Abstract
Up to now, efforts to crystallize the cataract-associated P23T mutant of human γD-crystallin have not been successful. Therefore, insights into the light scattering mechanism of this mutant have been exclusively obtained from solution work. Here we present the first crystal structure of the P23T mutant at 2.5Å resolution. The protein exhibits essentially the same overall structure as seen for the wild-type protein. Based on our structural data, we confirm that no major conformational changes are caused by the mutation, and that solution phase properties of the mutant appear exclusively associated with cataract formation.
Keywords: crystal structure, P23T crystallin mutant, cataract
INTRODUCTION
Human γD-crystallin (HGD) is the second most abundant member of the γ-crystallin family and mutations in HGD, such as R14C, P23T, R36S, R58H, and W42R, have been associated with a number of childhood cataracts.1-4 In general, these mutations exert various physiochemical effects on the protein and dramatic decreases in solubility were observed in all mutants, without major changes to the three-dimensional structure of the proteins. For the juvenile cataract associated R36S and R58H mutations, an increase in crystallization propensity was observed due to a decrease in the energy barrier for initiation of crystallization.5,6 In contrast, the introduction of a cysteine residue in the Arg14 position results in improper intermolecular disulfide bond formation, causing the protein to easily precipitate.7 The W42R mutation, on the other hand, changes the un/folding dynamics through a transient unfolding intermediate under physiological conditions and renders the protein as a mimic of photodamaged γD-crystallin that is involved in age-related cataract.8
The congenital cataract-associated P23T variant has been the subject of numerous investigations and is the hot spot of the crystallin family. The mutation is geographically widespread and appears phenotypically heterogeneous: at least ten different ADCC families of different ethnicity exhibit different phenotypes.4,9-17 The amino acid change dramatically lowers the solubility of the protein, causing protein aggregation and light scattering, and hence lens opacity. In contrast to the wild type protein, the P23T mutant protein exhibits retrograde solubility1 and Pande et al. proposed that net hydrophobic and anisotropic protein-protein interactions are responsible for the aggregation of P23T.18
High-resolution atomic structures are available for wild-type HGD19,20 as well as for several cataract causing mutants, such as R58H20, R36S5 and W42R8. However, efforts to crystallize the P23T protein were unsuccessful.1,2 The aggregation mechanism of this mutant was investigated in solution and it was revealed that only very minor structural differences from wild-type HGD are present that may initiate aggregation or polymerization of the protein.21 Here we provide an alternative view into the structural features of the mutant protein in the crystal. The structure was solved at 2.5 Å resolution, and, as already noted in the NMR solution structure, only differences in local conformational details are observed. The two most prominent ones pertain to the side chain of Trp42, one of the conserved Trp residues in the protein, that forms a different hydrogen bonding network in the mutant, and the side chain of His22 that exhibits a different conformation of the charged imidazole ring at the crystallization condition pH 4.6. Based on these crystallographic data, as well as our previous solution work, we can confirm with high confidence that no major structural changes of human γD-crystallin are present in the P23T mutant protein and that solution phase properties of this and other mutants may play critical roles in cataract formation.
MATERIALS AND METHODS
Protein Expression and Purification
Cloning, expression and purification of the P23T mutant protein were essentially carried out as described previously.21,22 In summary, the mutant γD-crystallin gene was cloned into the pET-14b vector with which E. coli BL21 (DE3) cells (EMD Chemicals) were transformed. For protein production, cells were grown at 37°C to an OD600 ~ 0.6, induced with 0.5 mM IPTG, and grown further at 18°C for 16 h. Cells were then harvested by centrifugation and the pellets were resuspended in 50 mM Tris buffer (pH 8.0), containing 1 mM EDTA, 1 mM DTT, 0.02% NaN3, and lyzed by passage through a microfluidizer (Microfluidics, Newton, MA). Cell debris was removed by centrifugation at 100,000 g for 1 hour and the supernatant was applied to a pre-equilibrated HiTrap QXL anion exchange column (GE Healthcare) in 50 mM Tris buffer (pH 8.0), 1 mM EDTA, 1 mM DTT, 0.02% NaN3. The flow-through fraction was collected and dialyzed overnight against 10 mM MES buffer (pH 6.2) containing 1 mM EDTA, 1 mM DTT, 0.02% NaN3 and 2% glycerol. The dialyzed sample was loaded on a HiTrap SP cation exchange column (GE Healthcare), equilibrated with the same buffer and eluted using a linear 0 to 1 M NaCl gradient over a 20-column volume. Fractions containing P23T were collected, concentrated, and further purified on a Superdex 75 26/60 column (GE Healthcare) in 20 mM sodium phosphate buffer (pH 6.2), 1 mM EDTA, 5 mM DTT, 0.02% NaN3.
