Abstract
Researchers have identified thousands of loci involved in complex traits and drug response. However, in most cases they only explain a small proportion of the heritability of the trait. Among different strategies conducted to identify this ‘missing heritability’, here we illustrate the importance of complex gene–environment interactions using findings regarding the role of leukotrienes on the bronchodilator response to albuterol in Latino asthmatics. Patients managing their asthma with leukotriene-modifying medication presented higher increases in the bronchodilator response to albuterol. Moreover, interactions between genes responsible for leukotriene production were associated with a decreased risk of asthma. Combining genetic and pharmacologic effects, leukotriene-modifying users carrying certain combinations of alleles presented higher improvements in lung function after bronchodilator administration. Genes and drugs act at different orders of interaction (from individual effects to gene–gene–drug–drug interactions) and population-specific effects have to be considered. These results may be extrapolated to other complex phenotypes.
Keywords: albuterol, asthma, bronchodilator drug response, drug–drug interaction, ethnic differences, gene–gene interaction, leukotriene modifier, leukotrienes
In recent decades, efforts in human genetics have identified many different loci associated with several complex phenotypes including drug response. These successes have recently been boosted with the advent of genome-wide association studies (GWAS) that allow the joint analysis of hundreds of thousands or millions of SNPs, capturing most, although not all, common variation across the genome. So far, more than 4000 loci have been associated (p ≤ 5 × 10−8) with several hundreds of different diseases or traits using GWAS [101]. Although there is some controversy about the accuracy of currently used methods for heritability estimation [1], all the identified genetic factors only explain a small proportion of the heritability of most common traits, despite rapid advances in human genetic studies. For example, more than 15 loci have so far been identified to be involved in Type 2 diabetes, but they explain less than 10% of its estimated heritability (for a review, see [2]).
Many different approaches have been suggested to overcome this limitation. Substantial efforts to identify the genetic determinants of this missing heritability have focused on the potential contribution of structural and rare variants. Structural variation, including copy number variants, inversions, and translocations, may be responsible for some of the unexplained heritability, although their variation beyond rare Mendelian disorders has been largely ignored [3]. Because of being unexplored, the global impact of these variants is still unknown but they have been involved in many different complex traits, such as Crohn’s disease, BMI or several neuropsychiatric disorders [4–7]. Rare variants, commonly defined as those with a minor allele frequency (MAF) lower than 1%, can have modest to high effect sizes. Many authors postulate that the increasing availability of ‘next-generation’ sequencing (NGS) technologies at affordable prices and the completion of NGS projects, such as the 1000 Genomes Project, will facilitate the detection of associations with rare variants [8,9,102]. However, the analysis of all rare variants is currently unlikely since sample size requirements are inversely proportional to MAF.
A more traditional strategy to increase the chances of identifying the genetic determinants of complex diseases or drug response has been to increase sample sizes in current GWAS [10] and to pool data via meta-analyses. Meta-analyses may clarify the role of previously identified loci, such as on the pharmacogenetics of lung cancer [11] or the genetic variants in the MTHFR gene involved in methotrexate toxicity [12]. It is well-established that increases in sample size increase the number of detected variants [13], but meta-analyses can also provide a comprehensive understanding of the differences in genetic risk patterns between different ethnic groups [14].
To clarify the heritability of complex traits across populations, recent efforts have focused on expanding studies to non-European populations because there are ethnic differences in the causes, expression and prevalence of various diseases [15,16]. Nonetheless, non-European groups have been and are still under-represented in clinical studies and biomedical research [17].
Beyond structural and rare variants, another potential explanation for the missing heritability of complex traits is the existence of genetic interactions among loci. In fact, genetic interactions may be responsible for a significant inflation of heritability estimates based on standard methods [1,18] highlighting the need for further investigation of the effects of gene–gene interactions. It is being increasingly recognized that genetic risk factors do not act in an isolated fashion or by combining their individual additive effects, but rather by interacting with other genes and the environment to determine overall risk in many different pathologies, such as high blood pressure, autoimmune disorders or multiple sclerosis [19–21]. Moreover, the role that more complex (i.e., third- or higher-order) interactions may play in complex outcomes is still not well understood.
