Abstract
Polarized total internal reflection fluorescence microscopy (polTIRFM) can be used to detect the spatial orientation and rotational dynamics of single molecules. polTIRFM determines the threed-imensional angular orientation and the extent of wobble of a fluorescent probe bound to the macromolecule of interest. This protocol describes how to label chicken calmodulin (CaM) with bifunctional rhodamine (BR) at two engineered cysteine (Cys) residues (P66C and A73C) so that it cross-links the two Cys sites. The resulting BR-CaM protein is then purified by high-performance liquid chromatography (HPLC) and concentrated by filter centrifugation. To confirm that the two Cys residues in the labeled CaM are actually cross-linked by BR, a sample of purified BR-CaM is digested by an endoproteinase and analyzed by mass spectrometry. The BR-CaM can then be used to label myosin V, which can in turn be used in a polTIRFM processive motility assay.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES: Please see the end of this article for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
-
Bisiodoacetamidorhodamine (bifunctional rhodamine, BR-I2; Corrie et al. 1998)
Alternatively, bis-[(N-iodoacetyl)piperazinyl]sulfonerhodamine (Invitrogen B10621) may be used.
Bovine serum albumin (BSA) standards and other reagents for Bradford assay (or other protein colorimetric assay)
-
Calmodulin (CaM) from chicken, genetically engineered to replace residues Pro-66 and Ala-73 with cysteines (P66C and A73C)
The protein should be bacterially expressed and purified as described by Putkey et al. (1983, 1985).
-
CaM, monofunctional acetamidotetramethylrhodamine (ATR)-labeled
This is used as a control. For more information, see Fig. 1 (bottom right) and the section Fluorescent Probes for Orientation Studies in the article Orientation and Rotational Motions of Single Molecules by Polarized Total Internal Reflection Fluorescence Microscopy (polTIRFM) (Beausang et al. 2012a).
Chromatography solvent A (0.1% w/v trifluoroacetic acid [TFA; Sigma-Aldrich 299537])
Chromatography solvent B (50% v/v acetonitrile [ACN; Fisher A9984] and 0.1% w/v TFA])
Dialysis buffer for calmodulin labeling <R>
Digestion buffer (50 mm NaHPO4 at pH 8.0)
Dithiothreitol (DTT; Fisher)
Endoproteinase Asp-N (sequencing grade; Roche Applied Science 11420488001)
Labeling buffer <R>
Sodium 2-mercaptoethanesulfonate (MESNA; Sigma-Aldrich M1511)
Tris(2-carboxyethyl)phosphine (TCEP; Invitrogen T2556)
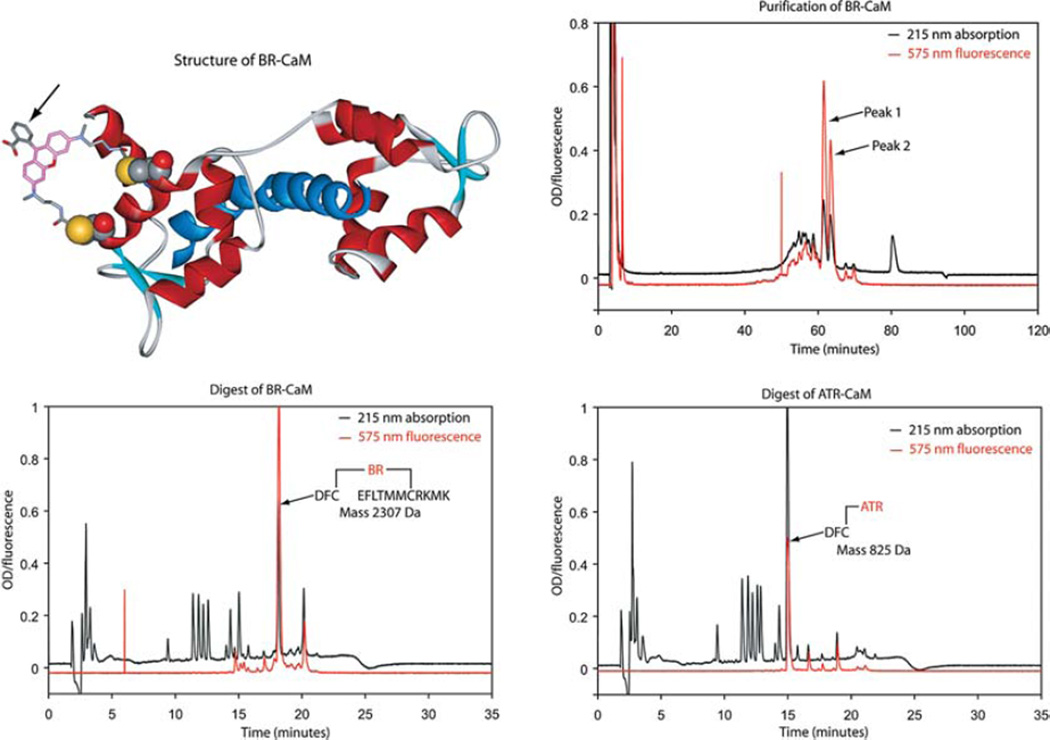
Figure 1.
