Abstract
Helicobacter pylori is arguably one of the most successful pathogens; it colonizes the stomachs of more than half of the human population. Colonization and persistence in such an inhospitable niche requires the presence of exquisite adaptive mechanisms. One of the proteins that contributes significantly to the remarkable adaptability of H. pylori is the ferric uptake regulator (Fur), which functions as a master regulator of gene expression. In addition to genes directly related to iron homeostasis, Fur controls expression of several enzymes that play a central role in metabolism and energy production. The absence of Fur leads to severe H. pylori colonization defects and, accordingly, several Fur-regulated genes have been shown to be essential for colonization. Moreover, proteins encoded by Fur-regulated genes have a strong impact on redox homeostasis in the stomach and are major determinants of inflammation. In this review, we discuss the main roles of Fur in the biology of H. pylori and highlight the importance of this regulatory protein in the infectious process.
Keywords: colonization, ferric uptake regulator, gastric cancer, Helicobacter pylori, inflammation, iron, iron acquisition, iron metabolism, virulence
Helicobacter pylori is a human pathogen with a unique capacity to efficiently colonize the hostile environment of the stomach. Gastric colonization by H. pylori induces chronic active gastritis in nearly all infected individuals, but most patients do not develop any apparent clinical signs of infection. However, in a subset of individuals, H. pylori infection progresses from gastritis to more severe upper gastrointestinal disorders such as peptic ulcers, mucosa-associated lymphoid tissue lymphoma or gastric adenocarcinoma. The reasons why certain individuals develop clinical disease while the majority of people remain asymptomatic are poorly understood. However, some progress has been made in the identification of factors that affect the wide range of disease states observed. Interestingly, these factors are multifactorial and include bacterial, host and environmental elements. Among the identified bacterial factors, the expression of the cytotoxin-associated protein CagA and the vacuolating cytotoxin VacA have been shown to be major contributors to disease severity. For example, CagA-positive H. pylori strains are at least twice as likely to cause cancer as H. pylori strains without CagA [1,2]. From the environmental perspective, diets rich in salt, pickled or smoked foods, or saturated fat, as well as the consumption of alcoholic beverages, exacerbate the severity of the infection symptoms. On the other hand, several studies examining host factors indicate that H. pylori is more likely to cause peptic ulcers in people with blood type O, while those with blood type A are more likely to develop gastric cancer [3].
According to the WHO, gastric cancer is the second most common cause of cancer-related death in the world, responsible for up to 736,000 deaths in 2008. Of note, it is estimated that H. pylori infection is responsible for 5.5% of all global cancers and 65% of gastric cancers worldwide [4]. The prevalence of H. pylori infection ranges from 20% in industrialized countries to more than 90% in the developing world, which makes H. pylori the most prevalent infection worldwide [5]. Currently, treatment of H. pylori infection involves triple therapy with a proton pump inhibitor or ranitidine, combined with clarithromycin and amoxicillin or metronidazole [6]. However, the rapid increase in antibiotic resistance may soon require the use of quadruple therapy. The complexity of the eradication therapy and the cost of these drugs is often excessive in nations where H. pylori is endemic, and thus often results in poor patient compliance [7]. Moreover, even successful eradication therapy does not protect the host from potential reinfection nor prevent asymptomatic infected individuals who do not realize that they need treatment from ultimately developing gastric cancer. Therefore, the identification of potential new drugs and new drug targets, along with the development of effective vaccines to prevent or cure chronic H. pylori infection, constitute a fundamental area of research. This endeavor demands a profound understanding of H. pylori pathogenesis and virulence factors.
As with many other organisms, H. pylori regulates gene expression in response to environmental change. To achieve this, H. pylori is equipped with a rather limited repertoire of response regulators and two component systems [8,9]. Control of iron homeostasis is mediated by the ferric uptake regulator (Fur), which essentially regulates transcription of genes involved in iron acquisition and storage in response to changes in iron availability [10,11]. However, Fur also regulates gene expression in response to low pH [12–15], oxidative stress [16,17] and salt [18]. Thus, this iron-sensing protein is actually a global regulator of gene expression in H. pylori that contributes significantly to the unique plasticity that is characteristic of this bacterium. In keeping with this, Fur has been shown to be important for survival under stressful conditions other than iron limitation [13]. Consequently, the Fur regulon includes genes involved in acid acclimation, resistance to oxygen reactive species and nitrogen metabolism. Therefore, collectively, Fur plays a key role in the adaptation of H. pylori to the hostile conditions that exist in the stomach. In H. pylori [13,19], as well as other pathogenic bacteria such as Staphylococcus aureus [20], Campylobacter jejuni [21] Listeria monocytogenes [22], Actinobacillus pleuropneumoniae [23], Bacillus cereus [24] and Vibrio cholerae [25], inactivation of the fur locus leads to reduced virulence of the corresponding mutants. Hence, Fur plays a critical role in bacterial pathogenesis. In the present review, we summarize the diverse means by which Fur maintains the intracellular iron balance in H. pylori, and discuss the importance of this regulatory protein in H. pylori colonization of the stomach.
Iron sources in the stomach, iron utilization mechanisms of H. pylori & association of H. pylori infection with iron-deficient anemia
Iron is an essential micronutrient for virtually all organisms, and H. pylori is no exception. In the stomach, ingested food provides iron in the form of heme and nonheme. The low pH and the digestive enzymes found in the stomach release iron from ligands to the gastric lumen. H. pylori and the host both compete for the free iron by deploying mechanisms specifically devised to sequester and facilitate the acquisition of this and other essential metals. Indeed, it is well known that sequestration of micro-nutrients represents one of the first lines of defense of the host against bacterial infection; this process is termed nutritional immunity. However, H. pylori seems particularly adept at competing for iron; it has been established that H. pylori successfully competes for iron in a murine host to such an extent that it can actually cause iron deficiency when the intake of dietary iron is poor [26]. This fact is evidence of the exquisite evolutive adaptation of this unique microorganism to its natural niche.
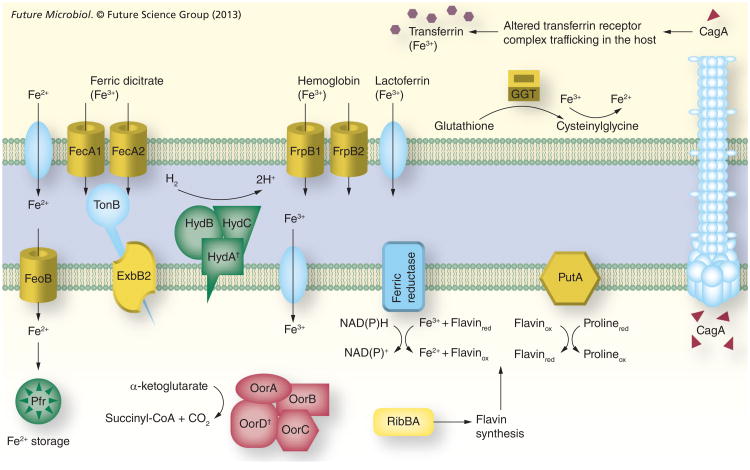
In order to guarantee a sufficient supply of iron from the environment, H. pylori cells display a repertoire of high-affinity iron-uptake systems (Figure 1). In the gastric lumen, released ferrous iron is kept in solution by means of the acidic pH and the low oxygen concentration characteristic of this niche. This in turn prevents the rapid conversion of the metal to ferric iron. Ferrous iron molecules pass freely through the outer membrane of the bacteria, but require high-affinity transporters in the cytoplasmic membrane to enter the cell; this function is carried out by FeoB in H. pylori [27]. Ferric citrate represents another important source of iron for H. pylori and, accordingly, three transporters implicated in the uptake of this molecule have been identified in this bacterium: FecA1, FecA2 and FecA3. Accumulation of ferric citrate into the periplasm via the Fec uptake systems requires energy, which is derived from the proton motive force mediated by the inner membrane protein complex composed of TonB, ExbB and ExbD [28,29].
Figure 1. The iron sources available in the stomach and the ferric uptake regulator-mediated responses aimed at maintaining the intracellular iron balance in Helicobacter pylori.