The purified protein was characterized by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry on a Voyager-DE PRO instrument, operated in a linear mode with external calibration. The experimentally obtained molecular mass for P23T was 20623.10 Da, in excellent agreement with the theoretical molecular mass of 20610.93 Da. An extinction coefficient of 42.86 mM−1 cm−1 was used to determine protein concentrations with a NanoDrop 1000 spectrophotometer (Thermo Scientific).
Crystallization, X-Ray data collection and refinement
Crystals of the P23T mutant were obtained from a solution containing 0.17 M CH3COONH4, 0.085 M C2H3NaO2*3H2O, pH 4.6, 25.5% (w/v) PEG 4000, and 15% (v/v) glycerol at 25°C by sitting drop vapor diffusion.
X-ray diffraction data were collected with a Rigaku FR-E generator on a Saturn 944 CCD detector at the wavelength corresponding to the copper edge (1.54 Å) on flash-cooled crystals (−180°C). The crystal of the P23T mutant diffracted to a resolution of 2.5 Å and initial indexing of the diffraction patterns indicated that P23T belonged to the P21 space group with unit cell parameters of a = 37.55 Å, b = 101.93 Å and c = 41.37 Å (α = γ = 90.00° and β = 96.53°). Two protein molecules are present per asymmetric unit with an estimated solvent content of ~35.56% (Vm=1.91 Å3/Da) as evaluated by Matthews coefficient.23 All diffraction data were processed, integrated, and scaled using the d*TREK software 24 and eventually converted to MTZ format using the CCP4 package.25
The structure of the P23T mutant was solved by molecular replacement, using the W42R structure (PDB ID: 4GR7) as the search model in PHASER.26 After generation of the initial model, iterative cycles of manual re-building using Coot,27 and refinement that involved alternating between manual re-building and automated refinement in REFMAC28 were carried out. The final P23T structure was refined to 2.50 Å resolution with an Rfactor of 22.8% and Rfree of 28.4%. 99.7% and 100% of all residues are located in the favored and allowed regions of the Ramachandran plot, respectively, as evaluated by MOLPROBITY.29 Data collection and refinement statistics are provided in Table I. The atomic coordinates and structure factors for the P23T mutant have been deposited in the Protein Data Bank under the accession code of 4JGF. All structure figures were prepared using the program PyMOL.30
Table I. Data Collection and Refinement Statistics for P23T.
