Abstract
The clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) system has been used as an efficient tool for genome editing. We report the application of CRISPR-Cas–mediated genome editing to wheat (Triticum aestivum), the most important food crop plant with a very large and complex genome. The mutations were targeted in the inositol oxygenase (inox) and phytoene desaturase (pds) genes using cell suspension culture of wheat and in the pds gene in leaves of Nicotiana benthamiana. The expression of chimeric guide RNAs (cgRNA) targeting single and multiple sites resulted in indel mutations in all the tested samples. The expression of Cas9 or sgRNA alone did not cause any mutation. The expression of duplex cgRNA with Cas9 targeting two sites in the same gene resulted in deletion of DNA fragment between the targeted sequences. Multiplexing the cgRNA could target two genes at one time. Target specificity analysis of cgRNA showed that mismatches at the 3′ end of the target site abolished the cleavage activity completely. The mismatches at the 5′ end reduced cleavage, suggesting that the off target effects can be abolished in vivo by selecting target sites with unique sequences at 3′ end. This approach provides a powerful method for genome engineering in plants.
Keywords: Cas, cgRNA, CRISPR, genome editing, indel, wheat genome editing
Specific and effective genome editing through nontransgenic approaches is an area of high-priority research for the improvement of food crops. Several genome editing technologies like zinc finger nuclease (ZFN) and transcription activator–like effector nuclease (TALEN) have been deployed for targeted genome modifications (Chen and Gao 2013; Zhang et al. 2010), but these are rather complicated in design and need protein engineering for each target sequence. Recently, a new technology based on the type II prokaryotic clustered, regularly interspaced, short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) system has been developed as an effective tool for genome engineering (Cong et al. 2013; Mali et al. 2013). It is highly specific, inexpensive, and easy to engineer. CRISPR consists of an array of repeat sequences separated by “spacer” sequences that belong to the targeted gene/genome. A long primary transcript transcribes from CRISPR arrays and gets processed into short CRISPR RNAs (crRNAs). The crRNA consists of a conserved repeat sequence and a variable spacer sequence (guide) complementary to the target gene sequence (Brouns et al. 2008; Hale et al. 2009). Trans activating crisper RNA (tracer RNA) is another important molecule that plays a critical role in the processing of pre-crRNA (Chylinski et al. 2013). It is a short RNA sequence and is complementary to the CRISPR repeat. It activates the processing and maturation of pre-crRNA into the short crRNA by RNAseIII and Cas9. The ribonucleoprotein complex formed by short crRNA and Cas9 proteins binds to the target sequence by base pairing (Jore et al. 2011) and causes sequence-specific dsDNA cleavage. A chimeric crRNA and tracer RNA hybrid has also been designed and has been reported to be as effective as those used individually (Mali et al. 2013). The presence of a conserved sequence motif (NGG) known as protospacer adjacent motif (PAM) at 3′ downstream of target spacer sequence is also reported as essential for cleavage (Gasiunas et al. 2012). The CRISPR-Cas system has been demonstrated to work efficiently for genome editing in bacterial, yeast, and animal systems (Cong et al. 2013; DiCarlo et al. 2013; Gasiunas et al. 2012; Jinek et al. 2013; Mali et al. 2013) and has been applied to plants recently (Li et al. 2013; Nekrasov et al. 2013; Shan et al. 2013). Although, some nonspecific editing has been reported (Fu et al. 2013), the CRISPR-Cas system is very simple to design, highly effective (Mali et al. 2013; Cong et al. 2013), and can be improved in specificity.
We report the application of the CRISPR-Cas–mediated genome editing in wheat (Triticum aestivum), the most important food crop plant, and Nicotiana benthamiana, a model plant species. Mutations in the inositol oxygenase (inox) and phytoene desaturase (pds) genes in cell suspension culture of wheat and the pds gene in leaves of N. benthamiana were targeted. The efficacy of multiplexed CRISPR RNA, targeting two different positions in two separate genes, was also studied.