Purification and characterization of BR-CaM. (Top left) The crystal structure of CaM (Houdusse et al. 1996) showing the location and orientation of the BR entity. The arrow identifies the carboxyphenyl ring of BR that produces the two diastereoisomer peaks appearing in the figure at the top right. Reverse-phase (C18) HPLC of labeled CaM (top right) and tryptic fragments (bottom left/right).
Equipment
Argon gas
Centrifugal filter unit (Centriplus YM-10, 10-kDa cutoff; Millipore 4421)
Centrifuge
Desalting column (PD-10 Sephadex-G25 column; Amersham Biosciences/GE Healthcare Life Sciences)
Dialysis tubing (12- to 14-kDa cutoff; Fisher Scientific132678)
-
HPLC columns
Analytical reverse-phase, C18 (Grace VYDAC 218TP5415)
Preparative reverse-phase, C18 (Grace VYDAC, 218TP510, with guard cartridge 218GD54)
Liquid nitrogen
Water baths, 37°C and boiling
METHOD
Labeling of Mutant CaM with BR
-
1.
Equilibrate a PD-10 desalting column with 15 mL of labeling buffer.
-
2.
Load 150 nmol mutant CaM onto the column. Elute it with 1.5 mL of labeling buffer.
-
3.
Determine the protein concentration in the eluate (e.g., by Bradford assay) and adjust the concentration of mutant CaM to 50 µm with labeling buffer.
-
4.
To reduce the disulfide bonds, incubate CaM in 50 µm TCEP for 40–60 min at room temperature.
For example, to 2 mL of 50 µm CaM, add 100 µL of 1 mm TCEP (pH 7.4).
-
5.
Add BR-I2 to a final concentration of 30 µm.
For example, add 4 µL of 15 mm BR-I2 to the 2.1-mL sample from Step 4.
-
6.
Incubate the solution at 20°C. After 12, 24, and 36 min, add 20-µm supplements of BR-I2 to a final concentration of 90 µm.
-
7.
After a total incubation time of 90 min, quench the labeling reaction by adding MESNA to a concentration of 3 mm.
Purification of BR-CaM
-
8.
Purify BR-CaM by HPLC on a preparative reverse-phase C18 column at room temperature.
Equilibrate the column with 30% solvent A, 70% solvent B at a flow rate of 4 mL/min.
-
Load the reaction mixture onto the column. Elute the BR-CaM with the following gradient profile:
10 min, isocratic 70% solvent B
40 min, linear gradient 70–80% solvent B
40 min, linear gradient 80–85% solvent B
1 min, linear gradient 85–70% solvent B
-
Collect 1.6-mL fractions starting at 50 min. Monitor the elution of the protein by absorption at λ = 215 nm and fluorescence at λex = 549 nm and λem = 560 nm.
The elution profile should show two distinct fluorescent protein peaks with 4–7 mL in each (Fig. 1, top right).
-
9.
During the HPLC run, deoxygenate 4 L of dialysis buffer by bubbling argon gas through it for at least 40 min at 4°C. After 30 min, add 5 mm DTT and continue bubbling. Because the DTT powder will not dissolve in dialysis buffer at 4°C, dissolve the DTT in 1 mL of room-temperature H2O before adding it to the dialysis buffer.
-
10.
Immediately after the HPLC run, transfer the two fluorescent protein peaks (purified diastereoisomers of BR-CaM) into separate dialysis tubes. Quickly place each of them into 500 mL of cold, deoxygenated dialysis buffer.
The immediate transfer of purified BR-CaM into dialysis and precautions against oxygen contamination are necessary to minimize the oxidation of methionine residues.
-
11.
Change the dialysis buffer after 2 and 4 h. Continue to dialyze the samples overnight with continual stirring and bubbling with argon.
-
12.
On the following day, transfer the samples into Centriplus YM-10 filter units. Concentrate the BR-CaM protein:
Centrifuge the samples at 4°C (3000 g) until the volume reaches 1.5 mL or less.
At 1-h intervals, stop the centrifuge and transfer the filtrate to a storage container.
Keep the filtrate on ice. After the BR-CaM protein is confirmed to be recovered in the retentate (in Step 16), discard the filtrate.
-
13.
To recover the protein, invert the filter and then centrifuge at 2000 g for 4 min at 4°C so that the protein falls into the tube cap.
-
14.
Flash-freeze 200-µL aliquots of the protein in liquid nitrogen. Store the aliquots for up to 1 yr at −80°C.
Analysis of Purified BR-CaM
-
15.