| Data Collection | ||
| Space group | P21 | |
| Cell dimensions | ||
| a, b, c (Å) | 37.55, 101.93, 41.37 | |
| α, β, γ (°) | 90.00, 96.53, 90.00 | |
| Resolution (Å) | 31.99 – 2.50 | (2.59 – 2.50) |
| Rmerge | 0.129 | (0.397) |
| <I/σI> | 9.2 | (3.4) |
| Completeness (%) | 100.0 | (99.8) |
| <Redundancy> | 8.39 | (6.69) |
| Refinement | ||
| Resolution (Å) | 32.01 -2.50 | (2.57-2.50) |
| No. reflections | 9664 | (720) |
| Rwork/Rfree | 0.228/0.284 | (0.331/0.426) |
| No. atoms | ||
| Protein | 2867 | |
| Water | 5 | |
| B-factors | ||
| Protein | 45.32 | |
| Water | 32.55 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.005 | |
| Bond angles (°) | 0.871 | |
RESULTS
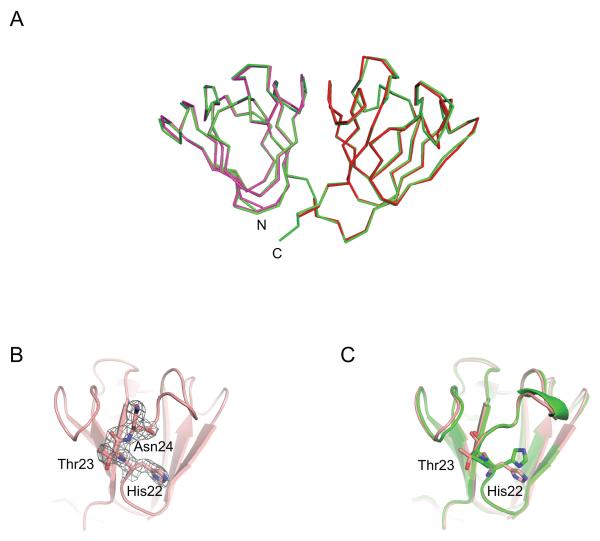
The solution structure of the P23T mutant had previously been determined in our laboratory by NMR spectroscopy.21 Based on this work, it was shown that only minor structural changes compared to the crystal structure of HGD were present. However, distinct differences in local conformational and dynamic behavior between the mutant and the wild-type proteins provided insights into the aggregation of the mutant protein.21 Here, we now report the P23T mutant crystal structure, determined at 2.5 Å resolution. Similar to the solution NMR P23T structure or the wild-type crystal HGD structure, the current structure solely exhibits local differences. Superposition of the N-terminal (N-td; residues 1-81) and C-terminal (C-td; residues 88-171) domains of the P23T mutant crystal structure (colored magenta and red in Fig. 1A, respectively) onto those of HGD (colored cyan in Fig. 1A) yielded pairwise backbone r.m.s.d. values of 0.433 Å and 0.518 Å, respectively. Slightly larger pairwise backbone atom r.m.s.d. values of 0.728 Å and 0.818 Å were seen to the NMR structure (PDB ID: 2KFB). This confirms that the individual domain structures in the P23T mutant crystal structure are essentially identical to either the wild-type HGD X-ray or the P23T solution structures. Note, that the two molecules of P23T in the asymmetric unit exhibit an average pairwise backbone atom r.m.s.d. value of 0.335 Å.
Figure 1. X-ray crystal structure of human P23T γD-crystallin mutant.
(A) Superposition of the P23T and HGD structures (cyan, PDB ID: 1HK0). The N-td (magenta, res. 1-81) and C-td (red, res. 82-171) coordinates of P23T were respectively fitted onto those of full length HGD (green). (B) Electron density for residue Thr23 in the P23T structure, contoured at 1.0 sigma. (C) Superposition of the N-tds of P23T (pink) and HGD (green), illustrating the side chain position of His22, Pro23 (HGD)/Thr23 (P23T).
Similar to our previous observation for the W42R mutant, the relative orientation between the N-td and C-td in the P23T mutant and HGD is slightly different, with a tilt angle of 8.9° between the domains. This explains the slightly higher backbone r.m.s.d. value of 0.644 Å between the P23T mutant and HGD crystal structures when the entire protein chain (residues 1-171) is superimposed, and most likely is the result of crystal packing.
The final refined electron density map of the P23T mutant reveals well-defined electron density for the threonine residue that replaces the proline ring in the mutant (Fig. 1B) and the backbone conformation of Thr23 is identical to that of Pro23 in the X-ray structure of HGD (Fig. 1C). The backbone carbonyl group of Thr23 (Pro23) accepts a hydrogen bond from the backbone amide group of Asn49, while the polar interaction between the hydroxyl group of Thr23 side chain and side chain of Tyr50 is only present in the mutant.