One difficulty in identifying potential gene–gene interactions is that we cannot follow the hypothesis-free approach that GWAS implement because of a problem of scale in the number of tests, and the subsequent lack of statistical power to detect most existing interactions. Testing all potential second-order (i.e., SNP–SNP) interactions in a dataset of, at least, several hundreds of thousands of SNPs would require strict significance thresholds to correct for multiple testing. For example, the significance threshold would become 5 × 10−14 for a study with one million markers and an initial α level of 0.05 applying a traditional Bonferroni correction. Consequently, we would need sample sizes some orders of magnitude higher than currently available and many relevant interactions would remain undetected. The detection of higher order interactions would increase exponentially with the number of tests performed.
A more feasible approach to detect interactions has been to analyze markers in smaller sets of genes that had previously been identified to influence the trait of interest or that are known to act in the same physiological or biochemical pathway. Using this approach we have identified complex second-, third- and fourth-order interactions between genes in the leukotriene pathway and asthma medications. These interactions modulate lung function and the risk of suffering from asthma in individuals of Hispanic/Latino descent and have clinical relevance.
Leukotriene pathway, bronchodilator response & asthma
Similar to most complex outcomes, the complexity in the diagnosis and treatment of asthma comes in part from the wide range of molecular pathways involved in the pathogenesis of asthma. Among these different mechanisms, leukotrienes (LTs) comprise a family of arachidonic acid metabolites that have a role in allergic and inflammatory diseases [22]. LT synthesis is initiated in airway leukocytes in response to a variety of stimuli, including allergens, and begins with ALOX and ALOX5AP producing leukotriene A4 (LTA4) from arachidonic acid. LTA4 is inactive and can be converted to leukotriene B4 (LTB4) by LTA4H or to cysteinyl leukotrienes by LTC4S. LTB4 and cysteinyl leukotrienes act on their specific receptors (BLT1 and BLT2 for LTB4, and CysLT1 and CysLT2 for cysteinyl leukotrienes) on various target cells within the respiratory tract (neutrophils and bronchial and vascular smooth muscle cells, among others) mediating bronchoconstriction, mucus production and airway edema. Several LT-modifying drugs have been approved for the treatment of asthma (e.g., montelukast, zafirlukast, pranlukast and zileuton), and guidelines for asthma management recommend them as primary therapies for disease control in patients with persistent asthma, along with the intermittent use of short-acting β2-agonists as rescue therapy for acute asthma exacerbations [103]. Despite these recommendations, it is estimated that 58–78% of all patients receiving LT modifiers do not respond to this class of medication [23,24].
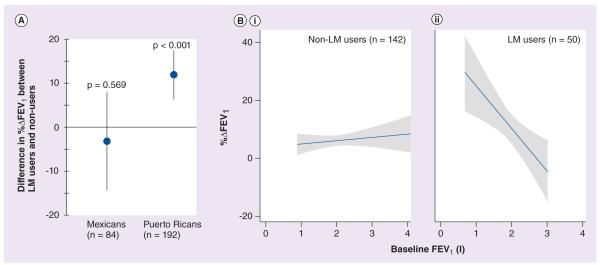
Since LTs attenuate the activity of β2-agonists on smooth muscle cells [25], we first postulated that LT modifiers may augment bronchodilator responsiveness to albuterol (a short-acting β2-agonist) by blocking this process. Because population-specific differences in drug responses have been well documented for short-acting β2-agonists [26,27], we conducted the analyses in the Mexican and Puerto Rican children with persistent asthma (Table 1) recruited for the GALA study [28]. We found that LT modifier use was associated with a clinically significant increase in lung function, assessed as the percentage change in forced expiratory volume in 1 s (FEV1) after the administration of albuterol, in Puerto Rican asthmatics (p < 0.001) but not in Mexicans (p = 0.57; Figure 1a). Among Puerto Ricans, asthmatics using LT-modifying drugs presented an average increase in FEV1 after the administration of albuterol (%ΔFEV1) of 11.8% (95% CI: 6.2–17.3) compared with patients that did not use LT modifiers. These results were independent of age, sex, concomitant corticosteroid use or recruitment site.
Table 1.
