Abstract
Evolutionary inferences are usually based on statistical models that compare mean genotypes and phenotypes (or their frequencies) among populations. An alternative is to use the actual distribution of genotypes and phenotypes to infer the “exchangeability” of individuals among populations. We illustrate this approach by using discriminant functions on principal components to classify individuals among paired lake and stream populations of threespine stickleback in each of six independent watersheds. Classification based on neutral and non-neutral microsatellite markers was highest to the population of origin and next-highest to populations in the same watershed. These patterns are consistent with the influence of historical contingency (separate colonization of each watershed) and subsequent gene flow (within but not between watersheds). In comparison to this low genetic exchangeability, ecological (diet) and morphological (trophic and armor traits) exchangeability was relatively high – particularly among populations from similar habitats. These patterns reflect the role of natural selection in driving parallel changes adaptive changes when independent populations colonize similar habitats. Importantly, however, substantial non-parallelism was also evident. Our results show that analyses based on exchangeability can confirm inferences based on statistical analyses of means or frequencies, while also refining insights into the drivers of – and constraints on – evolutionary diversification.
Keywords: adaptive radiation, ecological speciation, parallel evolution, convergent evolution, natural selection, Gasterosteus aculeatus
Introduction
Evolutionary landscape genetics uses spatial patterns of genetic variation to infer evolutionary processes. The usual route toward such inferences is to examine how genotypes and phenotypes traits differ among populations across a landscape. As one example, a role for deterministic natural selection is typically inferred when genotypes or phenotypes are similar for independent populations in similar environments: i.e., parallel or convergent evolution (Endler 1986; Schluter 2000; Langerhans and DeWitt 2004; Arendt and Reznick 2008; Losos 2011; Wake et al. 2011). As another example, specific causes of natural selection are typically inferred through correlations between genotypes or phenotypes and a particular ecological factor (Endler 1986; Wade and Kalisz 1990; MacColl 2011), such as diet (e.g., Schluter and McPhail 1992; Kaeuffer et al. 2012), structural habitat features (e.g., Losos 2009), predation (e.g., Reznick and Bryga 1996; Langerhans and DeWitt 2004), or water flow (e.g., Langerhans 2008). In addition, gene flow is often inferred to constrain adaptive evolution when divergence in non-neutral genotypes or phenotypes is negatively correlated with divergence in neutral markers (Hendry and Taylor 2004; Nosil and Crespi 2004; Bolnick and Nosil 2007; Räsänen and Hendry 2008).
Evolutionary inferences of the sort just described typically rely on the analysis of means or frequencies. For example, the first inference discussed above (parallel or convergent evolution) typically comes from statistical models (e.g., GLMs, ANOVAs, AMOVAs, chi-square tests) that consider the main effect of the environment term. The second two inferences (specific causal factors and the role of gene flow) typically come from statistical models (e.g., regressions, Mantel tests) that ask how population means or frequencies covary with potential drivers. At the most basic level, the essence of these approaches is to (1) distill populations down to estimates of their means, frequencies, and variances; (2) specify the precision of those estimates in the form of confidence intervals; (3) make statistical inferences based confidence interval overlap; and (4) infer effect sizes based on explained variance. We will jointly refer to all of these methods as the “significant effects on means” approach. Our goal is then to advance an alternative approach that might better capture the biological properties of populations and therefore allow improved evolutionary inference.
The alternative is to use the actual distribution of genotypes and phenotypes to infer how well individuals can be exchanged among populations without altering their ecological, phenotypic, or genetic properties. This consideration of the actual distribution of phenotypes and genotypes acknowledges the important role that individual variation can play in a variety of evolutionary and ecological processes (Bolnick et al. 2003; Hansen et al. 2006; Bolnick et al. 2011). To be more specific, this alternative approach evaluates the probability of classification of each individual into each sampled population, and then uses the distribution of these classification probabilities to inform exchangeability. We will henceforth use this “exchangeability” term when referring to the phenomenon in general, whereas we will add appropriate descriptors when our goal is to infer exchangeability with respect to a particular property (e.g., ecological, phenotypic, neutral genetic, non-neutral genetic, specific traits, etc.). This specificity is valuable because comparing exchangeability among the different properties aids evolutionary inference, as will be made apparent below.
Concepts of exchangeability have been mostly used in speciation and taxonomy (Templeton 1989, 2001; Stockman and Bond 2007; Bond and Stockman 2008) and in conservation biology (Funk and Fa 2006; Cano et al. 2008). In particular, Templeton (1989, 2001) argued that separate species are reciprocally monophyletic (genetic non-exchangeability) and differ in reproductive or ecological traits (ecological non-exchangeability). Crandall et al. (2000) then argued that the same criterion could be extended to the identification of evolutionarily significant units within species (see also Rader et al. 2005). Our invocation of exchangeability is conceptually related to these earlier ideas but it differs in two important ways. First, previous applications have relied on the “significant effects on means” approach rather than using the entire distribution to ask which populations could exchange individuals with the least alteration of those distributions. Second, previous applications have been concerned with determining if groups of organisms differ enough to be considered separate species or evolutionarily significant units, whereas we will be concerned with using conspecific populations to infer evolutionary processes.
We suggest that exchangeability can be best evaluated by using individual classification methods (details below). In particular, populations show high exchangeability when individuals are frequently “misclassified” between them: i.e., individuals originating from one source population are often classified to another population. Considering this exchangeability across a large number of populations in different locations and environments can be used to infer evolutionary processes. For instance, high exchangeability (high misclassification) of individuals between independent populations in similar environments, but low exchangeability (low misclassification) of individuals between independent populations in different environments, would suggest parallel (or convergent) evolution and thus imply the importance of deterministic selection. Similarly, high exchangeability between independent populations experiencing a particular environmental factor (e.g., temperature) would implicate that factor as a key selective force, and high exchangeability between nearby populations would imply an important role for gene flow.
Classification methods are already widely used and have proven powerful for generating inferences about population structure and evolutionary processes (Castric and Bernatchez 2004; Hauser et al. 2006; Waples and Gaggiotti 2006; Manel et al. 2006). For morphological traits, classification based on discriminant functions is often used to infer similarities and differences among populations (e.g., Lavin and McPhail 1993; Reznick et al. 1996; Hendry et al. 2002). For genetic markers, classification based on “assignment tests” is often used for similar inferences and to identify particular individuals that are migrants or hybrids (Pritchard et al. 2000; Hansen et al. 2001; Hauser et al. 2006; Hubisz et al. 2009). We extend these existing classification methods in several ways. First, we use identical sets of individuals to perform classification based on morphology (trophic and armor traits), genetic markers (neutral and non-neutral microsatellites), and ecology (diet as inferred by stomach contents and stable isotopes). Direct comparison of results among these different classes of variables is made possible by using the same classification method for each: discriminant functions on principal components (Jombart et al. 2010). Second, we consider not only the individuals originating from each population that are correctly classified to their home population versus “misclassified” to another population, but we also “cross-classify” all individuals. That is, we also ask which population, apart from the population of origin, is the best fit for each individual. This cross-classification approach complements misclassification because exchangeability then can be inferred based on all sampled individuals (as opposed to just those misclassified), which should increase power and precision.
Study system and predictions
Our illustrative analysis makes use of data collected in an earlier study of threespine stickleback (Gasterosteus aculeatus). Kaeuffer et al. (2012) examined parapatric population pairs of threespine stickleback in lake versus stream environments (henceforth “habitats”) within six different watersheds (one pair per watershed). Based on neutral microsatellites analyzed for these populations (Kaeuffer et al. 2012), FST between populations in different watersheds had a mean of 0.229, a standard deviation of 0.093, a maximum of 0.474, and a minimum of 0.060. Of 55 pair-wise comparisons, only three had FST < 0.100. These differences support the inference that lake-stream divergence was an independent evolutionary event within each watershed, and that any subsequent gene flow has been very low among watersheds (see also Hendry and Taylor 2004; Berner et al. 2009).