Materials and Methods
Synthesis of chimeric guide RNA encoding DNAs
We targeted the inox and pds genes of wheat and the pds gene of N. benthamiana to demonstrate RNA-guided genome editing in plants. Partial gene sequences were amplified from genomic DNA of the wild-type plants. Target sites (also known as protospacer) of 20 nucleotides were selected manually (Supporting Information, File S1) following the criteria described previously (Hwang et al. 2013; Mali et al. 2013). The presence of NGG trinucleotide protospacer adjacent motif (PAM) at the 3′ end of the target region was an essential criterion in target selection. The chimeric guide RNAs (cgRNAs) were designed to target one or two sites in the targeted genes (Hwang et al. 2013; Mali et al. 2013) (File S2). Overlapping primers were used for the synthesis of cgRNAs transcribing DNA. To develop duplex cgRNA for targeting two regions at one time, two cgRNAs were fused by using the SpeI restriction site. Assembled DNA was amplified by end primers, cloned, and sequenced.
Construction of expression vectors
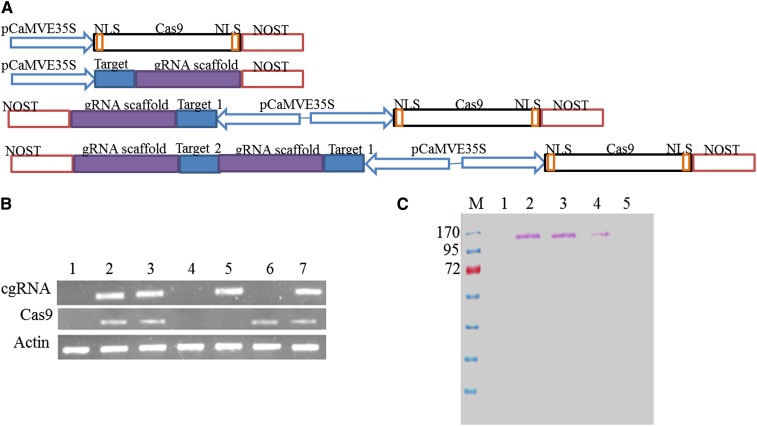
Four types of expression vectors were prepared by cloning cgRNA and Cas9 in different combinations (Figure 1A). Constitutive CaMVE35S promoter was used to drive the expression. We cloned cgRNA and Cas9 alone, as well as together, in plant expression vector pBI121. The Cas9 gene (4272 bp) including the FLAG tag and nuclear localization signal for eukaryotic expression was obtained from Addgene USA (Addgene plasmid 42229). The plant expression constructs and their target genes are provided in Table S1.
Figure 1.
Basic architecture of constructs used for CRISPR-Cas–mediated genome editing and expression analysis of cgRNA and Cas9. (A) Architecture of constructs used for expression. The four types of constructs developed for the expression of cgRNA and Cas9 are shown. (B) RT-PCR analysis of expression of cgRNA and Cas9 in wheat: untransformed (lane 1); pCinox1-transformed (lane 2) suspension cells; and pCwpds1-transformed (lane 3) suspension cells RT-PCR analysis of expression of cgRNA and Cas9 in. N. benthamiana: untransformed (lane 4); pTpds1 (lane 5); pCas9-transformed (lane 6) leaf; and pCtpds1-transformed (lane 7) leaf. (C) Western blot analysis of Cas9 using anti-Flag antibody (FLAG tag attached at N-terminus of Cas9). Lane M, molecular weight marker; lane 1, untransformed; lane 2, pCinox1-transformed suspension cells of wheat; lane 3, pCwpds1-transformed suspension cells of wheat; lane 4, pCtpds1 agro-infiltrated; lane 5, untransformed N. benthamiana leaves.
Plant transformation
Agrobacterium tumefaciens strain GV3101 was used for genetic transformation of N. benthamiana and wheat cells. Transient expression of cgRNA and Cas9 in different combinations in 3-wk-old to 4-wk-old N. benthamiana plant leaves was performed by agro-infiltration, as described by Bos et al. (2006). The infiltrated area was marked and used for DNA and RNA isolation 4 d after infiltration.