Yellow color indicates that expression is inactivated by iron-bound ferric uptake regulator (Fur). Green color indicates that expression is inactivated by apo-Fur. Red color indicates that expression is activated by iron-bound Fur. The subcellular location of GGT, PutA, HydABC and ferric reductase is only for illustration and has not been experimentally determined.
†Iron–sulfur proteins.
CagA: Cytotoxin-associated protein; GGT: γ-glutamyltranspeptidase; Pfr: Bacterial ferritin.
Although the presence of siderophores has not been described in H. pylori, it has been shown that this bacterium can bind and utilize several host iron-binding molecules as iron sources [30–32]. This is particularly relevant because H. pylori colonizes the gastric mucosa where free iron is readily sequestered by host iron-binding proteins such as lactoferrin and transferrin. The recent development of an iron-deficient chemically defined medium unequivocally demonstrated that H. pylori can acquire iron from fully saturated (holo) lactoferrin and transferrin [30]. Interestingly, however, the authors showed that H. pylori binds the apo-form of these two proteins with higher affinity than the holo-forms. Consequently, H. pylori cannot use iron from partially saturated (<75%) lactoferrin or transferrin and, thus, is unable to acquire iron from serum. Owing to this, the inability to acquire iron from serum has been suggested to block H. pylori virulence by preventing its growth in the bloodstream, where the presence of apo-lactoferrin and apo-transferrin is relatively high [30].
So far, little is known about how H. pylori cells acquire iron bound to host-binding proteins. A 70-kDa outer membrane protein with lactoferrin-binding properties was suggested several years ago, but the identity of this protein still remains unknown [33]. Similarly, the proteins responsible for transferrin binding in H. pylori have not yet been identified. By contrast, two outer membrane proteins, FrpB1 and FrpB2, have been implicated in hemoglobin binding [34,35]. In keeping with this, the ability of H. pylori to use hemoglobin as an iron source is well documented [30,32]. Several iron-repressible outer membrane proteins from H. pylori, including FrpB1, seem to be responsible for heme utilization [32,34]. It is noteworthy that the relative concentration of free heme can lead to opposite effects on H. pylori cells [30], while low concentrations of free heme stimulate growth, heme concentrations above 1 μM elicit toxicity. Even if heme and hemoglobin are released in the stomach from the ingested food, it seems that the use of hemoglobin may become most relevant when the bacterium is in contact with blood cells that are released as a result of ulceration.
Despite having previously generated controversy, epidemiological and clinical evidence suggest a relationship between iron deficiency anemia (IDA) and H. pylori infection [36–41]. Moreover, recent work has shown that H. pylori infection induces iron deficiency in insulingastrin mice [42]. Supporting this association, iron administration in combination with anti-H. pylori therapy has been demonstrated to be more effective than iron administration alone for the treatment of IDA [43]. H. pylori-induced anemia could result from iron loss due to microbleeding. However, H. pylori-induced hypoacidity may also contribute to IDA by impairing iron absorption. Furthermore, it seems that H. pylori strains isolated from patients with IDA show enhanced iron-uptake activity and thus may be more adept at competing with the host for iron [44]. Similarly, certain polymorphisms of the bacterioferritin Pfr and the high-affinity ferrous iron transporter FeoB are well correlated with IDA-associated H. pylori strains [45,46]. A comparative proteomic analysis of H. pylori strains from IDA and non-IDA patients revealed the existence of important differences between these two groups, and identified several proteins that are predominantly expressed in strains from IDA patients [47]. One of the proteins differentially expressed in these strains is AmiE, an aliphatic amidase implicated in ammonia production and acid adaptation. The gene encoding AmiE is upregulated under iron-restricted conditions and acidic pH [13,48–50]. This may suggest a deficiency in the regulatory circuitry that senses and responds to changes in iron availability and pH in the H. pylori strains isolated from IDA patients. Currently, despite all efforts, the specific role of H. pylori in the etiology of IDA, as well as the mechanism underlying the increased iron acquisition displayed by H. pylori from IDA patients remain unknown. Understanding iron uptake and trafficking in this bacterium as well as the regulation of these processes is critical to determine the molecular basis of this disorder.
Fur control of iron homeostasis involves activation & repression of genes with very diverse functions
Iron is required as a cofactor by several enzymes and as a catalyst in the electron transport processes. However, iron overload stimulates the formation of reactive oxygen species via the Fenton reaction. The resulting species damage DNA, proteins and membrane lipids [51]. To prevent this problem, bacteria have developed sophisticated mechanisms to sense and respond to fluctuations in iron availability as a means of maintaining an appropriate intracellular iron balance. In H. pylori, this control is carried out by the iron-sensing protein Fur. In addition to genes directly related to iron homeostasis, Fur also controls expression of very diverse genes that belong to different functional categories (Table 1 & Figure 1). However, the most widespread group of genes under the control of this metalloregulator is that of the iron uptake systems, including the previously mentioned high-affinity iron transporters FecA1, FecA2, FrpB1 and FeoB [11,13,52–54]. When the intracellular concentration of ferrous iron is low, Fur is unable to bind to the Fur box sequences within the promoters of these genes, transcription is derepressed and the capacity of H. pylori cells to acquire extracellular iron increases drastically. Similarly, recent experiments suggest that the lactoferrin and transferrin receptors from H. pylori are also upregulated under iron-deficient conditions [30].
Table 1.
List of Helicobacter pylori ferric uptake regulator-regulated genes.
| Gene | Function | Ref. |
|---|---|---|
| Repressed by iron-bound Fur | ||
| fecA1 | Iron transport | [13,54] |
| fecA2 | Iron transport | [13,54] |
| frpB1 | Iron transport | [13,54] |
| feoB | Iron transport | [11,60] |
| exbB2 | TonB-dependent energy transduction system | [13,54] |
| fur | Iron homeostasis | [102,103] |
| nikR | Nickel homeostasis | [104] |
| amiE | Aliphatic amidase | [13,54] |
| hpn2 | Nickel storage | [13,54] |
| pdxAJ | Vitamin B6 biosynthesis | [13,54] |
| ribBA | Riboflavin biosynthesis | [59] |
| putA | Proline metabolism | [60] |
| ggt | Synthesis and degradation of glutathione | [60] |
| ruvC | Recombination and repair | [13] |
| Activated by iron-bound Fur | ||
| cagA | Cytotoxicity | [60] |
| oorDABC | Metabolism | [13,70] |
| nifS | Iron–sulfur cluster biosynthesis | [68] |
| Repressed by apo-Fur | ||
| pfr | Iron storage | [13,54] |
| sodB | Oxidative stress defense | [17,54] |
| serB | Amino acid biosynthesis | [54] |
| hydABCDE | Hydrogenase | [54] |
| cheV2 | Chemotaxis | [54] |
| cytochrome c553 | Electron transport | [54] |
| Activated by apo-Fur | ||
| fur | Iron homeostasis | [102,103] |
Fur: Ferric uptake regulator.