Using the “significant effects on means” approach the following conclusions were reached by Kaeuffer et al. (2012). (1) Lake-stream diet divergence was in a similar direction in the six watersheds (lake fish forage more on zooplankton than do stream fish), implying that divergent selection on foraging related (trophic) traits was also in a similar direction. (2) Lake-stream divergence in body shape and gill raker structure was in a similar direction in the six watersheds (i.e., parallel evolution), suggesting an important role of deterministic natural selection. (3) By contrast, lake-stream divergence in armor traits differed in direction among the six watersheds (i.e., non-parallel evolution), suggesting a weak role of deterministic natural selection – at least that between generic “lake” and “stream” designations. (4) Lake-stream divergence in genetic (microsatellite) markers was sometimes in the same direction (parallel) and sometimes not (non-parallel), implying both deterministic and non-deterministic contributions of selection. (5) Lake-stream divergence in trophic traits and in some genetic markers was positively correlated with lake-stream divergence in diets, confirming intuition that diet divergence was a primarily determinant of selection.
Reframing this system in the context of exchangeability, we expect the following. (1). Misclassification and cross-classification based on diet should be higher between populations in similar habitats than between populations in different habitats. (2) Misclassification and cross-classification based on trophic traits should be higher between populations in similar habitats than between populations in different habitats. (3) Misclassification and cross-classification based on armor traits should not be closely associated with habitat type. (4) Misclassification and cross-classification based on genetic markers should be much lower between than within watersheds – because contingency increases differences between watersheds and gene flow decreases differences within watersheds. It is also possible that genetic misclassification and cross-classification should be more closely linked to habitat type for non-neutral than for neutral markers – if the non-neutral markers influenced by selection are similar in the different watersheds. (5) Misclassification and cross-classification patterns for diet should be predictive of patterns for trophic traits and perhaps also for non-neutral genetic markers – because diet is the main variable driving parallelism in divergent selection. The same should not be true, however, for armor traits and neutral markers.
Methods
Populations and sampling
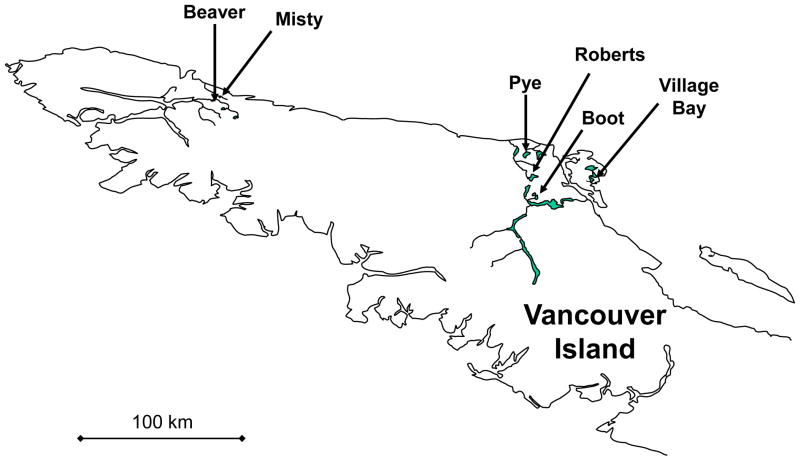
In May 2008, we collected threespine stickleback from paired lake and stream sites in each of six independent watersheds on Vancouver Island, British Columbia, Canada (12 sites in total). The specific pairs (Fig. 1; Appendix S1) were chosen based on prior knowledge of strong lake-stream divergence in morphology, and independent origins from marine ancestors (Hendry and Taylor 2004; Berner et al. 2008, 2009). At each site, we used unbaited minnow traps to capture and retain 36–40 fish (Appendix S1). The fish were euthanized with MS222 and their left side was photographed with a digital camera (Nikon D100). We then weighed each fish (±0.01 g) and preserved part of the pectoral fin in 95% ethanol. To enable analysis of stable isotopes (see below), white muscle tissue from the back of each fish was preserved on ice and later dried in an oven for 36 hours at 72 °C. Each fish was also dissected to determine its sex and to remove the stomach for diet analysis. The stomach (and its contents) and the remaining carcass were preserved separately in 95% ethanol.
Figure 1.
Map of the studied watersheds on northern Vancouver Island, British Columbia, Canada. Other watersheds and tributaries are omitted for clarity.
Ecology, morphology, and genetics
Our ecological inferences were based on diet, which is an excellent predictor of selection on trophic traits in threespine stickleback (for a detailed rationale, see Bolnick and Araujo 2011, and Kaeuffer et al. 2012). To consider short-term diets, food items from each stomach were categorized as either limnetic or benthic following Schluter and McPhail (1992). These counts were used to calculate the “proportion of limnetic prey” (PLP) for each individual as the number of limnetic prey items divided by the total number of identified prey items (limnetic plus benthic) (Schluter and McPhail 1992; Berner et al. 2008, 2009). To consider long-term diets and ecological niches, we used stable isotopes (Post 2002; Newsome et al. 2007). Following previous studies of threespine stickleback (Bolnick et al. 2008; Reimchen et al. 2008; Snowberg and Bolnick 2008; Matthews et al. 2010), we estimated (1) the relative importance of different sources of primary production for each individual (δ13C), and (2) the trophic position of each individual (δ15N). To generate these ratios, weighed and dried muscle samples were analyzed in the UC Davis Stable Isotope Facility, and the resulting δ13C and δ15N values were expressed relative to international standards: PDB (PeeDee Belemnite) for carbon and air for nitrogen. Kaeuffer et al. (2012) provides more details on these calculations.
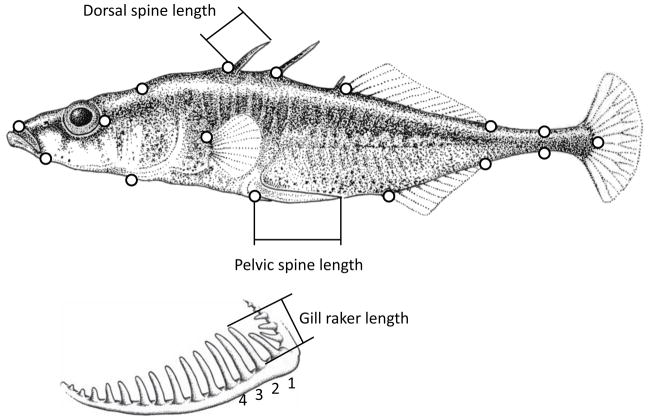
Our morphological inferences were based on trophic traits (body shape, gill raker number, and gill raker length) and defensive armor traits (plates and spines). More details on the functional significance of these traits, and their genetic basis, are provided in Kaeuffer et al. (2012). To analyze body shape, we used geometric morphometrics. TpsDig (Rohlf 2006) was used to place 16 homologous landmarks onto the digital image of each fish (Berner et al. 2009; Kaeuffer et al. 2012; Fig. 2). TpsRelw (Rohlf 2005) was used to estimate each fish’s centroid size (body size) and its affine (uniform) and non-affine (partial warp) components of body shape (Zelditch et al. 2004). TpsRelw was then used to extract relative warps (principal components) of shape variation across all individuals in the study. Lateral plates were counted on the left side of each fish and digital calipers were used to measure the lengths (± 0.01 mm) of the left pelvic spine and the first and second dorsal spines along the anterior side of the spine (Fig. 2). Gill rakers were counted on the ventral bone of the left gill arch from its base to its joint with the dorsal bone, and the lengths of the second to fourth gill raker were measured from the epibranchial-ceratobranchial joint on the ceratobranchial (Berner et al. 2009; Kaeuffer et al. 2012). These gill measurements were made at 45× magnification on a stereomicroscope with a micrometer (precision of 0.01 mm). Some of the measured traits were correlated with body size, and so allometric size standardizations were applied (see Kaeuffer et al. 2012).
Figure 2.