Clinical and demographic characteristics of the Mexican and the Puerto Rican subjects with asthma from the GALA study, stratified by ethnicity.
| Persistent asthmatics (analyzed in [28]) | All asthmatics (analyzed in [29,30]) | |||
|---|---|---|---|---|
| Mexicans | Puerto Ricans | Mexicans | Puerto Ricans | |
| n | 84 | 192 | 293 | 356 |
| Age, years | 11.4 (9.8–13.2) | 10.8 (9.3–12.8) | 13.2 (10.6–19.7) | 11.9 (9.5–14.9) |
| Male (%) | 61.9 | 58.1 | 53.6 | 55.9 |
| Baseline FEV1 | 2.11 (1.86–2.71) | 1.91 (1.52–2.31) | 2.34 (1.89–3.01) | 2.12 (1.67–2.73) |
| Baseline % predicted FEV1 | 93.0 (81.6–101.7) | 80.5 (72.0–90.7) | 89.7 (77.8–100.3) | 83.1 (73.9–93.4) |
| Serum IgE, IU/ml | 251 (106–623) | 273 (89–605) | 251 (95.7–600.0) | 258 (92.4–627.0) |
Figure 1. Association between the use of leukotriene modifiers and bronchodilator response to albuterol.
(A) LM use is associated with increased bronchodilator responsiveness in the whole Latino sample (data not shown) and in Puerto Ricans, but not in Mexicans. (B) Among Puerto Rican asthmatic patients, bronchodilator responsiveness to albuterol (%ΔFEV1) is independent of baseline lung function (baseline FEV1) in participants not taking leukotriene-modifying drugs (i); in leukotriene-modifying drug users responsiveness increases as baseline FEV1 decreases (ii).
%ΔFEV1: Percentage change in forced expiratory volume in 1 s; FEV1: Forced expiratory volume in 1 s; LM: Leukotriene modifier.
Data taken from [28].
The improvements in FEV1 after albuterol use in Puerto Rican participants were greatest in those patients who had the lowest baseline FEV1 values (Figure 1B), that is, in those patients with the most severe asthma. For every 200 ml decrease in baseline lung function, LT modifier use was associated with a mean increase of 2.9% (p = 0.003; 95% CI: 1.0–4.8) in bronchodilator responsiveness to albuterol. This is important given that Puerto Rican patients have lower baseline FEV1 values, lower bronchodilator responsiveness, and a more severe asthma phenotype than Mexicans [26].
In parallel, we sought to clarify the involvement of genes leading to the production of LTB4 (ALOX5AP and LTA4H) in asthma, since most previous genetic association studies had focused on ALOX5 and LTC4S genes. With this goal, we selected six SNPs in the ALOX5AP and LTA4H genes to be genotyped in the 687 Latino trios (asthmatic subject and both biological parents) collected for the GALA study (Table 1). Individual SNPs in both genes were associated with a decreased risk of asthma [29]. In the ALOX5AP gene, the minor allele in the intronic rs10507391 marker conferred protection from asthma in Puerto Ricans (p = 0.027; odds ratio [OR]: 0.78; 95% CI: 0.62–0.97) and in the combined Latino sample (p = 0.017). In the promoter of the LTA4H gene, the minor allele of the marker rs17525488 conferred a protective role for asthma in Mexicans (p = 0.035; OR: 0.62; 95% CI: 0.04–0.96) and in the combined Latino population (p = 0.007). In both cases, the associations that could not be replicated in one ethnicity showed the same trend and could be easily explained by a lower MAF of the SNPs limiting the statistical power. These SNPs significantly interacted to decrease the risk of asthma in the combined sample (OR: 0.47; 95% CI: 0.29–0.80; p = 0.003) and in Mexicans (OR: 0.44; 95% CI: 0.22–0.87; p = 0.01), and showed a similar trend in Puerto Ricans (OR: 0.54; 95% CI: 0.25–1.17; p = 0.10). This gene–gene interaction confers a decreased risk for asthma beyond what would have been expected from the main effects from each locus alone.
