Abstract
BACKGROUND:
Seasonal outbreaks of winter respiratory viruses are responsible for increases in morbidity and mortality in the community. Previous studies have used hospitalizations, intensive care unit and emergency department (ED) visits as indicators of seasonal influenza incidence.
OBJECTIVES:
To evaluate whether ED visits can be used as a proxy to detect respiratory viral disease outbreaks, as measured by laboratory confirmation.
METHODS:
An Emergency Department Syndromic Surveillance system was used to collect ED chief complaints in Eastern Ontario from 2006 to 2010. Comparable laboratory-confirmed cases of respiratory viral infections were collected from the Public Health Ontario Laboratory in Kingston, Ontario. Correlations between ED visits and laboratory-confirmed cases were calculated.
RESULTS:
Laboratory-confirmed cases of selected respiratory viruses were significantly correlated with ED visits for respiratory and fever/influenza-like illness. In particular, respiratory syncytial virus (Spearman’s rho = 0.593), rhinovirus (Spearman’s rho = 0.280), influenza A (Spearman’s rho = 0.528), influenza B (Spearman’s rho = 0.426) and pH1N1 (Spearman’s rho = 0.470) increased laboratory test levels were correlated with increased volume of ED visits across a number of age demographics. For the entire study population and all studied viruses, the Spearman’s rho was 0.702, suggesting a strong correlation with ED visits. Laboratory-confirmed cases lagged in reporting by between one and two weeks for influenza A and pH1N1 compared with ED visit volume.
CONCLUSION:
These findings support the use of an Emergency Department Syndromic Surveillance system to track the incidence of respiratory viral disease in the community. These methods are efficient and can be performed using automated electronic data entry versus the inherent delays in the primary care sentinel surveillance system, and can aid the timely implementation of preventive and preparatory health interventions.
Keywords: Emergency department surveillance, Respiratory, Syndromic surveillance, Viral disease
Abstract
HISTORIQUE :
Les éclosions saisonnières des virus respiratoires hivernaux sont responsables d’augmentations de la morbidité et de la mortalité dans la collectivité. Les études antérieures faisant appel aux hospitalisations, aux séjours aux unités de soins intensifs et aux consultations à l’urgence comme indicateurs de l’incidence d’influenza saisonnière.
OBJECTIFS :
Évaluer si les consultations à l’urgence pour déceler les éclosions de maladies respiratoires virales peuvent remplacer les confirmations mesurées en laboratoire.
MÉTHODOLOGIE :
Les chercheurs ont utilisé un système de surveillance syndromique des urgences pour colliger les principaux problèmes ayant suscité une consultation à l’urgence entre 2006 et 2010 dans l’est de l’Ontario. Ils ont colligé des cas comparables d’infections respiratoires virales confirmés en laboratoire auprès des Laboratoires de santé publique de l’Ontario situés à Kingston, en Ontario. Ils ont calculé les corrélations entre les consultations à l’urgence et les cas confirmés en laboratoire.
RÉSULTATS :
Les cas confirmés en laboratoire de certains virus respiratoires possédaient une corrélation significative avec les consultations à l’urgence en raison de maladies respiratoire, liées à la fièvre ou de type grippal. Notamment, l’augmentation en laboratoire des taux de virus respiratoire syncytial (Rho de Spearman = 0,593), de rhinovirus (Rho de Spearman = 0,280), d’influenza A (Rho de Spearman = 0,528), d’influenza B (Rho de Spearman = 0,426) et de grippe pH1N1 (Rho de Spearman = 0,470) était corrélée avec l’augmentation du volume de consultations à l’urgence dans plusieurs groupes d’âge. Dans l’ensemble de la population et des virus à l’étude, le Rho de Spearman s’établissait à 0,702, ce qui laisse supposer une forte corrélation avec les consultations à l’urgence. Les déclarations de cas d’influenza A et de grippe pH1N1 confirmés en laboratoire avaient de une à deux semaines de retard par rapport au volume de consultations à l’urgence.