Positions of landmarks used to quantify in body shape. Also shown are the spine measurements. The drawing is modified from Berner et al. (2008).
Genetic data were based on six putatively neutral microsatellite loci and six putatively non-neutral microsatellite loci (“neutral” and “selected” loci in Kaeuffer et al. 2012). The six neutral loci (Stn34, Stn67, Stn87, Stn159, Stn199, Stn234) were chosen because they are not tightly linked to any known quantitative trait locus (QTL). The six non-neutral loci (Stn45, Stn168, Stn232, Stn246, Stn321, Stn386) were chosen (out of 192 examined in bulk-segregant analyses) because they showed several signatures of divergent selection. These signatures are summarized here and are detailed in Kaeuffer et al. (2012). (1) Allele size distributions showed little overlap between the lake and stream fish within two or more watersheds, with the latter criterion decreasing the chance that divergence was due to drift or type 1 statistical error. (2) Previous work on stickleback found that five of the six loci were linked to QTL for adaptive traits, including morphology and gill raker number. (3) Lake-stream divergence in Stn321 was positively associated with lake-stream divergence in gill raker number, and the same was true for Stn45 and body shape. (4) The FST for each non-neutral loci was often outside the 95% CI of the neutral loci.
Classification
Exchangeability analyses can be based on any method that classifies individuals among different groups, such assignment tests for genetic data or discriminant functions for morphological data. Analyzing our data with several of these methods yielded qualitatively similar results, and so we will here present those based on only one method: Discriminant Analysis on Principal Components (DAPC, Jombart et al. 2010). DAPC has several advantages over other methods, including applicability to any data type (e.g., quantitative traits, metric traits, and multilocus genotypes). This property allowed us to directly compare results for different variables without any bias owing to different classification methods. Additional details regarding DAPC and its assumptions and advantages appear in Jombart et al. (2010).
We generated five different DAPCs using five different variable “classes”: diet (proportion of limnetic prey, δ13C, and δ15N), trophic traits (relative warp 1, gill raker length, and gill raker number), armor traits (plate number, pelvic spine length, and dorsal spine length), neutral markers (Stn34, Stn67, Stn87, Stn159, Stn199, and Stn234), and non-neutral markers (Stn45, Stn168, Stn232, Stn246, Stn321, and Stn386). Although some effects of sex were evident, these were minor in comparison to population differences and are ignored for the sake of simplicity. When information was not available for a particular individual (5.7% of individuals for gill raker number, 0.9% for PLP, and 1.91% for isotopes), we substituted the mean trait value estimated over all individuals (Jombart et al. 2010). Retaining too many principal components can lead to overfitting the discriminant analyses, and so we first used an a-score function to estimate the optimal number of PCs (Jombart et al. 2010), which was three for all variable classes except neutral markers (14) and non-neutral markers (13). In addition to these DAPC for variable classes, we also performed DAPC individually for each morphological variable: relative warp 1, gill raker size, gill raker length, lateral plate number, pelvic spine length, and dorsal spine length.
DAPC allowed us to determine the probability that a given individual from a given population of origin could be classified into the set of 12 candidate populations included in the analysis. In each case, we considered an individual to be classified to the population where its probability was highest excluding the population of origin in the case of cross-classification (see below). The population to which an individual fish was classified might represent, in reference to the population of origin, a similar or different habitat (lake or stream) between which gene flow would be likely (parapatric within a watershed) or unlikely (allopatric between watersheds). We could thus group individuals into four categories of classification: the population of origin (origin), a parapatric population in a different habitat (lake to stream or stream to lake within the same watershed), an allopatric population in a similar habitat (lake to lake or stream to stream between watersheds), and an allopatric population in a different habitat (lake to stream or stream to lake between watersheds). The above analysis yields patterns that we will analyze in the context of “misclassification.” For “cross-classification,” the analysis was the same except for one posthoc modification: for those individuals classified with the highest probability to their population of origin, we instead record the population to which that individual had the next-highest probability of classification. Thus, in the case of cross-classification, the analysis is based on only the last three of the above categories.
Interpretation of classification patterns requires standardizing for random expectations: i.e., how many individuals would be classified into each category if classification was equally probable into each of the 12 candidate populations (11 for cross-classification). We performed this standardization by dividing the number of individuals classified (or cross-classified) into a given category by the number of populations in that category. This standardization gives the mean number of individuals classified (or cross-classified) per population per category. These calculations were performed for each population of origin, yielding 12 standardized estimates that were used to calculate the mean and 95% confidence interval for classification into each category for each DAPC. Although more refined statistical approaches could be implemented (e.g., randomization, mixed models), confidence interval overlap will be used here to illustrate the general procedure in a simple and straightforward fashion.
Results
Classification to population of origin
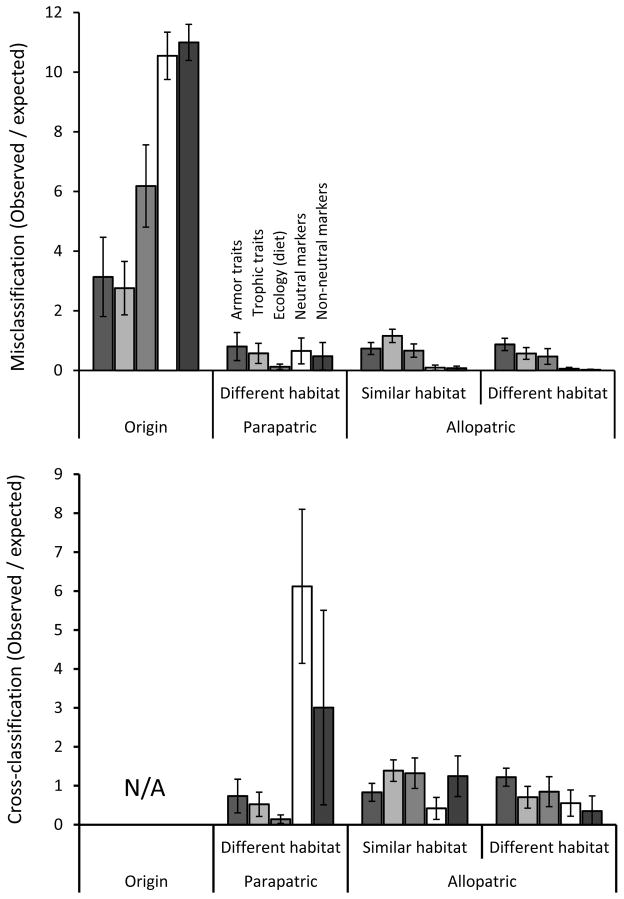
Classification to the population of origin was higher (2.7–11.0 times the random expectation) than to any other population for all classes of variables (Fig. 3). This result shows that all populations are unique to at least some extent for all variables– and, hence, are never fully exchangeable. This outcome presumably reflects the fact that each population has experienced a unique combination of colonization history, selection, drift, mutation, and gene flow.
Figure 3.
Ratio of the mean number of individuals classified into each category (origin, parapatric different habitat, allopatric similar habitat, and allopatric different habitat) to the mean number expected to be classified into those categories by chance (random expectation). The upper panel shows classification into the population of origin and misclassification into other populations, whereas the bottom panel shows cross-classification of all individuals away from the population of origin. In both panels, the values are means and 95% confidence intervals. Although the DAPCs are labeled for only one set of bars, the same labels apply in the same order to each set of bars.
For genetic markers, classification to the population of origin was very high: 10.5 times the random expectation for neutral markers and 11.0 times the random expectation for non-neutral markers (Fig. 3). In fact, 6 of the 24 population-by-marker type (neutral or non-neutral) combinations showed 100% classification to the population of origin, and only 1 of the 24 showed less than 75% (Appendix S2). These results highlight the fact that historical contingency, contemporary selection, and limited gene flow can cause very low genetic exchangeability even between adjacent populations.