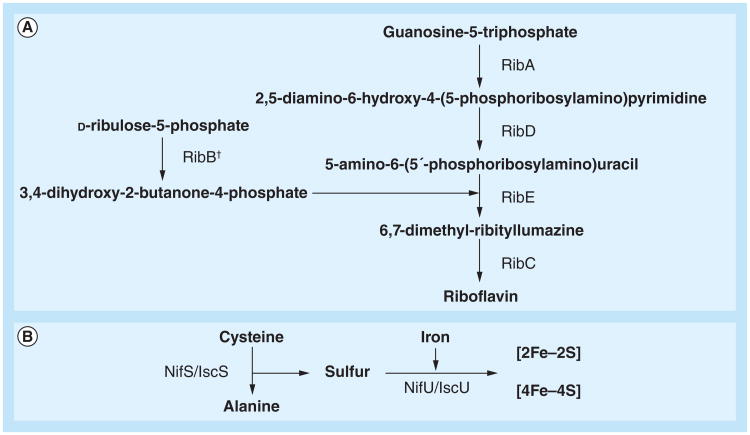
Flavin biosynthesis is also under the control of Fur in H. pylori as well as another gastrointestinal pathogen, C. jejuni [55,56]. Fur regulation of flavin biosynthesis is linked to the activity of the ferric iron reductases that catalyze the flavin-mediated reduction of extracellular ferric iron to the soluble ferrous form [57,58]. The activity of ferric iron reductases is believed to be important for the mobilization of insoluble ferric complexes in the almost neutral environment of the gastric epithelia. Given this, it is perhaps not surprising that expression of a key enzyme in the synthesis of flavins, RibBA, is regulated by Fur in H. pylori [55,59]. Indeed, RibBA plays a central role in the synthesis of riboflavin, the biochemical source of the flavin moiety of the flavin adenine dinucleotide and flavin mononucleotide coenzymes (Figure 2a). Of note, disruption of the ribBA gene eliminates the ferric iron reduction activity of H. pylori [55]. Transcriptional analysis revealed that the ribBA gene is repressed by iron-bound Fur. Thus, when iron is scarce, ribBA transcription is derepressed, which increases the production of flavins that are then ready to be used by ferric iron reductases. Similarly, the putA gene of H. pylori has recently been found to be repressed by Fur under iron-replete conditions [60]. PutA is a bifunctional enzyme that catalyzes the conversion of proline to glutamate. In the first step of this reaction, the proline dehydrogenase domain of PutA catalyzes the flavin adenine dinucleotide-dependent oxidation of proline to delta-1-pyrroline-5-carboxylate. Thus, proline catabolism generates reduced flavins that can be used by ferric iron reductases and, accordingly, the increase in putA transcript levels observed upon iron starvation in H. pylori is likely to be a Fur-mediated response aimed at increasing iron accessibility. Whether the expression of other flavoproteins of H. pylori is also under the control of Fur remains to be investigated.
Figure 2. Riboflavin and iron–sulfur cluster biosynthesis pathways.
(A) Riboflavin and (B) iron–sulfur clusters.
†RibB function in Helicobacter pylori is catalyzed by the bifunctional enzyme RibBA.
NifS/IscS: Cysteine desulfurase; NifU/IscU: (Iron–sulfur) cluster scaffold protein; RibA: GTP cyclohydrolase II; RibB: 3,4-dihydroxy-2-butanone 4-phosphate synthase; RibC: Riboflavin synthetase; RibD: Pyrimidine deaminase; RibE: 6,7-dimethyl-8-ribityllumazine synthase.
In a similar manner, the expression of the gene encoding γ-glutamyltranspeptidase (GGT) is Fur-dependent in H. pylori [60]. GGT is an enzyme that plays a crucial role in the utilization of extracellular glutamine and glutathione via conversion of these substrates to glutamate, which is subsequently transported into the H. pylori cell [61]. Remarkably, a recent study in Histoplasma capsulatum implicated this protein in the generation of extracellular reduced iron, hence revealing a new physiological role for GGT in iron acquisition [62]. It is noteworthy that the ferric reductase activity of this protein does not appear to be flavin cofactored. Perhaps for this reason, expression of the ggt gene in H. pylori is directly regulated by Fur. Despite the fact that the role of H. pylori GGT in iron assimilation has not yet been proven, Fur regulation of the ggt gene supports this idea. The activity of this enzyme may be extremely beneficial for H. pylori, allowing the reduction of ferric iron obtained from host binding proteins such as transferrin or hemin, as does its H. capsulatum counterpart [63].
Expression of iron-storage proteins is also under the control of Fur and, accordingly, this metalloregulator regulates the expression of the bacterial ferritin Pfr in H. pylori [11,13,54,64]. Pfr belongs to the nonheme ferritin subfamily and plays a significant role in protection against metal toxicity [65]. The intracellular levels of Pfr mimic those of iron: when iron is abundant, the levels of Pfr are high, and when iron is limited, pfr expression is downregulated. In this sense, pfr expression follows the opposite pattern of that of the iron-uptake genes, which are upregulated when iron is scarce. Interestingly, Pfr is constitutively expressed in a fur mutant, indicating that Fur functions as a repressor of pfr under iron-limited conditions [64]. To accomplish this role, H. pylori Fur binds to the pfr promoter in its iron-free (apo) form and prevents RNA polymerase binding. Since the classical view of Fur is that of an iron-bound repressor protein, this ability of apo-Fur to bind DNA has generated a significant amount of controversy. However, very recently, apo-Fur proteins from other organisms have been shown to bind and regulate transcription of Fur-regulated promoters [66,67]. Furthermore, the apo structure of C. jejuni has recently been determined [66]. Therefore, depending on the iron status, H. pylori Fur has the capacity to inactivate different sets of genes.
Increasing evidence suggests that the production of enzymes that require iron for their function is induced in H. pylori under iron-replete conditions. This control seems to be mediated by Fur at two different levels. First, Fur stimulates the synthesis of iron–sulfur (Fe–S) clusters. Fe–S clusters are small, inorganic, prosthetic groups that participate in a variety of biochemical processes, including electron transfer, substrate binding and activation, redox catalysis, DNA replication and repair, regulation of gene expression and tRNA modification. The positive effect of Fur on Fe–S biosynthesis is achieved by transcriptional activation of the nifS gene [68]. NifS is a cysteine desulfurase that releases sulfur or sulfide from L-cysteine and functions as a sulfur donor in the biogenesis of the Fe–S cluster (Figure 2b) [69]. Second, transcription of some genes encoding Fe–S-containing enzymes is directly regulated by Fur. For instance, the ferredoxin oxidoreductase gene cluster (oorDABC) was found to be activated by iron-bound Fur in the current authors' laboratory [13,70]. Similarly, a previous study revealed that the expression of the quinone-reactive Ni/Fe hydrogenase gene cluster (hydABCDE) was regulated by iron in a Fur-dependent manner [54]. However, in the latter case, transcriptional data indicate that regulation is achieved via apo-Fur, which down-regulates the expression of the hydABCDE cluster when iron is limiting for growth. Thus, Fur coordinates an iron-sparing response in H. pylori similar to that described in Bacillus subtilis [71,72]. This response is devised to guarantee a resourceful utilization of iron when this metal is limiting for growth. However, since deletion of the fur gene in H. pylori does not appear to have any impact on bacterial growth rate, differences in the iron status are not expected to prompt a major remodeling of the metabolic pathways in this bacterium. It is worth noting that, because of the unique features of the Fur protein from H. pylori, the iron-sparing response in this bacterium involves inactivation of gene expression by apo-Fur, as well as Fur-mediated activation of target genes when iron is abundant.
Expression of the iron-cofactored superoxide dismutase protein is also repressed by apo-Fur in H. pylori [17]. Superoxide dismutase constitutes a primary line of defense against superoxide radicals as it catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide (H2O2). Therefore, it is anticipated that sodB derepression under iron-replete conditions is aimed at alleviating the iron-induced oxidative stress previously described. Nevertheless, Fur regulation of sodB is not fully conserved across H. pylori strains and loss of regulation has been linked to nucleotide substitutions in the sodB promoter that lead to a marked decrease in the affinity of the metal-loregulator for the promoter region [73]. Interestingly, increased levels of superoxide dismutase have been associated with the development of metronidazole resistance [74]. Metronidazole is a prodrug that stimulates the formation of super-oxide radicals once it is absorbed by the cells and activated by reduction of its 5-nitro group [75]. It has been described that mutations in the Fur protein are responsible for the observed derepression of sodB in metronidazole-resistant strains [74]. At the present time, it is unknown whether these fur-mutant forms are deficient in regulation of other apo-Fur targets in addition to sodB.
Transcription of the gene encoding the cytotoxin-associated gene cagA was recently shown by our group to be activated by Fur [60]. CagA is an oncogenic protein that is delivered into the epithelial cells through a type IV secretion system. This delivery triggers a cascade of effects that interfere with signal transduction pathways in the host cell, which are known to be important for downstream disease development. Remarkably, a recent report also revealed an important role for CagA in iron acquisition [76]. Using a model polarized epithelium, Tan et al. demonstrated that, unlike the wild-type strain, CagA mutants were unable to colonize the apical surface of cell monolayers incubated with Dulbecco's modified Eagle medium [77]. This deficiency was later shown to be partially rescued by the addition of iron [76]. In uninfected monolayers, diferric transferrin binds to the transferrin receptor on the cell surface and the transferrin–receptor complexes are routed to the basolateral surface after endocytosis via clathrin-coated pits. However, colonization with H. pylori leads to mis-sorting of a subset of the transferrin–receptor complex and transcytosis of the complex from the basolateral to the apical surface. The virulence factors CagA and VacA were found to work in concert to alter the transferrin–receptor complex trafficking. In addition, mislocated transferrin–receptor complexes were found to accumulate at the sites of H. pylori colonization. Thus, CagA facilitates iron acquisition by H. pylori on the apical surface. The observed Fur-mediated activation of CagA may reflect an increased nutritional requirement of H. pylori cells when iron is abundant.