CONCLUSION :
Ces résultats appuient le recours à un système de surveillance syndromique des urgences pour suivre l’incidence de maladies respiratoires virales dans la collectivité. Ces méthodes sont efficaces et peuvent être effectuées au moyen de saisies de données électroniques automatisées plutôt que de s’associer aux délais inhérents au système de surveillance sentinelle en soins de première ligne, et elles peuvent contribuer à la mise en œuvre rapide d’interventions de santé préventives et préparatoires.
Respiratory infections are one of the leading causes of morbidity and mortality in Canada (1), with seasonal influenza (A or B) or influenza-related complications, such as pneumonia, accounting for the deaths of 4000 to 8000 Canadians per year (2). Other respiratory infections, such as respiratory syncytial virus (RSV), parainfluenza virus 1 to 4, and adenovirus and rhinovirus, also represent serious health risks, especially in young children, infants, elderly patients and the immunocompromised (3).
Outbreaks of influenza have been implicated in emergency department (ED) overcrowding and increased numbers of hospitalizations (4–7). It has been suggested that hospitalizations, intensive care unit admissions and ED visits could be used as indicators and predictors of seasonal influenza incidence and morbidity, as well as the effects of epidemics on public health resources at the community level (4,8,9). These findings indicate a role for syndromic surveillance in helping to detect and predict epidemiological patterns of disease outbreak, leading to timely and effective implementation of health policy (10). The WHO, under the new International Health Regulations, stresses the importance of detecting, reporting and confirming infections in new areas, testing severe cases and monitoring circulating viruses, as significant reasons for conducting virological surveillance in pandemic situations (11).
The present study describes several years in Eastern Ontario, from January 1, 2006, to February 29, 2010, and examines the validity of using ED visits as a proxy for the detection and prediction of actual laboratory-confirmed cases of respiratory viral disease. Surveillance could thus be used to track additional outbreaks of respiratory viral disease, aiding in the timely implementation of preventive and preparatory health interventions.
METHODS
The study region included three health units in Eastern Ontario: Hastings Prince Edward Counties, Leeds-Grenville and Lanark District and Kingston Frontenac Lennox and Addington. It represented 442,800 people (3.8% of the population of Ontario). The region had a slightly higher than average proportion of individuals ≥65 years of age compared with the overall population of the province, and a lower proportion of individuals 20 to 39 years of age. Forty-four per cent of the study population lived in communities of >10,000 people, and the region had a much smaller proportion of visible minorities and immigrants than the province as a whole (12). During the study period, there were 10 acute care facilities in the region, of which two were teaching hospitals. Health units were chosen as the geographical boundary for the present study because they are a common denominator for reporting for both streams of data used in the study (laboratory and ED).
The present study covered a period spanning January 1, 2006, to February 29, 2010. This included the abnormally high volume of ED visits and laboratory tests associated with the H1N1 pandemic (pH1N1) during the fall of 2009. While H1N1 first emerged in the study area in the spring of 2009, a pandemic level of cases did not occur until the fall of 2009. This high volume was included in the analysis, not only to use as a test case for a unique epidemic viral isolate, but also to ensure that other viral isolates could still be predicted and correlated while such an event was occurring.
Anonymized respiratory viral specimen data from tests performed by the Public Health Ontario Laboratory – Kingston (PHOL-K) during the study period was used for the analysis. Operated by the Ontario government, the PHOL-K processed specimens directly from physicians and acute care facilities across the three health units included in the present study. Tube cultures were used, with identification of specimens through monoclonal antibodies and immunofluorescence, except for pH1N1, for which real-time polymerase chain reaction was used. Specimens positive for influenza A were subtyped for seasonal influenza A strains (H1 and H3) and, during the 2009/2010 season, for pH1N1 influenza A (13). Subtyping was triaged if demand for laboratory confirmation exceeded testing capacity, favouring the identification of specimens obtained from high-risk and hospitalized patients. Due to this triage, H1 and H3 subtypes were not included in the present study. Influenza A specimens that were not subtyped in 2009 due to the high demand for laboratory services during the pH1N1 outbreak were considered to be pH1N1 cases because both Europe and North America reported <1% prevalence of seasonal H1N1 and H3N2 influenza A strains in 2009 (14,15).