For ecological (diet) variables, classification to the population of origin was 6.2 times the random expectation (Fig. 3), and 5 of the 12 populations showed greater than 50% classification to the population of origin (Appendix S2). These results highlight two important points. On the one hand, ecological exchangeability is much higher than genetic non-exchangeability (Fig. 1), which makes sense because diets overlap considerably among populations (Kaeuffer et al. 2012). On the other hand, ecological exchangeability is still somewhat limited, which suggests that diet differences could cause limited exchangeability for traits under divergent selection.
For morphological variables, classification to the population of origin was 2.8 times the random expectation for trophic traits and 3.1 times the random expectation for armor traits (Fig. 3). Classification to the population of origin was greater than 50% for only 1 of the 24 population-by-trait type (trophic or armor) combinations (Appendix S2). We see two major points here. First, selection on morphological traits increases exchangeability, relative to genetic markers, presumably because similar adaptations are suitable for multiple locations. This effect should be particularly strong when populations found in similar habitats, such as two streams or two lakes – a point we consider further below. Second, exchangeability is higher for trophic traits than for the selective factors (diet) thought to influence selection on those traits. This difference indicates that variation in morphological traits reflects patterns of selection that are not fully captured by our simple diet measures. That is, other environmental factors influence selection on stickleback morphology, and these other factors likely differ among populations of a given habitat “type.”
Misclassification
We now consider individuals from the above analysis that were not classified to their population of origin (Fig. 3; Appendix S2). A first striking pattern is that nearly all misclassifications for genetic markers were to the other population within the same watershed (6.1–18.3 times higher, relative to random expectations, within than between watersheds). This result reflects the large among-watershed genetic differences that arise owing to separate colonization from marine ancestors followed by low among-watershed gene flow (Hendry and Taylor 2004; Berner et al. 2009; Kaeuffer et al. 2012). The fact that this pattern held for both neutral and non-neutral markers suggests that the latter did not show strongly parallel lake-stream genetic divergence, at least not in the genomic regions where our markers were located (the strength of this statement will be tempered by results for cross-classification).
A second striking pattern is that misclassifications based on diet were less common between parapatric populations in different habitats than between allopatric populations in either habitat. Specifically, misclassifications were, relative to parapatric different habitats, 5.2 times more likely into allopatric similar habitats and 3.7 times more likely into allopatric different habitats – both relative to random expectations (Fig. 3). This result reflects the fact that diets always show low overlap between parapatric lake and stream habitats, whereas they sometimes show high overlap between allopatric populations, even from different habitats. That is, stickleback from a lake in one watershed might have a diet that is reasonably similar to stickleback from a stream in another watershed. As just one example, the proportion of limnetic prey in Beaver Lake was similar to that in Robert’s Stream (Berner et al. 2009; Kaeuffer et al. 2012).
A third striking pattern is that misclassification based on trophic traits was higher into allopatric populations of the same habitat type than into allopatric or parapatric populations of the other habitat type (Fig. 3). This result reflects the parallelism in lake-stream divergence reported in previous work on these populations (Hendry and Taylor 2004; Kaeuffer et al. 2012). That is, similar phenotypes evolve for population in similar habitats, which increases exchangeability for relevant adaptive traits. The difference was not large, however, which shows that parallelism was not especially strong – a point to which we will later return. By contrast, misclassification based on armor traits showed no association with habitat. This result reinforces previous assertions that armor traits do not show strong parallelism – because divergent selection acting on them is not consistent between lake and stream habitats in different watersheds (Hendry and Taylor 2004; Kaeuffer et al. 2012).
Cross-classification
As explained earlier, exchangeability inferences might be more powerful when based on cross-classification than when based on misclassification, because the former makes use of all individuals whereas the latter makes use only of individuals that are not classified to the population of origin. Fitting this expectation, cross-classification patterns (Fig. 3; Appendix S3) amplified many of the above inferences based on misclassification, while also revealing some additional nuances.
For genetic markers, many individuals were classified with 100% probability to their population of origin (Appendix S1), making them uninformative for cross-classification. However, some strong patterns were nevertheless evident (Fig. 3). In particular, cross-classification was much higher within than between watersheds, a result that further supports the important roles of contingency and gene flow in structuring genetic variation (see above). Some additional patterns were evident for non-neutral (but not neutral) markers: (1) high variation among focal populations meant that confidence intervals overlapped for cross-classification into parapatric versus allopatric populations, and (2) cross-classification into allopatric populations was 3.6 times higher for similar than different habitats. The second of these patterns suggests, in contrast to results for misclassification, that lake-stream divergence in the different watersheds might share at least some genetic basis: i.e., some parallelism at the genetic level (Kaeuffer et al. 2012; Roesti et al. 2012). However, a bias remains because we chose non-neutral markers based partly on this very criterion – see Methods.
As was the case for misclassification, ecological (diet) cross-classification was much lower into different habitats in the same watershed than into similar or different habitats in different watersheds – again likely reflecting diet differences in the former but some diet similarity in the latter. And, also as before, morphological cross-classification was higher into similar habitats than into different habitats for trophic traits but not for armor traits – again likely reflecting parallel selection pressures (diet) for the former but not for the latter.
Additional insights
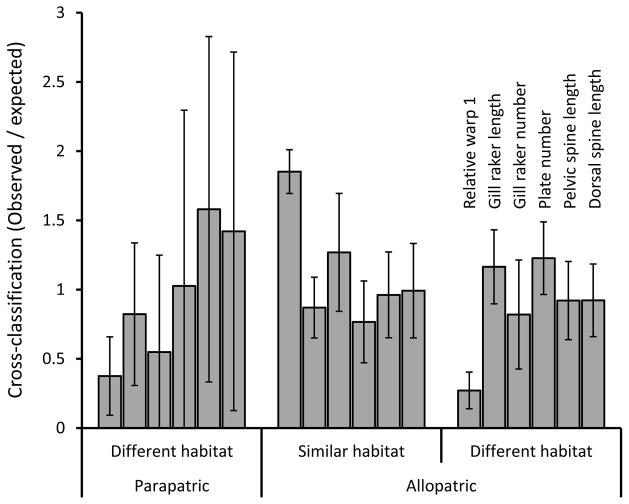
Cross-classification based on individual morphological variables (as opposed to variable “classes”) helps to refine inferences regarding the drivers of diversification (Fig. 4). In particular, relative warp 1, which mostly captures body depth (see also Kaeuffer et al. 2012), showed the strongest signature of parallelism (Fig. 4). First, cross-classification was higher into similar habitats in allopatry (1.85 times the random expectation) than into different habitats in parapatry (0.38 times) or allopatry (0.27 times). Second, cross-classification into similar habitats in allopatry was much higher for relative warp 1 (1.85 times) than for any of the other traits (0.77–1.27 times). These results confirm that traits showing the highest degree of parallelism (Kaeuffer et al. 2012) should also show the highest exchangeability among populations in similar habitats and the least exchangeability among populations in different habitats.
Figure 4.
Ratios for each morphological trait of the mean average number of individuals classified into each category (parapatric different habitat, allopatric similar habitat, allopatric different habitat) to the mean number expected to be classified into those categories by chance (random expectation). The values are means and 95% confidence intervals. Although the variables are labeled for only one set of bars, the same labels apply in the same order to each set of bars.