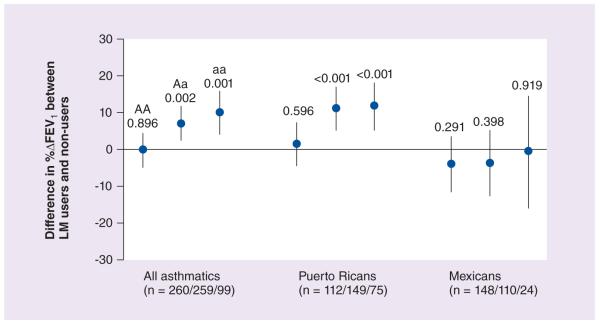
Given the results obtained from the drug–drug and the gene–gene analyses, we decided to pool the genetic and pharmacological data together and evaluate modulating effects of genetic variants in the ALOX5AP and LTA4H genes on the drug–drug interaction between LT modifiers and short-acting β2-agonists in Latinos with asthma recruited in the GALA study [30]. We did not find any effect of the SNPs analyzed in the ALOX5AP gene on the association between LT modifiers and bronchodilator responsiveness. On the other hand, two SNPs in the LTA4H gene had significant effects on this association. In patients carrying at least one copy of the minor allele of the LTA4H intronic rs2540491 SNP, the use of LT modifiers was associated with an augmentation of bronchodilator responsiveness, with a difference in mean %ΔFEV1 of 7.10% (95% CI: 2.61–11.59; p = 0.002) and 10.06% (95% CI: 4.29–15.82; p = 0.001) for heterozygotes and homozygotes, respectively (Figure 2). This same pattern of augmentation was followed by Puerto Rican participants, but not by Mexicans regardless of the allele present. At the LTA4H rs2540487 SNP in the promoter, the use of LT-modifying medication was associated with an augmentation of bronchodilator responsiveness to albuterol in Puerto Rican patients carrying at least one copy of the minor allele (4.93%; 95% CI: 0.11–9.74; p = 0.045) but not in Mexicans (p = 0.236). Because ALOX5AP and LTA4H had previously shown epistatic effects, we sought gene–gene interactions that altered the effect of LT4H SNPs on the augmentation of bronchodilator responsiveness by LT modifiers. Patients with genotypes containing the major allele of the ALOX5AP rs10507391 marker and a minor allele of either LTA4H rs2540487 or rs2540491 demonstrated augmentation of bronchodilator responsiveness by LT modifiers (all p < 0.01), with differences in mean %ΔFEV1 that ranged between 8.8 and 15.4%. Similar effects were observed for carriers of the minor allele of ALOX5AP rs9551963 with carriers of minor alleles at LTA4H rs2540487 or rs2540491.
Figure 2. Differences in mean percentage change in forced expiratory volume in 1 s between leukotriene modifier users and nonusers stratified by LTA4H rs2540491 genotype and ethnicity.
AA, Aa and aa represent homozygotes for the major allele, heterozygotes for the minor allele and homozygotes for the minor allele, respectively.
%ΔFEV1: Percentage change in forced expiratory volume in 1 s; FEV1: Forced expiratory volume in 1 s; LM: Leukotriene modifier.
Data taken from [30].
Conclusion
We have detected genetic and pharmacological interactive effects in the leukotriene pathway from second order (i.e., gene–gene and drug–drug) to higher order interactions (gene–drug–drug and gene–gene–drug–drug interactions). Many of these interactions have clinical implications, since the detected differences in %ΔFEV1 after the administration of albuterol can make the difference between adequate control of an acute asthma exacerbation at home and a run to an emergency room. Some of the SNPs in the ALOX5AP gene involved in these interactions were part of haplotypes previously associated with increased leukotriene production [31].
Additional findings keep increasing the list of genetic loci involved in the response to asthma controllers. For example, a recent report has identified several genes (e.g., the CHRM2 gene) associated with response to fluticasone, fluticasone/salmeterol and montelukast (a LT modifier) [32]. However, it is important to search for higher order gene–environment interactions and evaluate their clinical impact when exploring the heritability of complex traits, including drug response. Furthermore, it is also crucial to include the population perspective in the study of interactions. Although some differences between populations may be owing to our limited ability to detect or replicate our findings, population-specific interactions can be relevant and may direct differential prescriptions between populations, such as higher choice of LT modifiers as controller regimens for Puerto Rican patients with low baseline lung function.
Finally, it is important to remember that interaction networks do not explain the basis of the observed interactions and functional approaches are needed to understand their nature.
Future perspective
During the last 5 years, we have seen an exponential increase in the number of GWAS performed and in the number of samples that these studies include. Sample sizes of thousands or even tens of thousands of individuals are not exceptional anymore, including several studies that have identified and replicated genome-wide significant loci involved in the response to different drugs [33]. Furthermore, collaborative efforts to conduct meta-analyses and public initiatives to share data, such as the NCBI database of Genotypes and Phenotypes (dbGaP, [104]), facilitate the analysis of bigger sample sizes and the potential detection of loci of very small effects. In this sense, the number of studies searching for genetic interactions is likely to increase in the near future in an attempt to fill in the proportion of the heritability of complex traits that is not explained by individual genetic effects alone. Similarly, the increasing availability of larger tissue-specific expression datasets will identify the role of expression quantitative trait loci (eQTL) in drug response and other clinical quantitative traits [34] and, conversely, the effect of different drugs on transcriptional regulation [35]. These efforts will also enable the identification of potential eQTL–eQTL interactions relevant in pharmacogenomics.