To analyze the mutations in wheat, cell suspension culture developed from immature embryos was used for transformation following the protocol of Wu et al. (1998). Transformed cells were enriched by antibiotic selection and used for the isolation of DNA, RNA, and protein.
RT-PCR for expression analysis of cgRNA and Cas9
Total RNA was isolated from the transformed wheat suspension cells and agro-infiltrated area of N. benthamiana leaves, and was used to make cDNA. PCR amplification was performed with the primers listed in File S1 for the analysis of cgRNA expression. Cas9 expression was analyzed by using primers Cas9 forward 5′ tctgtgggctgggccgtgatc 3′ and Cas9 reverse 5′ cagatccggttcttccgtctgg 3′. Amplification of actin was used as control.
Western blotting
Total soluble protein was extracted from transformed wheat suspension cells and N. benthamiana leaves using the extraction buffer (20 mM Tris pH 8, 150 mM NaCl, 0.05% TritonX-100, and 100 µM PMSF), separated on 10% SDS-PAGE, and transferred onto PVDF membrane. Cas9 protein was detected by using anti-FLAG antibody (FLAG tag was attached at N-terminus of Cas9).
Mutation analysis
Mutations in targeted genes in plants were analyzed by PCR amplification and Sanger sequencing. Genomic DNA was isolated from transformed tissue and used as template in amplification. Primers from the vicinity of the targeted regions (highlighted in green in File S1) were used for amplification of control and transformed samples. Ten cycles of amplification were performed. The amplified DNA was cloned and used for sequencing. Sequences were aligned against wild-type, and average mutation percentage was calculated from three independent replicates. A minimum of 75 clones were sequenced for each transforming event and used for the analysis.
Specificity analysis of cgRNA
To analyze the specificity of cgRNA in binding to the target region, several sets of mutations were made in protospacer 1 binding site of inox cgRNA (File S1 and Table S2). Mutant cgRNA constructs were transformed with Cas9. Their editing potential was analyzed by amplification with the aforementioned primers and BsgI digestion (BsgI restriction site is present at target region in inox protospacer 1) (File S1). Mutated DNA could not be digested with BsgI. Average mutation percentage was estimated by densitometry.
Results
Target selection, construction of expression cassettes, and expression of Cas9 and cgRNAs
Despite using the expression of complete crRNA and tracer RNA, we designed cgRNA for gene targeting as described previously (Hwang et al. 2013; Mali et al. 2013). This reduces the complexity of the system. The cgRNA consisted of mature crRNA fused with partial tracer RNA, which mimics the natural architecture of crRNA and tracer RNA duplex. We used an 80-nucleotides-long guide RNA scaffold sequences fused with 20-nucleotide target (spacer) sequences at 5′ end (File S2).
We targeted two regions in each gene, separately and together. Therefore, several combinations of expression constructs were developed (Table S1); a representative architecture is shown in Figure 1A. Separate expression cassettes were developed for Cas9 (pCas9) and cgRNA (pTpds1 and pTpds2) to target the pds gene in N. benthamiana and transformed into leaves by agro-infiltration in different combinations (pCas9 or pTpds alone or together in 1:1 ratio). No mutation was detected when Cas9 and cgRNA were taken, one at a time, as expected. Combined expression of pCas9 and pTpds in 1:1 ratio provided mutations that increased significantly when Cas9 and cgRNA were coexpressed using the construct pCtpds1 (Table 1).