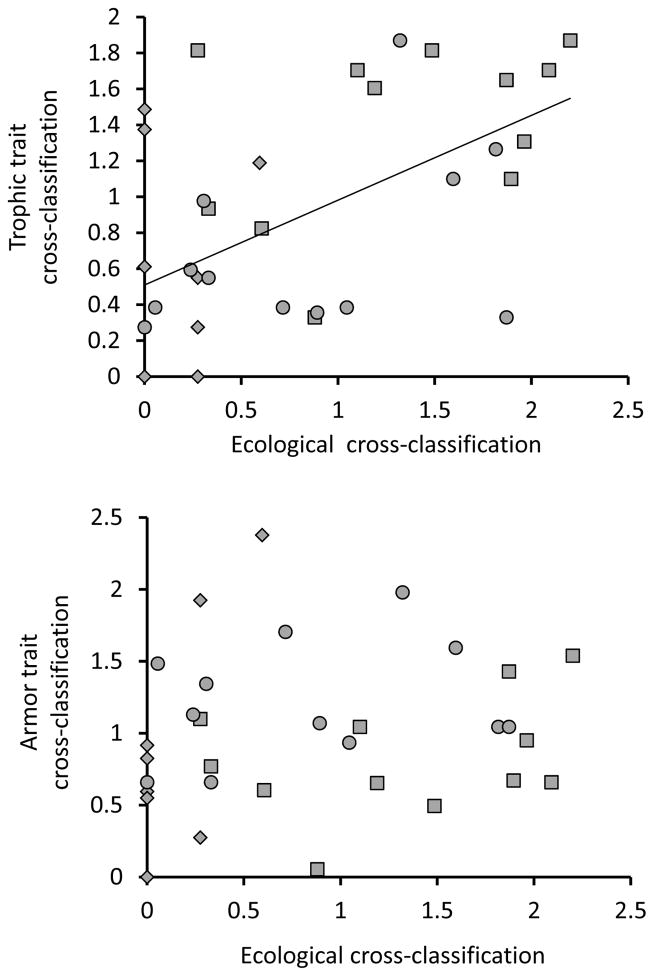
Patterns of cross-classification for ecological variables that shape selection should predict patterns of cross-classification for traits subject to that selection – as we have already noted. A test of this assertion is whether patterns of cross-classification for ecological (diet) variables are predictive of patterns of cross-classification for trophic traits but not for armor traits – because diet influences selection on the former much more than on the latter (Berner et al. 2008). This expectation was met for misclassification (not shown) and for cross-classification (Fig. 5), although the association was not very strong even for trophic traits. In particular, allopatric populations in different habitats showed higher cross-classification based on ecological variables than on trophic traits (Fig. 5), a result that suggests other factors influence divergence in trophic traits among watersheds.
Figure 5.
Relationships between ecological cross-classification and trophic (upper panel) and armor (lower panel) cross-classification. The ecological, trophic, and armor values were from DAPC and are the ratio of observed vales to random expectations (as in Fig. 3). Each population is represented by three data points corresponding to the three categories into which cross-classification could occur: parapatric different habitat (diamonds), allopatric similar habitat (squares), and allopatric different habitat (circles). The illustrated regression line is through all of the data together but the statistical test includes category as a random factor. This analysis revealed that ecological cross-classification was a significant predictor of cross-classification for foraging traits (r2 = 0.49 for the full model, F = 4.95, P = 0.033) but not armor traits (r2 = 0.48 for the full model, F = 2.67, P = 0.112).
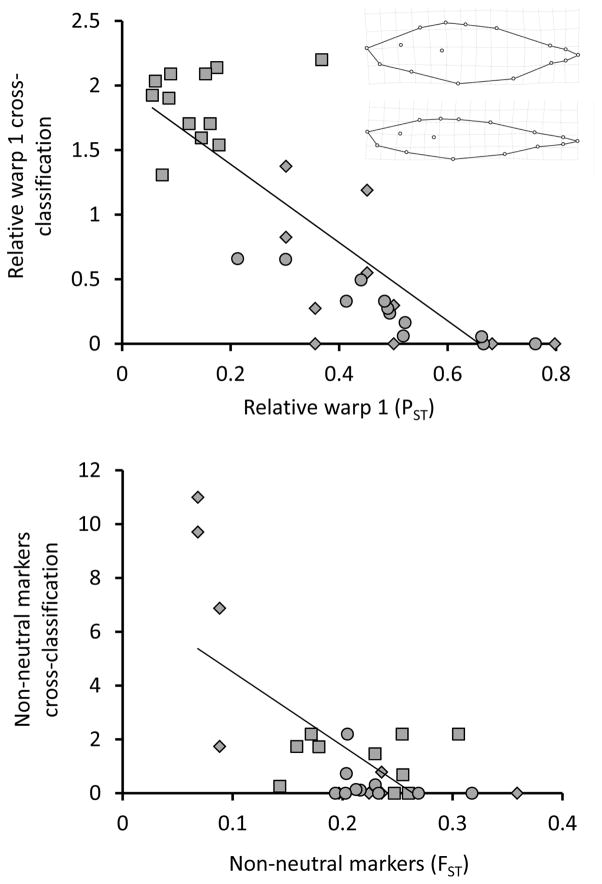
Results from cross-classification should – to at least some extent – parallel those based on significant effects on means. We explored this expectation by relating, for each variable, cross-classification patterns to differences in means and frequencies. For the former, we used cross-classification of individuals from each of the 12 focal populations into each of the three categories (parapatric different habitat, allopatric similar habitat, allopatric different habitat). For the latter, we used average differences between each focal population and the populations in each category, scaled by the phenotypic variance (i.e., mean PST, EST, and FST; see Kaeuffer et al. 2012). We used this variance scaling because the “significant effects on means” approach compares means relative to the variation with groups. The expectation that results from the two methods should be correlated was born out for relative warp 1, gill raker number, δ13C, δ15N, and non-neutral markers (Table 1; Fig. 6). Importantly, however, variance-scaled differences in means were not perfectly predictive of classification, confirming that the two approaches do not yield identical results.
Table 1.
Relationships for each variable between variance-standardized differences (PST, FST, EST) and cross-classification. Each of the 12 focal populations is represented by three data points corresponding to the three categories into which cross-classification could occur: parapatric different habitat, allopatric similar habitat, and allopatric different habitat. The predictor variable is the mean PST, FST, or EST between each focal population and populations in the different categories. The response variable is the mean cross-classification of each focal population into populations in the different categories. ‘Category’ is included as a random variable (but not shown in the table) so as to control for the three observations per population. All significant associations are negative (as expected) and r2 is for the full model.
| Variables | r2 | F | P |
|---|---|---|---|
| Relative warp 1 | 0.87 | 13.87 | 0.001 |
| Gill raker length | 0 | 0.00 | 0.973 |
| Gill raker number | 0.17 | 8.58 | 0.012 |
| Plate number | 0.02 | 1.93 | 0.174 |
| Pelvic spine length | 0.05 | 2.30 | 0.139 |
| Dorsal spine length | 0 | 0.120 | 1.000 |
| Proportion of limnetic prey | 0.52 | 1.31 | 0.260 |
| δ13C | 0.69 | 19.45 | < 0.001 |
| δ15N | 0.76 | 39.41 | < 0.001 |
| Neutral markers | 0.66 | 2.38 | 0.134 |
| Non-neutral markers | 0.46 | 18.91 | < 0.001 |
Figure 6.
Relative warp 1 (upper panel) and non-neutral markers (lower panel) were among the variables showing strong relationships between variance-standardized differences (PST and FST) and cross-classification. Each of the 12 focal populations is represented by three data points corresponding to the three categories into which cross-classification could occur: parapatric different habitat (diamonds), allopatric similar habitat (squares), and allopatric different habitat (circles). Note that a few focal populations do not appear for the non-neutral markers because all of their individuals were classified with 100% probability to the focal population (Appendix S3). The x-axis shows the mean PST or FST between each focal population and populations in the different categories. The y-axis shows mean cross-classification of each focal population into populations in the different categories. The inset in the upper panel shows the variation in body depth captured by relative warp 1: the deformations show the maximum (bottom: Pye Lake) and minimum (top: Village Bay Inlet Stream) population mean. The illustrated regression line is through all of the data but the statistical test included category as a random factor (Table 1).
Discussion
Our goal was to advance the idea that valuable evolutionary inferences can be derived from analyses that use classification methods to infer the exchangeability of individuals among populations. The basic premise is that population divergence might be best quantified by asking how well the genotypes/phenotypes of individuals from one population would “fit” into other populations. Or, in other words, the extent to which individuals can be swapped between populations without altering the ecological, morphological, or genetic properties of those populations. This consideration of populations as groups of potentially unique individuals, rather than means and variances, acknowledges the importance of individual variation to a variety of ecological and evolutionary processes (Bolnick et al. 2003; Hansen et al. 2006; Bolnick et al. 2011).