Moreover, the availability of NGS technologies at affordable prices will allow the inclusion in future studies of full sequence data. This will improve our knowledge of genetic interactions relevant to complex outcomes, especially in noncoding regions. Noncoding regions are increasingly showing previously unexpected roles in the regulation of gene expression through long-range elements and loops in the chromatin structure [36]. Finally, profiling epigenomic changes in the context of drug response is also a relatively unexplored but promising field [33]. Epigenomic variants may play a substantial role in drug response, both individually and interacting with additional genomic or epigenomic variation, and its characterization will likely shed light on the ‘dark matter’ of the missing heritability [2].
Executive summary.
▪ Despite recent advances in the identification of genetic factors underlying complex diseases and phenotypes, a substantial proportion of the heritability of most common traits is still unexplained.
▪ Several strategies have been proposed to overcome this limitation: analyzing structural and rare variants, increasing sample sizes, conducting meta-analyses, expanding to non-European populations and exploring gene–gene and gene–environment interactions.
▪ It is not feasible to explore all potential interactions in the genome and most efforts have analyzed small sets of genes and environmental factors in a common physiological or biochemical pathway, as we did with leukotrienes (LTs) and the bronchodilator response to albuterol in Latino asthmatics.
LT genes & drugs: bronchodilator response & asthma
▪ LT-modifying medications induced higher increases in the bronchodilator response to albuterol. Moreover, interactions between genes responsible for leukotriene production (ALOX5AP and LTA4H) were associated with a decreased risk of asthma.
▪ Combining genetic and pharmacologic effects, we observed that LT-modifying users carrying certain combinations of alleles presented higher improvements in lung function after bronchodilator administration.
Conclusion
▪ The effects of LT genes and asthma drugs occur at different orders of interaction: from individual effects to gene–gene–drug–drug interactions. Moreover, there are population-specific effects that have to be considered.
▪ The complex patterns of interactions observed may reflect processes involved in other complex phenotypes.
Acknowledgments
M Via was supported by the Beatriu de Pinós Program (2009 BP-B 00274). H Tcheurekdjian was funded by the Glaser Pediatric Research Network. EG Burchard was supported by the NIH (HL078885, AI077439 and HL088133), the Flight Attendant Medical Research Institute (FAMRI), American Asthma Foundation and the Sandler Foundation.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl Acad. Sci. USA. 2012;109(4):1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. ▪▪ Complete review regarding strategies to identify genetic risk factors underlying the unexplained proportion of the heritability of complex traits.
- 3.Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Via M, Gignoux C, Burchard EG. The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med. 2010;2(1):3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5(5):e1000477. doi: 10.1371/journal.pgen.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horgan AM, Yang B, Azad AK, et al. Pharmacogenetic and germline prognostic markers of lung cancer. J. Thorac. Oncol. 2011;6(2):296–304. doi: 10.1097/JTO.0b013e3181ffe909. [DOI] [PubMed] [Google Scholar]
- 12.Fisher MC, Cronstein BN. Metaana lysis of methylenetetrahydrofolate reductase (MTHFR) polymorphisms affecting methotrexate toxicity. J. Rheumatol. 2009;36(3):539–545. doi: 10.3899/jrheum.080576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torgerson DG, Ampleford EJ, Chiu GY, et al. Meta-ana lysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat. Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burchard EG, Ziv E, Coyle N, et al. The importance of race and ethnic background in biomedical research and clinical practice. N. Engl. J. Med. 2003;348(12):1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- 16.Bustamante CD, Burchard EG, de la Vega FM. Genomics for the world. Nature. 2011;475(7355):163–165. doi: 10.1038/475163a. ▪ Recent comment about the importance of expanding medical genomic studies to non-European populations.