Laboratory-confirmed infections were organized according to week into respiratory virus seasons (spanning September to August), as per the conventions of the Ontario Influenza Bulletin and the Public Health Agency of Canada’s FluWatch (which provide weekly summaries of influenza-like illness (ILI) activity in Canada and definitions for declaring seasonal outbreaks) (16,17).
ED visit volume data for respiratory infections and/or fever/ILI, hereafter referred to as RESP/ILI, were collected using archived data from a real-time ED Syndromic Surveillance (EDSS) system, which monitored ED volumes at the triage level in acute care facilities throughout the study region. The EDSS was programmed to identify syndromes based on predefined, validated keyword lists used to scan the chief complaint field of the triage records of all patients entering the ED (10). The classification of ED visits as RESP/ILI visits was based on the patients’ chief complaints (eg, RESP: cough, sore throat, upper respiratory tract infection and sinus infection; ILI: fever- or influenza-related).
RESP/ILI ED visits and laboratory data were aggregated weekly over the course of the study time period. Overall, ED data were broken down into three age classes: 0 to 17, 18 to 64 and ≥65 years of age. Age classes were not used for the laboratory isolate data due to the low sample sizes apparent when age limitations were applied. Data from all health units were analyzed collectively. SPSS version 21 (IBM Corporation, USA) was used for statistical analysis.
The frequency distribution of the data was analyzed to select the most appropriate regression model for analysis. The data were found to be overdispersed with zero values, making a standard linear or Poisson distribution model inappropriate. This was due to the fact that tests often show a negative result (identified by a zero). The testing regimen was completed in accordance with industry standard techniques for viral testing by an accredited laboratory. Although laboratory biases do exist (see limitations), for the present analysis, zero values were assumed to be accurate and, thus did not represent false negatives (errors) from laboratory testing or missed positive results from samples not submitted. For this reason, a non-zero-inflated, negative binomial regression model was chosen to analyze the data (18). Model validity was determined using overdispersion, which was defined as the Pearson χ2 value divided by the degrees of freedom and be less than or equal to (positive or negative) two. Models were statistically significant at P<0.05.
Negative binomial correlations between the different virus isolates and specific combinations were calculated. Because the data were right-skewed, the nonparametric Spearman’s rho was used.
In an effort to detect the potential delay between ED visits for RESP/ILI and laboratory confirmation, a cross-correlation with differencing was calculated between the weekly ED RESP/ILI visits and weekly aggregated virus data.
RESULTS
There were 1299 unique laboratory-confirmed cases of respiratory virus infections identified by PHOL-K from January 1, 2006 to February 29, 2010. Approximately 1.3% of the laboratory tests were from an individual who also tested positive for at least one of the other virus isolates. No patients tested positive for more than two viral isolates.
Approximately 15% (172,094 of 1,147,324) of all ED visits during the study period and within the study region were by patients presenting with symptoms belonging to the RESP/ILI categories.
Model and correlation results
In the all-ages analysis, the negative binomial regression showed that the model was valid and statistically significant for rhinovirus, influenza A, influenza B, RSV and pH1N1 (Table 1), as well as for combined influenza, RSV and combined influenza as well as all viral isolates. The same viruses and virus combinations were valid and statistically significant in the 0 to 17 years of age model and the 18 to 64 years of age model. In the ≥65 years age group, the negative binomial regression showed the model was valid only for parainfluenza, adenovirus, RSV and rhinovirus, of which only RSV and rhinovirus were statistically significant.
TABLE 1.