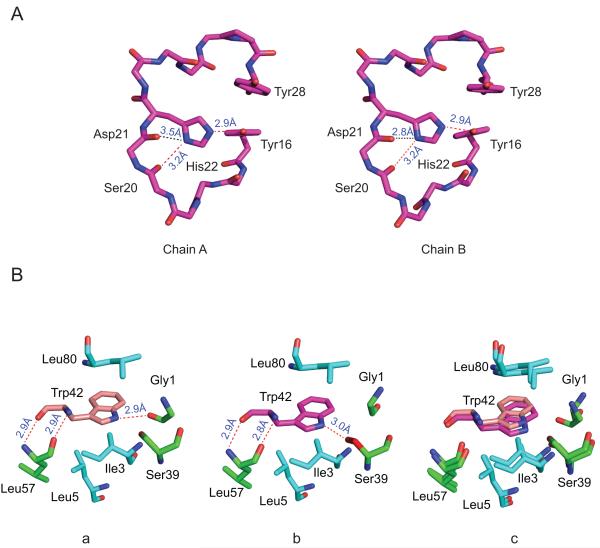
In the solution NMR studies it was noted that the His22 imidazole ring exists predominantly as the Nδ1 tautomer in the P23T mutant, different from the predominant Nε2 tautomer in the wild type protein.21 We therefore investigated the side chain of His22 in our X-ray crystal structure of P23T mutant. Since the pKa of the histidine side-chain is ~ 6 and the P23T crystals were grown at pH 4.6, only the charged species will be present, and only one conformation of His22 side chain is seen in the two molecules in the asymmetric unit (Fig. 2A), with the Nε2 group of the imidazole ring of His22 forming a hydrogen bond with Tyr16 side chain and Nδ1 interacting with backbone carbonyl group of Ser20 through a hydrogen bond.
Figure 2. Local conformations in the P23T and HGD γD-crystallin structures.
(A) No difference in His22 side chain conformations is observed in the two independent protein molecules in the asymmetric unit. (B) Contacts around the Trp42 side chain in the HGD (PDB ID: 1HK0) (a) and the P23T (b) structures and their best-fit superposition (c).
Further detailed analysis of the P23T and the HGD X-ray structures revealed minor structural readjustments around the Trp42 side chain. In the P23T structure a hydrogen bond is formed between the side chain of Trp42 and the backbone carbonyl group of Ser39 (Fig. 2B (b), colored in red) while in the HGD structure, the equivalent hydrogen bond is formed between Trp42 and the backbone carbonyl group of Gly1 (Fig. 2B (a)). The switch in this hydrogen bonding of Trp42 results in a small rotation of its aromatic ring (Fig. 2B (c)). In previous studies we noted that Trp42 played a significant role in maintaining the stability and integrity of the solution structure of HGD.8 Testing the P23T mutant by trypsin digestion we did not observe any cleavage products (data not shown), as previously found for the W42R mutant.8 Therefore, it is unlikely that the minor structural changes around Trp42 in the P23T mutant significantly influence its global structure.
We also evaluated whether the Thr substitution alters the solvent accessible surface of the protein. No significant differences are apparent, with the wild-type HGD and the P23T structures possessing 8894.32 and 8560.03 Å2 solvent accessible area, respectively, as calculated by VMD31. Therefore, at present, no features can be detected in the globular structure of the P23T mutant protein can explain its involvement in cataract formation, and other properties need to be considered and examined.