Fur plays an important role in the colonization of the stomach
The fur gene of H. pylori is not essential for in vitro growth, and isogenic fur mutants have been successfully constructed [13,50,64]. The loss of Fur function in H. pylori is likely, at least in part, compensated for by the action of other regulatory proteins encoded for in the chromosome [8,9]. Despite the fact that Fur is dispensable for survival in vitro, several laboratories have reported the existence of severe colonization defects associated with the inactivation of the H. pylori fur locus [13,19,50,78].
Initially, Bury-Moné and coworkers found that H. pylori fur mutants recovered from mice 1 month postinfection were 1–2 logs less abundant than wild-type bacteria recovered at the same time point [50]. In agreement with this, a transposon-based mutagenesis study identified fur as one of several loci that contribute to colonization of the stomach in a murine infection model [78]. In an attempt to better understand the infection dynamics of the fur mutant, the current authors' laboratory temporally monitored the bacterial loads seen in animals infected with either the wild-type or a Δfur strain [13,19]. These studies were carried out using the Mongolian gerbil model of H. pylori infection, which has been shown to be well suited to study the development of H. pylori-induced carcinoma [79,80]. Remarkably, the number of bacteria required to establish an infection was 2 logs higher for the fur mutant than for the wild-type strain, reinforcing the role of Fur in colonization [13]. In addition, the obtained results corroborated a decrease in the number of recoverable Δfur bacteria at days 3 and 7 postinfection. However, bacterial loads of the wild-type strain and the fur mutant were virtually identical at later time points. The discrepancies between these results and those of the previous studies are probably due to the different animal models employed; differences in gastric physiology in the different models probably affect the pool of genes necessary for various stages of colonization, as well as impact the dynamics of infection. Nevertheless, our findings revealed that the role of Fur in colonization is more important at early time points. Perhaps the initial steps of infection are when bacteria are particularly challenged to adapt to the new environment, and thus this period is the critical point when changes in gene expression are most needed to ensure rapid acclimation to the gastric niche.
The role for Fur during the earliest stages of establishment of infection was confirmed by experiments in which gerbils were first infected with the Δfur strain and subsequently super-infected with the wild-type strain at various times postinfection [19]. Consistently, the wild-type strain efficiently displaced the Δfur bacteria during the first week of infection, but this ability progressively diminished over time. When the converse experiment was performed, the Δfur mutant was unable to displace the wild-type strain at any time point tested [19]. Furthermore, competition assays, wherein mutant and wild-type bacteria were coinfected into the same animals, further confirmed the colonization defects seen upon the loss of Fur [19] – the number of mutant bacteria recovered from the stomachs was 1 log lower at day 1 and 2 logs lower at later time points [13,19]. Thus, on the whole, the presence of Fur increases the colonization potential of H. pylori and is important for the process of establishing an infection, but it is not crucial for maintaining the infection once it has occurred.
Given the large number of genes regulated by Fur, the defect of the fur mutant in the colonization of the stomach is probably the deregulation of numerous Fur-regulated loci rather than the specific activity of a single gene. Accordingly, many Fur-regulated genes act in concert to facilitate survival of H. pylori cells under different environments. In keeping with this, many genes under the control of Fur have been shown to play important roles in colonization (Table 2). Therefore, the Fur-orchestrated expression of these genes is critical for H. pylori to efficiently colonize the gastric mucosa.
Table 2.
Helicobacter pylori ferric uptake regulator-regulated genes that have been shown to play a role in colonization.
| Gene name | Function | Animal model of infection | Ref. |
|---|---|---|---|
| feoB | Iron transport | Mouse | [27] |
| fec A1 | Iron transport | Gerbil | [105] |
| pfr | Iron storage | Gerbil | [106] |
| pdxJ | Vitamin B6 biosynthesis | Mouse | [107] |
| putA | Proline metabolism | Mouse | [101] |
| ggt | Synthesis and degradation of glutathione | Mouse/piglet | [108,109] |
| faB† | Motility | Piglet | [110] |
| sodB | Oxidative stress defense | Mouse | [111] |
| hydABCDE | Hydrogenase | Mouse | [112] |
| cagA‡ | Cytotoxicity | Gerbil | [76] |
| nikR | Nickel homeostasis | Mouse | [50] |
Colonization of a double flaA/flaB mutant was analyzed.
Mongolian gerbils were maintained on an iron-deficient diet.
Fur determines the distribution of bacteria in the stomach
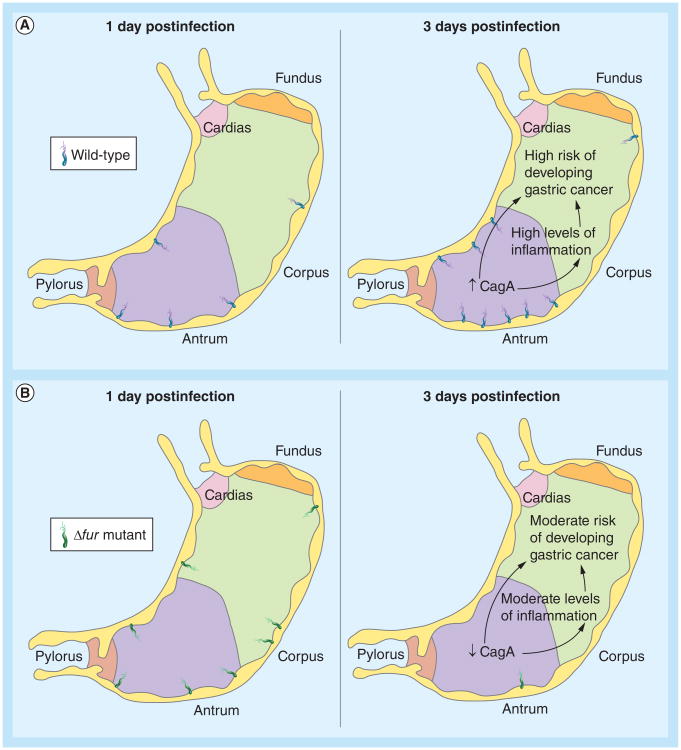
It is well documented that H. pylori colonizes both the corpus and the antrum of the human stomach. Despite this, the bacterium predominantly localizes to the antral region, where acid production is lower than in the corpus [81,82]. Therefore, local acid production is a major determinant of colonization. This distinct distribution has been shown to be preserved in the Mongolian gerbil model of infection (Figure 3) [19]. However, the distribution of H. pylori in the stomach is altered in gerbils infected with the Δfur mutant (Figure 3) [19]. In this sense, single-strain infection experiments revealed a dramatic increase in the number of Δfur bacteria localized to the corpus of the stomach at 1 day postinfection. This is in acute contrast to the low levels of colonization of the corpus featured by the wild-type strain at this and later time points. Remarkably, wild-type cells multiply in the antrum immediately and the bacterial load increases. On the contrary, by day 3 postinfection, Δfur bacteria have been rapidly cleared from both the corpus and the antrum, causing a concomitant decrease in the overall bacterial load, which then slowly recovers over the next 2 weeks. The increased tropism of the fur mutant towards the corpus of the stomach may be due, at least in part, to the altered expression of genes coding for components of the chemotaxis apparatus. Transcriptional data suggest that Fur regulates the expression of the gene coding for the chemotaxis protein CheV2 [54]. Furthermore, nonchemotactic mutants of H. pylori display an anomalous distribution in the stomach of infected mice; they are not predominantly localized to the antrum as normal [83]. Interestingly, it has been reported that H. pylori mutants lacking the type IV secretion protein CagY or the effector protein CagA colonize the antral region of the stomach more prominently than the corpus [84]. As mentioned earlier, transcription of cagA was recently found by the current authors' group to be activated by Fur [60]. Thus, the lower levels of CagA expression expected for the fur mutant may facilitate the higher colonization of the antrum observed at the very initial steps of colonization. Whether or not disregulation of cheV2 and cagA is responsible for the atypical localization of H. pylori in the stomach, or whether this can be attributed to the altered expression of other Fur-regulated factors that serve as adhesins, remains to be determined.