To determine the dye-to-protein ratio, use a protein colorimetric assay (e.g., a Bradford assay with BSA standards). The rhodamine chromophore absorption is ε547 = 89,250 m−1 cm−1 (Corrie et al. 1998).
The conjugate should contain one chromophore per 17-kDa CaM peptide.
-
16.
To analyze the purity of the labeled protein, use analytical reverse-phase HPLC with a flow rate of 1 mL/min.
Load 15 µL (or 3 pmol) of protein sample onto the column, equilibrated in solvent A.
Wash the column with solvent A for 2 min.
Elute the protein with a 0–100% solvent B linear gradient over 35 min. Monitor the absorption and fluorescence as in Step 8.iii.
Wash the column with solvent A between samples.
-
17.
Confirm that the two Cys residues in the labeled CaM are actually cross-linked by BR.
Dialyze 4 nmol of BR-CaM (or monofunctional ATR-labeled CaM) into digestion buffer.
Add 3.2 µg of endoproteinase Asp-N, and digest for 19 h at 37°C.
Quench the digestion reaction by incubating it at a rolling boil for 5 min.
-
Separate the digested peptide fragments by analytical HPLC. Using a flow rate of 1mL/min, load the digested sample onto the column in 70% solvent A, 30% solvent B. Elute the protein with the following profile:
2 min, 30% solvent B
17.5 min, linear gradient 30–100% solvent B
Monitor the eluate as in Step 8.iii. Collect 200-µL fractions.
-
Determine the mass of the fluorescent fragments by electrospray time-of-flight mass spectrometry (e.g., at the Keck Foundation Biotechnology Resource Laboratory at Yale University).
A two-site labeled peptide, cleaved between the two fluorophore linkers, retains the mass of both flanking peptide fragments (Fig. 1, bottom left).
RELATED INFORMATION
The BR-CaM produced in this protocol is used in Fluorescent Labeling of Myosin V for Polarized Total Internal Reflection Fluorescence Microscopy (polTIRFM) Motility Assays (Beausang et al. 2012b). A method is also available for Preparation of Filamentous Actin for Polarized Total Internal Reflection Fluorescence Microscopy (polTIRFM) Motility Assays (Beausang et al. 2012c).
RECIPES
Dialysis Buffer for Calmodulin Labeling
10 mm Tris-HCl
30 mm KCl
5 mm dithiothreitol (DTT; add fresh just prior to use)
Adjust pH to 7.0 with KOH.
Labeling Buffer
-
25 mm Tris-Cl at pH 7.4
100 mm NaCl
2 mm EDTA (Sigma-Aldrich)
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health grants AR26846 and AR51174 to the Pennsylvania Muscle Institute and by the National Science Foundation (NSF) NSEC Nano/Bio Interface Center (NBIC) DMR 04-25780. J.F.B. is supported by an NSF IGERT fellowship through the NBIC DGE 0221664. We thank Drs. Philip C. Nelson and Jody A. Dantzig-Brody for review and useful comments on the manuscript.
REFERENCES
- Beausang JF, Sun Y, Quinlan ME, Forkey JN, Goldman YE. Orientation and rotational motions of single molecules by polarized total internal reflection fluorescence microscopy (polTIRFM) Cold Spring Harb Protoc. 2012a doi: 10.1101/pdb.top069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausang JF, Sun Y, Quinlan ME, Forkey JN, Goldman YE. Fluorescent labeling of myosin V for polarized total internal reflection fluorescence microscopy (polTIRFM) motility assays. Cold Spring Harb Protoc. 2012b doi: 10.1101/pdb.prot069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausang JF, Sun Y, Quinlan ME, Forkey JN, Goldman YE. Preparation of filamentous actin for polarized total internal reflection fluorescence microscopy (polTIRFM) motility assays. Cold Spring Harb Protoc. 2012c doi: 10.1101/pdb.prot069377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrie JET, Craik JS, Munasinghe VRN. A homo-bifunctional rhodamine for labeling proteins with defined orientations of a fluorophore. Bioconjug Chem. 1998;9:160–167. doi: 10.1021/bc970174e. [DOI] [PubMed] [Google Scholar]
- Houdusse A, Silver M, Cohen C. A model of Ca2+-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch. Structure. 1996;4:1475–1490. doi: 10.1016/s0969-2126(96)00154-2. [DOI] [PubMed] [Google Scholar]
- Putkey JA, Ts’ui KF, Tanaka T, Lagacé L, Stein JP, Lai EC, Means AR. Chicken calmodulin genes. A species comparison of cDNA sequences and isolation of a genomic clone. J BiolChem. 1983;258:11864–11870. [PubMed] [Google Scholar]
- Putkey JA, Slaughter GR, Means AR. Bacterial expression and characterization of proteins derived from the chicken calmodulin cDNA and a calmodulin processed gene. J Biol Chem. 1985;260:4704–4712. [PubMed] [Google Scholar]