Table 1. Average mutation rate at protospacer 1 of phytoene desaturase gene of Nicotiana benthamiana after agro-infiltration with different constructs.
| Construct Used | Average Mutation (%) at Target Sites |
|---|---|
| pCas9 | 0 |
| pTpds1 | 0 |
| pCas9 and pTpds1 (1:1 ratio) | 1.8 (2/108) |
| pCtpds1 | 12.7 (12/94) |
RT-PCR of total RNA from transformed cells showed the expression of cgRNA and Cas9 in cells of both plants (Figure 1B). Cas9 protein was also detected by Western blotting using anti-FLAG antibody against the FLAG tag attached at N-terminus (Figure 1C).
cgRNA-Cas9 guided genome editing in N. benthamiana
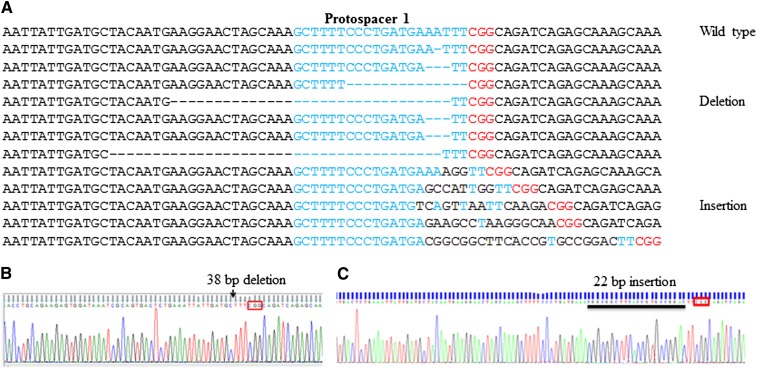
As described, we targeted two sites in the pds gene of N. benthamiana leaves by agro-infiltration (File S1). We detected mutations (deletions and insertions) in N. benthamiana in 1.8% of the clones obtained from leaves infiltrated with the two Agrobacterium strains (containing pCas9 or pTpds1) mixed in 1:1 ratio. However, 12.7% indels were detected in N. benthamiana when Cas9 and cgRNA were present within a single plasmid construct pCtpds1 (Table 1) and, hence, codelivered. In protospacer 2 region, 2.4% and 13.8% indels were detected with pCas9:pTpds2 (1:1) and pCtpds2 treatments, respectively. The results established that the co-delivery of cgRNA and Cas9 improved efficacy of the system. Therefore, in case of wheat only, co-delivery was used to examine the application of the CRISPR-Cas system. A maximum of 38 bp deletion and of 22 bp addition were detected (Figure 2). The indel pattern was similar to that reported in other studies (Hwang et al. 2013; Li et al. 2013; Mali et al. 2013; Nekrasov et al. 2013; Shan et al. 2013). Maximum indels were noticed approximately 2–3 bp away from the 5′ end of PAM.
Figure 2.
Editing at protospacer 1 of phytoene desaturase (pds) gene of N. benthamiana by the CRISPR-Cas system. (A) Alignment of wild-type and sequences with indel at protospacer 1 of the pds gene. (B) and (C) Sanger sequencing of selected deletion and insertion mutants, respectively.
cgRNA-Cas9 guided genome editing in T. aestivum
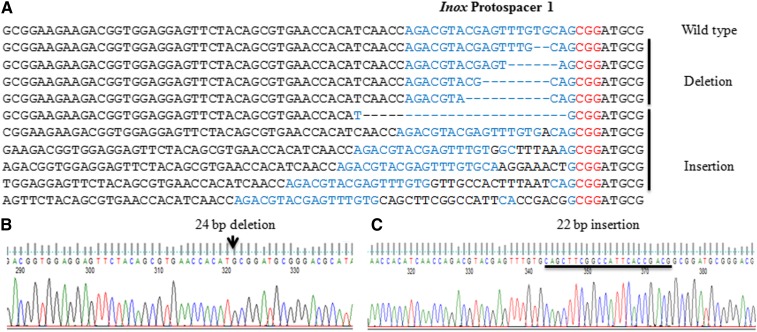
Four regions of the two genes (inox and pds) were targeted for editing in suspension cells of wheat (File S1). The rate of mutations observed in wheat was higher as compared to that in N. benthamiana. We observed 18–22% mutations in different protospacers (Table 2) in wheat. Both deletion and insertion mutations were detected. A maximum of 24 bp deletion and of 22 bp insertion were detected in inox protospacer 1 (Figure 3). Indels were located at similar positions (2–3 bp away from the 5′ end of PAM) in wheat, as observed in N. benthamiana and as reported in other studies (Hwang et al. 2013; Li et al. 2013; Mali et al. 2013; Nekrasov et al. 2013; Shan et al. 2013).
