutionary biology. Annu. Rev. Ecol. Syst. 31: 163–196. [Google Scholar]
- Liang J., Yu L., Yin J., Savage-Dunn C., 2007. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev. Biol. 305: 714–725. [DOI] [PubMed] [Google Scholar]
- Liu S., Madiai F., Hackshaw K. V., Allen C. E., Carl J., et al. , 2011. The large zinc finger protein ZAS3 is a critical modulator of osteoclastogenesis. PLoS One 6: e17161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G. G., Ovcharenko I., Pachter L., Dubchak I., Rubin E. M., 2002. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12: 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Sakura H., Sudo T., Ishii S., 1989. Putative metal finger structure of the human immunodeficiency virus type 1 enhancer binding protein HIV-EP1. J. Biol. Chem. 264: 14591. [PubMed] [Google Scholar]
- Marty T., Müller B., Basler K., Affolter M., 2000. Schnurri mediates Dpp-dependent repression of brinker transcription. Nat. Cell Biol. 2: 745–749. [DOI] [PubMed] [Google Scholar]
- Matthews B., Harmon L. J., M’Gonigle L., Marchinko K. B., Schaschl H., 2010. Sympatric and allopatric divergence of MHC genes in threespine stickleback. PLoS One 5: e10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., et al. , 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Meyer A., Van de Peer Y., 2005. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). Bioessays 27: 937–945. [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos J. I., 2011. Patterns of positive selection and neutral evolution in the protein-coding genes of Tetraodon and Takifugu. PLoS One 6: e24800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Seeler J. S., Nirula A., Shurland D. L., Gaynor R. B., 1992. Regulation of human immunodeficiency virus enhancer function by PRDII-BF1 and c-rel gene products. J. Virol. 66: 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschick M., Barluenga M., Salzburger W., Meyer A., 2011. Adaptive phenotypic plasticity in the Midas cichlid fish pharyngeal jaw and its relevance in adaptive radiation. BMC Evol. Biol. 11: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschick M., Indermaur A., Salzburger W., 2012. Convergent evolution within an adaptive radiation of cichlid fishes. Curr. Biol. 22: 2362–2368. [DOI] [PubMed] [Google Scholar]
- Møller A. P., Erritzøe J., 1998. Host immune defence and migration in birds. Evol. Ecol. 12: 945–953. [Google Scholar]
- Ng P., Henikoff S. T., 2003. SIFT: Predicting amino acid changes that affect protein functions. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A. G., Glanowski S., Sackton T. B., et al. , 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., 2008. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- Press C. M., Evensen O., 1999. The morphology of the immune system in teleost fishes. Fish Shellfish Immun. 9: 309–318. [Google Scholar]
- Roberts R. B., Ser J. R., Kocher T. D., 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326: 998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, R. M., A. López-Vázquez, and C. López-Larrea, 2012 Immune systems evolution, pp. 237–251 in Sensing in Nature: Advances in experimental medicine and biology, edited by C. López-Larrea. Landes Bioscience and Springer Science and Business Media. New York, USA. [DOI] [PubMed] [Google Scholar]
- Rombout J. H. W. M., Huttenhuis H. B. T., Picchietti S., Scapigliati G., 2005. Phylogeny and ontogeny of fish leucocytes. Fish Shellfish Immunol. 19: 441–455. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Roth O., Scharsack J. P., Keller I., Reusch T. B. H., 2011. Bateman’s principle and immunity in a sex-role reversed pipefish. J. Evol. Biol. 24: 1410–1420. [DOI] [PubMed] [Google Scholar]
- Roth O., Keller I., Landis S. H., Salzburger W., Reusch T. B. H., 2012. Hosts are ahead in a marine host-parasite coevolutionary arms race: Innate immune system adaptation in pipefish syngnathus typhle against Vibrio phylotypes. Evolution 66: 2528–2539. [DOI] [PubMed] [Google Scholar]
- Rothmeier A. S., Ruf W., 2012. Protease-activated receptor 2 signaling in inflammation. Semin. Immunopathol. 34: 133–149. [DOI] [PubMed] [Google Scholar]