Negative binomial regression for respiratory infections and/or fever/influenza-like illness (RESP/ILI) emergency department (ED) visits from all age groups
| Isolate | Pearson χ2,* | n | B† | P |
|---|---|---|---|---|
| Rhinovirus | 334.5 | 216 | 0.001 | 0.001 |
| Influenza A | 332.5 | 216 | 0.004 | <0.001 |
| Parainfluenza | NV | NV | NV | NV |
| Influenza B | 302.6 | 216 | 0.005 | <0.001 |
| Adenovirus | NV | NV | NV | NV |
| RSV | 258.7 | 216 | 0.004 | <0.001 |
| pH1N1 | 67.3 | 43 | 0.003 | <0.001 |
| Isolate groups | ||||
| All influenza | 367.6 | 216 | 0.004 | <0.001 |
| Influenza + RSV | 295.1 | 216 | 0.005 | <0.001 |
| All viruses | 205.0 | 216 | 0.003 | <0.001 |
Pearson χ2 values can be used to determine the relative weight each isolate or group of isolates has on the model. A lower χ2 means a stronger fit of the model comparing ED RESP/ILI visits with laboratory cases;
The negative bionomial test statistic B can be used to determine the rise in laboratory cases for each additional ED RESP/ILI visit. For example, for each additional RESP/ILI visit, there was a 0.5% increase in laboratory cases for influenza B. NV (not valid) represents nonsignificant model results; pH1N1 Pandemic H1N1; RSV Respiratory syncytial virus
Goodness of fit measures in the all ages, the 0 to 17 years and the 18 to 64 years age group analyses suggested that for individual viral isolates, pH1N1 levels were most closely associated with ED visit volume. For all ages and the 0 to 17 years age group, RSV was the second most closely associated, while for the 18 to 64 years age group rhinovirus was the second. However, in all cases this was followed closely by the remaining viruses. When combined, all viruses collectively showed the closest association with ED visit volume in the above three analyses.
For all ages, when each individual isolate was present and circulating in the population, for each additional RESP/ILI visit, there was a corresponding increase in laboratory-confirmed viral isolates for rhinovirus (0.1%), influenza A (0.4%), influenza B (0.5%), RSV (0.4%), pH1N1 (0.3%), all influenza (0.4%), all influenza plus RSV (0.5%) and all viruses (0.3%). According to the 0 to 17 years age group analysis, for each additional RESP/ILI ED visit, there was a corresponding increase in viral isolates in the following manner: influenza B (0.8%), influenza A (0.6%), RSV (0.7%), all influenza (0.7%), influenza plus RSV (0.7%) and all viruses (0.5%) (Table 2). For the 18 to 64 years age group analysis, the increases in isolates were: influenza B (0.8%), influenza A (0.8%), RSV (0.7%), rhinovirus (0.3%), pH1N1 (0.6%), all influenza (0.9%), influenza plus RSV (0.9%) and all viruses (0.7%). Finally, in the ≥65 years age group analysis, for each additional RESP/ILI ED visit, the model indicated an increase in RSV (0.27%) and rhinovirus (0.1%).
TABLE 2.