DISCUSSION
P23T is a mutant of human γD-crystallin and is implicated in several different autosomal dominant cataract phenotypes, including cerulean cataract,9 lamellar cataract,4 coralliform cataract,12,32 fasciculiform cataract,10 bilateral aculeiform cataract17 and an unclassified silica-like nuclear cataract13. The P23T protein is significantly less soluble (<10 mg/mL) than wild-type HGD (>200 mg/mL) at 37°C.21 This behavior is not associated with a structural change in the P23T mutant, as evidenced by CD, FT-IR, and NMR studies.1,2,21 Even though a slightly increased β-sheet content of P23T was proposed by Mackay et al,12 this is not observed here and no direct structural difference can currently explain the dramatically lowered solubility of this mutant. Given the present P23T X-ray crystal structure, which exhibits no significant differences from that of HGD, as pointed out earlier, the lowered solubility of the P23T protein must arise predominantly from solution properties that increase net protein-protein interactions. Interestingly, the fact that the native structure does not always reveal the underlying features of reactions that originate from a non-native, frequently transient, conformer is not unique to HGD. One example is QHF-lithostathine, a protein found in human brain and pancreas, whose structure appears unchanged in the insoluble fibers.33 Similarly, the folded structure of sickle-cell hemoglobin34 does not provide any clues as to its aggregation behavior.
Pande’s group has extensively and systematically examined the solubility changes caused by the P23T change, using solution NMR and chemical probes. They observed that the surface hydrophobicities of P23T and other mutants such as P23V and P23S are inversely related to their solubility and that the P23T mutant protein exhibits retrograde solubility, i.e. is less soluble at elevated temperature.35 Therefore, they proposed that net hydrophobic and anisotropic protein-protein interactions dominate the aggregation of P23T.18,35 However, in the crystal structure of P23T, no direct structural evidence for this suggestion can be seen. Additionally, we also observed that the P23T mutant exhibited a slight increase in solubility at higher ionic strength (between 0 and 0.5 M NaCl; data not shown), indicating that the presence of high concentrations of salt may prevent random protein-protein ionic interactions, thereby reducing aggregation to some degree.
The local difference in the tautomeric form of the His22 side chain observed in solution between P23T and HGD by Jung et al21 cannot be assessed in the X-ray structure of P23T reported here since the protein was crystallized at pH 4.6. Thus the His22 imidazole ring is protonated and no distinction between the Nε2 and the Nδ1 atom is possible. With respect to crystal contacts around the mutation site, His22 in P23T is not involved in any protein-protein contacts, similar to what is seen in HGD (PDB ID: 1HK0) and the R58H mutant (PDB ID: 1H4A) and different from R36S (PDB ID: 2G98) in which His22 contacts Arg116 of the neighboring molecule. Tyr28, which is in the vicinity of Thr23, however, engages in a contact with Asp155 of the C-td of a neighboring molecule, similar to the situation seen in bovine γD-crystallin (PDB ID: 1ELP), where Tyr28 interacts with Ile102 of the neighboring molecule. Since bovine γD-crystallin is highly soluble, such interactions cannot be responsible for the solubility difference between P23T and HGD. Therefore, as pointed out above, it is important to investigate the solution behavior of aggregating proteins and, for the cataract associated P23T crystallin, further studies are necessary to explain its dramatically lowered solubility and aggregation behavior.
CONCLUSION
Here, we report the X-ray crystal structure of the most commonly occurring HGD mutant, P23T, which up to now has been refractory to crystallization. Only minor conformational differences were observed when compared to the crystal structure of wild-type HGD. Having now the P23T mutant crystal structure available further reinforces the notion that this single mutation mainly affects the solution behavior of the protein that causes aggregation and/or precipitation or may change its interactions with other proteins or factors in the eye lens, and not its 3D structure. In addition, since the P23T mutant is associated with more than ten cataract phenotypes, the structural analysis and comparison between its crystal structure and solution structure provides insights into the mechanism of its dramatically lowered solubility and aggregation behavior.
Acknowledgments
Grant sponsor: NIH; Grant number: EY021193.