We illustrated an exchangeability approach through the analysis of empirical data for lake and stream stickleback. The populations were mostly non-exchangeable at the genetic level, presumably because each carries its own unique signature of historical and contemporary events and processes. When genetic exchangeability did occur, it was typically between populations within the same watershed, which are more likely to share these events and processes and are more likely to be tied together by gene flow. Relative to this rare genetic exchangeability, ecological (diet) exchangeability was much higher – because diets overlapped considerably among populations. This ecological overlap suggested that exchangeability also should be high for morphological traits, which proved to be the case. In addition, morphological exchangeability was highest among populations that experienced similar selection: trophic traits were most exchangeable among populations of a similar habitat type. That is, parallel evolution increases exchangeability among populations in similar habitats – while decreasing it among populations in different habitats.
Most of the inferences drawn from these exchangeability analyses match those from the previous “significant effects on means” analyses of the same samples (Kaeuffer et al. 2012). This correspondence is reassuring because it shows that exchangeability analyses can replicate conclusions more traditional analyses. (Of course, basic caveats about inferring parallel evolution from population samples – see references in the Introduction – will attend both approaches.) The question remains, however, as to whether or not any new insight was gained through the exchangeability approach. For starters, it is important to note that results of the two approaches were not identical; i.e., population differences in variance-standardized means were not perfectly predictive of classification patterns. Moreover, exchangeability analyses led to clarified and improved inferences. For instance, we previously inferred, from significant effects on means (Kaeuffer et al. 2012), that parallelism in lake-stream divergence was very high for trophic traits because it was in the same direction in every watershed, and because the habitat term explained more than half of the total variation. Yet the present exchangeability analysis showed that classification based on these same traits was always highest to the population of origin and that classification away from the population of origin was only modestly predicted by habitat type. These findings indicate that habitat-specific processes (e.g., divergent selection that is similar between all lakes and streams) can be overwhelmed by location-specific processes (e.g., selection that is unique to a particular lake or stream). The exchangeability thus forces the new realization that parallelism is not really that high – even for the traits previously inferred to be very parallel. We suspect that reliance on the “significant effects on means” approach might generally over-emphasize the importance of parallelism in nature.
Extensions
Our exposition of how evolutionary inferences can be derived from exchangeability was based on only a few trait types, one species, and a particular form of geographical structure (allopatric pairs of parapatric populations). However, the basic approach should be relevant and informative for any type of trait, organism, or geographical structure. In addition, they should be amenable to a wider range of questions. We here summarize some of these possibilities.
First, it would also be useful to include more populations within each allopatric group, such as multiple stream and lake populations within each watershed. These additional populations would aid inferences regarding interactions between selection and gene flow. In particular, the ecological theory of adaptive radiation predicts that increasing adaptive divergence reduces gene flow: i.e., ecological speciation (Schluter 2000; Crispo et al. 2006; Räsänen and Hendry 2008; Thibert-Plante and Hendry 2010). If so, exchangeability at neutral genetic markers should be lower between populations in different habitats than between populations in similar habitats.
Second, we considered only two ecotypes (lake and stream), whereas exchangeability analyses might be equally useful for more ecotypes, such as phytophagous insects that occupy a range of different host plants (e.g., Ferrari et al. 2006). In this case, one might first follow the above-described procedure and record the extent to which cross-classification occurs to a similar or different ecotype when gene flow is present (parapatry/sympatry) or absent (allopatry). This broad division into similar versus different ecotypes would inform the overall action of divergent selection across the entire group. One might then examine classification between particular ecotype pairs so as to inform which particular ecological contrasts (different host plant species) drive the greatest divergence. Comparative analyses of this sort could make a useful contribution to current discussions regarding factors that determine the degree of progress toward ecological speciation (Hendry 2009; Nosil et al. 2009).
Third, we considered classification among populations within a species, whereas similar analyses could be applied to variation among ed species. For example, Darwin’s finches specialize on different food types at different locations on the same islands (parapatry/sympatry) and on different islands (allopatry) (Grant 1999). These species hybridize in nature and the role of historical events and contemporary gene flow remains an active area of research (Grant and Grant 1994; Grant 1999; Grant et al. 2004; Petren et al. 2005). Exchangeability analyses could be implemented by sampling multiple species at each of multiple locations on each of multiple islands. Individuals could then be cross-classified within species (i.e., the same “environment”) or between species (i.e., a different “environment”) on the same island (parapatry/sympatry) or on different islands (allopatry). Similar analyses might prove useful for groups such as Anolis lizard ecomorphs (Losos 2009) or whitefish species in postglacial lakes (Siwertsson et al. 2010; Vonlanthen et al. 2012). Results from these analyses would be directly relevant to discussions about the collective evolution of species comprised of multiple populations (Morjan and Rieseberg 2004; Futuyma 2010) and to debates about the reality of species designations (Zink 2002).
Fourth, our analyses were based on the dichotomous decision to classify each individual into a particular population, which seemed appropriate given our focus on how individuals could be “exchanged” between populations. The reality, however, is that probabilities of classification of each individual into a given population were highly variable among individuals, suggesting that analyzing the raw probabilities might increase power and precision. For example, one could use the probabilities of classification for each individual from each focal population into each other population, and average these values across individuals. We performed a preliminary analysis along these lines, and found that results based on the dichotomous analysis were always correlated with results based on the raw probabilities (Table 2). The two alternatives should therefore yield similar conclusions; although the imperfect correlations suggest the value of further considering the accuracy and power of each.
Table 2.
Relationships for DAPCs between cross-classification based on the dichotomous approach (an individual is or is not assigned to a given population) versus raw probabilities of classification of each individual into each population. Each of the 12 focal populations is represented by three data points corresponding to the three categories into which cross-classification could occur: parapatric different habitat, allopatric similar habitat, and allopatric different habitat. The predictor variable is the mean probability of assignment of individuals from each focal population into populations in each of the three categories. The response variable is the mean cross-classification of individuals in each focal population into populations in the other categories. ‘Category’ is included as a random variable (but not shown in the table) so as to control for the three observations per population. All associations are positive (as expected) and r2 is for the full model.
| DAPC variables | r2 | F | P |
|---|---|---|---|
| Ecology (diet) | 0.73 | 37.49 | < 0.001 |
| Foraging traits | 0.73 | 50.05 | < 0.001 |
| Armor traits | 0.21 | 4.02 | 0.031 |
| Neutral markers | 0.62 | 33.83 | 0.017 |
| Non-neutral markers | 0.72 | 78.46 | < 0.001 |
Fifth, we considered morphological variation that has a strong genetic basis. For traits with stronger environmental contributions, exchangeability analyses could be used to assess the extent to which plastic differences represent deterministic responses to particular environments, as well as the effect that gene flow has on plastic divergence (Crispo 2008). An intriguing further possibility is that classification might be based on the genetic and plastic components of the phenotype as estimated in “animal model” analyses of natural populations (e.g. Kruuk 2004; Hadfield et al. 2009). That is, breeding values (evolution) and environmental deviations (plasticity) could be analyzed separately for their exchangeability.
Sixth, classification-based analyses allow the identification of particular individuals that show the highest exchangeability among particular populations, as well as the traits that make them so. These individuals might be recent migrants or they might be hybrids between the populations – as is already commonly inferred from genetic “assignment” tests (Manel et al. 2005). Whether or not these individuals are also the most morphologically exchangeable can then inform whether or not dispersal is “phenotype-biased” (Edelaar et al. 2008; Bolnick et al. 2009; Shine et al. 2011), whether or not “pre-adaptation” to alternative conditions is a primary criterion for successful gene flow (Quinn et al. 2001), and the extent to which dispersal between populations is likely to have maladaptive consequences (Hendry et al. 2001; Yeaman and Guillaume 2009).