Figure 3. Colonization pattern of Helicobacter pylori at early stages of infection.
Distribution of Helicobacter pylori in the stomach of Mongolian gerbils infected with either the (A) wild-type strain or a (B) Δfur mutant. The expected levels of CagA expression and the associated risk of developing inflammation and gastric cancer are indicated.
Fur impacts the level of inflammation & the degree of disease
The absence of the fur locus was also found to correlate with lower levels of inflammation of both the corpus and the antrum of infected gerbils [19]. This is important because the level of inflammation has been identified as one of the factors associated with an elevated risk of developing gastric cancer. Inflammation probably plays a significant role in nutrient acquisition by H. pylori, since mucosal/cellular damage could lead to leakage of serum and other cellular contents. This in turn provides an excellent source of nutrients and urea. The main bacterial factors that contribute to H. pylori-induced gastric inflammation are the type IV secretion system and CagA. Several studies have shown that serum antibody response to CagA correlates with severity of gastric inflammation [85–87]. To this end, CagA-positive strains induce higher levels of IL-1β and IL-8, which may induce further inflammation [88]. In addition, it has been reported that the conserved repeat responsible for phosphorylation-independent activity motif in the C-terminal region of CagA may stimulate NF-κB-mediated inflammation by interacting with the HGF receptor [89]. NF-κB regulates a variety of genes whose products are involved in cell growth, inflammation and immune responses. Taken together with the fact that Fur functions as an activator of cagA expression, the lower levels of inflammation elicited by the fur mutant are not unexpected [60]. In addition, CagA-positive strains have been shown to correlate with a higher risk of developing gastric cancer. Therefore, one should expect to find milder disease outcomes associated with H. pylori fur mutants. This concept is supported by the finding that gerbils infected with the fur mutant developed invasive adenocarcinoma much later and with less frequency than gerbils infected with the wild-type strain [19].
In addition to regulation of CagA, Fur also regulates the expression of VacA, a potent exotoxin that causes progressive vacuolation of host cells as well as gastric injury. Fur regulation of VacA expression is indirect and, accordingly, Fur does not bind to the vacA promoter [13]. It has been reported that oral administration of VacA to mice causes degeneration of the gastric mucosa and acute inflammation, followed by gastric ulcer disease [90]. Of note, VacA seems to be important for H. pylori during the initial steps of colonization [91]. Thus, the defects in early-stage colonization demonstrated by the fur mutant may be due to alterations in VacA. Clearly the necessity for Fur-dependent regulation of CagA and VacA explains why fur mutants demonstrate changes in the rate of disease development and overall induced disease severity.
The expression of two other genes coding for proteins that have been shown to induce higher levels of inflammation, GGT and PutA, is also under the control of Fur in H. pylori. [60]. In the case of GGT, it has been postulated that glutathione hydrolysis by this enzyme induces H2O2 production [92]. The increase in H2O2 production by host cells in the presence of GGT has been linked to NF-κB activation and upregulation of IL-8 production [93]. In addition, elevated levels of H2O2 generate oxidative stress, which has been associated with the development of severe gastric diseases in H. pylori infections [94]. In fact, epidemiological studies suggest that gastric cancer and precancerous lesions may be caused by reactive oxygen species and that dietary antioxidants can prevent stomach cancer [95]. Supporting this hypothesis, previous data suggest that H2O2 in combination with NH3 and NH2Cl hinders the healing process of peptic ulcers [96]. In the presence of cloride ions, H2O2 generates hypochlorus anions that subsequently react with ammonia to lead to the formation of monochloramine. Previous reports have demonstrated that monochloramine is a potent inductor of mucosal toxicity [97].
Proline oxidation by PutA, an enzyme that plays a central role in proline metabolism, also generates reactive oxygen species, such as H2O2 [98,99]. Based on studies that demonstrate that H. pylori can use proline as a respiratory substrate [100], it has been suggested that proline obtained from damaged host epithelia may support persistent colonization and growth of Helicobacter species. This hypothesis is supported by the inability of an H. pylori putA mutant to colonize the stomach of a murine animal model [101]. Interestingly, Helicobacter hepaticus mutants defective in the putA gene display wild-type levels of infection in mice [98]. Therefore, putA appears essential for the colonization of the gastric mucosa, but dispensable for the colonization of other niches. Despite this, in agreement with the role of PutA in the generation of reactive oxygen species, liver samples from mice infected with the putA mutant show less inflammation than liver samples from mice infected with the wild-type strain. Therefore, in summary, Fur-regulated genes play a major role in the development of the inflammatory response, and deregulation derived from the absence of the Fur protein has a strong impact on redox homeostasis in the stomach.
Future perspective
One area of H. pylori research that has not been fully explored is transmission of the bacterium from one host to the next. In this sense, despite the fact that several animal models have been successfully employed to study the pathogenesis of H. pylori infection, none of the available animal models have yet proven to be useful to study the process of transmission of this bacterium. The lack of a transmission model for H. pylori limits a full understanding of the role of virulence genes in disease development. Among the genes that could reasonably play a role in transmission, we believe those responsible for adaptation to stressful conditions, including fur, would likely be important. Thus, during the process of transmission, it is expected that bacteria must survive outside the host gastric mucosa, at least temporarily; the conditions will probably be substantially different from those existing in the stomach or the digestive tract. Given the important role of Fur in adaptation, it would be interesting to determine whether animals infected with an H. pylori fur mutant could transmit this infection. This information could give us insight into the role of Fur in the ultimate dissemination of the bacterium. Thus, in the coming years, we expect that the increasing interest of the scientific community to understand H. pylori pathogenesis and the need to identify new therapeutic targets will lead to the development of reliable models to study H. pylori transmission.
In addition, we expect that the role of Fur in the activation of cagA will be studied in detail. Since individuals infected with cagA-positive strains show increased risk of developing gastric cancer, it is reasonable to think that Fur-mediated upregulation of cagA will also contribute to such an association. Thus, it is expected that fur-positive strains, showing higher levels of CagA expression, should be associated with increased toxicity and risk of developing gastric cancer. Similarly, we expect that the role in colonization and pathogenesis of particular Fur-regulated genes that have not been previously analyzed, such as ribBA or hpn2, will be studied using one of the available animal models of infection. Since Fur plays a role in colonization and disease, we believe that understanding the role of each particular Fur-regulated gene during the infection process is pivotal to identifying the best targets to develop live vaccines.
Finally, we also expect that the role of Fur in IDA etiology will be explored further. Since Fur is the main regulator of iron homeostasis in H. pylori and it inhibits the expression of high-affinity iron transporters, malfunction of this protein may lead to increased iron uptake from the host. Chronic infection by fur-mutant H. pylori strains that constitutively express these transport proteins could lead to iron deficiency. To our knowledge, the iron concentration in the blood of animals infected with a fur mutant has never been assessed. Therefore, it would be interesting to determine whether there is a decrease of iron in the blood of animals infected with a fur-mutant strain.
Executive summary.
The complexity and elevated cost of the eradication therapy used to treat Helicobacter pylori infections requires the identification of potential new drugs and new drug targets, along with the development of effective vaccines to prevent or cure chronic H. pylori infection.
Pathogenic bacteria often use iron as an environmental signal to regulate virulence genes; thus, the study of the mechanisms devised to sense and respond to changes in iron availability are paramount to identify and understand virulence factors.
H. pylori seems particularly adept at competing for iron with the host, and it can actually cause iron deficiency when the intake of dietary iron is poor.
The ferric uptake regulator (Fur) controls iron homeostasis and functions as a master regulator of gene expression in H. pylori. Fur regulates genes implicated in iron homeostasis as well as genes coding for enzymes that play a central role in metabolism and energy production.
The absence of Fur leads to severe colonization defects. Consistently, several Fur-regulated genes have been shown to be essential for colonization. In addition, Fur plays a role in the distribution of H. pylori in the stomach.
Proteins encoded by Fur-regulated genes have a strong impact on redox homeostasis in the stomach and are major determinants of inflammation.