REFERENCES
- 1.Pande A, Annunziata O, Asherie N, Ogun O, Benedek GB, Pande J. Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human gamma D-crystallin. Biochemistry. 2005;44:2491–2500. doi: 10.1021/bi0479611. [DOI] [PubMed] [Google Scholar]
- 2.Evans P, Wyatt K, Wistow GJ, Bateman OA, Wallace BA, Slingsby C. The P23T cataract mutation causes loss of solubility of folded gammaD-crystallin. J Mol Biol. 2004;343:435–444. doi: 10.1016/j.jmb.2004.08.050. [DOI] [PubMed] [Google Scholar]
- 3.Hejtmancik JF. The genetics of cataract: our vision becomes clearer. American journal of human genetics. 1998;62:520–525. doi: 10.1086/301774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santhiya ST, Shyam Manohar M, Rawlley D, Vijayalakshmi P, Namperumalsamy P, Gopinath PM, Loster J, Graw J. Novel mutations in the gamma-crystallin genes cause autosomal dominant congenital cataracts. J Med Genet. 2002;39:352–358. doi: 10.1136/jmg.39.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kmoch S, Brynda J, Asfaw B, Bezouska K, Novak P, Rezacova P, Ondrova L, Filipec M, Sedlacek J, Elleder M. Link between a novel human gammaD-crystallin allele and a unique cataract phenotype explained by protein crystallography. Human molecular genetics. 2000;9:1779–1786. doi: 10.1093/hmg/9.12.1779. [DOI] [PubMed] [Google Scholar]
- 6.Pande A, Pande J, Asherie N, Lomakin A, Ogun O, King J, Benedek GB. Crystal cataracts: human genetic cataract caused by protein crystallization. Proc Natl Acad Sci U S A. 2001;98:6116–6120. doi: 10.1073/pnas.101124798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pande A, Pande J, Asherie N, Lomakin A, Ogun O, King JA, Lubsen NH, Walton D, Benedek GB. Molecular basis of a progressive juvenile-onset hereditary cataract. Proc Natl Acad Sci U S A. 2000;97:1993–1998. doi: 10.1073/pnas.040554397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji F, Jung J, Koharudin LM, Gronenborn AM. The human W42R gammaD-crystallin mutant structure provides a link between congenital and age-related cataracts. The Journal of biological chemistry. 2013;288:99–109. doi: 10.1074/jbc.M112.416354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nandrot E, Slingsby C, Basak A, Cherif-Chefchaouni M, Benazzouz B, Hajaji Y, Boutayeb S, Gribouval O, Arbogast L, Berraho A, Abitbol M, Hilal L. Gamma-D crystallin gene (CRYGD) mutation causes autosomal dominant congenital cerulean cataracts. Journal of medical genetics. 2003;40:262–267. doi: 10.1136/jmg.40.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shentu X, Yao K, Xu W, Zheng S, Hu S, Gong X. Special fasciculiform cataract caused by a mutation in the gammaD-crystallin gene. Mol Vis. 2004;10:233–239. [PubMed] [Google Scholar]
- 11.Xu WZ, Zheng S, Xu SJ, Huang W, Yao K, Zhang SZ. Autosomal dominant coralliform cataract related to a missense mutation of the gammaD-crystallin gene. Chinese medical journal. 2004;117:727–732. [PubMed] [Google Scholar]
- 12.Mackay DS, Andley UP, Shiels A. A missense mutation in the gammaD crystallin gene (CRYGD) associated with autosomal dominant “coral-like” cataract linked to chromosome 2q. Mol Vis. 2004;10:155–162. [PubMed] [Google Scholar]
- 13.Burdon KP, Wirth MG, Mackey DA, Russell-Eggitt IM, Craig JE, Elder JE, Dickinson JL, Sale MM. Investigation of crystallin genes in familial cataract, and report of two disease associated mutations. Br J Ophthalmol. 2004;88:79–83. doi: 10.1136/bjo.88.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang LY, Gong B, Tong JP, Fan DS, Chiang SW, Lou D, Lam DS, Yam GH, Pang CP. A novel gammaD-crystallin mutation causes mild changes in protein properties but leads to congenital coralliform cataract. Mol Vis. 2009;15:1521–1529. [PMC free article] [PubMed] [Google Scholar]