Conclusion
We have built a case for deriving evolutionary inferences from the determination of individual exchangeability among populations. We see three principal advantages over the more typical “significant effects on means” approach. One is conceptual: populations are composed of a diversity of individuals that might not be well represented by simple means, variances, and frequencies. Individual classification, by contrast approaches acknowledges and exploits the actual distribution of genotypes and phenotypes, and therefore facilitates consideration of the many unique consequences of individual variation for ecology and evolution. A second advantage is methodological: discriminant functions on principal components places all variables on the same scale, and therefore makes classification patterns comparable across populations and traits, including morphological measurements, discrete counts, proportions, and multilocus genotypes. A third major advantage comes from the identification of specific individuals that are most exchangeable among populations, thus enabling further analysis of the properties of those individuals, as well as their consequences for gene flow and adaptive divergence. Increased appreciation and application of exchangeability-inspired classification-based methods is likely to improve inferences in evolutionary biology, landscape genetics, and the interface between them: evolutionary landscape genetics.
Supplementary Material
Acknowledgments
Field work was assisted by J.-S. Moore. Morphological measurements were assisted by S. Barrette, C. Macnaughton, and S. Muttalib. Genetic data collection was assisted by members of the Peichel lab. Additional comments and advice were provided by E. Taylor and X. Thibert-Plante. Financial support was provided by an FQRNT postdoctoral fellowship (RK), an NIH grant P50 HG002568 (CLP), a David and Lucille Packard Foundation Fellowship (DIB), the Howard Hughes Medical Institute (DIB), and the Natural Sciences and Engineering Research Council of Canada (APH).
BIBLIOGRAPHY
- Arendt J, Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Berner D, Grandchamp AC, Hendry AP. Variable progress toward ecological speciation in parapatry: stickleback across eight lake-stream transitions. Evolution. 2009;63:1740–1753. doi: 10.1111/j.1558-5646.2009.00665.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Araújo MS. Partitioning the relative fitness effects of diet and trophic morphology in the threespine stickleback. Evol Ecol Res. 2011;13:439–459. [Google Scholar]
- Berner D, Adams DC, Grandchamp AC, Hendry AP. Natural selection drives patterns of lake-stream divergence in stickleback foraging morphology. J Evol Biol. 2008;21:1653–1665. doi: 10.1111/j.1420-9101.2008.01583.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Nosil P. Natural selection in populations subject to a migration load. Evolution. 2007;61:2229–2243. doi: 10.1111/j.1558-5646.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Caldera EJ, Matthews B. Evidence for asymmetric migration load in a pair of ecologically divergent stickleback populations. Biol J Linn Soc. 2008;94:273–287. [Google Scholar]
- Bolnick DI, Snowberg LK, Patenia C, Stutz WE, Ingram T, Lau OL. Phenotype-dependent native habitat preference facilitates divergence between parapatric lake and stream stickleback. Evolution. 2009;63:2004–2016. doi: 10.1111/j.1558-5646.2009.00699.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Svanback R, Fordyce JA, Yang LH, Davis JM, Hulsey CD, Forister ML. The ecology of individuals: incidence and implications of individual specialization. Am Nat. 2003;161:1–28. doi: 10.1086/343878. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Amarasekare P, Araujo MS, Bürger R, Levine JM, Novak M, Rudolf VHW, Schreiber SJ, Urban MC, Vasseur DA. Why intraspecific trait variation matters in community ecology. Trends Ecol Evol. 2011;26:183–192. doi: 10.1016/j.tree.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond JE, Stockman AK. An integrative method for delimiting cohesion species: finding the population-species interface in a group of Californian trapdoor spiders with extreme genetic divergence and geographic structuring. Sys Biol. 2008;57:628–646. doi: 10.1080/10635150802302443. [DOI] [PubMed] [Google Scholar]
- Cano JM, Mäkinen HS, Leinonen T, Freyhof J, Merilä J. Extreme neutral genetic and morphological divergence supports classification of Adriatic three-spined stickleback (Gasterosteus aculeatus) populations as distinct conservation units. Biol Cons. 2008;141:1055–1066. [Google Scholar]
- Castric V, Bernatchez L. Individual assignment test reveals differential restriction to dispersal between two salmonids despite no increase of genetic differences with distance. Mol Ecol. 2004;13:1299–1312. doi: 10.1111/j.1365-294X.2004.02129.x. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends Ecol Evol. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Crispo E. Modifying effects of phenotypic plasticity in interactions among natural selection, adaptation and gene flow. J Evol Biol. 2008;21:1460–1469. doi: 10.1111/j.1420-9101.2008.01592.x. [DOI] [PubMed] [Google Scholar]
- Crispo E, Bentzen P, Reznick DN, Kinnison MT, Hendry AP. The relative influence of natural selection and geography on gene flow in guppies. Mol Ecol. 2006;15:49–62. doi: 10.1111/j.1365-294X.2005.02764.x. [DOI] [PubMed] [Google Scholar]
- Doughty P, Reznick DN. Patterns and analysis of adaptive phenotypic plasticity in animals. In: DeWitt TJ, Scheiner SM, editors. Phenotypic plasticity: functional and conceptual approaches. Oxford University Press; Oxford, UK: 2004. pp. 126–150. [Google Scholar]
- Edelaar P, Siepielski AM, Clobert J. Matching habitat choice causes directed gene flow: a neglected dimension in evolution and ecology. Evolution. 2008;62:2462–2472. doi: 10.1111/j.1558-5646.2008.00459.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Natural Selection in the Wild. Princeton University Press; Princeton, NJ: 1986. [Google Scholar]
- Ferrari J, Godfray HCJ, Faulconbridge AS, Prior K, Via S. Population differentiation and genetic variation in host choice among pea aphids from eight host plant genera. Evolution. 2006;60:1574–1584. [PubMed] [Google Scholar]
- Funk SM, Fa JE. Phylogeography of the endemic St. Lucia whiptail lizard Cnemidophorus vanzoi: conservation genetics at the species boundary. Cons Genet. 2006;7:651–663. [Google Scholar]
- Futuyma DJ. Evolutionary constraint and ecological consequences. Evolution. 64:1865–1884. doi: 10.1111/j.1558-5646.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- Grant PR. Ecology and evolution of Darwin’s finches. Princeton University Press; Princeton, NJ: 1999. [Google Scholar]
- Grant PR, Grant BR. Phenotypic and genetic effects of hybridization in Darwin’s finches. Evolution. 1994;48:297–316. doi: 10.1111/j.1558-5646.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR, Markert JA, Keller LF, Petren K. Convergent evolution of Darwin’s finches caused by introgressive hybridization and selection. Evolution. 2004;58:1588–1599. doi: 10.1111/j.0014-3820.2004.tb01738.x. [DOI] [PubMed] [Google Scholar]
- Hadfield JD, Wilson AJ, Garant D, Sheldon BC, Kruuk LEB. The misuse of BLUP in ecology and evolution. Am Nat. 2010;175:116–125. doi: 10.1086/648604. [DOI] [PubMed] [Google Scholar]
- Hansen MM, Nielsen EE, Bekkevold D, Mensberg KLD. Admixture analysis and stocking impact assessment in brown trout (Salmo trutta), estimated with incomplete baseline data. Can J Fish Aquat Sci. 2001;58:1853–1860. [Google Scholar]
- Hansen TF, Carter AJR, Pelabon C. On adaptive accuracy and precision in natural populations. Am Nat. 2006;168:168–181. doi: 10.1086/505768. [DOI] [PubMed] [Google Scholar]
- Hauser L, Seamons TR, Dauer M, Naish KA, Quinn TP. An empirical verification of population assignment methods by marking and parentage data: hatchery and wild steelhead (Oncorhynchus mykiss) in Forks Creek, Washington, USA. Mol Ecol. 2006;15:3157–3173. doi: 10.1111/j.1365-294X.2006.03017.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP. Ecological speciation! Or the lack thereof? Can. J Fish Aquat Sci. 2009;66:1383–1398. [Google Scholar]