Table 2. Average indel percentage in T. aestivum at different protospacers with related expression cassettes.
| Expression Cassette | Indel (%) | |
|---|---|---|
| Protospacer 1 | Protospacer 2 | |
| pCinox1 | 17.9 (14/78) | NA |
| pCinox2 | NA | 20.7 (17/82) |
| pCwpds1 | 22.3 (19/85) | NA |
| pCwpds2 | NA | 18.4 (14/76) |
| pCinox12 | 12.7 (12/94) | 11.2 (9/80) |
| pCpin1 | 10.2 (8/78) | 8.6 (7/81) |
NA, not applicable.
Figure 3.
CRISPR-cas–guided editing at protospacer 1 of inositol oxygenase (inox) gene of T. aestivum. (A) Alignment of wild-type and sequences with indel at protospacer 1 of inox gene. (B) and (C) Sanger sequencing of selected deletion and insertion mutants, respectively.
Duplex cgRNA-Cas9 induced deletion in inox gene
To analyze the efficacy of duplex cgRNA targeting two sites simultaneously in one gene, we fused the two cgRNA (pCinox12) targeting the inox gene in wheat. The expression of duplex cgRNA caused indels at both the sites. However, the efficiency was lower as compared to that of the single cgRNA (Table 2). Approximately 11–12% of mutations were detected at each site. Complete deletion of the region between two targeted sites was observed in 2.8% cases (Figure 4).
Figure 4.
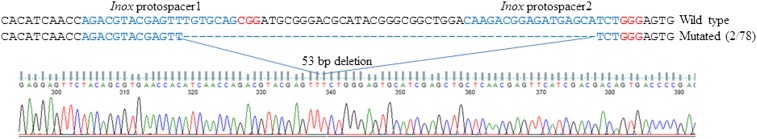
Duplex cgRNA-Cas9–induced deletion in inox gene. The figure shows that the targeting of two protospacers in one gene resulted into the deletion of the complete fragment between targeted regions.
Multiple gene targeting with cgRNA-Cas9
To detect the editing potential of cgRNA-Cas9 for more than one gene at a time, we fused cgRNA targeting the protospacer 1 of pds and inox genes of wheat (pCpin1). Expression of this cassette in suspension cells caused indels at both the target sites, although with different efficiency (Table 2). Approximately 10% of indels were detected in pds and 8% were detected in inox gene.
Target specificity of cgRNA
To evaluate the off-target effects of CRISPR, several sets of random mutations in protospacer 1 binding site of inox cgRNA were developed (Table S2) and transformed in suspension cells of wheat for coexpression with Cas9. The mutations at the 3′ end of the target binding region abolished the genome cleavage activity completely in the presence of a single base mismatch up to base 12. Multiple mutations up to the base 16 position also prevented cleavage (Figure S1), whereas single base mismatches at the 5′ end were tolerated to some extent and provided 1–3% mutation.
Discussion
The study demonstrates that the CRISPR-Cas system can be applied successfully to genome editing in very large genomes such as the 17 Gb hexaploid genome of wheat. The application of the CRISPR-Cas system has been demonstrated in bacteria, yeast, and animal cells with a high percentage of indels (Cong et al. 2013; Mali et al. 2013; DiCarlo et al. 2013; Jinek et al. 2013). Application of this system in plants has also been reported using transient expression by protoplast transformation and agro-infiltration (Li et al. 2013; Nekrasov et al. 2013; Shan et al. 2013).
We have demonstrated that CRISPR-Cas–guided genome editing can be achieved by using wheat suspension cells. This makes the application of this technique simpler to crop plants that can be regenerated from cell cultures. We also showed that the deletion of a gene fragment can be achieved in wheat by simultaneous cleavage at two targeted sites within one gene. Similar deletion has been reported in the case of Arabidopsis (Li et al. 2013; Mao et al. 2013), although its genome size is much smaller than the wheat genome. This result suggests that the method can be useful in deletion of target regions from the genome and the development of knockout mutants.