Acknowledgments
Research in the laboratory of DS Merrell is made possible by grant AI065529 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Disclaimer: The contents of this report are the sole responsibility of the authors and do not necessarily represent the official views of the US Department of Defense or the NIH.
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
•of interest
••of considerable interest
- 1.Blaser MJ, Perez-Perez GI, Kleanthous H, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111–2115. [PubMed] [Google Scholar]
- 2.Gwack J, Shin A, Kim CS, et al. CagA-producing Helicobacter pylori and increased risk of gastric cancer: a nested case–control study in Korea. Br J Cancer. 2006;95(5):639–641. doi: 10.1038/sj.bjc.6603309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkout AM, Blackwell CC, Weir DM. Increased inflammatory responses of persons of blood group O to Helicobacter pylori. J Infect Dis. 2000;181(4):1364–1369. doi: 10.1086/315375. [DOI] [PubMed] [Google Scholar]
- 4.Malfertheiner P, Sipponen P, Naumann M, et al. Helicobacter pylori eradication has the potential to prevent gastric cancer: a state-of-the-art critique. Am J Gastroenterol. 2005;100(9):2100–2115. doi: 10.1111/j.1572-0241.2005.41688.x. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard TG, Eisenberg JC, Matsumoto Y. Clearance of Helicobacter pylori infection through immunization: the site of T cell activation contributes to vaccine efficacy. Vaccine. 2004;22(7):888–897. doi: 10.1016/j.vaccine.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Malfertheiner P, Megraud F, O'Morain C, et al. Current concepts in the management of Helicobacter pylori infection – the Maastricht 2–2000 Consensus Report. Aliment Pharmacol Ther. 2002;16(2):167–180. doi: 10.1046/j.1365-2036.2002.01169.x. [DOI] [PubMed] [Google Scholar]
- 7.Ruggiero P, Peppoloni S, Rappuoli R, Del Giudice G. The quest for a vaccine against Helicobacter pylori: how to move from mouse to man? Microbes Infect. 2003;5(8):749–756. doi: 10.1016/s1286-4579(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 8.Baltrus DA, Amieva MR, Covacci A, et al. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191(1):447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter BM, Whitmire JM, Merrell DS. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect Immun. 2009;77(7):2590–2601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Vliet AH, Stoof J, Vlasblom R, et al. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter. 2002;7(4):237–244. doi: 10.1046/j.1523-5378.2002.00088.x. [DOI] [PubMed] [Google Scholar]
- 12.Bijlsma JJ, Waidner B, Vliet AH, et al. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect Immun. 2002;70(2):606–611. doi: 10.1128/iai.70.2.606-611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Gancz H, Censini S, Merrell DS. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect Immun. 2006;74(1):602–614. doi: 10.1128/IAI.74.1.602-614.2006. Describes the identification of genes regulated by iron and pH in a ferric uptake regulator (Fur)-dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Vliet AH, Kuipers EJ, Stoof J, Poppelaars SW, Kusters JG. Acid-responsive gene induction of ammonia-producing enzymes in Helicobacter pylori is mediated via a metal-responsive repressor cascade. Infect Immun. 2004;72(2):766–773. doi: 10.1128/IAI.72.2.766-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Vliet AH, Stoof J, Poppelaars SW, et al. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori fur repressor. J Biol Chem. 2003;278(11):9052–9057. doi: 10.1074/jbc.M207542200. [DOI] [PubMed] [Google Scholar]
- 16.Delany I, Spohn G, Rappuoli R, Scarlato V. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol Microbiol. 2001;42(5):1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x. [DOI] [PubMed] [Google Scholar]
- 17.Ernst FD, Homuth G, Stoof J, et al. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J Bacteriol. 2005;187(11):3687–3692. doi: 10.1128/JB.187.11.3687-3692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gancz H, Merrell DS. The Helicobacter pylori ferric uptake regulator (Fur) is essential for growth under sodium chloride stress. J Microbiol. 2011;49(2):294–298. doi: 10.1007/s12275-011-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Miles S, Piazuelo MB, Semino-Mora C, et al. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect Immun. 2010;78(7):3073–3082. doi: 10.1128/IAI.00190-10. Provides the most comprehensive study on the role of Fur in the colonization of the stomach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun. 2001;69(6):3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186(14):4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rea RB, Gahan CG, Hill C. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect Immun. 2004;72(2):717–727. doi: 10.1128/IAI.72.2.717-727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen I, Gerstenberger J, Gruber AD, et al. Deletion of the ferric uptake regulator Fur impairs the in vitro growth and virulence of Actinobacillus pleuropneumoniae. Infect Immun. 2005;73(6):3740–3744. doi: 10.1128/IAI.73.6.3740-3744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvie DR, Vilchez S, Steggles JR, Ellar DJ. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology. 2005;151(Pt 2):569–577. doi: 10.1099/mic.0.27744-0. [DOI] [PubMed] [Google Scholar]
- 25.Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect Immun. 2005;73(12):8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keenan JI, Peterson RA, Fraser R, et al. The effect of Helicobacter pylori infection and dietary iron deficiency on host iron homeostasis: a study in mice. Helicobacter. 2004;9(6):643–650. doi: 10.1111/j.1083-4389.2004.00278.x. [DOI] [PubMed] [Google Scholar]
- 27.Velayudhan J, Hughes NJ, Mccolm AA, et al. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol Microbiol. 2000;37(2):274–286. doi: 10.1046/j.1365-2958.2000.01987.x. [DOI] [PubMed] [Google Scholar]
- 28.Braun V, Braun M. Active transport of iron and siderophore antibiotics. Curr Opin Microbiol. 2002;5(2):194–201. doi: 10.1016/s1369-5274(02)00298-9. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson AD, Deisenhofer J. Metal import through microbial membranes. Cell. 2004;116(1):15–24. doi: 10.1016/s0092-8674(03)01030-4. [DOI] [PubMed] [Google Scholar]
- 30•.Senkovich O, Ceaser S, Mcgee DJ, Testerman TL. Unique host iron utilization mechanisms of Helicobacter pylori revealed with iron-deficient chemically defined media. Infect Immun. 2010;78(5):1841–1849. doi: 10.1128/IAI.01258-09. Analyzes the iron binding proteins from the host that Helicobacter pylori can use as an iron source. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Husson MO, Legrand D, Spik G, Leclerc H. Iron acquisition by Helicobacter pylori: importance of human lactoferrin. Infect Immun. 1993;61(6):2694–2697. doi: 10.1128/iai.61.6.2694-2697.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worst DJ, Otto BR, De Graaff J. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect Immun. 1995;63(10):4161–4165. doi: 10.1128/iai.63.10.4161-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhaenens L, Szczebara F, Husson MO. Identification, characterization, and immunogenicity of the lactoferrin-binding protein from Helicobacter pyloris. Infect Immun. 1997;65(2):514–518. doi: 10.1128/iai.65.2.514-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrizo-Chavez MA, Cruz-Castaneda A, Olivares-Trejo Jde J. The frpB1 gene of Helicobacter pylori is regulated by iron and encodes a membrane protein capable of binding haem and haemoglobin. FEBS Lett. 2012;586(6):875–879. doi: 10.1016/j.febslet.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Lopez MA, Olivares-Trejo JJ. The gene frpB2 of Helicobacter pylori encodes an hemoglobin-binding protein involved in iron acquisition. Biometals. 2009;22(6):889–894. doi: 10.1007/s10534-009-9240-5. [DOI] [PubMed] [Google Scholar]
- 36.Cardenas VM, Mulla ZD, Ortiz M, Graham DY. Iron deficiency and Helicobacter pylori infection in the United States. Am J Epidemiol. 2006;163(2):127–134. doi: 10.1093/aje/kwj018. [DOI] [PubMed] [Google Scholar]