Age-stratified results summary*
| Isolates |
Age, years
|
|||||
|---|---|---|---|---|---|---|
|
0 to 17
|
18 to 64
|
≥65
|
||||
| Spearman’s | Spearman’s | Spearman’s | ||||
| B | rho | B | rho | B | rho | |
| RSV | 0.007 | 0.566 | 0.007 | 0.481 | 0.027 | 0.416 |
| Adenovirus | NV | NV | NV | NV | NV | NV |
| Rhinovirus | 0.001 | NV | 0.003 | 0.280 | 0.010 | 0.194 |
| Parainfluenza | NV | NV | NV | NV | NV | NV |
| Influenza | ||||||
| Influenza A | 0.006 | 0.473 | 0.008 | 0.511 | NV | NV |
| Influenza B | 0.008 | 0.442 | 0.008 | 0.330 | NV | NV |
| pH1N1 | 0.004 | 0.370 | 0.003 | 0.470 | NV | NV |
| Isolate groups | ||||||
| All | 0.007 | 0.521 | 0.009 | 0.566 | NV | NV |
| Influenza | ||||||
| Flu + RSV | 0.007 | 0.626 | 0.009 | 0.653 | NV | NV |
| All viruses | 0.005 | 0.644 | 0.007 | 0.677 | NV | NV |
Includes test statistics from negative binomial regression of respiratory infections and/or fever/influenza-like illness (RESP/ILI) emergency department (ED) visits and correlations between isolates and RESP/ILI ED visits. Spearman’s rho values ≥0.5 are considered to be highly correlated; 0.25 to 0.5 are considered to be moderately correlated and <0.25 are considered to be weakly correlated. Flu Influenza; NV Results which were not valid due to model significance or over dispersion of the data within the model; pH1N1 Pandemic H1N1; RSV Respiratory syncytial virus
RSV was found to have the highest correlation between ED visits and the number of positive laboratory isolates for the all ages (Table 3), the 0 to 17 years age group and the ≥65 years age group analyses. For the first two, it was found to be highly correlated, while for the latter it was moderately correlated. In the 18 to 64 years age group, influenza A was found to have the highest correlation. Influenza A was also classified as highly correlated with ED visits in the all-ages analysis, while influenza B and pH1N1 were moderately correlated. For the 0 to 17 years age group analysis, influenza A, influenza B and pH1N1 were moderately correlated while for the 18 to 64 years age group analysis, RSV, influenza B and rhinovirus were moderately correlated. In the all ages group, the 0 to 17 years age group and the 18 to 64 years age group analyses, all combined isolate groups were found to have high correlations with increased ED visit volumes.
TABLE 3.
Correlations between isolates and respiratory infections and/or fever/influenza-like illness emergency department visits for all age groups
| Isolate | n | Spearman’s rho | P |
|---|---|---|---|
| RSV | 216 | 0.593 | <0.001 |
| Influenza A | 216 | 0.528 | <0.001 |
| Influenza B | 216 | 0.426 | <0.001 |
| pH1N1 | 216 | 0.413 | 0.006 |
| Rhinovirus | 216 | 0.192 | 0.005 |
| Parainfluenza | NV | NV | NV |
| Adenovirus | NV | NV | NV |
| Isolate groups | |||
| Influenza + RSV | 216 | 0.709 | <0.001 |
| All viruses | 216 | 0.702 | <0.001 |
| All influenza | 216 | 0.585 | <0.001 |
Spearman’s rho values ≥0.5 are considered to be highly correlated; 0.25 to 0.5 are considered to be moderately correlated and <0.25 are considered to be weakly correlated. NV Nonsignificant correlations or failure to achieve significance in the regression model; pH1N1 Pandemic H1N1; RSV Respiratory syncytial virus
Figures 1 and 2 illustrate the close association between influenza A, influenza B, RSV, rhinovirus and pH1N1 (Figure 1) and without the large amplitude of pH1N1 (Figure 2) with RESP/ILI ED visit volume for all ages.
Figure 1).
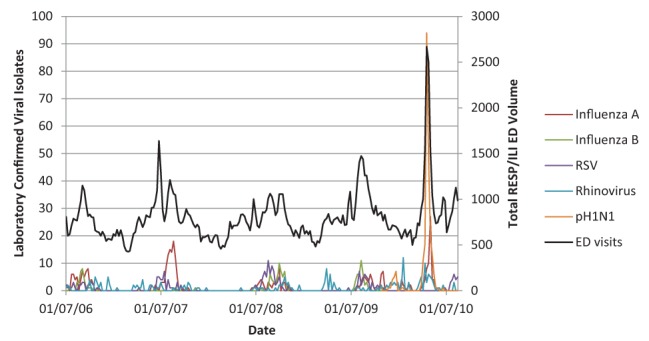
Respiratory infections and/or fever/influenza-like illness (RESP/ILI) emergency department (ED) visit volume compared with specific viral isolates for all age classes including pandemic H1N1 (pH1N1). RSV Respiratory syncytial virus
Figure 2).