We have also analyzed the efficacy of multiplex cgRNA targeting two genes at one time. Significant indels were detected in both the target genes in wheat. Because the experiment was performed with suspension cells, we cannot comment on whether both the target sites were mutated in the same cell or in different cells. However, the results establish that the method introduces mutations in multiple genes using the same expression cassette. Such results have been reported in the case of animal systems (Cong et al. 2013) but have not been reported in plants.
The CRISPR-Cas system is much simpler than the other reported systems such as ZFN and TALEN (Chen and Gao 2013; Zhang et al. 2010). High-efficiency editing can be achieved even in large and complex plant genomes at each of the multiple targeted locations using the CRISPR-Cas system. Unlike ZFNs, it is not necessary to screen several cgRNAs to target the sites. Certain off-target mutations have also been reported (Fu et al. 2013) in the case of mismatch at the 5′ end of the target binding region. We observed that even single mismatch toward the 3′ end until base 12 abolished the binding of CRISPR completely. Similar results have been reported in the case of animal and bacterial systems (Cong et al. 2013). Although our experiments for the detection of off-target binding do not include extensive analysis, the results suggested that off-target mutations can be avoided by careful selection of the target sequences. Instead of targeting the conserved and semiconserved regions, unique sequences should be chosen during CRISPR design, especially toward the 3′ side.
Because of the simplicity in designing cgRNA (Cong et al. 2013) and the applicability to a wide variety of plants and animals, the CRISPR-Cas system will be a very useful tool for reverse genetics as well as functional genomics. The necessity of NGG (PAM) at the 3′ of the target region is the only limitation for the target site.
Supplementary Material
Acknowledgments
We thank Dr. Siddharth Tiwari for providing wheat cell suspension culture. We thank National Agri-Food Biotechnology Institute (Department of Biotechnology, Government of India) for providing research support. S.K.U. thanks the Department of Science and Technology (DST), India, for a DST-INSPIRE Faculty Fellowship. J.K. is grateful to Council of Scientific and Industrial Research, India, for a Senior Research Fellowship. R.T. thanks DST for a JC Bose Fellowship.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Brouns S. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., et al. , 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. I. B., Kanneganti T. D., Young C., Cakir C., Huitema E., et al. , 2006. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48: 165–176. [DOI] [PubMed] [Google Scholar]
- Chen K., Gao C., 2013. TALENs: Customizable molecular DNA scissors for genome engineering of plants. J. Genet. Genomics 40: 271–279. [DOI] [PubMed] [Google Scholar]
- Chylinski K., Le R. A., Charpentier E., 2013. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., et al. , 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., et al. , 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31: 822–826. DOI: 10.1038/nbt.2623. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V., 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109. 109: E2579– E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., et al. , 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore M. M., Lundgren M., Duijn E. V., Bultema J. B., Westra E. R., et al. , 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 18: 529–536. [DOI] [PubMed] [Google Scholar]
- Li J. F., Norville J. E., Aach J., McCormack M., Zhang D., et al. , 2013. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31: 688–691. DOI: 10.1038/nbt.2654. 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Staskawicz B., Weigel D., Jones J. D. J., Kamoun S., 2013. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 691–693. DOI: 10.1038/nbt.2655. 10.1038/nbt.2655 [DOI] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., et al. , 2013. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31: 686–688. DOI: 10.1038/nbt.2650. 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- Wu H., McCormac A. C., Elliott M. C., Chen D. F., 1998. Agrobacterium-mediated stable transformation of cell suspension cultures of barley (Hordeum vulgare). Plant Cell Tissue Organ Cult. 54: 161–171. [Google Scholar]
- Zhang F., Maeder M. L., Unger-Wallace E., Hoshaw J. P., Reyon D., et al. , 2010. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107: 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.