- 37.Qu XH, Huang XL, Xiong P, et al. Does Helicobacter pylori infection play a role in iron deficiency anemia? A meta-analysis. World J Gastroenterol. 2010;16(7):886–896. doi: 10.3748/wjg.v16.i7.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Annibale B, Capurso G, Delle Fave G. The stomach and iron deficiency anaemia: a forgotten link. Dig Liver Dis. 2003;35(4):288–295. doi: 10.1016/s1590-8658(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 39.Barabino A. Helicobacter pylori-related iron deficiency anemia: a review. Helicobacter. 2002;7(2):71–75. doi: 10.1046/j.1083-4389.2002.00073.x. [DOI] [PubMed] [Google Scholar]
- 40.Konno M, Muraoka S, Takahashi M, Imai T. Iron-deficiency anemia associated with Helicobacter pylori gastritis. J Pediatr Gastroenterol Nutr. 2000;31(1):52–56. doi: 10.1097/00005176-200007000-00012. [DOI] [PubMed] [Google Scholar]
- 41.Digirolamo AM, Perry GS, Gold BD, et al. Helicobacter pylori, anemia, and iron deficiency: relationships explored among Alaska native children. Pediatr Infect Dis J. 2007;26(10):927–934. doi: 10.1097/INF.0b013e31812e52cd. [DOI] [PubMed] [Google Scholar]
- 42.Thomson MJ, Pritchard DM, Boxall SA, et al. Gastric Helicobacter infection induces iron deficiency in the INS-GAS mouse. PLoS ONE. 2012;7(11):e50194. doi: 10.1371/journal.pone.0050194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X, Qu X, Yan W, et al. Iron deficiency anaemia can be improved after eradication of Helicobacter pylori. Postgrad Med J. 2010;86(1015):272–278. doi: 10.1136/pgmj.2009.089987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokota S, Konno M, Mino E, Sato K, Takahashi M, Fujii N. Enhanced Fe ion-uptake activity in Helicobacter pylori strains isolated from patients with iron-deficiency anemia. Clin Infect Dis. 2008;46(4):e31–e33. doi: 10.1086/526784. [DOI] [PubMed] [Google Scholar]
- 45.Choe YH, Hwang TS, Kim HJ, Shin SH, Song SU, Choi MS. A possible relation of the Helicobacter pylori pfr gene to iron deficiency anemia? Helicobacter. 2001;6(1):55–59. doi: 10.1046/j.1523-5378.2001.00007.x. [DOI] [PubMed] [Google Scholar]
- 46.Jeon BH, Oh YJ, Lee NG, Choe YH. Polymorphism of the Helicobacter pylori feoB gene in Korea: a possible relation with iron-deficiency anemia? Helicobacter. 2004;9(4):330–334. doi: 10.1111/j.1083-4389.2004.00239.x. [DOI] [PubMed] [Google Scholar]
- 47.Park SA, Lee HW, Hong MH, et al. Comparative proteomic analysis of Helicobacter pylori strains associated with iron deficiency anemia. Proteomics. 2006;6(4):1319–1328. doi: 10.1002/pmic.200500293. [DOI] [PubMed] [Google Scholar]
- 48.Merrell DS, Thompson LJ, Kim CC, et al. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect Immun. 2003;71(11):6510–6525. doi: 10.1128/IAI.71.11.6510-6525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect Immun. 2003;71(6):3529–3539. doi: 10.1128/IAI.71.6.3529-3539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bury-Moné S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53(2):623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 51.Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373(1):1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 52.Danielli A, Romagnoli S, Roncarati D, Costantino L, Delany I, Scarlato V. Growth phase and metal-dependent transcriptional regulation of the fecA genes in Helicobacter pylori. J Bacteriol. 2009;191(11):3717–3725. doi: 10.1128/JB.01741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delany I, Pacheco AB, Spohn G, Rappuoli R, Scarlato V. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J Bacteriol. 2001;183(16):4932–4937. doi: 10.1128/JB.183.16.4932-4937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Ernst FD, Bereswill S, Waidner B, et al. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expressio. Microbiology. 2005;151(Pt 2):533–546. doi: 10.1099/mic.0.27404-0. Provides an extensive list of genes from H. pylori that are under the control of iron, either in a Fur-dependent or -independent manner. [DOI] [PubMed] [Google Scholar]
- 55.Worst DJ, Gerrits MM, Vandenbroucke-Grauls CM, Kusters JG. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J Bacteriol. 1998;180(6):1473–1479. doi: 10.1128/jb.180.6.1473-1479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crossley RA, Gaskin DJ, Holmes K, et al. Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl Environ Microbiol. 2007;73(24):7819–7825. doi: 10.1128/AEM.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cowart RE. Reduction of iron by extracellular iron reductases: implications for microbial iron acquisition. Arch Biochem Biophys. 2002;400(2):273–281. doi: 10.1016/S0003-9861(02)00012-7. [DOI] [PubMed] [Google Scholar]
- 58.Schroder I, Johnson E, De Vries S. Microbial ferric iron reductases. FEMS Microbiol Rev. 2003;27(2–3):427–447. doi: 10.1016/S0168-6445(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 59.Fassbinder F, van Vliet AH, Gimmel V, Kusters JG, Kist M, Bereswill S. Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp) FEMS Microbiol Lett. 2000;184(2):225–229. doi: 10.1111/j.1574-6968.2000.tb09018.x. [DOI] [PubMed] [Google Scholar]
- 60•.Pich OQ, Carpenter BM, Gilbreath JJ, Merrell DS. Detailed analysis of Helicobacter pylori Fur-regulated promoters reveals a Fur box core sequence and novel Fur-regulated genes. Mol Microbiol. 2012;84(5):921–941. doi: 10.1111/j.1365-2958.2012.08066.x. Identifies novel genes under the control of Fur in H pylori. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibayama K, Wachino J, Arakawa Y, Saidijam M, Rutherford NG, Henderson PJ. Metabolism of glutamine and glutathione via gamma-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: possible significance in the pathophysiology of the organism. Mol Microbiol. 2007;64(2):396–406. doi: 10.1111/j.1365-2958.2007.05661.x. [DOI] [PubMed] [Google Scholar]
- 62.Zarnowski R, Cooper KG, Brunold LS, Calaycay J, Woods JP. Histoplasma capsulatum secreted gamma-glutamyltransferase reduces iron by generating an efficient ferric reductant. Mol Microbiol. 2008;70(2):352–368. doi: 10.1111/j.1365-2958.2008.06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Timmerman MM, Woods JP. Potential role for extracellular glutathione-dependent ferric reductase in utilization of environmental and host ferric compounds by Histoplasma capsulatum. Infect Immun. 2001;69(12):7671–7678. doi: 10.1128/IAI.69.12.7671-7678.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bereswill S, Greiner S, van Vliet AH, et al. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J Bacteriol. 2000;182(21):5948–5953. doi: 10.1128/jb.182.21.5948-5953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bereswill S, Waidner U, Odenbreit S, et al. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology. 1998;144(Pt 9):2505–2516. doi: 10.1099/00221287-144-9-2505. [DOI] [PubMed] [Google Scholar]
- 66.Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci USA. 2012;109(25):10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng X, Sun F, Ji Q, et al. Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J Bacteriol. 2012;194(7):1753–1762. doi: 10.1128/JB.06582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alamuri P, Mehta N, Burk A, Maier RJ. Regulation of the Helicobacter pylori Fe–S cluster synthesis protein NifS by iron, oxidative stress conditions, and Fur. J Bacteriol. 2006;188(14):5325–5330. doi: 10.1128/JB.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olson JW, Agar JN, Johnson MK, Maier RJ. Characterization of the NifU and NifS FeS cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry. 2000;39(51):16213–16219. doi: 10.1021/bi001744s. [DOI] [PubMed] [Google Scholar]
- 70.Gilbreath JJ, West AL, Pich OQ, Carpenter BM, Michel S, Merrell DS. Fur activates expression of the 2-oxoglutarate oxidoreductase genes (oorDABC) in Helicobacter pylori. J Bacteriol. 2012;194(23):6490–6497. doi: 10.1128/JB.01226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smaldone GT, Revelles O, Gaballa A, Sauer U, Antelmann H, Helmann JD. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J Bacteriol. 2012;194(10):2594–2605. doi: 10.1128/JB.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaballa A, Antelmann H, Aguilar C, et al. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci USA. 2008;105(33):11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carpenter BM, Gancz H, Gonzalez-Nieves RP, et al. A single nucleotide change affects Fur-dependent regulation of sodB in H. pylori. PLoS ONE. 2009;4(4):e5369. doi: 10.1371/journal.pone.0005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsugawa H, Suzuki H, Satoh K, et al. Two amino acids mutation of ferric uptake regulator determines Helicobacter pylori resistance to metronidazole. Antioxid Redox Signal. 2011;14(1):15–23. doi: 10.1089/ars.2010.3146. [DOI] [PubMed] [Google Scholar]