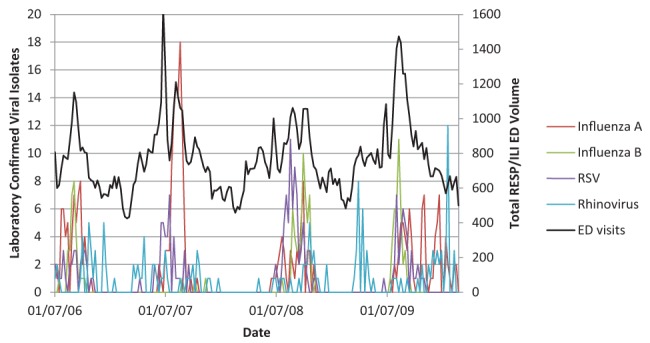
Respiratory infections and/or fever/influenza-like illness (RESP/ILI) emergency department (ED) visit volume compared with specific viral isolates for all age classes, not including pandemic H1N1. RSV Respiratory syncytial virus
When the ED visit volume for RESP/ILI was analyzed against the reporting week laboratory results, a lagged effect was noted. Only influenza A and pH1N1 showed strong lags, with influenza A showing a lag within the second week (cross-correlation function = 0.533) and pH1N1 showing a lag of within the first week and also within the second week (cross-correlation function = 0.751 and 0.476, respectively) (Figure 3).
Figure 3.
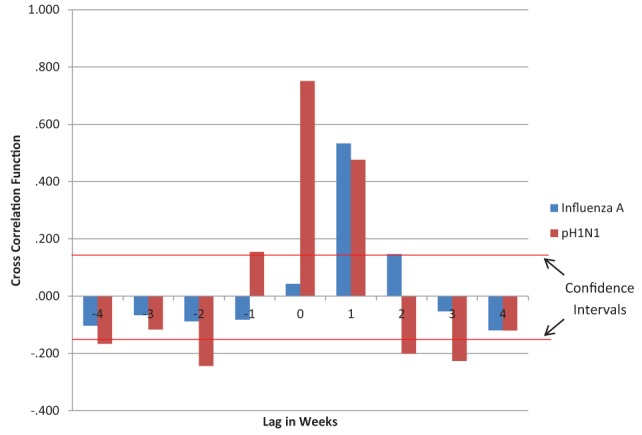
Time series lag analysis of respiratory infections and/or fever/influenza-like illness emergency department (ED) visits compared with laboratory reporting of influenza A and pandemic H1N1 (pH1N1) specimen confirmation. The red horizontal lines represent the 95% CIs for a lack of trend (0.145). Values above or below these lines represent time-lagged trends. Only pH1N1 showed negative cross correlation values greater than the CIs. This may be due to large fluctuation in ED and laboratory data due to the pandemic levels; as such, only the pH1N1 cross-correlation values that were much greater than the CIs during lags of 0 and 1 will be used for discussion. A cross-correlation function greater than the CI indicates a probable connection between the timing of ED visits and laboratory case reporting. A lag of 0 indicates that the laboratory virus case reporting increase lagged the ED visit volume increase by up to one week. A lag of 1 indicates the laboratory virus case reporting increase lagged the ED visits increase by up to two weeks
DISCUSSION
Laboratory-confirmed infections of influenza and other respiratory viruses are generally regarded to be the best measure of true prevalence in the population (6,7); however, few other studies have examined the relationship between laboratory-confirmed cases of respiratory viruses and ED-based respiratory syndromic surveillance.