- Hendry AP, Taylor EB. How much of the variation in adaptive divergence can be explained by gene flow? An evaluation using lake-stream stickleback pairs. Evolution. 2004;58:2319–2331. doi: 10.1111/j.0014-3820.2004.tb01606.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Day T, Taylor EB. Population mixing and the adaptive divergence of quantitative traits in discrete populations: a theoretical framework for empirical tests. Evolution. 2001;55:459–466. doi: 10.1554/0014-3820(2001)055[0459:pmatad]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resources. 2000;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010:1194. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeuffer R, Peichel CL, Bolnick DI, Hendry AP. Parallel and nonparallel aspects of ecological, phenotypic, and genetic divergence across replicate population pairs of lake and stream stickleback. Evolution. 2012;66:402–418. doi: 10.1111/j.1558-5646.2011.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk LEB. Estimating genetic parameters in natural populations using the ‘animal model’. Phil Trans R Soc Lond B Biol Sci. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerhans RB, DeWitt TJ. Shared and unique features of evolutionary diversification. Am Nat. 2004;164:335–349. doi: 10.1086/422857. [DOI] [PubMed] [Google Scholar]
- Langerhans RB. Predictability of phenotypic differentiation across flow regimes in fishes. Integr Comp Biol. 2008;48:750–768. doi: 10.1093/icb/icn092. [DOI] [PubMed] [Google Scholar]
- Lavin PA, McPhail JD. Parapatric lake and stream sticklebacks on northern Vancouver Island: disjunct distribution or parallel evolution? Can J Zool. 1993;71:11–17. [Google Scholar]
- Losos JB. Lizards in an evolutionary tree: ecology and adaptive radiation of Anoles. University of California Press; Berkeley, CA: 2009. [Google Scholar]
- Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- MacColl ADC. The ecological causes of evolution. Trends Ecol Evol. 2011;26:514–522. doi: 10.1016/j.tree.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Manel S, Gaggiotti OE, Waples RS. Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol. 2005;20:136–142. doi: 10.1016/j.tree.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Matthews B, Marchinko KB, Bolnick DI, Mazumder A. Specialization of trophic position and habitat use by sticklebacks in an adaptive radiation. Ecology. 2010;91:1025–1034. doi: 10.1890/09-0235.1. [DOI] [PubMed] [Google Scholar]
- Morjan CL, Rieseberg LH. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol Ecol. 2004;13:1341–1356. doi: 10.1111/j.1365-294X.2004.02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome SD, del Rio CM, Bearhop S, Phillips DL. A niche for isotopic ecology. Front Ecol Environ. 2007;5:429–436. [Google Scholar]
- Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol Evol. 2009;24:145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ. Does gene flow constrain adaptive divergence or vice versa? A test using ecomorphology and sexual isolation in Timema cristinae walking-sticks. Evolution. 2004;58:102–112. doi: 10.1111/j.0014-3820.2004.tb01577.x. [DOI] [PubMed] [Google Scholar]
- Petren K, Grant PR, Grant BR, Keller LF. Comparative landscape genetics and the adaptive radiation of Darwin’s finches: the role of peripheral isolation. Mol Ecol. 2005;14:2943–2957. doi: 10.1111/j.1365-294X.2005.02632.x. [DOI] [PubMed] [Google Scholar]
- Post DM. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology. 2002;83:703–718. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotypes. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TP, Kinnison MT, Unwin MJ. Evolution of chinook salmon (Oncorhynchus tshawytscha) populations in New Zealand: pattern, rate, and process. Genetica. 2001;112–113:493–513. [PubMed] [Google Scholar]
- Rader RB, Belk MC, Shiozawa DK, Crandall KA. Empirical tests for ecological exchangeability. Anim Cons. 2005;8:239–247. [Google Scholar]
- Räsänen K, Hendry AP. Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecol Lett. 2008;11:624–636. doi: 10.1111/j.1461-0248.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- Reimchen TE, Ingram T, Hansen SC. Assessing niche differences of sex, armour and asymmetry phenotypes using stable isotope analyses in Haida Gwaii sticklebacks. Behaviour. 2008;145:561–577. [Google Scholar]
- Reznick DN, Rodd FH, Cardenas M. Life-history evolution in guppies (Poecilia reticulata: Poeciliidae). 4. Parallelism in life-history phenotypes. Am Nat. 1996;147:319–338. [Google Scholar]
- Reznick DN, Bryga HA. Life-history evolution in guppies (Poecilia reticulata: Poeciliidae). 5. Genetic basis of parallelism in life histories. Am Nat. 1996;147:339–359. [Google Scholar]
- Roesti M, Hendry AP, Salzburger W, Berner D. Genome divergence during evolutionary diversification as revealed in replicate lake-stream stickleback population pairs. Mol Ecol. 2012;12:2852–2862. doi: 10.1111/j.1365-294X.2012.05509.x. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. tpsRelw. 2005 http://life.bio.sunysb.edu/morph/
- Rohlf FJ. tpsDIG2. 2006 http://life.bio.sunysb.edu/morph/
- Schluter D. The ecology of adaptive radiation. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- Schluter D, McPhail JD. Ecological character displacement and speciation in sticklebacks. Am Nat. 1992;140:85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- Shine R, Brown GP, Phillips BL. An evolutionary process that assembles phenotypes through space rather than through time. Proc Natl Acad Sci USA. 2011;108:5708–5711. doi: 10.1073/pnas.1018989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwertsson A, Knudsen R, Kahilainen KK, Præbel K, Primicerio R, Amundsen PA. Sympatric diversification as influenced by ecological opportunity and historical contingency in a young species lineage of whitefish. Evol Ecol Res. 2010;12:929–947. [Google Scholar]
- Snowberg LK, Bolnick DI. Assortative mating by diet in a phenotypically unimodal but ecologically variable population of stickleback. Am Nat. 2008;172:733–739. doi: 10.1086/591692. [DOI] [PubMed] [Google Scholar]
- Stockman AK, Bond JE. Delimiting cohesion species: extreme population structuring and the role of ecological interchangeability. Mol Ecol. 2007;16:3374–3392. doi: 10.1111/j.1365-294X.2007.03389.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. In: The meaning of species and speciation: a genetic perspective. Otte D, Endler JA, editors. Speciation and its consequensesSinauer Associates Inc; Sunderland, MA: 1989. pp. 3–27. [Google Scholar]
- Templeton AR. Using phylogeographic analyses of gene trees to test species status and processes. Mol Ecol. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. [DOI] [PubMed] [Google Scholar]
- Thibert-Plante X, Hendry AP. When can ecological speciation be detected with neutral loci? Mol Ecol. 2010;19:2301–2314. doi: 10.1111/j.1365-294X.2010.04641.x. [DOI] [PubMed] [Google Scholar]
- Vonlanthen P, Bittner D, Hudson AG, Young KA, Müller R, Lundsgaard-Hansen B, Roy D, Di Piazza S, Largiader CR, Seehausen O. Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature. 2012;482:357–362. doi: 10.1038/nature10824. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Kalisz S. The causes of natural selection. Evolution. 1990;44:1947–1955. doi: 10.1111/j.1558-5646.1990.tb04301.x. [DOI] [PubMed] [Google Scholar]
- Wake DB, Wake MH, Specht CD. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science. 2011;331:1032–1035. doi: 10.1126/science.1188545. [DOI] [PubMed] [Google Scholar]
- Waples RS, Gaggiotti O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- Yeaman S, Guillaume F. Predicting adaptation under migration load: the role of genetic skew. Evolution. 2009;63:2926–2938. doi: 10.1111/j.1558-5646.2009.00773.x. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Swiderski DL, Sheets HD, Fink WL. Geometric morphometrics for biologists: a primer. Elsevier Academic Press; New York: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.