- Sackton T. B., Lazzaro B. P., Schlenke T. A., Evans J. D., Hultmark D., et al. , 2007. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39: 1461–1468. [DOI] [PubMed] [Google Scholar]
- Saita Y., Takagi T., Kitahara K., Usui M., Miyazono K., et al. , 2007. Lack of Schnurri-2 expression associates with reduced bone remodeling and osteopenia. J. Biol. Chem. 282: 12907–12915. [DOI] [PubMed] [Google Scholar]
- Salzburger W., 2009. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol. Ecol. 18: 169–185. [DOI] [PubMed] [Google Scholar]
- Salzburger W., Meyer A., Baric S., Verheyen E., Sturmbauer C., 2002. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Syst. Biol. 51: 113–135. [DOI] [PubMed] [Google Scholar]
- Salzburger W., Meyer A., 2004. The species flocks of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften 91: 277–290. [DOI] [PubMed] [Google Scholar]
- Salzburger W., Mack T., Verheyen E., Meyer A., 2005. Out of Tanganyika: Genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol. Biol. 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W., Braasch I., Meyer A., 2007. Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 5: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M. E., Salzburger W., 2012. How cichlids diversify. Science 338: 619–621. [DOI] [PubMed] [Google Scholar]
- Savan R., Igawa D., Sakai M., 2003. Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus carpio L. Eur. J. Biochem. 270: 4647–4654. [DOI] [PubMed] [Google Scholar]
- Sawyer S. L., Emerman M., Malik H. S., 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2: e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharsack J. P., Kalbe M., Harrod C., Rauch G., 2007. Habitat-specific adaptation of immune responses of stickleback (Gasterosteus aculeatus) lake and river ecotypes. P. Roy. Soc. B: Biol. Sci. 274: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke T. A., Begun D. J., 2003. Natural selection drives drosophila immune system evolution. Genetics 164: 1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter, D., 2000 The Ecology of Adaptive Radiation, edited by P. H. Harvey, R. M. May. Oxford: Oxford University Press. [Google Scholar]
- Seehausen O., 2006. African cichlid fish: a model system in adaptive radiation research. Proc. Biol. Sci. 273: 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler J. S., Muchardt C., Suessle A., Gaynor R. B., 1994. Transcription factor PRDII-BF1 activates human immunodeficiency virus type 1 gene expression. J. Virol. 68: 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A., 1996. Gen(om)e duplications in the evolution of early vertebrates. Curr. Opin. Genet. Dev. 6: 715–722. [DOI] [PubMed] [Google Scholar]
- Swofford D. L., 2002 Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Massachusetts.
- Takagi T., Jin W., Taya K., Watanabe G., Mori K., et al. , 2006. Schnurri-2 mutant mice are hypersensitive to stress and hyperactive. Brain Res. 1108: 88–97. [DOI] [PubMed] [Google Scholar]
- Takahashi T., 2003. Systematics of Tanganyikan cichlid fishes (Teleostei: Perciformes). Ichthyol. Res. 50: 367–382. [Google Scholar]
- Taylor J. S., Braasch I., Frickey T., Meyer A., Van de Peer Y., 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 13: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y., Morikawa N., Kawakami K., Okada N., 2002a Accelerated evolution of the surface amino acids in the WD-repeat domain encoded by the hagoromo gene in an explosively speciated lineage of east African cichlid fishes. Mol. Biol. Evol. 19: 574–578. [DOI] [PubMed] [Google Scholar]
- Terai Y., Morikawa N., Okada N., 2002b The evolution of the pro-domain of bone morphogenetic protein 4 (Bmp4) in an explosively speciated lineage of East African cichlid fishes. Mol. Biol. Evol. 19: 1628–1632. [DOI] [PubMed] [Google Scholar]
- Terai Y., Morikawa N., Kawakami K., Okada N., 2003. The complexity of alternative splicing of hagoromo mRNAs is increased in an explosively speciated lineage in East African cichlids. Proc. Natl. Acad. Sci. USA 100: 12798–12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis A., Salzburger W., Egger B., 2012. The function of anal fin egg-spots in the cichlid fish Astatotilapia burtoni. PLoS One 7: e29878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. N., 1988. Variation in interspecific interactions. Annu. Rev. Ecol. Syst. 19: 65–87. [Google Scholar]