The present study investigated the efficacy of RESP/ILI ED visit volume to gauge the level of activity of specific respiratory viruses in the underlying population. When all age groups were considered for ED visit volume, there was a moderate to strong correlation between the levels of RSV, influenza A, influenza B, pH1N1 and rhinovirus. This suggests that RESP/ILI ED visit volume may be an appropriate proxy for the levels of these isolates when they are confirmed to be circulating within a population. With the exception of rhinovirus, this continued to be the case within the age classes of 0 to 17 years. RESP/ILI ED visits did not appear to be a good proxy for the levels of adenovirus, rhinovirus and parainfluenza circulating within this age class. For those 18 to 64 years of age, results were similar in strength to the all-ages category, with only parainfluenza and adenovirus showing difficulty in being indicated by ED visit volume. For individuals ≥65 years of age, RESP/ILI appeared to be a potential proxy only for the level of RSV and rhinovirus circulating within the population.
The models used in the present study may also be able to predict potential increases in specific respiratory virus levels within the underlying population, when the virus is already present and circulating – most notably RSV, influenza A, influenza B and pH1N1. This can be particularly important for planning possible public health interventions. It can also be useful for preparing for surge conditions within acute care centres and testing laboratories. Currently, increases in viral testing are expected during respiratory season in Canada; however, linking this expectation to a real-time EDSS may enable public health laboratories to better time the requirements for their services and to allocate resources in a more efficient manner. A prospective investigation linking an EDSS to a public health laboratory in real time is warranted.
The length of lag between a rise in ED visit volume for RESP/ILI visits and a corresponding increase in laboratory confirmation of circulating viral isolates for influenza A and pH1N1 was interesting in that RESP/ILI visits appeared to increase approximately two weeks before a rise in reporting from the laboratory. Although this is highly likely due to the length of time required to report on laboratory results, it illustrates the potential utility of using ED visit volume surveillance as an earlier indicator of the levels of viruses that are known to be circulating within the local population.
The present study had a number of limitations. Laboratory confirmation is deemed the best measure of the true prevalence of respiratory viral illness in the community; however, these methods are associated with case ascertainment and reporting biases. The inclusion of most viruses implicated in causing seasonal respiratory illness similar to influenza reduced possible confounding effects that could arise due to their omission. However, the present study also did not account for environmental causes of respiratory illness, and further work is warranted to determine the contribution of these nonmicrobiological causes to ED visits. Of key note is the fact that certain viruses included in the present study did not show strong correlations or did not fit well within the statistical models. Although there may be a number of factors involved, it is hypothesized that symptom severity may be playing a leading role in whether an individual infected with a particular virus will seek treatment in an ED, seek treatment at his or her family physician or simply convalesce at home. Finally, the models used gave a measure of predicted increases in viral isolates as ED visit volume increased. It is important to note that this was only a potential increase and was only valid when a particular isolate was known and confirmed to be circulating within the population.
We conclude that rises in circulating viral isolates may be indicated and potentially predicted using RESP/ILI ED visit volume from a real-time EDSS system when the particular viral isolates are known to be circulating within an underlying population. EDSS showed efficacy at indicating increased circulation of RSV, influenza A, influenza B and pH1N1 and is timelier than laboratory confirmation. ED surveillance may complement laboratory surveillance, especially in the face of finite laboratory capacity (such as during a pandemic).
SUMMARY
Seasonal outbreaks of winter respiratory viruses are responsible for increases in morbidity and mortality in the community. Traditionally, laboratory-based testing has been used to determine the levels of circulating seasonal viruses within an underlying population. In the present study, surveillance of the volume of patients visiting local EDs was used as a proxy for determining the potential levels of circulating seasonal viruses as detected by laboratory-confirmed cases. RSV-, rhinovirus-, influenza A-, influenza B- and pH1N1-positive laboratory test levels were shown to be predicted by ED visit volume across a number of age demographics.
REFERENCES
- 1.Public Health Agency of Canada. Leading causes of death and hospitalization in Canada. Public Health Agency of Canada; 2008 (Cited March 23, 2010). <www.phac-aspc.gc.ca/publicat/lcd-pcd97/table1-eng.php> (Accessed January 2012).