- 17.Gifford AL, Cunningham WE, Heslin KC, et al. Participation in research and access to experimental treatments by HIV-infected patients. N. Engl. J. Med. 2002;346:1373–1382. doi: 10.1056/NEJMsa011565. [DOI] [PubMed] [Google Scholar]
- 18.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era – concepts and misconceptions. Nat. Rev. Genet. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 19.Kelly TN, He J. Genomic epidemiology of blood pressure salt sensitivity. J. Hypertens. 2012;30(5):861–873. doi: 10.1097/HJH.0b013e3283524949. [DOI] [PubMed] [Google Scholar]
- 20.Rose AM, Bell LC. Epistasis and immunity: the role of genetic interactions in autoimmune diseases. Immunology. 2012;137(2):131–138. doi: 10.1111/j.1365-2567.2012.03623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berlanga-Taylor AJ, Disanto G, Ebers GC, Ramagopalan SV. Vitamin D-gene interactions in multiple sclerosis. J. Neurol. Sci. 2011;311(1–2):32–36. doi: 10.1016/j.jns.2011.08.041. [DOI] [PubMed] [Google Scholar]
- 22.Peters-Golden M, Henderson WR., Jr Leukotrienes. N. Engl. J. Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 23.Malmstrom K, Rodriguez-Gomez G, Guerra J, et al. Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. A randomized, controlled trial. Montelukast/Beclomethasone Study Group. Ann. Intern. Med. 1999;130:487–495. doi: 10.7326/0003-4819-130-6-199903160-00005. [DOI] [PubMed] [Google Scholar]
- 24.Szefler SJ, Phillips BR, Martinez FD, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J. Allergy Clin. Immunol. 2005;115:233–242. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Rovati GE, Baroffio M, Citro S, et al. Cysteinyl-leukotrienes in the regulation of β2-adrenoceptor function: an in vitro model of asthma. Respir. Res. 2006;7:103. doi: 10.1186/1465-9921-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burchard EG, Avila PC, Nazario S, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am. J. Respir. Crit. Care Med. 2004;169:386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 27.Choudhry S, Ung N, Avila PC, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am. J. Respir. Crit. Care Med. 2005;171:563–570. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 28.Tcheurekdjian H, Thyne SM, Williams LK, et al. Augmentation of bronchodilator responsiveness by leukotriene modifiers in Puerto Rican and Mexican children. Ann. Allergy Asthma Immunol. 2009;102(6):510–517. doi: 10.1016/S1081-1206(10)60126-3. ▪ Description of drug–drug interactions between leukotriene (LT)-modifying medication and short-acting β2-agonists.
- 29.Via M, De Giacomo A, Corvol H, et al. The role of LTA4H and ALOX5AP genes in the risk for asthma in Latinos. Clin. Exp. Allergy. 2010;40(4):582–589. doi: 10.1111/j.1365-2222.2009.03438.x. ▪ Identification of associations between genetic markers in the ALOX5AP and LTA4H genes and their interactions and the risk of asthma.
- 30.Tcheurekdjian H, Via M, de Giacomo A, et al. ALOX5AP and LTA4H polymorphisms modify augmentation of bronchodilator responsiveness by leukotriene modifiers in Latinos. J. Allergy Clin. Immunol. 2010;126(4):853–858. doi: 10.1016/j.jaci.2010.06.048. ▪▪ Description of third and fourth-order interactions between LT genes and asthma drugs in Latino asthmatic patients.
- 31.Helgadottir A, Manolescu A, Thorleifsson G, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat. Genet. 2004;36(3):233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 32.Mougey EB, Chen C, Tantisira KG, et al. Pharmacogenetics of asthma controller treatment. Pharmacogenomics J. 2012 doi: 10.1038/tpj.2012.5. doi:10.1038/tpj.2012.5. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harper AR, Topol EJ. Pharmacogenomics in clinical practice and drug development. Nat. Biotechnol. 2012;30(11):1117–1124. doi: 10.1038/nbt.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min JL, Taylor JM, Richards JB, et al. The use of genome-wide eQTL associations in lymphoblastoid cell lines to identify novel genetic pathways involved in complex traits. PLoS ONE. 2011;6(7):e22070. doi: 10.1371/journal.pone.0022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maranville JC, Luca F, Richards AL, et al. Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet. 2011;7(7):e1002162. doi: 10.1371/journal.pgen.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishiro T, Ishihara K, Hino S, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28(9):1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101. [Accessed 17 October 2012];A catalog of published genome-wide association studies. www.genome.gov/gwastudies.
- 102.The 1000 Genomes Project. www.1000genomes.org.
- 103.Global Initiative for Asthma (GINA) [Accessed 3 December 2012];Global Strategy for Asthma Management and Prevention. 2011 www.ginasthma.org.
- 104.NCBI database of Genotypes and Phenotypes (dbGaP) doi: 10.1093/nar/gkt1211. www.ncbi.nlm.nih.gov/gap. [DOI] [PMC free article] [PubMed]