- 75.Perez-Reyes E, Kalyanaraman B, Mason RP. The reductive metabolism of metronidazole and ronidazole by aerobic liver microsomes. Mol Pharmacol. 1980;17(2):239–244. [PubMed] [Google Scholar]
- 76•.Tan S, Noto JM, Romero-Gallo J, Peek RM, Jr, Amieva MR. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 2011;7(5):e1002050. doi: 10.1371/journal.ppat.1002050. Describes the unanticipated role of the cytotoxins CagA and VacA in iron acquisition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5(5):e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baldwin DN, Shepherd B, Kraemer P, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75(2):1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franco AT, Israel DA, Washington MK, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102(30):10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe T, Tada M, Nagai H, Sasaki S, Nakao M. Helicobacter pylori infection induces gastric cancer in Mongolian gerbils. Gastroenterology. 1998;115(3):642–648. doi: 10.1016/s0016-5085(98)70143-x. [DOI] [PubMed] [Google Scholar]
- 81.Hackelsberger A, Gunther T, Schultze V, Labenz J, Roessner A, Malfertheiner P. Prevalence and pattern of Helicobacter pylori gastritis in the gastric cardia. Am J Gastroenterol. 1997;92(12):2220–2224. [PubMed] [Google Scholar]
- 82.Stolte M, Eidt S, Ohnsmann A. Differences in Helicobacter pylori associated gastritis in the antrum and body of the stomach. Z Gastroenterol. 1990;28(5):229–233. [PubMed] [Google Scholar]
- 83.Terry K, Williams SM, Connolly L, Ottemann KM. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun. 2005;73(2):803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rieder G, Merchant JL, Haas R. Helicobacter pylori cag-type IV secretion system facilitates corpus colonization to induce precancerous conditions in Mongolian gerbils. Gastroenterology. 2005;128(5):1229–1242. doi: 10.1053/j.gastro.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 85.Peek RM, Jr, Miller GG, Tham KT, et al. Heightened inflammatory response and cytokine expression in vivo to cagA+ Helicobacter pylori strains. Lab Invest. 1995;73(6):760–770. [PubMed] [Google Scholar]
- 86.Kuipers EJ, Perez-Perez GI, Meuwissen SG, Blaser MJ. elicobacter pylori and atrophic gastritis: importance of the cagA status. J Natl Cancer Inst. 1995;87(23):1777–1780. doi: 10.1093/jnci/87.23.1777. [DOI] [PubMed] [Google Scholar]
- 87•.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40(3):297–301. doi: 10.1136/gut.40.3.297. Identifies CagA as a major determinant of cancer development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88••.Yamaoka Y, Kita M, Kodama T, Sawai N, Kashima K, Imanishi J. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut. 1997;41(4):442–451. doi: 10.1136/gut.41.4.442. Identifies CagA as one of the factors contributing to the inflamatory response elicited during H. pylori infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suzuki M, Mimuro H, Kiga K, et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5(1):23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 90.Telford JL, Ghiara P, Dell'orco M, et al. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179(5):1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salama NR, Otto G, Tompkins L, Falkow S. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect Immun. 2001;69(2):730–736. doi: 10.1128/IAI.69.2.730-736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Busiello I, Acquaviva R, Di Popolo A, et al. Helicobacter pylori gamma-glutamyltranspeptidase upregulates COX-2 and EGF-related peptide expression in human gastric cells. Cell Microbiol. 2004;6(3):255–267. doi: 10.1046/j.1462-5822.2004.00366.x. [DOI] [PubMed] [Google Scholar]
- 93.Gong M, Ling SS, Lui SY, Yeoh KG, Ho B. Helicobacter pylori gamma-glutamyl transpeptidase is a pathogenic factor in the development of peptic ulcer disease. Gastroenterology. 2010;139(2):564–573. doi: 10.1053/j.gastro.2010.03.050. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Q, Dawodu JB, Etolhi G, Husain A, Gemmell CG, Russell RI. Relationship between the mucosal production of reactive oxygen radicals and density of Helicobacter pylori in patients with duodenal ulcer. Eur J Gastroenterol Hepatol. 1997;9(3):261–265. doi: 10.1097/00042737-199703000-00008. [DOI] [PubMed] [Google Scholar]
- 95.Correa P, Piazuelo MB, Camargo MC. The future of gastric cancer prevention. Gastric Cancer. 2004;7(1):9–16. doi: 10.1007/s10120-003-0265-0. [DOI] [PubMed] [Google Scholar]
- 96.Sato K, Watanabe S, Yoshizawa T, Hirose M, Murai T, Sato N. Ammonia, hydrogen peroxide, and monochloramine retard gastric epithelial restoration in rabbit cultured cell model. Dig Dis Sci. 1999;44(12):2429–2434. doi: 10.1023/a:1026670518567. [DOI] [PubMed] [Google Scholar]
- 97.Kodama M, Tsukada H, Ooya M, et al. Gastric mucosal damage caused by monochloramine in the rat and protective effect of taurine: endoscopic observation through gastric fistula. Endoscopy. 2000;32(4):294–299. doi: 10.1055/s-2000-7383. [DOI] [PubMed] [Google Scholar]
- 98.Krishnan N, Doster AR, Duhamel GE, Becker DF. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect Immun. 2008;76(7):3037–3044. doi: 10.1128/IAI.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krishnan N, Becker DF. Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. J Bacteriol. 2006;188(4):1227–1235. doi: 10.1128/JB.188.4.1227-1235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagata K, Nagata Y, Sato T, Fujino MA, Nakajima K, Tamura T. L-Serine, D- and L-proline and alanine as respiratory substrates of Helicobacter pylori: correlation between in vitro and in vivo amino acid levels. Microbiology. 2003;149(Pt 8):2023–2030. doi: 10.1099/mic.0.26203-0. [DOI] [PubMed] [Google Scholar]
- 101.Nakajima K, Inatsu S, Mizote T, et al. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res. 2008;29(1):9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- 102.Delany I, Spohn G, Pacheco AB, et al. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol Microbiol. 2002;46(4):1107–1122. doi: 10.1046/j.1365-2958.2002.03227.x. [DOI] [PubMed] [Google Scholar]
- 103.Delany I, Spohn G, Rappuoli R, Scarlato V. An antirepression Fur operator upstream of the promoter is required for iron-mediated transcriptional autoregulation in Helicobacter pylori. Mol Microbiol. 2003;50(4):1329–1338. doi: 10.1046/j.1365-2958.2003.03757.x. [DOI] [PubMed] [Google Scholar]
- 104.Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V. In vitro analysis of protein–operator interactions of the NikR and Fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J Bacteriol. 2005;187(22):7703–7715. doi: 10.1128/JB.187.22.7703-7715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsugawa H, Suzuki H, Matsuzaki J, Hirata K, Hibi T. FecA1, a bacterial iron transporter, determines the survival of Helicobacter pylori in the stomach. Free Radic Biol Med. 2012;52(6):1003–1010. doi: 10.1016/j.freeradbiomed.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 106.Waidner B, Greiner S, Odenbreit S, et al. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect Immun. 2002;70(7):3923–3929. doi: 10.1128/IAI.70.7.3923-3929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grubman A, Phillips A, Thibonnier M, et al. Vitamin B6 is required for full motility and virulence in Helicobacter pylori. MBio. 2010;1(3):pii:e00112–10. doi: 10.1128/mBio.00112-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chevalier C, Thiberge JM, Ferrero RL, Labigne A. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol Microbiol. 1999;31(5):1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 109.Mcgovern KJ, Blanchard TG, Gutierrez JA, Czinn SJ, Krakowka S, Youngman P. Gamma-glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect Immun. 2001;69(6):4168–4173. doi: 10.1128/IAI.69.6.4168-4173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64(7):2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seyler RW, Jr, Olson JW, Maier RJ. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect Immun. 2001;69(6):4034–4040. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298(5599):1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]