- 2.Health Canada. Influenza (the “flu”). Health Canada; 2009 (Cited March 23, 2010). <www.hc-sc.gc.ca/hl-vs/alt_formats/pacrb-dgapcr/pdf/iyh-vsv/diseases-maladies/flu-grippe-eng.pdf> (Accessed January 2012).
- 3.Kesson AM. Respiratory virus infections. Paediatr Respir Rev. 2007;8:240–8. doi: 10.1016/j.prrv.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schull MJ, Mamdani MM, Fang JM. Community influenza outbreaks and emergency department ambulance diversion. Ann Emerg Med. 2004;44:61–7. doi: 10.1016/j.annemergmed.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Menec VH, Black C, MacWilliam L, Aoki FY. The impact of influenza-associated respiratory illnesses on hospitalizations, physician visits, emergency room visits, and mortality. Can J Public Health-Rev Can Sante Publ. 2003;94:59–63. doi: 10.1007/BF03405054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upshur RE, Knight K, Goel V. Time-series analysis of the relation between influenza virus and hospital admissions of the elderly in Ontario, Canada, for pneumonia, chronic lung disease, and congestive heart failure. Am J Epidemiol. 1999;149:85–92. doi: 10.1093/oxfordjournals.aje.a009731. [DOI] [PubMed] [Google Scholar]
- 7.Bourgeois FT, Olson KL, Brownstein JS, McAdam AJ, Mandl KD. Validation of syndromic surveillance for respiratory infections. Ann Emerg Med. 2006;47:265–71. doi: 10.1016/j.annemergmed.2005.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullooly JP, Bridges CB, Thompson WW, et al. Influenza- and RSV-associated hospitalizations among adults. Vaccine. 2007;25:846–55. doi: 10.1016/j.vaccine.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Moineddin R, Nie JX, Domb G, Leong AM, Upshur RE. Seasonality of primary care utilization for respiratory diseases in Ontario: A time-series analysis. BMC Health Serv Res. 2008;8:6. doi: 10.1186/1472-6963-8-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore KM, Edgar BL, McGuinness D. Implementation of an automated, real-time public health surveillance system linking emergency departments and health units: Rationale and methodology. Can J Emerg Med Care. 2008;10:114–9. doi: 10.1017/s1481803500009817. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization Human infection with pandemic (H1N1) 2009 virus: updated interim WHO guidance on global surveillance. World Health Organization, 2009 July 10, 2009. < www.who.int/csr/disease/swineflu/WHO_case_definition_swine_flu_2009_04_29.pdf> (Accessed January 2012)
- 12.Bains N, Dall K, Hay C, Pacey M, Sarkella J, Ward M. 2004. Population Health Profile: South East LHIN,
- 13.Ontario Agency for Health Protection and Promotion. SOP-SD-MO-019-000: Real-time RT-PCR (rRT-PCR) for sub-typing of pandemic influenza A (H1N1) virus and seasonal influenza virus A (H1 and H3).
- 14.Centers for Disease Control and Prevention Surveillance for the 2009 pandemic influenza A (H1N1) virus and seasonal influenza viruses – New Zealand, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:918–21. [PubMed] [Google Scholar]
- 15.World Health Organization. Pandemic H1N1 2009 – update 74. 2009.
- 16.FluWatch. Public Health Agency of Canada; 2009 (cited 2009–2010). <www.phac-aspc.gc.ca/fluwatch/09-10/index-eng.php> (Accessed January 2012).
- 17.Ontario Influenza Bulletins. Ontario Ministry of Health and Long-Term Care; 2012 (cited 2009–2010). <www.health.gov.on.ca/english/providers/program/pubhealth/flu/flu_09/flubul_mn.html> (Accessed January 2012).
- 18.Hilbe JE. Negative Binomial Regression. Cambridge: Cambridge University Press; 2007. p. 251. [Google Scholar]