- Toll-Riera M., Laurie S., Alba M. M., 2011. Lineage-specific variation in intensity of natural selection in mammals. Mol. Biol. Evol. 28: 383–398. [DOI] [PubMed] [Google Scholar]
- Torres-Vazquez J., Park S., Warrior R., Arora K., 2001. The transcription factor Schnurri plays a dual role in mediating Dpp signaling during embryogenesis. Development 128: 1657–1670. [DOI] [PubMed] [Google Scholar]
- Tschirren B., Raberg L., Westerdahl H., 2011. Signatures of selection acting on the innate immunity gene Toll-like receptor 2 (TLR2) during the evolutionary history of rodents. J. Evol. Biol. 24: 1232–1240. [DOI] [PubMed] [Google Scholar]
- Vamathevan J. J., Hasan S., Emes R. D., Amrine-Madsen H., Rajagopalan D., et al. , 2008. The role of positive selection in determining the molecular cause of species differences in disease. BMC Evol. Biol. 8: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff J. N., 2005. Genome evolution and biodiversity in teleost fish. Heredity 94: 280–294. [DOI] [PubMed] [Google Scholar]
- Watano K., Iwabuchi K., Fujil S., Ishimori N., Mitsuhashi S., et al. , 2001. Allograft inflammatory factor-1 augments production of interleukin-6, -10 and -12 by a mouse macrophage line. Immunogenetics 104: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte S. K., 2007. The innate immune response of finfish–A review of current knowledge. Fish Shellfish Immunol. 23: 1127–1151. [DOI] [PubMed] [Google Scholar]
- Wickler W., 1962. “Egg-dummies” as natural releasers in mouth-breeding cichlids. Nature 194: 1092–1093. [Google Scholar]
- William R., Leisha E., Alexander I., Melissa B., Ichiro I., et al. , 1995. The Myc intron-binding polypeptide associates with RFX1 in vivo and binds to the Major Histocompatibility Complex Class II promoter region, to the Hepatitis B virus enhancer, and to regulatory regions of several distinct viral genes. Mol. Cell. Biol. 15: 3041–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley S., Johnson J., Smith M. J., Crandall K. A., McClellan D. A., 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19: 671–672. [DOI] [PubMed] [Google Scholar]
- Wu C.-I., Ting C.-T., 2004. Genes and speciation. Nat. Rev. Genet. 5: 114–122. [DOI] [PubMed] [Google Scholar]
- Wu L.-C., 2002. ZAS: C2H2 zinc finger proteins involved in growth and development. Gene Expr. 10: 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-C., Mak C. H., Dear N., Boehm T., Foroni L., et al. , 1993. Molecular cloning of a zinc finger protein which binds to the heptamer of the signal sequence for V (D) J recombination. Nucleic Acids Res. 21: 5067–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-C., Yiling L., Strandtmann J., Chi-Ho M., Lee B., et al. , 1996. The mouse DNA binding protein Rc for the Kappa B motif of transcription and for the V(D)J recombination signal sequences contains composite DNA–protein interaction domains and belongs to a new family of large transcriptional proteins. Genomics 35: 415–424. [DOI] [PubMed] [Google Scholar]
- Yang Z., 1997. PAML, A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z., Bielawski J. P., 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wong W., Nielsen R., 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Yang Z., 2006. Computational molecular evolution, Oxford University Press, Oxford. [Google Scholar]
- Yang Z., 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yao L. C., 2006. Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development 133: 4025–4034. [DOI] [PubMed] [Google Scholar]
- Yin K., Yu L., Savage-Dunn C., 2010. Alternative trans-splicing of Caenorhabditis elegans sma-9/schnurri generates a short transcript that provides tissue-specific function in BMP signaling. BMC Mol. Biol. 11: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Lee V., Finn A., Senger K., Zarrin A. A., et al. , 2012. Origin of immunoglobulin isotype switching. Curr. Biol. 22: 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.