- 15.Khan AO, Aldahmesh MA, Ghadhfan FE, Al-Mesfer S, Alkuraya FS. Founder heterozygous P23T CRYGD mutation associated with cerulean (and coralliform) cataract in 2 Saudi families. Mol Vis. 2009;15:1407–1411. [PMC free article] [PubMed] [Google Scholar]
- 16.Yang G, Xiong C, Li S, Wang Y, Zhao J. A recurrent mutation in CRYGD is associated with autosomal dominant congenital coralliform cataract in two unrelated Chinese families. Mol Vis. 2011;17:1085–1089. [PMC free article] [PubMed] [Google Scholar]
- 17.Vanita V, Singh D. A missense mutation in CRYGD linked with autosomal dominant congenital cataract of aculeiform type. Mol Cell Biochem. 2012;368:167–172. doi: 10.1007/s11010-012-1355-2. [DOI] [PubMed] [Google Scholar]
- 18.Pande A, Ghosh KS, Banerjee PR, Pande J. Increase in surface hydrophobicity of the cataract-associated P23T mutant of human gammaD-crystallin is responsible for its dramatically lower, retrograde solubility. Biochemistry. 2010;49:6122–6129. doi: 10.1021/bi100664s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chirgadze YN, Driessen HP, Wright G, Slingsby C, Hay RE, Lindley PF. Structure of bovine eye lens gammaD (gammaIIIb)-crystallin at 1.95 A. Acta crystallographica Section D, Biological crystallography. 1996;52:712–721. doi: 10.1107/S0907444996000352. [DOI] [PubMed] [Google Scholar]
- 20.Basak A, Bateman O, Slingsby C, Pande A, Asherie N, Ogun O, Benedek GB, Pande J. High-resolution X-ray crystal structures of human gammaD crystallin (1.25 A) and the R58H mutant (1.15 A) associated with aculeiform cataract. J Mol Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 21.Jung J, Byeon IJ, Wang Y, King J, Gronenborn AM. The structure of the cataract-causing P23T mutant of human gammaD-crystallin exhibits distinctive local conformational and dynamic changes. Biochemistry. 2009;48:2597–2609. doi: 10.1021/bi802292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji F, Jung J, Gronenborn AM. Structural and biochemical characterization of the childhood cataract-associated R76S mutant of human gammaD-crystallin. Biochemistry. 2012;51:2588–2596. doi: 10.1021/bi300199d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 24.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 25.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. (1994/09/01) 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delano WL. The PyMOL molecular graphics system. 2002 Available at: http://www.pymol.org.
- 31.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 32.Xu WZ, Zheng S, Xu SJ, Huang W, Yao K, Zhang SZ. [Localization and screening of autosomal dominant coralliform cataract associated gene] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2004;21:19–22. [PubMed] [Google Scholar]
- 33.Laurine E, Gregoire C, Fandrich M, Engemann S, Marchal S, Thion L, Mohr M, Monsarrat B, Michel B, Dobson CM, Wanker E, Erard M, Verdier JM. Lithostathine quadruple-helical filaments form proteinase K-resistant deposits in Creutzfeldt-Jakob disease. The Journal of biological chemistry. 2003;278:51770–51778. doi: 10.1074/jbc.M306767200. [DOI] [PubMed] [Google Scholar]
- 34.Eaton WA, Hofrichter J. Sickle cell hemoglobin polymerization. Advances in protein chemistry. 1990;40:63–279. doi: 10.1016/s0065-3233(08)60287-9. [DOI] [PubMed] [Google Scholar]
- 35.Pande A, Zhang J, Banerjee PR, Puttamadappa SS, Shekhtman A, Pande J. NMR study of the cataract-linked P23T mutant of human gammaD-crystallin shows minor changes in hydrophobic patches that reflect its retrograde solubility. Biochem Biophys Res Commun. 2009;382:196–199. doi: 10.1016/j.bbrc.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]